Circulating Serum Cell-Free Mitochondrial DNA in Amyotrophic Lateral Sclerosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Standard Protocol Approvals, Registrations, and Patient Consents
2.3. Clinical Measures
2.4. Sample Collection and Laboratory Assays
2.5. DNA Extraction and mtDNA Quantification Through Digital Droplet PCR
2.6. Statistical Methods
3. Results
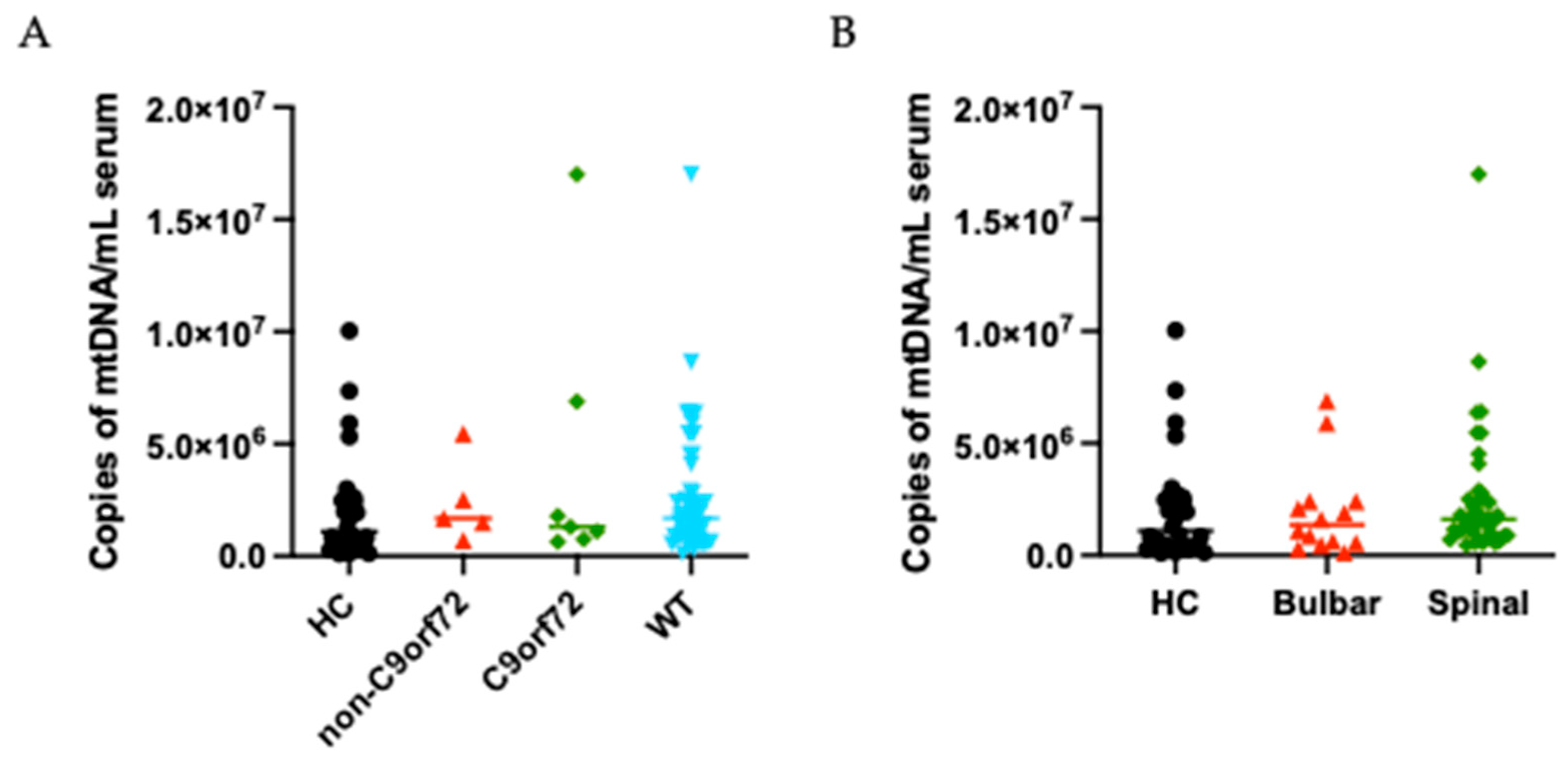
3.1. Biomarkers’ Distribution in ALS Patients and Healthy Controls
3.2. Cf-mtDNA Association with Clinical Indicators of Progression in ALS
3.3. Correlation of Cf-mtDNA with ALS Biomarkers
3.4. Cf-mtDNA and Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| AISI | Aggregate systemic inflammation index |
| ALS | Amyotrophic lateral sclerosis |
| ALS-FRSr | Amyotrophic Lateral Sclerosis Functional Rating Scale—revised |
| AUC | Area under the curve |
| C9ORF72 | Chromosome 9 open reading frame 72 |
| CHI3L1 | Chitinase-3-like protein 1 |
| CI | Confidence interval |
| COPD | Chronic obstructive pulmonary disease |
| DPR | Disease progression rate |
| FTD | Fronto-temporal dementia |
| FVC | Forced vital capacity |
| HC | Healthy controls |
| HR | Hazard ratio |
| IV | Invasive ventilation |
| MLR | Monocyte-to-lymphocyte ratio |
| NF | Neurofilament |
| NfL | Neurofilament light chain |
| NIV | Non-invasive ventilation |
| NLR | Neutrophil-to-lymphocyte ratio |
| PD | Parkinson’s disease |
| PEG | Percutaneous endoscopic gastrostomy |
| pNfH | Neurofilament heavy chain |
| ROC | Receiver operating characteristic |
| SD | Standard deviation |
| SII | Systemic–immune–inflammation index |
| SIRI | Systemic inflammation response index |
| WBC | White blood cell |
References
- Genin, E.C.; Abou-Ali, M.; Paquis-Flucklinger, V. Mitochondria, a Key Target in Amyotrophic Lateral Sclerosis Pathogenesis. Genes 2023, 14, 1981. [Google Scholar] [CrossRef]
- Smith, E.F.; Shaw, P.J.; De Vos, K.J. The Role of Mitochondria in Amyotrophic Lateral Sclerosis. Neurosci. Lett. 2019, 710, 132933. [Google Scholar] [CrossRef]
- Newman, L.E.; Shadel, G.S. Mitochondrial DNA Release in Innate Immune Signaling. Annu. Rev. Biochem. 2023, 92, 299–332. [Google Scholar] [CrossRef]
- Xu, X.; Pang, Y.; Fan, X. Mitochondria in Oxidative Stress, Inflammation and Aging: From Mechanisms to Therapeutic Advances. Signal Transduct. Target. Ther. 2025, 10, 190. [Google Scholar] [CrossRef]
- Gambardella, S.; Limanaqi, F.; Ferese, R.; Biagioni, F.; Campopiano, R.; Centonze, D.; Fornai, F. cCf-mtDNA as a Potential Link Between the Brain and Immune System in Neuro-Immunological Disorders. Front. Immunol. 2019, 10, 1064. [Google Scholar] [CrossRef] [PubMed]
- Pinti, M.; Cevenini, E.; Nasi, M.; De Biasi, S.; Salvioli, S.; Monti, D.; Benatti, S.; Gibellini, L.; Cotichini, R.; Stazi, M.A.; et al. Circulating Mitochondrial DNA Increases with Age and Is a Familiar Trait: Implications for “Inflamm-Aging”. Eur. J. Immunol. 2014, 44, 1552–1562. [Google Scholar] [CrossRef]
- Zanini, G.; Selleri, V.; Lopez Domenech, S.; Malerba, M.; Nasi, M.; Mattioli, A.V.; Pinti, M. Mitochondrial DNA as Inflammatory DAMP: A Warning of an Aging Immune System? Biochem. Soc. Trans. 2023, 51, 735–745. [Google Scholar] [CrossRef]
- De Gaetano, A.; Solodka, K.; Zanini, G.; Selleri, V.; Mattioli, A.V.; Nasi, M.; Pinti, M. Molecular Mechanisms of mtDNA-Mediated Inflammation. Cells 2021, 10, 2898. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhang, H.; Xu, X.; Chen, H.; Qi, B. Association between Circulating Cell-Free Mitochondrial DNA and Inflammation Factors in Noninfectious Diseases: A Systematic Review. PLoS ONE 2024, 19, e0289338. [Google Scholar] [CrossRef] [PubMed]
- Kunze, R.; Fischer, S.; Marti, H.H.; Preissner, K.T. Brain Alarm by Self-Extracellular Nucleic Acids: From Neuroinflammation to Neurodegeneration. J. Biomed. Sci. 2023, 30, 64. [Google Scholar] [CrossRef]
- Giordano, L.; Ware, S.A.; Lagranha, C.J.; Kaufman, B.A. Mitochondrial DNA Signals Driving Immune Responses: Why, How, Where? Cell Commun. Signal. 2025, 23, 192. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.Y.; Yong, Y.K.; Xue, Y.C.; Liu, H.; Furihata, T.; Shankar, E.M.; Ng, C.S. cGAS and DDX41-STING Mediated Intrinsic Immunity Spreads Intercellularly to Promote Neuroinflammation in SOD1 ALS Model. iScience 2022, 25, 104404. [Google Scholar] [CrossRef]
- Yu, C.-H.; Davidson, S.; Harapas, C.R.; Hilton, J.B.; Mlodzianoski, M.J.; Laohamonthonkul, P.; Louis, C.; Low, R.R.J.; Moecking, J.; De Nardo, D.; et al. TDP-43 Triggers Mitochondrial DNA Release via mPTP to Activate cGAS/STING in ALS. Cell 2020, 183, 636–649.e18. [Google Scholar] [CrossRef] [PubMed]
- Park, S.S.; Jeong, H.; Andreazza, A.C. Circulating Cell-Free Mitochondrial DNA in Brain Health and Disease: A Systematic Review and Meta-Analysis. World J. Biol. Psychiatry 2022, 23, 87–102. [Google Scholar] [CrossRef] [PubMed]
- Risi, B.; Imarisio, A.; Cuconato, G.; Padovani, A.; Valente, E.M.; Filosto, M. Mitochondrial DNA (mtDNA) as Fluid Biomarker in Neurodegenerative Disorders: A Systematic Review. Eur. J. Neurol. 2025, 32, e70014. [Google Scholar] [CrossRef]
- Verde, F.; Otto, M.; Silani, V. Neurofilament Light Chain as Biomarker for Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Front. Neurosci. 2021, 15, 679199. [Google Scholar] [CrossRef]
- Poesen, K.; De Schaepdryver, M.; Stubendorff, B.; Gille, B.; Muckova, P.; Wendler, S.; Prell, T.; Ringer, T.M.; Rhode, H.; Stevens, O.; et al. Neurofilament Markers for ALS Correlate with Extent of Upper and Lower Motor Neuron Disease. Neurology 2017, 88, 2302–2309. [Google Scholar] [CrossRef]
- Simonini, C.; Zucchi, E.; Bedin, R.; Martinelli, I.; Gianferrari, G.; Fini, N.; Sorarù, G.; Liguori, R.; Vacchiano, V.; Mandrioli, J. CSF Heavy Neurofilament May Discriminate and Predict Motor Neuron Diseases with Upper Motor Neuron Involvement. Biomedicines 2021, 9, 1623. [Google Scholar] [CrossRef]
- Zucchi, E.; Bonetto, V.; Sorarù, G.; Martinelli, I.; Parchi, P.; Liguori, R.; Mandrioli, J. Neurofilaments in Motor Neuron Disorders: Towards Promising Diagnostic and Prognostic Biomarkers. Mol. Neurodegener. 2020, 15, 58. [Google Scholar] [CrossRef]
- Sturmey, E.; Malaspina, A. Blood Biomarkers in ALS: Challenges, Applications and Novel Frontiers. Acta Neurol. Scand. 2022, 146, 375–388. [Google Scholar] [CrossRef]
- Simonini, C.; Zucchi, E.; Martinelli, I.; Gianferrari, G.; Lunetta, C.; Sorarù, G.; Trojsi, F.; Pepe, R.; Piras, R.; Giacchino, M.; et al. Neurodegenerative and Neuroinflammatory Changes in SOD1-ALS Patients Receiving Tofersen. Sci. Rep. 2025, 15, 11034. [Google Scholar] [CrossRef]
- Brooks, B.R.; Miller, R.G.; Swash, M.; Munsat, T.L.; World Federation of Neurology Research Group on Motor Neuron Diseases. El Escorial Revisited: Revised Criteria for the Diagnosis of Amyotrophic Lateral Sclerosis. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 2000, 1, 293–299. [Google Scholar] [CrossRef]
- Shefner, J.M.; Al-Chalabi, A.; Baker, M.R.; Cui, L.-Y.; de Carvalho, M.; Eisen, A.; Grosskreutz, J.; Hardiman, O.; Henderson, R.; Matamala, J.M.; et al. A Proposal for New Diagnostic Criteria for ALS. Clin. Neurophysiol. 2020, 131, 1975–1978. [Google Scholar] [CrossRef]
- Benatar, M.; Granit, V.; Andersen, P.M.; Grignon, A.-L.; McHutchison, C.; Cosentino, S.; Malaspina, A.; Wuu, J. Mild Motor Impairment as Prodromal State in Amyotrophic Lateral Sclerosis: A New Diagnostic Entity. Brain J. Neurol. 2022, 145, 3500–3508. [Google Scholar] [CrossRef] [PubMed]
- Chio, A.; Calvo, A.; Moglia, C.; Mazzini, L.; Mora, G.; PARALS Study Group. Phenotypic Heterogeneity of Amyotrophic Lateral Sclerosis: A Population Based Study. J. Neurol. Neurosurg. Psychiatry 2011, 82, 740–746. [Google Scholar] [CrossRef]
- Gianferrari, G.; Martinelli, I.; Zucchi, E.; Simonini, C.; Fini, N.; Vinceti, M.; Ferro, S.; Gessani, A.; Canali, E.; Valzania, F.; et al. Epidemiological, Clinical and Genetic Features of ALS in the Last Decade: A Prospective Population-Based Study in the Emilia Romagna Region of Italy. Biomedicines 2022, 10, 819. [Google Scholar] [CrossRef]
- Kimura, F.; Fujimura, C.; Ishida, S.; Nakajima, H.; Furutama, D.; Uehara, H.; Shinoda, K.; Sugino, M.; Hanafusa, T. Progression Rate of ALSFRS-R at Time of Diagnosis Predicts Survival Time in ALS. Neurology 2006, 66, 265–267. [Google Scholar] [CrossRef] [PubMed]
- Mandrioli, J.; Biguzzi, S.; Guidi, C.; Sette, E.; Terlizzi, E.; Ravasio, A.; Casmiro, M.; Salvi, F.; Liguori, R.; Rizzi, R.; et al. Heterogeneity in ALSFRS-R Decline and Survival: A Population-Based Study in Italy. Neurol. Sci. 2015, 36, 2243–2252. [Google Scholar] [CrossRef]
- Martinelli, I.; Zucchi, E.; Simonini, C.; Gianferrari, G.; Bedin, R.; Biral, C.; Ghezzi, A.; Fini, N.; Carra, S.; Mandrioli, J. SerpinA1 Levels in Amyotrophic Lateral Sclerosis Patients: An Exploratory Study. Eur. J. Neurol. 2024, 31, e16054. [Google Scholar] [CrossRef]
- Li, J.; Gao, C.; Wang, Q.; Liu, J.; Xie, Z.; Zhao, Y.; Yu, M.; Zheng, Y.; Lv, H.; Zhang, W.; et al. Elevated Serum Circulating Cell-free Mitochondrial DNA in Amyotrophic Lateral Sclerosis. Eur. J. Neurol. 2024, 31, e16493. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, R.J.; Lee, J.H.; Kefford, R.F.; Rizos, H. Evaluation of Commercial Kits for Purification of Circulating Free DNA. Cancer Genet. 2018, 228–229, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Higgins, C.M.J.; Jung, C.; Ding, H.; Xu, Z. Mutant Cu, Zn Superoxide Dismutase That Causes Motoneuron Degeneration Is Present in Mitochondria in the CNS. J. Neurosci. 2002, 22, RC215. [Google Scholar] [CrossRef]
- Eirin, A.; Saad, A.; Tang, H.; Herrmann, S.M.; Woollard, J.R.; Lerman, A.; Textor, S.C.; Lerman, L.O. Urinary Mitochondrial DNA Copy Number Identifies Chronic Renal Injury in Hypertensive Patients. Hypertension 2016, 68, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Nasi, M.; Bianchini, E.; De Biasi, S.; Gibellini, L.; Neroni, A.; Mattioli, M.; Pinti, M.; Iannone, A.; Mattioli, A.V.; Simone, A.M.; et al. Increased Plasma Levels of Mitochondrial DNA and Pro-Inflammatory Cytokines in Patients with Progressive Multiple Sclerosis. J. Neuroimmunol. 2020, 338, 577107. [Google Scholar] [CrossRef] [PubMed]
- Ying, C.; Li, Y.; Zhang, H.; Pang, S.; Hao, S.; Hu, S.; Zhao, L. Probing the Diagnostic Values of Plasma Cf-nDNA and Cf-mtDNA for Parkinson’s Disease and Multiple System Atrophy. Front. Neurosci. 2024, 18, 1488820. [Google Scholar] [CrossRef]
- Gorham, I.K.; Reid, D.M.; Sun, J.; Zhou, Z.; Barber, R.C.; Phillips, N.R. Blood-Based mtDNA Quantification Indicates Population-Specific Differences Associated with Alzheimer’s Disease-Related Risk. J. Alzheimers Dis. 2024, 97, 1407–1419. [Google Scholar] [CrossRef]
- Huang, J.; Song, Z.; Wei, B.; Li, Q.; Lin, P.; Li, H.; Dong, K. Immunological Evaluation of Patients with Alzheimer’s Disease Based on Mitogen-Stimulated Cytokine Productions and Mitochondrial DNA Indicators. BMC Psychiatry 2023, 23, 145. [Google Scholar] [CrossRef]
- Aydın, Ş.; Özdemir, S.; Adıgüzel, A. The Potential of CfDNA as Biomarker: Opportunities and Challenges for Neurodegenerative Diseases. J. Mol. Neurosci. 2025, 75, 34. [Google Scholar] [CrossRef]

| Variable | Patients (n = 54), n (%), Mean [SD] Median [IQR] |
|---|---|
| Age at onset, years | 52.03 [13.15] 50.47 [40.74–62.70] |
| Weight loss at sampling, Kg | 2.70 [6.09] 0.50 [0.00–3.75] |
| Mutational status † | |
| C9ORF72/other/WT | 7 (14.3%)/5 (10.2%)/40 (74.1%) |
| Site of onset | |
| Bulbar, spinal | 14 (25.9%)/40 (74.1%) |
| Phenotype | |
| Flail, UMNp, bulbar, classic | 8 (14.8%)/3 (5.6%)/9 (16.7%)/34 (63.0%) |
| ALSFRS-r total score at sampling, points | 40.87 [4.39] 42.00 [38.25–44.00] |
| DPR at sampling, points/month | 1.25 [2.16] 0.65 [0.36–1.21] |
| MiToS score at sampling | 0.15 [0.36] 0.00 [0.00–0.00] |
| King’s staging at sampling | 1.72 [0.79] 2.00 [1.00–2.00] |
| FVC at sampling, % | 94.24 [20.02] 91.50 [79.75–106.00] |
| NIV | 34 (63.0%) |
| PEG | 29 (53.7%) |
| IV | 22 (40.7%) |
| ALSFRS-r total score at last observation, points | 14.85 [10.58] 14.00 [6.00-20-00] |
| DPR at last observation, points/month | 1.12 [0.90] 0.84 [0.70–1.26] |
| MiToS score at sampling | 2.60 [1.25] 2.00 [2.00–4.00] |
| King’s staging at sampling | 3.49 [0.72] 4.00 [3.00–4.00] |
| Comorbidities | |
| Depression/psychosis | 15 (27.8%)/2 (3.7%) |
| COPD/other respiratory disease | 4 (7.4%)/5 (9.3%) |
| Diabetes | 4 (7.4%) |
| Hypertension | 20 (37.0%) |
| Cardiopathies | 6 (11.1%) |
| Dyslipidemia | 16 (29.6%) |
| Autoimmune diseases | 4 (7.4%) |
| Oncological history | 5 (9.3%) |
| Variable | Association Measure (95% CI) | p-Value |
|---|---|---|
| FVC at sampling | MD = 0.074 (−0.900, 1.048) | 0.8822 |
| ALSFRS-R at sampling | MD = −0.040 (−0.253, 0.174) | 0.7166 |
| DPR at sampling | MD = −0.045 (−0.150, 0.059) | 0.3993 |
| MiTos at sampling | MR = 0.989 (0.862, 1.135) | 0.8750 |
| King’s staging at sampling | MR = 1.007 (0.973, 1.042) | 0.7020 |
| ALSFRS-R at last observation | MD = −0.200 (−0.718, 0.318) | 0.4529 |
| DPR at last observation | MD = −0.002 (−0.047, 0.042) | 0.9144 |
| MiTos at last observation | MR = 1.010 (0.982, 1.038) | 0.5006 |
| King’s staging at last observation | MR = 1.004 (0.979, 1.029) | 0.7830 |
| Variable | Spearman Correlation | p-Value |
|---|---|---|
| NfLserum | 0.022 | 0.8774 |
| pNfHserum | −0.054 | 0.7022 |
| SerpinA1serum | −0.110 | 0.4438 |
| TREM2serum | 0.185 | 0.2078 |
| CHIT3L1serum | −0.053 | 0.7109 |
| NLR | −0.043 | 0.7625 |
| MLR | 0.074 | 0.5967 |
| SIRI | 0.038 | 0.7879 |
| AISI | 0.043 | 0.7603 |
| SII | 0.001 | 0.9961 |
| Total Cholesterol | −0.046 | 0.7493 |
| HDL Cholesterol | −0.166 | 0.2592 |
| LDL Cholesterol | −0.080 | 0.5884 |
| Triglycerides | −0.015 | 0.9164 |
| Creatinine | −0.280 | 0.0442 |
| Outcome | HR (95% CI) | p-Value |
|---|---|---|
| NIV | 1.005 (0.935–1.079) | 0.8991 |
| PEG | 1.020 (0.970–1.074) | 0.4332 |
| IV | 1.021 (0.967–1.078) | 0.4485 |
| Death | 1.016 (0.967–1.067) | 0.5279 |
| Tracheostomy-free survival | 1.016 (0.974–1.059) | 0.4665 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zanini, G.; Martinelli, I.; Sinigaglia, G.; Zucchi, E.; Banchelli, F.; Simonini, C.; Gianferrari, G.; Ghezzi, A.; Mandrioli, J.; Pinti, M. Circulating Serum Cell-Free Mitochondrial DNA in Amyotrophic Lateral Sclerosis. Cells 2025, 14, 1433. https://doi.org/10.3390/cells14181433
Zanini G, Martinelli I, Sinigaglia G, Zucchi E, Banchelli F, Simonini C, Gianferrari G, Ghezzi A, Mandrioli J, Pinti M. Circulating Serum Cell-Free Mitochondrial DNA in Amyotrophic Lateral Sclerosis. Cells. 2025; 14(18):1433. https://doi.org/10.3390/cells14181433
Chicago/Turabian StyleZanini, Giada, Ilaria Martinelli, Giorgia Sinigaglia, Elisabetta Zucchi, Federico Banchelli, Cecilia Simonini, Giulia Gianferrari, Andrea Ghezzi, Jessica Mandrioli, and Marcello Pinti. 2025. "Circulating Serum Cell-Free Mitochondrial DNA in Amyotrophic Lateral Sclerosis" Cells 14, no. 18: 1433. https://doi.org/10.3390/cells14181433
APA StyleZanini, G., Martinelli, I., Sinigaglia, G., Zucchi, E., Banchelli, F., Simonini, C., Gianferrari, G., Ghezzi, A., Mandrioli, J., & Pinti, M. (2025). Circulating Serum Cell-Free Mitochondrial DNA in Amyotrophic Lateral Sclerosis. Cells, 14(18), 1433. https://doi.org/10.3390/cells14181433









