CRISPR-Powered Liquid Biopsies in Cancer Diagnostics
Abstract
1. Introduction
2. CRISPR—From Gene Editing Tool to Point-of-Care Diagnostics
3. Overview of CRISPR-Cas Systems Used in Diagnostics
3.1. Cas9
3.2. Cas13
3.3. Cas12
3.4. Cas14
3.5. CasΦ/Cas12j
3.6. Strategies for Result Readouts
4. CRISPR-Dx for Precise and Rapid Cancer Screening
4.1. Biomarkers Enabling CRISPR-Dx Detection
4.1.1. ctDNA
4.1.2. RNA
4.1.3. DNA Methylation
4.1.4. Proteins
4.1.5. Other Non-Nucleic-Acid Biomarkers
4.2. Cancer Associated Viruses
5. Comparison of Performance Among CRISPR-Dx Platforms
5.1. Biomarker Capacity
5.2. POC Capacity: Trade-Offs in LOD and Time-to-Result
5.3. Clinical Sensitivity and Specificity
5.4. ctDNA Sensitivity: Measure of Variant Allelic Fraction
| CRISPR-Dx Platform | Tested Biomarkers | Limit of Detection | Time-to-Result | Point-of-Care Capacity |
|---|---|---|---|---|
| NASBACC [31] | RNA | ≤10 fM | >60 min | ✔ |
| CAS-EXPAR [32] | DNA | ≤10 aM | ≤60 min | ✔ |
| CAS-EXPAR [32] | DNA methylation | ≤10 aM | – | – |
| FLASH [33] | DNA | ≤10 aM | – | – |
| LEOPARD [34] | RNA | ≤10 aM | – | ✔ |
| PC reporter [35] | DNA | >10 fM | >60 min | ✔ |
| RCA-CRISPR-split-HRP [36] | mRNA | ≤10 fM | >60 min | ✔ |
| HOLMES [37] | DNA/RNA | ≤10 aM | ≤60 min | – |
| DETECTR [38] | DNA | ≤10 aM | ≤60 min | ✔ |
| E-CRISPR [39] | DNA | >10 fM | ≤30 min | ✔ |
| E-CRISPR [39] | Protein | >10 fM | >60 min | ✔ |
| Wax-CRISPR [40] | DNA | ≤10 aM | ≤30 min | ✔ |
| CDetection [41] | DNA | ≤10 aM | ≤30 min | ✔ |
| CDetection [41] | ctDNA | 1% VAF 1 | ≤30 min | ✔ |
| HOLMESv2 [42] | DNA/RNA | ≤10 aM | ≤30 min | ✔ |
| HOLMESv2 [42] | DNA methylation | – | – | – |
| SHERLOCK [43] | RNA | ≤10 aM | ≤60 min | ✔ |
| SHERLOCK [43] | DNA | ≤10 aM | >60 min | ✔ |
| SHERLOCK [43] | ctDNA | 0.1% VAF 1 | >60 min | ✔ |
| HUDSON-SHERLOCK [44] | RNA | ≤10 aM | >60 min | ✔ |
| SHERLOCKv2 [45] | RNA/DNA | ≤10 aM | ≤30 min | ✔ |
| SHERLOCKv2 [45] | ctDNA | 0.6% VAF 1 | ≤30 min | ✔ |
| Cas14-DETECTR [27] | DNA | – | >60 min | – |
| EXP-J [28] | mRNA | ≤10 fM | ≤60 min | – |
6. Challenges and Future Directions in CRISPR-Dx for Cancer Diagnosis
6.1. Challenges in Detecting Elusive ctDNA
6.2. Point-of-Care Deployment Challenges
6.3. Clinical and Regulatory Challenges
6.4. Future Directions for CRISPR-Dx
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| AFP | Alpha-foetoprotein |
| APML | Acute promyelocytic leukaemia |
| Cas | CRISPR associated |
| CAS-EXPAR | CRISPR associated EXPAR |
| CDetection | Cas12b-mediated DNA detection |
| cfDNA | Cell-free DNA |
| COVID-19 | Coronavirus disease 2019 |
| CRISPR | Clustered regularly interspaced palindromic repeats |
| CRISPR-Dx | CRISPR diagnostics |
| crRNA | CRISPR RNA |
| dCas9 | Nuclease-deficient Cas9 |
| ddPCR | Digital droplet PCR |
| DETECTR | DNA endonuclease-targeted CRISPR trans reporter |
| DNA | Deoxyribonucleic acid |
| dPCR | Digital polymerase chain reaction |
| dPCR | Digital PCR |
| dsDNA | Double-stranded DNA |
| EBV | Epstein–Barr virus |
| EUA | Emergency Use Authorization |
| EXPAR | Exponential amplification reaction |
| EXP-J | Exponential amplification reaction (Cas12j) |
| FEN1 | Flap endonuclease 1 |
| FLASH | Finding low abundance sequences by hybridization |
| HBV | Hepatitis B virus |
| HCV | Hepatitis C virus |
| HER2 | Tyrosine-protein kinase erbB-2 |
| HOLMES | A one-hour low-cost multipurpose highly efficient system |
| HPV | Human papilloma virus |
| HRP | horseradish peroxidase |
| HUDSON | Heating unextracted diagnostic samples to obliterate nucleases |
| KLK3 | Kallikrein related peptidase 3 |
| LAMP | Loop-mediated isothermal amplification |
| LEOPARD | Leveraging engineered tracrRNAs and on-target DNAs for parallel RNA detection |
| LOD | Limit of detection |
| miRNA | Micro RNA |
| mRNA | Messenger RNA |
| NAA | Nucleic acid amplification |
| NASBACC | Nucleic acid sequence-based amplification CRISPR |
| NGS | Next-generation sequencing |
| NSCLC | Non-small-cell lung cancer |
| PAM | Protospacer adjacent motif |
| PC | Paired dCas9 |
| PCA3 | Prostate cancer antigen 3 |
| PCR | Polymerase chain reaction |
| PFS | Protospacer flanking site |
| PSA | Prostate-specific antigen |
| qPCR | Quantitative polymerase chain reaction |
| RAT | Rapid antigen test |
| RCA | Rolling circle amplification |
| RNA | Ribonucleic acid |
| RPA | Recombinase polymerase amplification |
| Rptr | Reprogrammed tracrRNA |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| SHERLOCK | Specific high-sensitivity enzymatic reporter unlocking |
| ssDNA | Single-stranded DNA |
| tracrRNA | Trans-activating CRISPR RNA |
| WT | Wild-type |
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthi, B.V.S.K.; Nepal, S.; Varambally, S. Genomic and epigenomic alterations in cancer. Am. J. Pathol. 2016, 186, 1724–1735. [Google Scholar] [CrossRef] [PubMed]
- Richards, M.A.; Hiom, S.; Hamilton, W. Diagnosing cancer earlier: What progress is being made? Br. J. Cancer 2023, 128, 441–442. [Google Scholar] [CrossRef]
- Rituraj; Pal, R.S.; Wahlang, J.; Pal, Y.; Chaitanya, M.; Saxena, S. Precision oncology: Transforming cancer care through personalized medicine. Med. Oncol. 2025, 42, 246. [Google Scholar] [CrossRef]
- Australian Institute of Health and Welfare Cancer Data in Australia. Available online: https://www.aihw.gov.au/reports/cancer/cancer-data-in-australia/contents/cancer-incidence-and-survival-by-stage-data-visual (accessed on 12 August 2025).
- Borrero, L.J.H.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188556. [Google Scholar] [CrossRef]
- Cocco, E.; Lopez, S.; Santin, A.D.; Scaltriti, M. Prevalence and role of HER2 mutations in cancer. Pharmacol. Ther. 2019, 199, 188–196. [Google Scholar] [CrossRef]
- Jin, X.; Zhou, Y.-F.; Ma, D.; Zhao, S.; Lin, C.-J.; Xiao, Y.; Fu, T.; Liu, C.-L.; Chen, Y.-Y.; Xiao, W.-X.; et al. Molecular classification of hormone receptor-positive HER2-negative breast cancer. Nat. Genet. 2023, 55, 1696–1708. [Google Scholar] [CrossRef]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef]
- Ashworth, T.R. A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Australas. Med. J. 1869, 14, 146–149. [Google Scholar]
- Mandel, P.; Metais, P. Les acides nucléiques du plasma sanguin chez l’homme. CR Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Christie, E.L.; Dawson, S.-J.; Bowtell, D.D.L. Blood worth bottling: Circulating tumor DNA as a cancer biomarker. Cancer Res. 2016, 76, 5590–5591. [Google Scholar] [CrossRef]
- Neal, R.D.; Tharmanathan, P.; France, B.; Din, N.U.; Cotton, S.; Fallon-Ferguson, J.; Hamilton, W.; Hendry, A.; Hendry, M.; Lewis, R.; et al. Is increased time to diagnosis and treatment in symptomatic cancer associated with poorer outcomes? Systematic review. Br. J. Cancer 2015, 112 (Suppl. 1), S92–S107. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, K. Earlier diagnosis: The importance of cancer symptoms. Lancet Oncol. 2020, 21, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Waarts, M.R.; Stonestrom, A.J.; Park, Y.C.; Levine, R.L. Targeting mutations in cancer. J. Clin. Investig. 2022, 132, e154943. [Google Scholar] [CrossRef] [PubMed]
- Dawson, S. Characterizing the cancer genome in blood. Cold Spring Harb. Perspect. Med. 2019, 9, a026880. [Google Scholar] [CrossRef]
- Low, S.J.; O’Neill, M.T.; Kerry, W.J.; Krysiak, M.; Papadakis, G.; Whitehead, L.W.; Savic, I.; Prestedge, J.; Williams, L.; Cooney, J.P.; et al. Rapid detection of monkeypox virus using a CRISPR-Cas12a mediated assay: A laboratory validation and evaluation study. Lancet Microbe 2023, 4, e800–e810. [Google Scholar] [CrossRef]
- Puig-Serra, P.; Casado-Rosas, M.C.; Martinez-Lage, M.; Olalla-Sastre, B.; Alonso-Yanez, A.; Torres-Ruiz, R.; Rodriguez-Perales, S. CRISPR approaches for the diagnosis of human diseases. Int. J. Mol. Sci. 2022, 23, 1757. [Google Scholar] [CrossRef]
- Land, K.J.; Boeras, D.I.; Chen, X.-S.; Ramsay, A.R.; Peeling, R.W. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 2019, 4, 46–54. [Google Scholar] [CrossRef]
- Low, S.J.; O’Neill, M.; Kerry, W.J.; Wild, N.; Krysiak, M.; Nong, Y.; Azzato, F.; Hor, E.; Williams, L.; Taiaroa, G.; et al. PathoGD: An integrative genomics approach to primer and guide RNA design for CRISPR-based diagnostics. Commun. Biol. 2025, 8, 147. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Al-Shayeb, B.; Sachdeva, R.; Chen, L.-X.; Ward, F.; Munk, P.; Devoto, A.; Castelle, C.J.; Olm, M.R.; Bouma-Gregson, K.; Amano, Y.; et al. Clades of huge phages from across Earth’s ecosystems. Nature 2020, 578, 425–431. [Google Scholar] [CrossRef]
- Mali, P.; Esvelt, K.M.; Church, G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods 2013, 10, 957–963. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, J.E.; Segerink, L.I. Building the future of clinical diagnostics: An analysis of potential benefits and current barriers in CRISPR/Cas diagnostics. ACS Synth. Biol. 2025, 14, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-Espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; Doudna, J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 2018, 362, 839–842. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-E.; Kim, H.; Lee, Y.-H.; Lee, H.-Y.; Park, Y.; Jang, H.; Kim, J.-R.; Lee, M.-Y.; Jeong, B.-H.; Byun, J.-Y.; et al. Unveiling Cas12j trans-cleavage activity for CRISPR diagnostics: Application to miRNA detection in lung cancer diagnosis. Adv. Sci. 2024, 11, e2402580. [Google Scholar] [CrossRef]
- Katti, A.; Diaz, B.J.; Caragine, C.M.; Sanjana, N.E.; Dow, L.E. CRISPR in cancer biology and therapy. Nat. Rev. Cancer 2022, 22, 259–279. [Google Scholar] [CrossRef]
- Redman, M.; King, A.; Watson, C.; King, D. What is CRISPR/Cas9? Arch. Dis. Child. Educ. Pract. Ed. 2016, 101, 213–215. [Google Scholar] [CrossRef]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef]
- Huang, M.; Zhou, X.; Wang, H.; Xing, D. Clustered regularly interspaced short palindromic repeats/Cas9 triggered isothermal amplification for site-specific nucleic acid detection. Anal. Chem. 2018, 90, 2193–2200. [Google Scholar] [CrossRef]
- Quan, J.; Langelier, C.; Kuchta, A.; Batson, J.; Teyssier, N.; Lyden, A.; Caldera, S.; McGeever, A.; Dimitrov, B.; King, R.; et al. FLASH: A next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Res. 2019, 47, e83. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.; Sharma, S.; Dugar, G.; Peeck, N.L.; Bischler, T.; Wimmer, F.; Yu, Y.; Barquist, L.; Schoen, C.; Kurzai, O.; et al. Noncanonical crRNAs derived from host transcripts enable multiplexable RNA detection by Cas9. Science 2021, 372, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qian, L.; Wei, W.; Wang, Y.; Wang, B.; Lin, P.; Liu, W.; Xu, L.; Li, X.; Liu, D.; et al. Paired design of dCas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains. ACS Synth. Biol. 2017, 6, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.-Y.; Zhu, L.-Y.; Zhu, C.-S.; Ma, J.-X.; Hou, T.; Wu, X.-M.; Xie, S.-S.; Min, L.; Tan, D.-A.; Zhang, D.-Y.; et al. Highly effective and low-cost microRNA detection with CRISPR-Cas9. ACS Synth. Biol. 2018, 7, 807–813. [Google Scholar] [CrossRef]
- Li, S.; Cheng, Q.; Wang, J.; Li, X.; Zhang, Z.; Gao, S.; Cao, R.; Zhao, G.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Dai, Y.; Somoza, R.A.; Wang, L.; Welter, J.F.; Li, Y.; Caplan, A.I.; Liu, C.C. Exploring the trans-cleavage activity of CRISPR-Cas12a (cpf1) for the development of a universal electrochemical biosensor. Angew. Chem. Int. Ed. 2019, 58, 17399–17405. [Google Scholar] [CrossRef]
- Zhou, H.; Cai, Y.; He, L.; Li, T.; Wang, Z.; Li, L.; Hu, T.; Li, X.; Zhuang, L.; Huang, X.; et al. Phase transition of wax enabling crispr diagnostics for automatic at-home testing of multiple sexually transmitted infection pathogens. Small 2025, 21, e2407931. [Google Scholar] [CrossRef]
- Teng, F.; Guo, L.; Cui, T.; Wang, X.-G.; Xu, K.; Gao, Q.; Zhou, Q.; Li, W. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol. 2019, 20, 132. [Google Scholar] [CrossRef]
- Li, L.; Li, S.; Wu, N.; Wu, J.; Wang, G.; Zhao, G.; Wang, J. HOLMESv2: A CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 2019, 8, 2228–2237. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K.; et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 2015, 60, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Bandaru, S.; Tsuji, M.H.; Shimizu, Y.; Usami, K.; Lee, S.; Takei, N.K.; Yoshitome, K.; Nishimura, Y.; Otsuki, T.; Ito, T. Structure-based design of gRNA for Cas13. Sci. Rep. 2020, 10, 11610. [Google Scholar] [CrossRef]
- Zetsche, B.; Heidenreich, M.; Mohanraju, P.; Fedorova, I.; Kneppers, J.; DeGennaro, E.M.; Winblad, N.; Choudhury, S.R.; Abudayyeh, O.O.; Gootenberg, J.S.; et al. Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat. Biotechnol. 2017, 35, 31–34. [Google Scholar] [CrossRef]
- Li, S.; Cheng, Q.; Liu, J.; Nie, X.; Zhao, G.; Wang, J. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018, 28, 491–493. [Google Scholar] [CrossRef]
- Karvelis, T.; Bigelyte, G.; Young, J.K.; Hou, Z.; Zedaveinyte, R.; Budre, K.; Paulraj, S.; Djukanovic, V.; Gasior, S.; Silanskas, A.; et al. PAM recognition by miniature CRISPR–Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res. 2020, 48, 5016–5023. [Google Scholar] [CrossRef]
- Pausch, P.; Al-Shayeb, B.; Bisom-Rapp, E.; Tsuchida, C.A.; Li, Z.; Cress, B.F.; Knott, G.J.; Jacobsen, S.E.; Banfield, J.F.; Doudna, J.A. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 2020, 369, 333–337. [Google Scholar] [CrossRef]
- Dai, F.; Zhang, T.; Pang, F.; Jiao, T.; Wang, K.; Zhang, Z.; Wang, N.; Xie, Z.; Zhang, Y.; Wang, Z.; et al. A compact, palm-sized isothermal fluorescent diagnostic intelligent IoT device for personal health monitoring and beyond via one-tube/one-step LAMP-CRISPR assay. Biosens. Bioelectron. 2025, 270, 116945. [Google Scholar] [CrossRef]
- Nagpal, M.; Singh, S.; Singh, P.; Chauhan, P.; Zaidi, M.A. Tumor markers: A diagnostic tool. Natl. J. Maxillofac. Surg. 2016, 7, 17–20. [Google Scholar] [CrossRef]
- Bosch, X.; Molina, R.; Marrades, R.; Augé, J.M.; Pellicé, M.; López-Soto, A. Tumour markers with clinically controlled cut-offs for suspected cancer. Eur. J. Clin. Investig. 2021, 51, e13523. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Hsieh, C.-H.; Wen, C.-N.; Wen, Y.-H.; Chen, C.-H.; Lu, J.-J. Cancers screening in an asymptomatic population by using multiple tumour markers. PLoS ONE 2016, 11, e0158285. [Google Scholar] [CrossRef] [PubMed]
- Ossowski, S.; Neeman, E.; Borden, C.; Stram, D.; Giraldo, L.; Kotak, D.; Thomas, S.; Suga, J.M.; Lin, A.; Liu, R. Improving time to molecular testing results in patients with newly diagnosed, metastatic non–small-cell lung cancer. JCO Oncol. Pract. 2022, 18, e1874–e1884. [Google Scholar] [CrossRef]
- IJzerman, M.J.; de Boer, J.; Azad, A.; Degeling, K.; Geoghegan, J.; Hewitt, C.; Hollande, F.; Lee, B.; To, Y.H.; Tothill, R.W.; et al. Towards routine implementation of liquid biopsies in cancer management: It is always too early, until suddenly it is too late. Diagnostics 2021, 11, 103. [Google Scholar] [CrossRef] [PubMed]
- Yeh, P.; Hunter, T.; Sinha, D.; Ftouni, S.; Wallach, E.; Jiang, D.; Chan, Y.-C.; Wong, S.Q.; Silva, M.J.; Vedururu, R.; et al. Circulating tumour DNA reflects treatment response and clonal evolution in chronic lymphocytic leukaemia. Nat. Commun. 2017, 8, 14756. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, Y.; Dang, L.; Liu, Y.; Huang, S.; Wu, S.; Ma, P.; Jiang, H.; Li, Y.; Pan, Y.; et al. EasyCatch, a convenient, sensitive and specific CRISPR detection system for cancer gene mutations. Mol. Cancer 2021, 20, 157. [Google Scholar] [CrossRef]
- Tsou, J.-H.; Leng, Q.; Jiang, F. A CRISPR Test for detection of circulating nuclei acids. Transl. Oncol. 2019, 12, 1566–1573. [Google Scholar] [CrossRef]
- Chen, W.; Wu, S.; Li, G.; Duan, X.; Sun, X.; Li, S.; Zhao, Y.; Gu, D.; Zeng, G.; Liu, H. Accurate diagnosis of prostate cancer with CRISPR-based nucleic acid test strip by simultaneously identifying PCA3 and KLK3 genes. Biosens. Bioelectron. 2023, 220, 114854. [Google Scholar] [CrossRef] [PubMed]
- Maity, A.; Sathyanarayanan, A.; Kumar, R.; Vora, J.; Gawde, J.; Jain, H.; Bagal, B.; Subramanian, P.G.; Sengar, M.; Khattry, N.; et al. RAPID-CRISPR: Highly sensitive diagnostic assay for detection of PML::RARA isoforms in acute promyelocytic leukemia. Blood Adv. 2025, 9, 463–472. [Google Scholar] [CrossRef]
- Inoue, J.; Johji, I. Cancer-associated miRNAs and their therapeutic potential. J. Hum. Genet. 2021, 66, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhou, F.; Yang, D.; Chen, Y.; Li, M.; Wang, P. miRNA detection for prostate cancer diagnosis by miRoll-Cas: miRNA rolling circle transcription for CRISPR-Cas assay. Anal. Chem. 2023, 95, 13220–13226. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Tang, J.; Tan, Q.; Xie, X.; Zhao, X.; Xing, D. CRISPR/Cas13a-triggered Cas12a biosensing method for ultrasensitive and specific miRNA detection. Talanta 2023, 260, 124582. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Wan, Y.; Li, M.; Xu, J.; Wang, Q.; Wu, D. Stimuli-responsive incremental DNA machine auto-catalyzed CRISPR-Cas12a feedback amplification permits ultrasensitive molecular diagnosis of esophageal cancer-related microRNA. Talanta 2024, 271, 125675. [Google Scholar] [CrossRef]
- Boulias, K.; Greer, E.L. Means, mechanisms and consequences of adenine methylation in DNA. Nat. Rev. Genet. 2022, 23, 411–428. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Swarts, D.C. Making the cut(s): How Cas12a cleaves target and non-target DNA. Biochem. Soc. Trans. 2019, 47, 1499–1510. [Google Scholar] [CrossRef]
- van Dongen, J.E.; Berendsen, J.T.W.; Eijkel, J.C.T.; Segerink, L.I. A CRISPR/Cas12a-assisted in vitro diagnostic tool for identification and quantification of single CpG methylation sites. Biosens. Bioelectron. 2021, 194, 113624. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Y.; Zhang, Y.; Zhong, Q.; Zhu, X.; Wu, Q. A functionalized magnetic nanoparticle regulated CRISPR-Cas12a sensor for the ultrasensitive detection of alpha-fetoprotein. Analyst 2022, 147, 3186–3192. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Lu, Z.; Huang, Q.; Li, H.; Wang, Y.; Lai, Y.; He, Y.; Deng, M.; Liu, W. An ultrasensitive hybridization chain reaction-amplified CRISPR-Cas12a aptasensor for extracellular vesicle surface protein quantification. Theranostics 2020, 10, 10262–10273. [Google Scholar] [CrossRef]
- Guan, X.; Zhao, J.; Sha, Z.; Liang, Y.; Huang, J.; Zhang, J.; Sun, S. CRISPR/Cas12a and aptamer-chemiluminescence based analysis for the relative abundance determination of tumor-related protein positive exosomes for breast cancer diagnosis. Biosens. Bioelectron. 2024, 259, 116380. [Google Scholar] [CrossRef]
- Song, S.-F.; Zhang, X.-W.; Chen, S.; Shu, Y.; Yu, Y.-L.; Wang, J.-H. CRISPR-based dual-aptamer proximity ligation coupled hybridization chain reaction for precise detection of tumor extracellular vesicles and cancer diagnosis. Talanta 2024, 280, 126780. [Google Scholar] [CrossRef]
- Tan, W.; Zhang, C.; Cheng, S.; Hu, X.; Wang, M.; Xian, Y. DNA gate-based CRISPR-Cas exponential amplification system for ultrasensitive small extracellular vesicle detection to enhance breast cancer diagnosis. Anal. Chem. 2024, 96, 1328–1335. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Wu, Y.; Liu, L.-E.; He, L.; Yu, S.; Effah, C.Y.; Liu, X.; Qu, L.; Wu, Y. Universal DNAzyme walkers-triggered CRISPR-Cas12a/Cas13a bioassay for the synchronous detection of two exosomal proteins and its application in intelligent diagnosis of cancer. Biosens. Bioelectron. 2023, 219, 114827. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zhao, W.; Chen, D.; Ren, D.; Qian, T.; Xia, X.; Wang, X.; Li, Q.; Yang, J.; Gu, Y.; et al. Ultrasensitive detection of FEN1 activity for cancer diagnosis using a CRISPR/Cas13a-based triple cascade amplification system. Adv. Healthc. Mater. 2025, 14, e2404411. [Google Scholar] [CrossRef]
- Hao, L.; Zhao, R.T.; Welch, N.L.; Tan, E.K.W.; Zhong, Q.; Harzallah, N.S.; Ngambenjawong, C.; Ko, H.; Fleming, H.E.; Sabeti, P.C.; et al. CRISPR-Cas-amplified urinary biomarkers for multiplexed and portable cancer diagnostics. Nat. Nanotechnol. 2023, 18, 798–807. [Google Scholar] [CrossRef]
- Guo, L.; Zhang, S.; Du, X.; Zhou, M.; Gu, H. Fusing allosteric ribozymes with CRISPR-Cas12a for efficient diagnostics of small molecule targets. Small Methods 2024, 9, e2401236. [Google Scholar] [CrossRef]
- De Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef]
- Damian, D. The role of viruses in cellular transformation and cancer. Cancer Rep. 2025, 8, e70150. [Google Scholar] [CrossRef]
- Wen, Y.; Zhu, Y.; Zhang, C.; Yang, X.; Gao, Y.; Li, M.; Yang, H.; Liu, T.; Tang, H. Chronic inflammation, cancer development and immunotherapy. Front. Pharmacol. 2022, 13, 1040163. [Google Scholar] [CrossRef]
- Bavor, C.; Brotherton, J.M.; Smith, M.A.; Prang, K.-H.; McDermott, T.; Rankin, N.M.; Zammit, C.M.; Jennett, C.J.; Sultana, F.; Machalek, D.A.; et al. The early impacts of primary HPV cervical screening implementation in Australia on the pathology sector: A qualitative study. BMC Health Serv. Res. 2023, 23, 1073. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liu, S.-X.; Wang, F.; Zeng, M.-S. Room temperature detection of plasma Epstein-Barr virus DNA with CRISPR-Cas13. Clin. Chem. 2019, 65, 591–592. [Google Scholar] [CrossRef]
- Jiang, C.; Zheng, X.; Lin, L.; Li, X.; Li, X.; Liao, Y.; Jia, W.; Shu, B. CRISPR Cas12a-mediated amplification-free digital DNA assay improves the diagnosis and surveillance of nasopharyngeal carcinoma. Biosens. Bioelectron. 2023, 237, 115546. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, S.R.; Wilson, D.F. Evidence of Epstein-Barr virus association with head and neck cancers: A review. J. Can. Dent. Assoc. 2016, 82, 1–11. [Google Scholar]
- Carpén, T.; Syrjänen, S.; Jouhi, L.; Randen-Brady, R.; Haglund, C.; Mäkitie, A.; Mattila, P.S.; Hagström, J. Epstein–Barr virus (EBV) and polyomaviruses are detectable in oropharyngeal cancer and EBV may have prognostic impact. Cancer Immunol. Immunother. 2020, 69, 1615–1626. [Google Scholar] [CrossRef]
- Wang, S.; Shen, X.; Chen, G.; Zhang, W.; Tan, B. Application and development of CRISPR-Cas12a methods for the molecular diagnosis of cancer: A review. Anal. Chim. Acta 2025, 1341, 343603. [Google Scholar] [CrossRef] [PubMed]
- Pormohammad, A.; Mohtavinejad, N.; Gholizadeh, P.; Dabiri, H.; Salimi Chirani, A.; Hashemi, A.; Nasiri, M.J. Global estimate of gastric cancer in Helicobacter pylori–infected population: A systematic review and meta-analysis. J. Cell Physiol. 2019, 234, 1208–1218. [Google Scholar] [CrossRef]
- Bernardo, C.; Cunha, M.C.; Santos, J.H.; Da Costa, J.M.C.; Brindley, P.J.; Lopes, C.; Amado, F.; Ferreira, R.; Vitorino, R.; Santos, L.L. Insight into the molecular basis of Schistosoma haematobium-induced bladder cancer through urine proteomics. Tumor Biol. 2016, 37, 11279–11287. [Google Scholar] [CrossRef]
- Forootan, A.; Sjöback, R.; Björkman, J.; Sjögreen, B.; Linz, L.; Kubista, M. Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR). Biomol. Detect. Quantif. 2017, 12, 1–6. [Google Scholar] [CrossRef]
- Deprez, L.; Corbisier, P.; Kortekaas, A.-M.; Mazoua, S.; Beaz Hidalgo, R.; Trapmann, S.; Emons, H. Validation of a digital PCR method for quantification of DNA copy number concentrations by using a certified reference material. Biomol. Detect. Quantif. 2016, 9, 29–39. [Google Scholar] [CrossRef]
- The Global Fund New Pricing for Cepheid GeneXpert Tuberculosis Testing. Available online: https://www.theglobalfund.org/media/13442/operational_2023-10-cepheid-genexpert-tb-testing_briefingnote_en.pdf (accessed on 9 September 2025).
- Perez-Toralla, K.; Pereiro, I.; Garrigou, S.; Di Federico, F.; Proudhon, C.; Bidard, F.-C.; Viovy, J.-L.; Taly, V.; Descroix, S. Microfluidic extraction and digital quantification of circulating cell-free DNA from serum. Sens. Actuators B Chem. 2019, 286, 533–539. [Google Scholar] [CrossRef]
- Hou, Y.; Chen, S.; Zheng, Y.; Zheng, X.; Lin, J.-M. Droplet-based digital PCR (ddPCR) and its applications. Trends Anal. Chem. 2023, 158, 116897. [Google Scholar] [CrossRef]
- Australian Institute of Health and Welfare Rural and Remote Health. Available online: https://www.aihw.gov.au/reports/rural-remote-australians/rural-and-remote-health (accessed on 10 July 2025).
- Trenti, T. Synergy between point-of-care testing and laboratory consolidations. Electron. J. Int. Fed. Clin. Chem. Lab. Med. 2021, 32, 328–336. [Google Scholar]
- Cingam, S.R.; Koshy, N.V. Acute Promyelocytic Leukemia. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Centers for Disease Control and Prevention Malaria Diagnostic Tests. Available online: https://www.cdc.gov/malaria/hcp/diagnosis-testing/malaria-diagnostic-tests.html (accessed on 9 September 2025).
- Ghouneimy, A.; Mahas, A.; Marsic, T.; Aman, R.; Mahfouz, M. CRISPR-based diagnostics: Challenges and potential solutions toward point-of-care applications. ACS Synth. Biol. 2023, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- United States Food and Drug Administration. DETECTR BOOST SARS-CoV-2 Reagent Kit; United States Food and Drug Administration: Silver Spring, MD, USA, 2022.
- United States Food and Drug Administration. SherlockTM CRISPR SARS-CoV-2 Kit; United States Food and Drug Administration: Silver Spring, MD, USA, 2022.
- Hu, F.; Zhang, Y.; Yang, Y.; Peng, L.; Cui, S.; Ma, Q.; Wang, F.; Wang, X. A rapid and ultrasensitive RPA-assisted CRISPR-Cas12a/Cas13a nucleic acid diagnostic platform with a smartphone-based portable device. Biosens. Bioelectron. 2025, 280, 117428. [Google Scholar] [CrossRef]
- Ibrahim, A.U.; Al-Turjman, F.; Sa’id, Z.; Ozsoz, M. Futuristic CRISPR-based biosensing in the cloud and internet of things era: An overview. Multimed. Tools Appl. 2022, 81, 35143–35171. [Google Scholar] [CrossRef] [PubMed]
- Shokr, A.; Pacheco, L.G.C.; Thirumalaraju, P.; Kanakasabapathy, M.K.; Gandhi, J.; Kartik, D.; Silva, F.S.R.; Erdogmus, E.; Kandula, H.; Luo, S.; et al. Mobile health (mHealth) viral diagnostics enabled with adaptive adversarial learning. ACS Nano 2021, 15, 665–673. [Google Scholar] [CrossRef]
- Ansari, M.A.; Verma, D.; Hamizan, M.-A.; Mukherjee, M.D.; Mohd-Naim, N.F.; Ahmed, M.U. Trends in aptasensing and the enhancement of diagnostic efficiency and accuracy. ACS Synth. Biol. 2025, 14, 21–40. [Google Scholar] [CrossRef]
- Futane, A.; Jadhav, P.; Mustafa, A.H.; Srinivasan, A.; Narayanamurthy, V. Aptamer-functionalized MOFs and AI-driven strategies for early cancer diagnosis and therapeutics. Biotechnol. Lett. 2024, 46, 1–17. [Google Scholar] [CrossRef] [PubMed]


| CRISPR-Dx Platform | Year | Cas | Primary Target 1 | NAA 2 | Multiplexing | Readout |
|---|---|---|---|---|---|---|
| NASBACC [31] | 2016 | Cas9 | RNA | NASBA 3 | No | Colourimetric |
| CAS-EXPAR [32] | 2018 | Cas9 | ssDNA | EXPAR 4 | No | Fluorescence |
| FLASH [33] | 2019 | Cas9 | dsDNA | PCR 5 | Yes | Illumina sequencing |
| LEOPARD [34] | 2021 | Cas9 | RNA | PCR | Yes | Gel Electrophoresis |
| PC reporter [35] | 2017 | dCas9 | dsDNA | PCR | No | Luminescence |
| RCA-CRISPR-split-HRP [36] | 2018 | dCas9 | miRNA | RCA 6 | No | Colourimetric |
| HOLMES [37] | 2018 | Cas12a | DNA | PCR | No | Fluorescence |
| DETECTR [38] | 2018 | Cas12a | DNA | RPA 7 | No | Fluorescence |
| E-CRISPR [39] | 2019 | Cas12a | dsDNA | None | No | Electrical |
| Wax-CRISPR [40] | 2025 | Cas12a | dsDNA | RPA | Yes | Fluorescence |
| CDetection [41] | 2019 | Cas12b | DNA | RPA | No | Fluorescence |
| HOLMESv2 [42] | 2019 | Cas12b | DNA | LAMP 8 | No | Fluorescence |
| SHERLOCK [43] | 2017 | Cas13a | RNA | PCR | No | Fluorescence |
| HUDSON-SHERLOCK [44] | 2018 | Cas13a | RNA | RPA | No | Lateral flow strip |
| SHERLOCKv2 [45] | 2018 | Cas13a Cas13b Cas12a | RNA | RPA | Limited | Lateral flow strip |
| Cas14-DETECTR [27] | 2018 | Cas14a | ssDNA | PCR | No | Fluorescence |
| EXP-J [28] | 2024 | CasΦ | DNA | EXPAR | No | Fluorescence |
| Cas Protein | Cas9 | Cas12 | Cas13 | Cas14 | CasΦ |
|---|---|---|---|---|---|
| Figure |  |  |  | 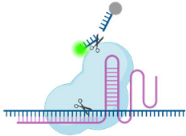 |  |
| Example platform | LEOPARD [34] | DETECTR [38] | SHERLOCK [43] | Cas14- DETECTR [27] | EXP-J [28] |
| Target | dsDNA | dsDNA | RNA | ss/dsDNA | ss/dsDNA |
| tracrRNA required | Yes | No | No | Yes | No |
| PAM 3/PFS 4 restricted | PAM (NGG) 1 | PAM (TTTV) 1 | PFS (H) 1 | None/PAM (TTTA) 1,2 | PAM (AAA) 1 |
| Trans-cleavage | No | ssDNA | RNA | ssDNA | ssDNA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Slattery, J.R.; Naung, N.Y.; Kalinna, B.H.; Pal, M. CRISPR-Powered Liquid Biopsies in Cancer Diagnostics. Cells 2025, 14, 1539. https://doi.org/10.3390/cells14191539
Slattery JR, Naung NY, Kalinna BH, Pal M. CRISPR-Powered Liquid Biopsies in Cancer Diagnostics. Cells. 2025; 14(19):1539. https://doi.org/10.3390/cells14191539
Chicago/Turabian StyleSlattery, Joshua R., Noel Ye Naung, Bernd H. Kalinna, and Martin Pal. 2025. "CRISPR-Powered Liquid Biopsies in Cancer Diagnostics" Cells 14, no. 19: 1539. https://doi.org/10.3390/cells14191539
APA StyleSlattery, J. R., Naung, N. Y., Kalinna, B. H., & Pal, M. (2025). CRISPR-Powered Liquid Biopsies in Cancer Diagnostics. Cells, 14(19), 1539. https://doi.org/10.3390/cells14191539







