Tumor Tissue Microbiota in Colorectal Cancer: PCR Profile of FFPE Blocks in Associations with Metastatic Status
Abstract
1. Introduction
2. Materials and Methods
2.1. Characteristics of Patients with Colorectal Cancer (CRC)
2.2. Assessment of Quantitative and Qualitative Composition of the Microbiota by PCR
2.3. Statistical Data Analysis
2.4. Ethical Approval and Informed Consent Statements
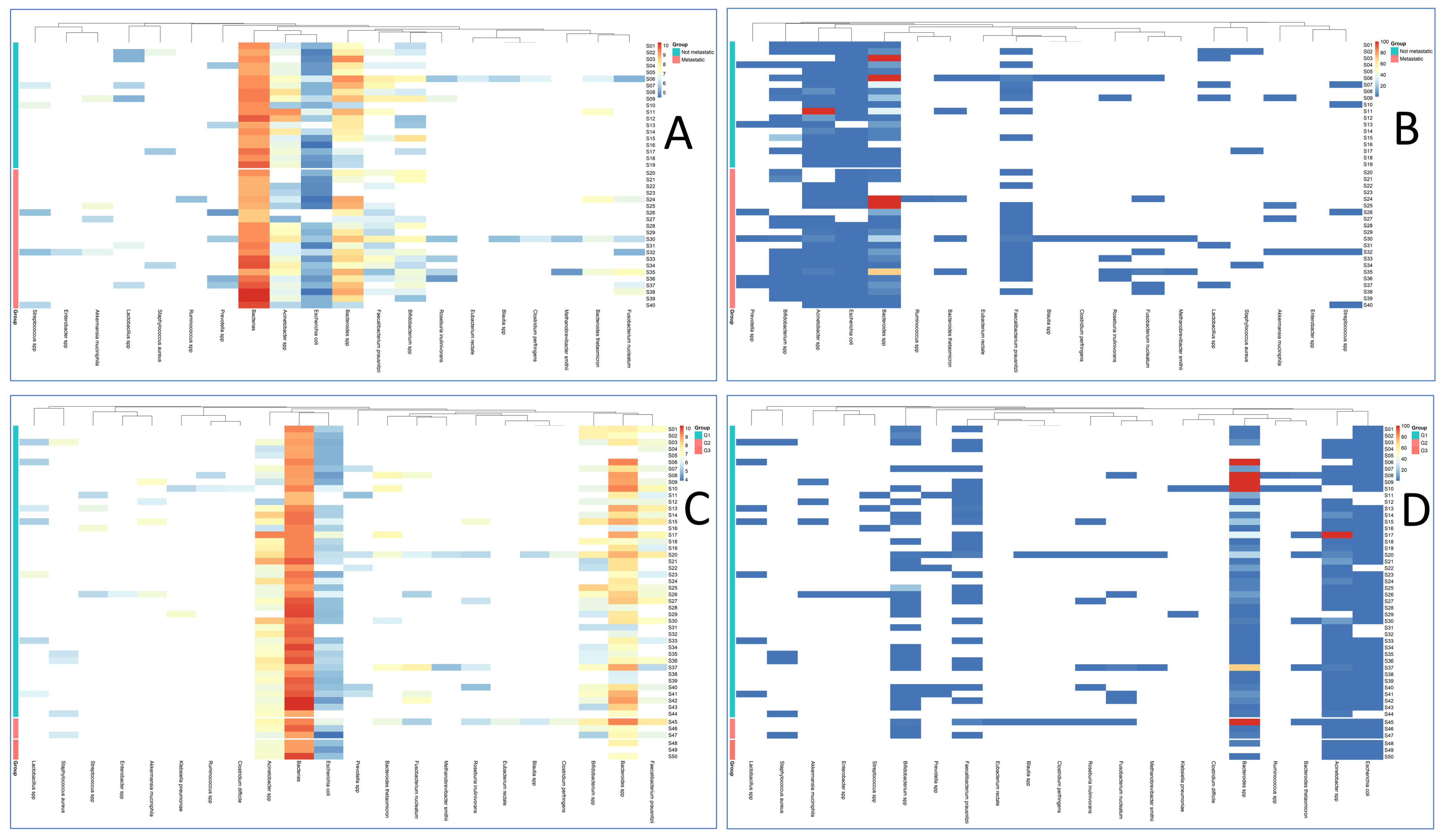
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AHR | Aryl hydrocarbon receptor (AHR/β-catenin signaling) |
| AKT | AKT serine/threonine kinase (PI3K–AKT pathway) |
| AUC | Area under the ROC curve |
| CRC | Colorectal cancer |
| CV | Cross-validation (five-fold, in LASSO) |
| FDR | False discovery rate (Benjamini–Hochberg control) |
| FFPE | Formalin-fixed paraffin-embedded (tumor tissue). |
| IHC | Immunohistochemistry |
| IQR | Interquartile range |
| LASSO | Least Absolute Shrinkage and Selection Operator (L1-regularized logistic regression) |
| Lg | Decimal (base-10) logarithm (log10) |
| MAPK | Mitogen-activated protein kinase (pathway) |
| MMR | Mismatch repair (tumor MMR status by IHC) |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| PCR | Polymerase chain reaction. |
| PI3K | Phosphoinositide 3-kinase (PI3K–AKT pathway) |
| ROC | Receiver operating characteristic (curve) analysis |
| SD | Standard deviation. |
| TNM | Tumor–Node–Metastasis classification (8th ed., 2017) |
| U-test | Mann–Whitney U test (nonparametric pairwise test) |
| χ2 | Chi-squared statistic/test (Pearson’s χ2) |
References
- Roshandel, G.; Ghasemi-Kebria, F.; Malekzadeh, R. Colorectal Cancer: Epidemiology, Risk Factors, and Prevention. Cancers 2024, 16, 1530. [Google Scholar] [CrossRef] [PubMed]
- Partyka, O.; Pajewska, M.; Czerw, A.; Deptała, A.; Mękal, D.; Sygit, K.; Kowalczyk, D.; Cipora, E.; Kaczmarski, M.; Gazdowicz, L.; et al. Current Progress in Clinical Research in Secondary Prevention and Early Detection of Colorectal Cancer. Cancers 2025, 17, 367. [Google Scholar] [CrossRef] [PubMed]
- Mąkosza, K.M.; Muc-Wierzgoń, M.; Dzięgielewska-Gęsiak, S. Nutrition and Selected Lifestyle Elements as a Tertiary Prevention in Colorectal Cancer Patients. Nutrients 2024, 16, 3129. [Google Scholar] [CrossRef]
- Rebersek, M. Gut microbiome and its role in colorectal cancer. BMC Cancer 2021, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fong, W.; Li, Q.; Yu, J. Gut microbiota modulation: A novel strategy for prevention and treatment of colorectal cancer. Oncogene 2020, 39, 4925–4943. [Google Scholar] [CrossRef]
- Kim, J.; Lee, H.K. Potential Role of the Gut Microbiome in Colorectal Cancer Progression. Front. Immunol. 2022, 12, 807648. [Google Scholar] [CrossRef]
- Shakhpazyan, N.K.; Mikhaleva, L.M.; Bedzhanyan, A.L.; Gioeva, Z.V.; Mikhalev, A.I.; Midiber, K.Y.; Pechnikova, V.V.; Biryukov, A.E. Exploring the Role of the Gut Microbiota in Modulating Colorectal Cancer Immunity. Cells 2024, 13, 1437. [Google Scholar] [CrossRef]
- Shakhpazyan, N.; Mikhaleva, L.; Bedzhanyan, A.; Gioeva, Z.; Mikhalev, A.; Pechnikova, V.; Midiber, K.; Orekhov, A. Altered proinflammatory cytokine response in colorectal cancer patients: Insights into immune dysregulation. Clin. Exp. Morphol. 2024, 13, 29–35. [Google Scholar] [CrossRef]
- Shakhpazyan, N.; Mikhaleva, L.; Midiber, K. The role of myeloid-derived suppressor cells in antitumor immune response. Clin. Exp. Morphol. 2025, 14, 5–15. [Google Scholar] [CrossRef]
- Grellier, N.; Severino, A.; Archilei, S.; Kim, J.; Gasbarrini, A.; Cammarota, G.; Porcari, S.; Benech, N. Gut microbiota in inflammation and colorectal cancer: A potential Toolbox for Clinicians. Best Pract. Res. Clin. Gastroenterol. 2024, 72, 101942. [Google Scholar] [CrossRef] [PubMed]
- Avuthu, N.; Guda, C. Meta-Analysis of Altered Gut Microbiota Reveals Microbial and Metabolic Biomarkers for Colorectal Cancer. Microbiol. Spectr. 2022, 10, e0001322. [Google Scholar] [CrossRef]
- Van-Wehle, T.; Vital, M. Investigating the response of the butyrate production potential to major fibers in dietary intervention studies. npj Biofilms Microbiomes 2024, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Li, F.; Yang, R. The Roles of Gut Microbiota Metabolites in the Occurrence and Development of Colorectal Cancer: Multiple Insights for Potential Clinical Applications. Gastro Hep Adv. 2024, 3, 855–870. [Google Scholar] [CrossRef]
- Wang, Y.; Wan, X.; Wu, X.; Zhang, C.; Liu, J.; Hou, S. Eubacterium rectale contributes to colorectal cancer initiation via promoting colitis. Gut Pathog. 2021, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef]
- Huangfu, S.-C.; Zhang, W.-B.; Zhang, H.-R.; Li, Y.; Zhang, Y.-R.; Nie, J.-L.; Chu, X.-D.; Chen, C.-S.; Jiang, H.-P.; Pan, J.-H. Clinicopathological and prognostic significance of Fusobacterium nucleatum infection in colorectal cancer: A meta-analysis. J. Cancer 2021, 12, 1583–1591. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, Y.; Li, J.; Zhao, L.; Yan, W.; Lin, B.; Guo, X.; Wei, Y. Fusobacterium nucleatum Acts as a Pro-carcinogenic Bacterium in Colorectal Cancer: From Association to Causality. Front. Cell Dev. Biol. 2021, 9, 710165. [Google Scholar] [CrossRef]
- Obuya, S.; Elkholy, A.; Avuthu, N.; Behring, M.; Bajpai, P.; Agarwal, S.; Kim, H.-G.; El-Nikhely, N.; Akinyi, P.; Orwa, J.; et al. A signature of Prevotella copri and Faecalibacterium prausnitzii depletion, and a link with bacterial glutamate degradation in the Kenyan colorectal cancer patients. J. Gastrointest. Oncol. 2022, 13, 2282–2292. [Google Scholar] [CrossRef]
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium prausnitzii: From microbiology to diagnostics and prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef]
- Dikeocha, I.J.; Al-Kabsi, A.M.; Chiu, H.-T.; Alshawsh, M.A. Faecalibacterium prausnitzii Ameliorates Colorectal Tumorigenesis and Suppresses Proliferation of HCT116 Colorectal Cancer Cells. Biomedicines 2022, 10, 1128. [Google Scholar] [CrossRef]
- Giacconi, R.; Donghia, R.; Arborea, G.; Savino, M.T.; Provinciali, M.; Lattanzio, F.; Caponio, G.R.; Coletta, S.; Bianco, A.; Notarnicola, M.; et al. Plasma Bacterial DNA Load as a Potential Biomarker for the Early Detection of Colorectal Cancer: A Case–Control Study. Microorganisms 2023, 11, 2360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ji, Q.; Chen, Q.; Wei, Z.; Liu, S.; Zhang, L.; Zhang, Y.; Li, Z.; Liu, H.; Sui, H. Akkermansia muciniphila inhibits tryptophan metabolism via the AhR/β-catenin signaling pathway to counter the progression of colorectal cancer. Int. J. Biol. Sci. 2023, 19, 4393–4410. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via its FadA Adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Zalila-Kolsi, I.; Dhieb, D.; Osman, H.A.; Mekideche, H. The Gut Microbiota and Colorectal Cancer: Understanding the Link and Exploring Therapeutic Interventions. Biology 2025, 14, 251. [Google Scholar] [CrossRef] [PubMed]
- Bundgaard-Nielsen, C.; Baandrup, U.T.; Nielsen, L.P.; Sørensen, S. The presence of bacteria varies between colorectal adenocarcinomas, precursor lesions and non-malignant tissue. BMC Cancer 2019, 19, 1–13. [Google Scholar] [CrossRef]
- da Costa, C.P.; Vieira, P.; Mendes-Rocha, M.; Pereira-Marques, J.; Ferreira, R.M.; Figueiredo, C. The Tissue-Associated Microbiota in Colorectal Cancer: A Systematic Review. Cancers 2022, 14, 3385. [Google Scholar] [CrossRef]
- Song, D.; Wang, X.; Ma, Y.; Liu, N.-N.; Wang, H. Beneficial insights into postbiotics against colorectal cancer. Front. Nutr. 2023, 10, 1111872. [Google Scholar] [CrossRef]
- Yuan, D.; Tao, Y.; Wang, H.; Wang, J.; Cao, Y.; Cao, W.; Pan, S.; Yu, Z. A comprehensive analysis of the microbiota composition and host driver gene mutations in colorectal cancer. Investig. New Drugs 2022, 40, 884–894. [Google Scholar] [CrossRef]
- Abed, J.; Emgård, J.E.; Zamir, G.; Faroja, M.; Almogy, G.; Grenov, A.; Sol, A.; Naor, R.; Pikarsky, E.; Atlan, K.A.; et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 2016, 20, 215–225. [Google Scholar] [CrossRef]
| Taxon Combinations | Support | Confidence | Lift | p-Value | Empirical p |
|---|---|---|---|---|---|
| Bacteroides spp., Fusobacterium nucleatum | 0.175 | 0.29 | 1.632653 | 0.095 | 0.094 |
| Bacteroides spp., Acinetobacter spp., Fusobacterium nucleatum | 0.175 | 0.29 | 1.632653 | 0.095 | 0.094 |
| Acinetobacter spp., Fusobacterium nucleatum | 0.175 | 0.29 | 1.632653 | 0.095 | 0.094 |
| Escherichia coli, Acinetobacter spp., Fusobacterium nucleatum | 0.175 | 0.29 | 1.63 | 0.095 | 0.094 |
| Fusobacterium nucleatum | 0.175 | 0.29 | 1.63 | 0.095 | 0.094 |
| Escherichia coli, Fusobacterium nucleatum | 0.175 | 0.29 | 1.63 | 0.095 | 0.094 |
| Bacteroides spp., Escherichia coli, Fusobacterium nucleatum | 0.175 | 0.29 | 1.63 | 0.095 | 0.094 |
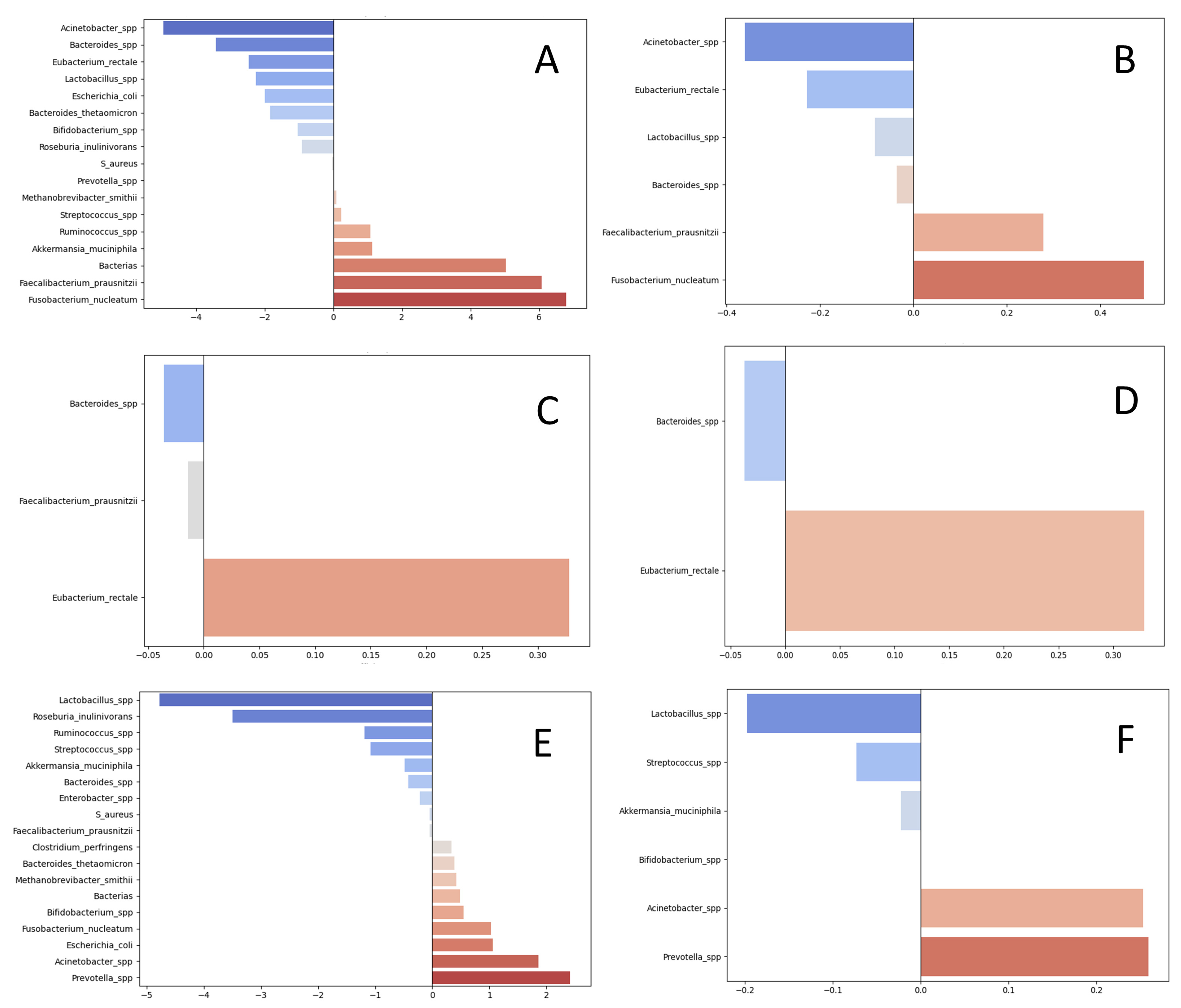
| Groups | Taxons | Regression Coefficients for Absolute Abundances (LASSO) | Regression Coefficients for Relative Measures (LASSO) |
|---|---|---|---|
| pro-metastatic | Fusobacterium nucleatum | 6.79 | 0.49 |
| Faecalibacterium prausnitzii | 6.07 | 0.27 | |
| Total bacterial load | 5.03 | 0 | |
| Akkermansia muciniphila | 1.14 | 0 | |
| Ruminococcus spp. | 1.08 | 0 | |
| Streptococcus spp. | 0.23 | 0 | |
| Methanobrevibacter smithii | 0.09 | 0 | |
| Prevotella spp. | 0.01 | 0 | |
| anti-metastatic | Acinetobacter spp. | −4.95 | −0.36 |
| Bacteroides spp. | −3.43 | −0.04 | |
| Eubacterium rectale | −2.47 | −0.23 | |
| Lactobacillus spp. | −2.27 | −0.08 | |
| Escherichia coli | −2.00 | 0 | |
| Bacteroides thetaomicron | −1.84 | 0 | |
| Bifidobacterium spp. | −1.04 | 0 | |
| Roseburia inulinivorans | −0.91 | 0 | |
| Staphylococcus aureus | −0.03 | 0 | |
| associated with higher grade | Eubacterium rectale | 0.33 | 0.33 |
| associated with lower grade | Bacteroides spp. | −0.04 | −0.04 |
| Faecalibacterium prausnitzii | −0.01 | ||
| associated with mutations | Prevotella spp. | 2.41 | 0.26 |
| Acinetobacter spp. | 1.86 | 0.25 | |
| Escherichia coli | 1.06 | 0 | |
| Fusobacterium nucleatum | 1.03 | 0 | |
| Bifidobacterium spp. | 0.54 | 0 | |
| Total bacterial load | 0.48 | 0 | |
| Methanobrevibacter smithii | 0.42 | 0 | |
| Bacteroides thetaomicron | 0.39 | 0 | |
| Clostridium perfringens | 0.34 | 0 | |
| associated with wild-type status | Lactobacillus_spp. | −4.77 | −0.2 |
| Roseburia_inulinivorans | −3.5 | 0 | |
| Ruminococcus_spp. | −1.19 | 0 | |
| Streptococcus_spp. | −1.08 | −0.07 | |
| Akkermansia_muciniphila | −0.48 | −0.02 | |
| Bacteroides_spp. | −0.42 | 0 | |
| Enterobacter_spp. | −0.22 | 0 | |
| S_aureus | −0.05 | 0 | |
| Faecalibacterium_prausnitzii | −0.04 | 0 |
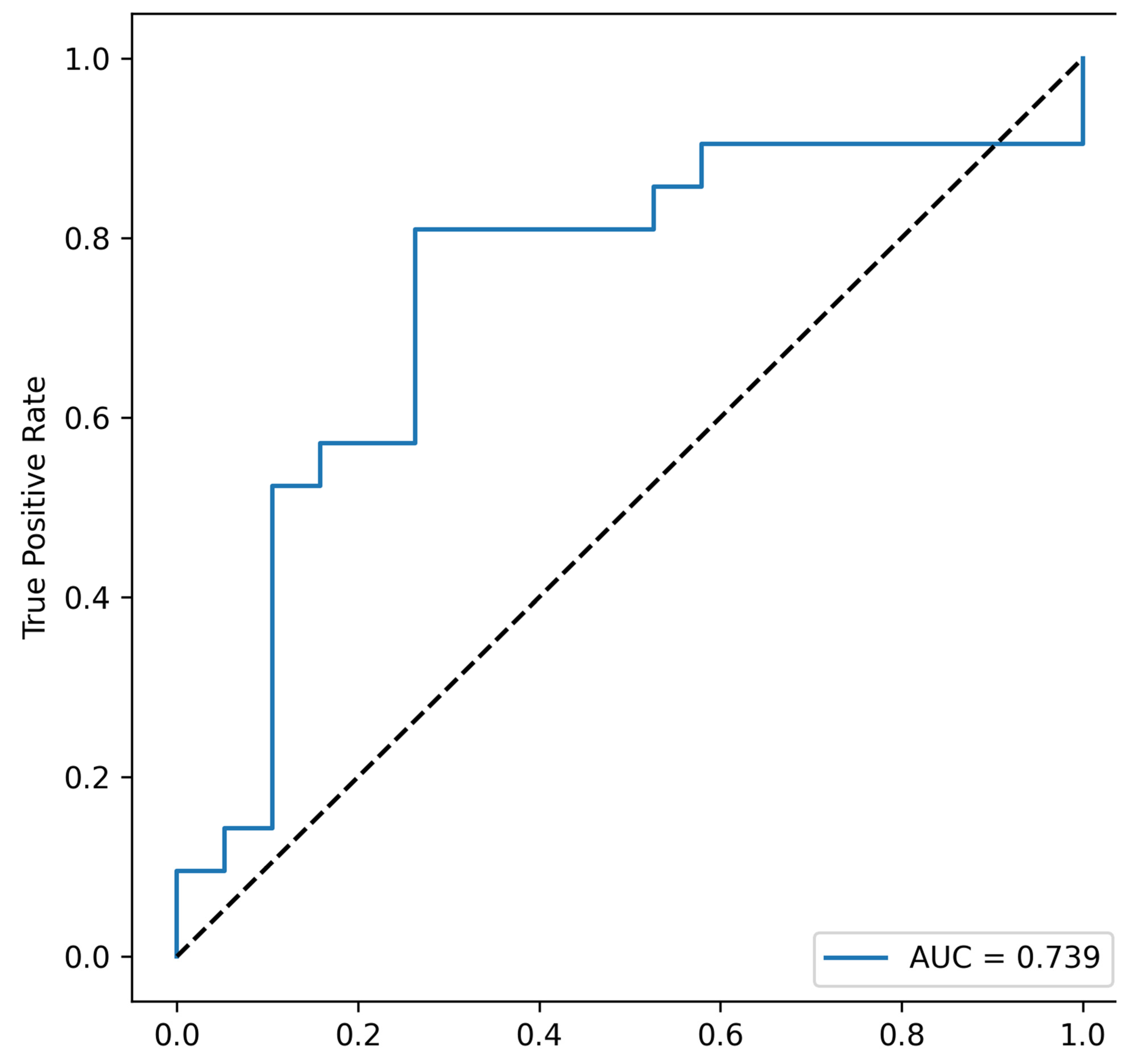
| Combination | AUC (Area Under Curve) | p Value |
|---|---|---|
| Fusobacterium_nucleatum, Faecalibacterium_prausnitzii, Total bacterial load, Akkermansia_muciniphila | 0.739 | 0.047 |
| Fusobacterium_nucleatum, Faecalibacterium_prausnitzii, Total bacterial load, Ruminococcus_spp. | 0.721805 | 0.047 |
| Fusobacterium_nucleatum, Faecalibacterium_prausnitzii, Akkermansia_muciniphila, Ruminococcus_spp. | 0.699248 | 0.047 |
| Fusobacterium_nucleatum, Total bacterial load, Akkermansia_muciniphila, Ruminococcus_spp. | 0.677945 | 0.047 |
| Fusobacterium_nucleatum, Faecalibacterium_prausnitzii, Total bacterial load | 0.709273 | 0.047 |
| Fusobacterium_nucleatum, Faecalibacterium_prausnitzii, Akkermansia_muciniphila | 0.694236 | 0.047 |
| Fusobacterium_nucleatum, Faecalibacterium_prausnitzii, Ruminococcus_spp. | 0.674185 | 0.047 |
| Fusobacterium_nucleatum, Total bacterial load, Akkermansia_muciniphila | 0.672932 | 0.047 |
| Fusobacterium_nucleatum, Total bacterial load, Ruminococcus_spp. | 0.610276 | 0.047 |
| Fusobacterium_nucleatum, Akkermansia_muciniphila, Ruminococcus_spp. | 0.645363 | 0.047 |
| Fusobacterium_nucleatum, Faecalibacterium_prausnitzii | 0.671679 | 0.047 |
| Fusobacterium_nucleatum, Total bacterial load | 0.607769 | 0.047 |
| Fusobacterium_nucleatum, Akkermansia_muciniphila | 0.642857 | 0.047 |
| Fusobacterium_nucleatum, Ruminococcus_spp. | 0.622807 | 0.047 |
| Fusobacterium_nucleatum | 0.622807 | 0.047 |
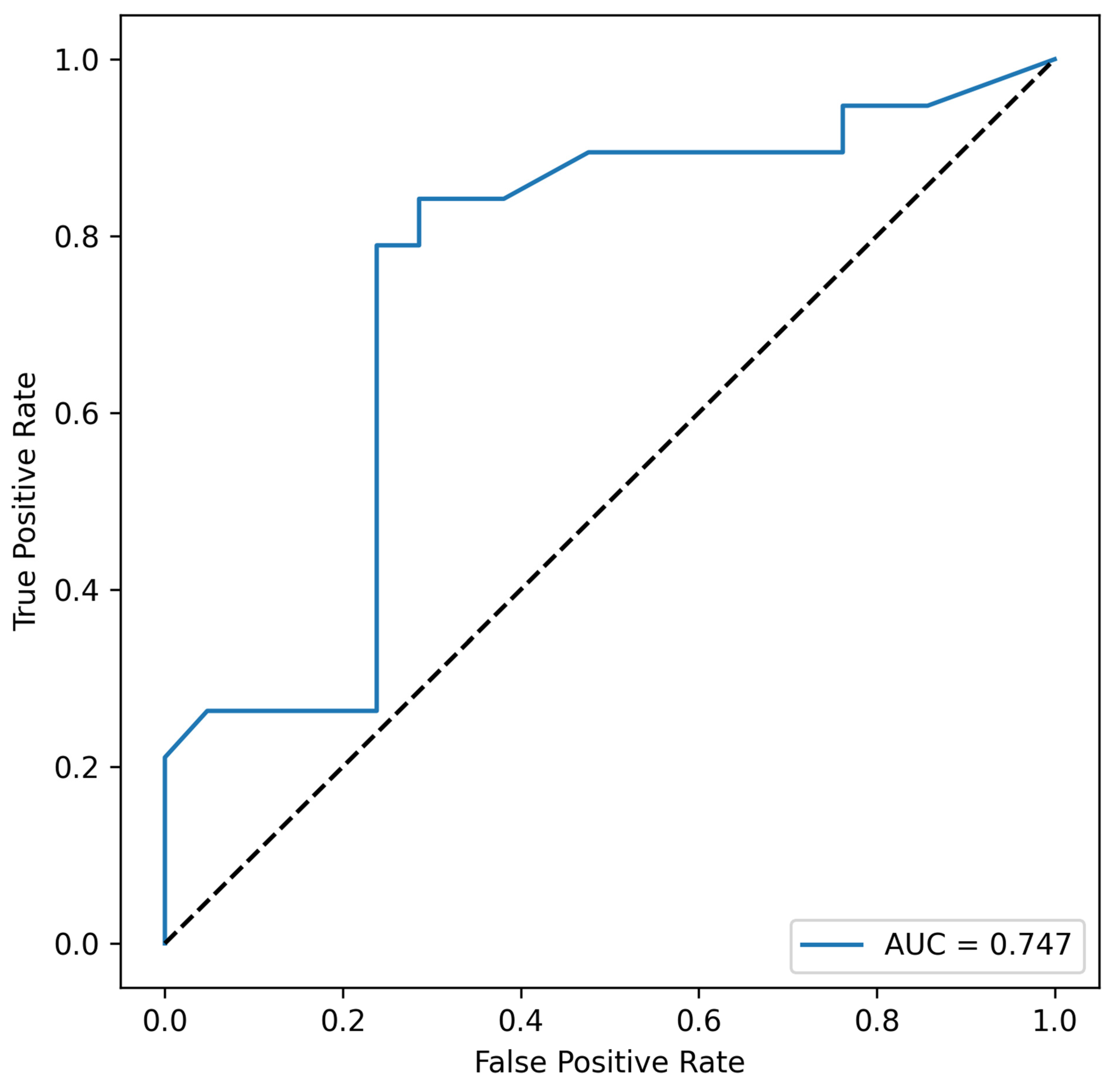
| Combination | AUC | min_p_Value |
|---|---|---|
| Eubacterium_rectale, Acinetobacter_spp. | 0.747 | 0.0104 |
| Acinetobacter_spp. | 0.738 | 0.0104 |
| Lactobacillus_spp., Eubacterium_rectale, Acinetobacter_spp. | 0.724 | 0.0104 |
| Lactobacillus_spp., Acinetobacter_spp. | 0.711 | 0.0104 |
| Bacteroides_spp., Lactobacillus_spp., Eubacterium_rectale, Acinetobacter_spp. | 0.642 | 0.0104 |
| Bacteroides_spp., Eubacterium_rectale, Acinetobacter_spp. | 0.634 | 0.0104 |
| Bacteroides_spp., Lactobacillus_spp., Acinetobacter_spp. | 0.632 | 0.0104 |
| Bacteroides_spp., Acinetobacter_spp. | 0.629 | 0.0104 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shakhpazyan, N.K.; Mikhaleva, L.M.; Sadykhov, N.K.; Midiber, K.Y.; Buchaka, A.S.; Gioeva, Z.V.; Mikhalev, A.I.; Bedzhanyan, A.L. Tumor Tissue Microbiota in Colorectal Cancer: PCR Profile of FFPE Blocks in Associations with Metastatic Status. Cells 2025, 14, 1508. https://doi.org/10.3390/cells14191508
Shakhpazyan NK, Mikhaleva LM, Sadykhov NK, Midiber KY, Buchaka AS, Gioeva ZV, Mikhalev AI, Bedzhanyan AL. Tumor Tissue Microbiota in Colorectal Cancer: PCR Profile of FFPE Blocks in Associations with Metastatic Status. Cells. 2025; 14(19):1508. https://doi.org/10.3390/cells14191508
Chicago/Turabian StyleShakhpazyan, Nikolay K., Liudmila M. Mikhaleva, Nikolay K. Sadykhov, Konstantin Y. Midiber, Anton S. Buchaka, Zarina V. Gioeva, Alexander I. Mikhalev, and Arkady L. Bedzhanyan. 2025. "Tumor Tissue Microbiota in Colorectal Cancer: PCR Profile of FFPE Blocks in Associations with Metastatic Status" Cells 14, no. 19: 1508. https://doi.org/10.3390/cells14191508
APA StyleShakhpazyan, N. K., Mikhaleva, L. M., Sadykhov, N. K., Midiber, K. Y., Buchaka, A. S., Gioeva, Z. V., Mikhalev, A. I., & Bedzhanyan, A. L. (2025). Tumor Tissue Microbiota in Colorectal Cancer: PCR Profile of FFPE Blocks in Associations with Metastatic Status. Cells, 14(19), 1508. https://doi.org/10.3390/cells14191508





