Barriers in the Nervous System: Challenges and Opportunities for Novel Biomarkers in Amyotrophic Lateral Sclerosis
Abstract
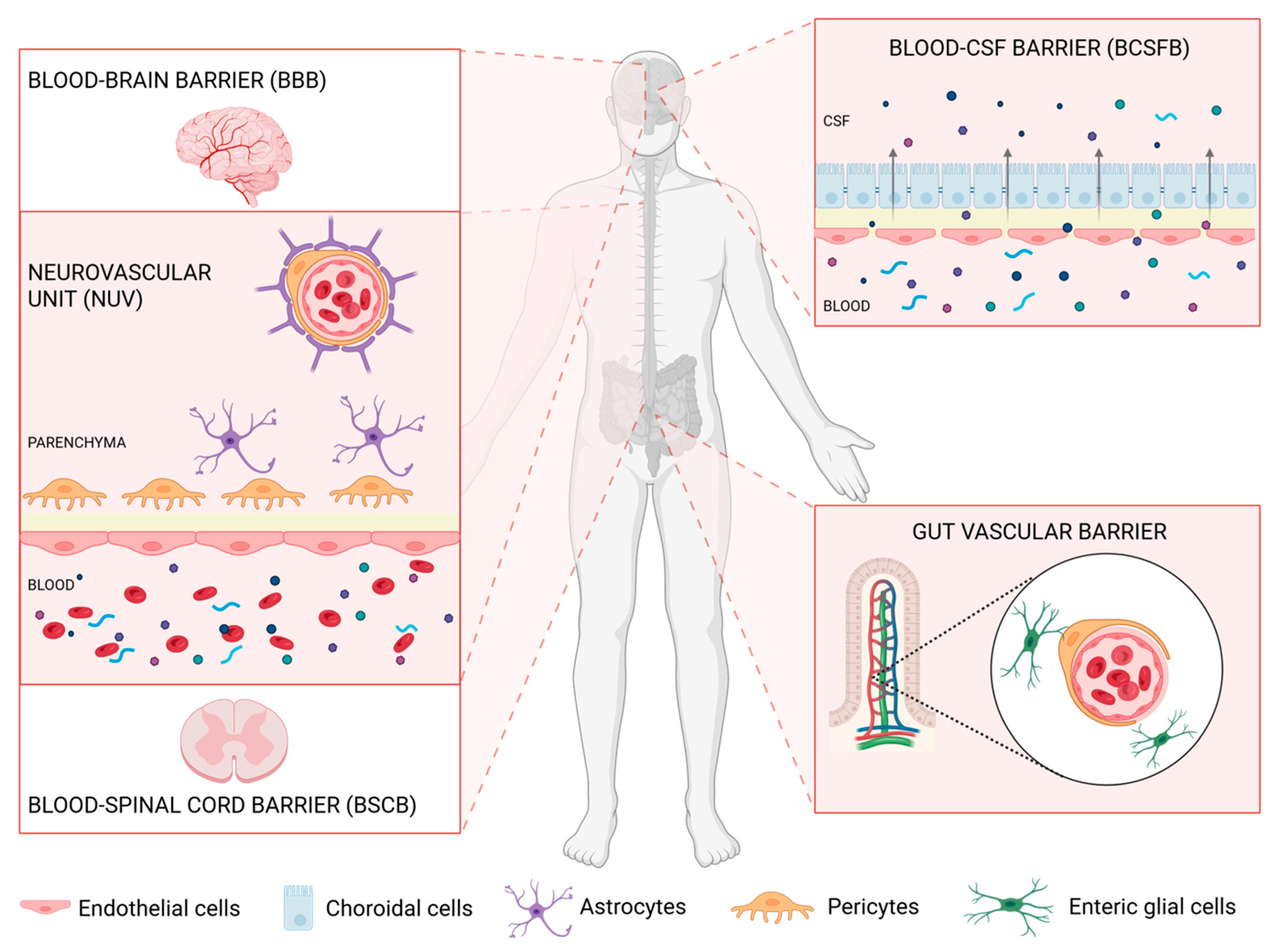
1. Barriers Delimiting and Protecting the Nervous System
1.1. The Blood–CSF Barrier (BCSFB)
1.2. The Blood–Brain Barrier (BBB)
1.3. The Blood–Spinal Cord Barrier (BSCB)
1.4. Defects in the CNS Barriers in Amyotrophic Lateral Sclerosis
1.5. Extra-Neuronal Barriers Affecting the CNS: The Gut–Vascular Barrier
2. Does the Damage to the Barriers Offer Opportunities to Search for Biomarkers in Circulation?
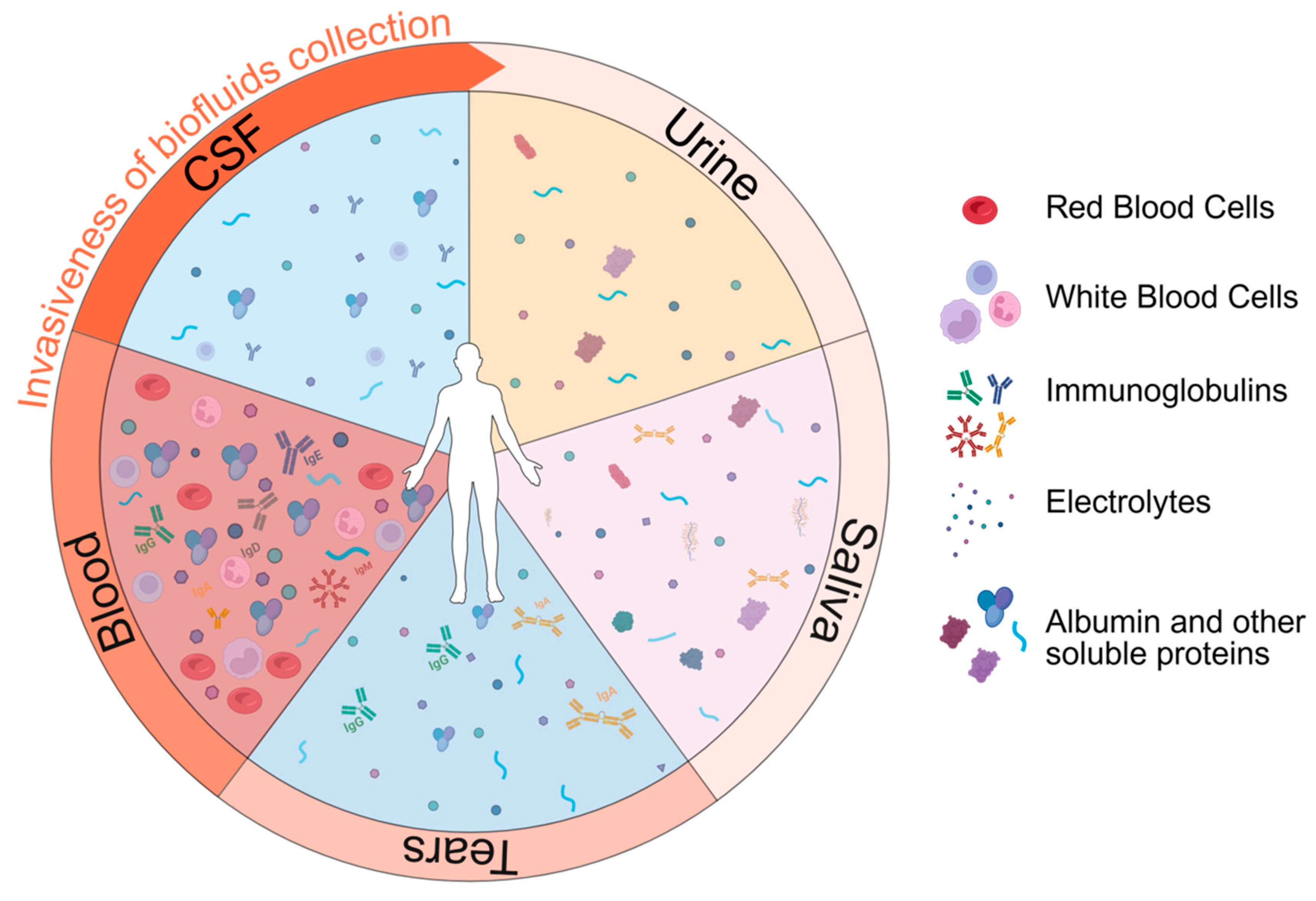
Biofluids for Biomarker Detection
- The cerebrospinal fluid and the interstitial fluid
- The blood
- Urine
- Saliva
- Tears
| Biofluid | ALS Markers |
|---|---|
| CSF | NFL [126] |
| neurofilament high (NFH and phosphorylated NFH) [80] | |
| tau and its phosphorylated form [82] | |
| myeloid protein chitotriosidase-1 (CHIT1) [127] | |
| glial protein YKL-40, also known as chitinase-3-like protein 1 (CHI3L1) [128] | |
| YKL-39 [79] | |
| inserting cryptic exons of the HDGFL2 [83] | |
| Blood | NFL [79] |
| phosphorylated tau [78] | |
| GFAP [91,92] | |
| cardiac troponin T [93] | |
| Urine | neurotrophin receptor p75 extracellular domain (p75ECD) [79] |
| Saliva | no saliva biomarkers have been associated with ALS yet |
| Tears | no tear biomarkers have been associated with ALS yet |
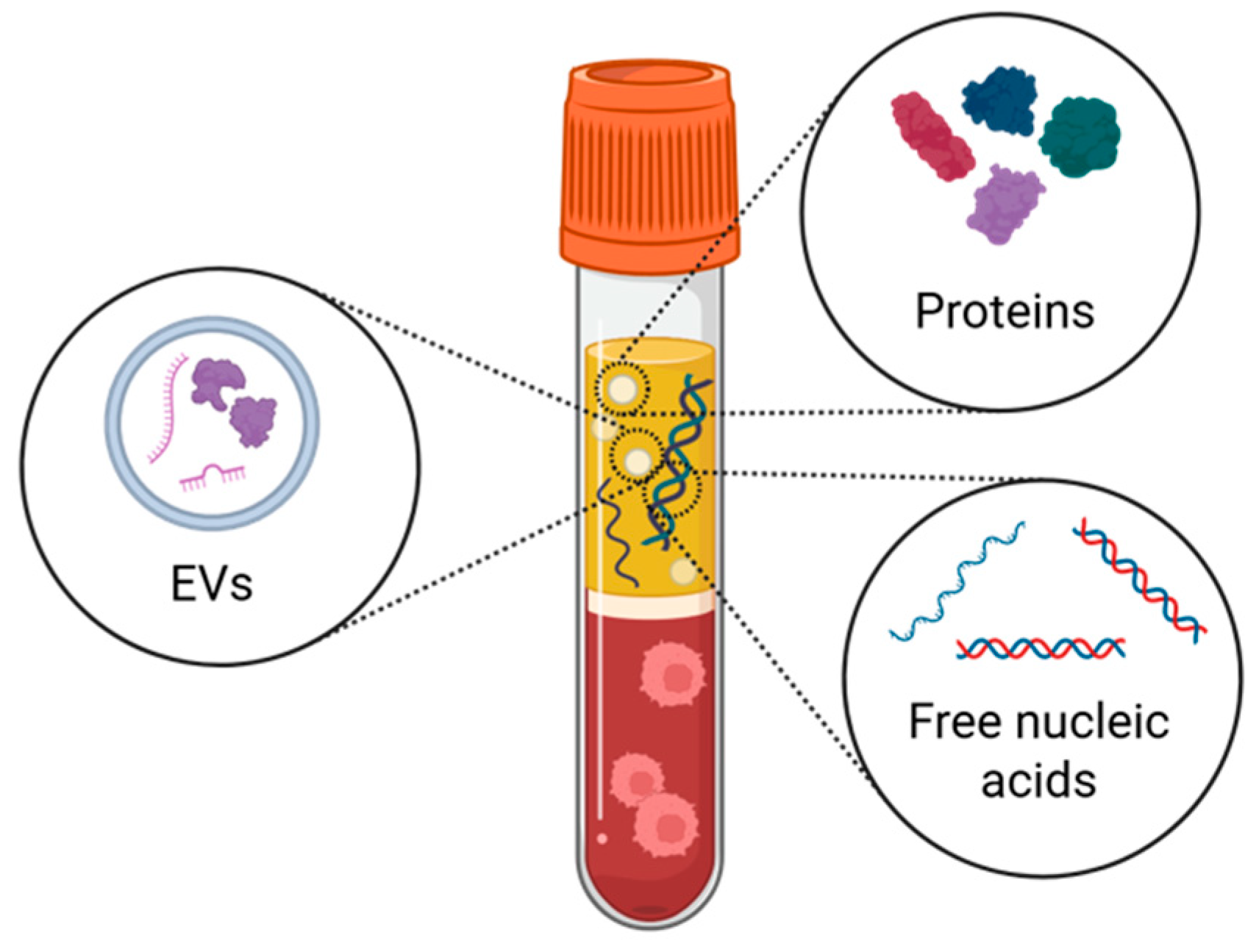
3. Beyond Free Circulating Proteins: Extracellular Vesicles and Cell-Free DNA
- Extracellular vesicles (EVs)
- Circulating cell-free DNA (cfDNA)
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CNS | central nervous system |
| BCSFB | blood–cerebrospinal fluid barrier |
| BBB | blood–brain barrier |
| BSCB | blood–spinal cord barrier |
| GVB | gut–vascular barrier |
| NUV | neurovascular unit |
| BCRP | ATP-binding cassettes, breast cancer resistance protein |
| ABCG2 | ATP-binding cassette subfamily G member 2 |
| CYP1B1 | cytochrome P450 family, with cytochrome P450 family 1 subfamily B |
| PDGF-β | platelet-derived growth factor |
| AQP4 | aquaporin 4 |
| AD | Alzheimer’s disease |
| PD | Parkinson’s disease |
| HD | Huntington’s disease |
| ALS | Amyotrophic Lateral Sclerosis |
| MS | multiple sclerosis |
| TDP-43 | TAR DNA-binding protein 43 |
| COL6A1 | collagen type VI alpha 1 chain |
| SPP1 | secreted phosphoprotein 1 |
| JAM-1 | junctional adhesion molecule 1 |
| GFAP | glial fibrillary acidic protein |
| EGC | enteric glial cell |
| PSD95 | postsynaptic density protein 95 |
| FMT | fecal microbiota transplantation |
| SCFAs | short-chain fatty acids |
| CSF | cerebrospinal fluid |
| ANP | Atrial Natriuretic Peptide |
| CHIT1 | myeloid protein chitotriosidase-1 |
| CHI3L1 | chitinase-3-like protein 1 |
| HDGFL2 | hepatoma-derived growth factor-like protein 2 |
| EVs | extracellular vesicles |
| cfDNA | cell-free DNA |
| L1CAM | L1 cell adhesion molecule |
| NCAM | Neural Cell Adhesion Molecule |
| GluR2/3 | glutamate ionotropic receptor AMPA type subunits 2 and 3 |
| NMDAR2A | glutamate receptor, ionotropic, N-methyl D-aspartate 2A |
| ATP1A3 | ATPase, Na+/K+ transporting, and alpha 3 polypeptide |
| BMECs | brain microvascular endothelial cells |
| TNF-α | Tumor Necrosis Factor-alpha |
| LPS | Lipopolysaccharide |
| WGA | wheat germ agglutinin |
| M6P | mannose 6-phosphate |
| cfRNA | cell-free RNA |
| cf-mtDNA | circulating cell-free mitochondrial DNA |
| MCI | mild cognitive impairment |
| ddPCR | droplet digital PCR |
| NFL | neurofilament light polypeptide |
| NFH | neurofilament heavy polypeptide |
| BMECs | brain microvascular endothelial cells |
| p75ECD | neurotrophin receptor p75 extracellular domain |
| HSC | hemopoietic stem cell |
| CHI3L2 | Chitinase-3-like protein 2 |
References
- Engelhardt, B. Cluster: Barriers of the Central Nervous System. Acta Neuropathol. 2018, 135, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Carloni, S.; Rescigno, M. The Gut-Brain Vascular Axis in Neuroinflammation. Semin. Immunol. 2023, 69, 101802. [Google Scholar] [CrossRef]
- Lun, M.P.; Monuki, E.S.; Lehtinen, M.K. Development and Functions of the Choroid Plexus–Cerebrospinal Fluid System. Nat. Rev. Neurosci. 2015, 16, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Sakka, L.; Coll, G.; Chazal, J. Anatomy and Physiology of Cerebrospinal Fluid. Eur. Ann. Otorhinolaryngol. Head. Neck Dis. 2011, 128, 309–316. [Google Scholar] [CrossRef]
- Spector, R.; Johanson, C.E. The Mammalian Choroid Plexus. Sci. Am. 1989, 261, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Ayre, J.R.; Bazira, P.J.; Abumattar, M.; Makwana, H.N.; Sanders, K.A. A New Classification System for the Anatomical Variations of the Human Circle of Willis: A Systematic Review. J. Anat. 2022, 240, 1187–1204. [Google Scholar] [CrossRef]
- Jones, J.D.; Castanho, P.; Bazira, P.; Sanders, K. Anatomical Variations of the Circle of Willis and Their Prevalence, with a Focus on the Posterior Communicating Artery: A Literature Review and Meta-analysis. Clin. Anat. 2021, 34, 978–990. [Google Scholar] [CrossRef]
- Wälchli, T.; Bisschop, J.; Carmeliet, P.; Zadeh, G.; Monnier, P.P.; De Bock, K.; Radovanovic, I. Shaping the Brain Vasculature in Development and Disease in the Single-Cell Era. Nat. Rev. Neurosci. 2023, 24, 271–298. [Google Scholar] [CrossRef]
- Winkler, E.A.; Bell, R.D.; Zlokovic, B.V. Central Nervous System Pericytes in Health and Disease. Nat. Neurosci. 2011, 14, 1398–1405. [Google Scholar] [CrossRef]
- Kadry, H.; Noorani, B.; Cucullo, L. A Blood–Brain Barrier Overview on Structure, Function, Impairment, and Biomarkers of Integrity. Fluids Barriers CNS 2020, 17, 69. [Google Scholar] [CrossRef]
- Komarova, Y.A.; Kruse, K.; Mehta, D.; Malik, A.B. Protein Interactions at Endothelial Junctions and Signaling Mechanisms Regulating Endothelial Permeability. Circ. Res. 2017, 120, 179–206. [Google Scholar] [CrossRef] [PubMed]
- Dauchy, S.; Dutheil, F.; Weaver, R.J.; Chassoux, F.; Daumas-Duport, C.; Couraud, P.; Scherrmann, J.; De Waziers, I.; Declèves, X. ABC Transporters, Cytochromes P450 and Their Main Transcription Factors: Expression at the Human Blood–Brain Barrier. J. Neurochem. 2008, 107, 1518–1528. [Google Scholar] [CrossRef]
- Bell, R.D.; Winkler, E.A.; Sagare, A.P.; Singh, I.; LaRue, B.; Deane, R.; Zlokovic, B.V. Pericytes Control Key Neurovascular Functions and Neuronal Phenotype in the Adult Brain and during Brain Aging. Neuron 2010, 68, 409–427. [Google Scholar] [CrossRef] [PubMed]
- Armulik, A.; Genové, G.; Mäe, M.; Nisancioglu, M.H.; Wallgard, E.; Niaudet, C.; He, L.; Norlin, J.; Lindblom, P.; Strittmatter, K.; et al. Pericytes Regulate the Blood–Brain Barrier. Nature 2010, 468, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Peppiatt, C.M.; Howarth, C.; Mobbs, P.; Attwell, D. Bidirectional Control of CNS Capillary Diameter by Pericytes. Nature 2006, 443, 700–704. [Google Scholar] [CrossRef]
- Iliff, J.J.; Wang, M.; Liao, Y.; Plogg, B.A.; Peng, W.; Gundersen, G.A.; Benveniste, H.; Vates, G.E.; Deane, R.; Goldman, S.A.; et al. A Paravascular Pathway Facilitates CSF Flow Through the Brain Parenchyma and the Clearance of Interstitial Solutes, Including Amyloid β. Sci. Transl. Med. 2012, 4, 147ra111. [Google Scholar] [CrossRef]
- Ray, L.A.; Heys, J.J. Fluid Flow and Mass Transport in Brain Tissue. Fluids 2019, 4, 196. [Google Scholar] [CrossRef]
- Murlidharan, G.; Crowther, A.; Reardon, R.A.; Song, J.; Asokan, A. Glymphatic Fluid Transport Controls Paravascular Clearance of AAV Vectors from the Brain. JCI Insight 2016, 1, e88034. [Google Scholar] [CrossRef]
- Nielsen, S.; Arnulf Nagelhus, E.; Amiry-Moghaddam, M.; Bourque, C.; Agre, P.; Petter Ottersen, O. Specialized Membrane Domains for Water Transport in Glial Cells: High-Resolution Immunogold Cytochemistry of Aquaporin-4 in Rat Brain. J. Neurosci. 1997, 17, 171–180. [Google Scholar] [CrossRef]
- Rash, J.E.; Yasumura, T.; Hudson, C.S.; Agre, P.; Nielsen, S. Direct Immunogold Labeling of Aquaporin-4 in Square Arrays of Astrocyte and Ependymocyte Plasma Membranes in Rat Brain and Spinal Cord. Proc. Natl. Acad. Sci. USA 1998, 95, 11981–11986. [Google Scholar] [CrossRef]
- Bartanusz, V.; Jezova, D.; Alajajian, B.; Digicaylioglu, M. The Blood–Spinal Cord Barrier: Morphology and Clinical Implications. Ann. Neurol. 2011, 70, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.-Y.; Kwon, H.-B.; Ahn, J.-C.; Kang, D.; Kwon, S.-H.; Park, J.A.; Kim, K.-W. Functional and Developmental Analysis of the Blood–Brain Barrier in Zebrafish. Brain Res. Bull. 2008, 75, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Chopra, N.; Menounos, S.; Choi, J.P.; Hansbro, P.M.; Diwan, A.D.; Das, A. Blood-Spinal Cord Barrier: Its Role in Spinal Disorders and Emerging Therapeutic Strategies. NeuroSci 2021, 3, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Winkler, E.A.; Sengillo, J.D.; Bell, R.D.; Wang, J.; Zlokovic, B.V. Blood–Spinal Cord Barrier Pericyte Reductions Contribute to Increased Capillary Permeability. J. Cereb. Blood Flow. Metab. 2012, 32, 1841–1852. [Google Scholar] [CrossRef]
- Sweeney, M.D.; Zhao, Z.; Montagne, A.; Nelson, A.R.; Zlokovic, B.V. Blood-Brain Barrier: From Physiology to Disease and Back. Physiol. Rev. 2019, 99, 21–78. [Google Scholar] [CrossRef]
- Chen, T.; Dai, Y.; Hu, C.; Lin, Z.; Wang, S.; Yang, J.; Zeng, L.; Li, S.; Li, W. Cellular and Molecular Mechanisms of the Blood–Brain Barrier Dysfunction in Neurodegenerative Diseases. Fluids Barriers CNS 2024, 21, 60. [Google Scholar] [CrossRef]
- Riva, N.; Domi, T.; Pozzi, L.; Lunetta, C.; Schito, P.; Spinelli, E.G.; Cabras, S.; Matteoni, E.; Consonni, M.; Bella, E.D.; et al. Update on Recent Advances in Amyotrophic Lateral Sclerosis. J. Neurol. 2024, 271, 4693–4723. [Google Scholar] [CrossRef]
- Bendotti, C.; Bonetto, V.; Pupillo, E.; Logroscino, G.; Al-Chalabi, A.; Lunetta, C.; Riva, N.; Mora, G.; Lauria, G.; Weishaupt, J.H.; et al. Focus on the Heterogeneity of Amyotrophic Lateral Sclerosis. Amyotroph. Lateral Scler. Front. Degener. 2020, 21, 485–495. [Google Scholar] [CrossRef]
- Schreiber, S.; Bernal, J.; Arndt, P.; Schreiber, F.; Müller, P.; Morton, L.; Braun-Dullaeus, R.C.; Valdés-Hernández, M.D.C.; Duarte, R.; Wardlaw, J.M.; et al. Brain Vascular Health in ALS Is Mediated through Motor Cortex Microvascular Integrity. Cells 2023, 12, 957. [Google Scholar] [CrossRef]
- Garbuzova-Davis, S.; Saporta, S.; Haller, E.; Kolomey, I.; Bennett, S.P.; Potter, H.; Sanberg, P.R. Evidence of Compromised Blood-Spinal Cord Barrier in Early and Late Symptomatic SOD1 Mice Modeling ALS. PLoS ONE 2007, 2, e1205. [Google Scholar] [CrossRef]
- Nicaise, C.; Mitrecic, D.; Demetter, P.; De Decker, R.; Authelet, M.; Boom, A.; Pochet, R. Impaired Blood–Brain and Blood–Spinal Cord Barriers in Mutant SOD1-Linked ALS Rat. Brain Res. 2009, 1301, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Deane, R.; Ali, Z.; Parisi, M.; Shapovalov, Y.; O’Banion, M.K.; Stojanovic, K.; Sagare, A.; Boillee, S.; Cleveland, D.W.; et al. ALS-Causing SOD1 Mutants Generate Vascular Changes Prior to Motor Neuron Degeneration. Nat. Neurosci. 2008, 11, 420–422. [Google Scholar] [CrossRef]
- Meister, S.; Storck, S.E.; Hameister, E.; Behl, C.; Weggen, S.; Clement, A.M.; Pietrzik, C.U. Expression of the ALS-Causing Variant hSOD1G93A Leads to an Impaired Integrity and Altered Regulation of Claudin-5 Expression in an in Vitro Blood–Spinal Cord Barrier Model. J. Cereb. Blood Flow. Metab. 2015, 35, 1112–1121. [Google Scholar] [CrossRef]
- Tang, J.; Kang, Y.; Zhou, Y.; Li, X.; Lan, J.; Wu, L.; Feng, X.; Peng, Y. ALS-Causing SOD1 Mutants Regulate Occludin Phosphorylation/Ubiquitination and Endocytic Trafficking via the ITCH/Eps15/Rab5 Axis. Neurobiol. Dis. 2021, 153, 105315. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Kagawa, Y.; Sun, J.; Turner, B.J.; Huang, C.; Shah, A.D.; Schittenhelm, R.B.; Nicolazzo, J.A. Altered Blood–Brain Barrier Dynamics in the C9orf72 Hexanucleotide Repeat Expansion Mouse Model of Amyotrophic Lateral Sclerosis. Pharmaceutics 2022, 14, 2803. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, K.; Ohta, Y.; Nagai, M.; Morimoto, N.; Kurata, T.; Takehisa, Y.; Ikeda, Y.; Matsuura, T.; Abe, K. Disruption of Neurovascular Unit Prior to Motor Neuron Degeneration in Amyotrophic Lateral Sclerosis. J. Neurosci. Res. 2011, 89, 718–728. [Google Scholar] [CrossRef]
- Nicaise, C.; Soyfoo, M.S.; Authelet, M.; De Decker, R.; Bataveljic, D.; Delporte, C.; Pochet, R. Aquaporin-4 Overexpression in Rat ALS Model. Anat. Rec. 2009, 292, 207–213. [Google Scholar] [CrossRef]
- Watanabe-Matsumoto, S.; Moriwaki, Y.; Okuda, T.; Ohara, S.; Yamanaka, K.; Abe, Y.; Yasui, M.; Misawa, H. Dissociation of Blood-Brain Barrier Disruption and Disease Manifestation in an Aquaporin-4-Deficient Mouse Model of Amyotrophic Lateral Sclerosis. Neurosci. Res. 2018, 133, 48–57. [Google Scholar] [CrossRef]
- Månberg, A.; Skene, N.; Sanders, F.; Trusohamn, M.; Remnestål, J.; Szczepińska, A.; Aksoylu, I.S.; Lönnerberg, P.; Ebarasi, L.; Wouters, S.; et al. Altered Perivascular Fibroblast Activity Precedes ALS Disease Onset. Nat. Med. 2021, 27, 640–646. [Google Scholar] [CrossRef]
- Aragón-González, A.; Shaw, A.C.; Kok, J.R.; Roussel, F.S.; Santos Souza, C.D.; Granger, S.M.; Vetter, T.; De Diego, Y.; Meyer, K.C.; Beal, S.N.; et al. C9ORF72 Patient-Derived Endothelial Cells Drive Blood-Brain Barrier Disruption and Contribute to Neurotoxicity. Fluids Barriers CNS 2024, 21, 34. [Google Scholar] [CrossRef]
- Garbuzova-Davis, S.; Haller, E.; Saporta, S.; Kolomey, I.; Nicosia, S.V.; Sanberg, P.R. Ultrastructure of Blood–Brain Barrier and Blood–Spinal Cord Barrier in SOD1 Mice Modeling ALS. Brain Res. 2007, 1157, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Garbuzova-Davis, S.; Hernandez-Ontiveros, D.G.; Rodrigues, M.C.O.; Haller, E.; Frisina-Deyo, A.; Mirtyl, S.; Sallot, S.; Saporta, S.; Borlongan, C.V.; Sanberg, P.R. Impaired Blood–Brain/Spinal Cord Barrier in ALS Patients. Brain Res. 2012, 1469, 114–128. [Google Scholar] [CrossRef]
- Henkel, J.S.; Beers, D.R.; Wen, S.; Bowser, R.; Appel, S.H. Decreased mRNA expression of tight junction proteins in lumbar spinal cords of patients with ALS. Neurology 2009, 72, 1614–1616. [Google Scholar] [CrossRef]
- Saul, J.; Hutchins, E.; Reiman, R.; Saul, M.; Ostrow, L.W.; Harris, B.T.; Van Keuren-Jensen, K.; Bowser, R.; Bakkar, N. Global Alterations to the Choroid Plexus Blood-CSF Barrier in Amyotrophic Lateral Sclerosis. Acta Neuropathol. Commun. 2020, 8, 92. [Google Scholar] [CrossRef]
- Winkler, E.A.; Sengillo, J.D.; Sullivan, J.S.; Henkel, J.S.; Appel, S.H.; Zlokovic, B.V. Blood–Spinal Cord Barrier Breakdown and Pericyte Reductions in Amyotrophic Lateral Sclerosis. Acta Neuropathol. 2013, 125, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Yamadera, M.; Fujimura, H.; Inoue, K.; Toyooka, K.; Mori, C.; Hirano, H.; Sakoda, S. Microvascular Disturbance with Decreased Pericyte Coverage Is Prominent in the Ventral Horn of Patients with Amyotrophic Lateral Sclerosis. Amyotroph. Lateral Scler. Front. Degener. 2015, 16, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-Y.; Cai, Z.-Y.; Sun, X.-H.; Shen, D.; Yang, X.-Z.; Liu, M.-S.; Cui, L.-Y. Blood–Brain Barrier Dysfunction and Myelin Basic Protein in Survival of Amyotrophic Lateral Sclerosis with or without Frontotemporal Dementia. Neurol. Sci. 2022, 43, 3201–3210. [Google Scholar] [CrossRef]
- Verde, F.; Ferrari, I.; Maranzano, A.; Ciusani, E.; Torre, S.; Milone, I.; Colombo, E.; Doretti, A.; Peverelli, S.; Ratti, A.; et al. Relationship between Cerebrospinal Fluid/Serum Albumin Quotient and Phenotype in Amyotrophic Lateral Sclerosis: A Retrospective Study on 328 Patients. Neurol. Sci. 2023, 44, 1679–1685. [Google Scholar] [CrossRef]
- Brescia, P.; Rescigno, M. The Gut Vascular Barrier: A New Player in the Gut–Liver–Brain Axis. Trends Mol. Med. 2021, 27, 844–855. [Google Scholar] [CrossRef]
- Savidge, T.C.; Newman, P.; Pothoulakis, C.; Ruhl, A.; Neunlist, M.; Bourreille, A.; Hurst, R.; Sofroniew, M.V. Enteric Glia Regulate Intestinal Barrier Function and Inflammation Via Release of S-Nitrosoglutathione. Gastroenterology 2007, 132, 1344–1358. [Google Scholar] [CrossRef]
- Bush, T.G.; Savidge, T.C.; Freeman, T.C.; Cox, H.J.; Campbell, E.A.; Mucke, L.; Johnson, M.H.; Sofroniew, M.V. Fulminant Jejuno-Ileitis Following Ablation of Enteric Glia in Adult Transgenic Mice. Cell 1998, 93, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Bonaz, B.; Bazin, T.; Pellissier, S. The Vagus Nerve at the Interface of the Microbiota-Gut-Brain Axis. Front. Neurosci. 2018, 12, 49. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut Microbiota in Human Metabolic Health and Disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef]
- Gan, Y.; Chen, Y.; Zhong, H.; Liu, Z.; Geng, J.; Wang, H.; Wang, W. Gut Microbes in Central Nervous System Development and Related Disorders. Front. Immunol. 2024, 14, 1288256. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Murdock, M.H.; Jing, D.; Won, T.H.; Chung, H.; Kressel, A.M.; Tsaava, T.; Addorisio, M.E.; Putzel, G.G.; Zhou, L.; et al. The Microbiota Regulate Neuronal Function and Fear Extinction Learning. Nature 2019, 574, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Heijtz, R.D.; Wang, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal Gut Microbiota Modulates Brain Development and Behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef]
- Braniste, V.; Al-Asmakh, M.; Kowal, C.; Anuar, F.; Abbaspour, A.; Tóth, M.; Korecka, A.; Bakocevic, N.; Ng, L.G.; Kundu, P.; et al. The Gut Microbiota Influences Blood-Brain Barrier Permeability in Mice. Sci. Transl. Med. 2014, 6, 263ra158. [Google Scholar] [CrossRef]
- Yan, J.; Chen, H.; Zhang, Y.; Peng, L.; Wang, Z.; Lan, X.; Yu, S.; Yang, Y. Fecal Microbiota Transplantation Significantly Improved Respiratory Failure of Amyotrophic Lateral Sclerosis. Gut Microbes 2024, 16, 2353396. [Google Scholar] [CrossRef]
- Vázquez-Liébanas, E.; Mocci, G.; Li, W.; Laviña, B.; Reddy, A.; O’Connor, C.; Hudson, N.; Elbeck, Z.; Nikoloudis, I.; Gaengel, K.; et al. Mosaic Deletion of Claudin-5 Reveals Rapid Non-Cell-Autonomous Consequences of Blood-Brain Barrier Leakage. Cell Rep. 2024, 43, 113911. [Google Scholar] [CrossRef]
- Li, K.; Wei, S.; Hu, L.; Yin, X.; Mai, Y.; Jiang, C.; Peng, X.; Cao, X.; Huang, Z.; Zhou, H.; et al. Protection of Fecal Microbiota Transplantation in a Mouse Model of Multiple Sclerosis. Mediat. Inflamm. 2020, 2020, 2058272. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T.R.; Debelius, J.W.; Thron, T.; Janssen, S.; Shastri, G.G.; Ilhan, Z.E.; Challis, C.; Schretter, C.E.; Rocha, S.; Gradinaru, V.; et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 2016, 167, 1469–1480.E12. [Google Scholar] [CrossRef]
- Xie, J.; Bruggeman, A.; De Nolf, C.; Vandendriessche, C.; Van Imschoot, G.; Van Wonterghem, E.; Vereecke, L.; Vandenbroucke, R.E. Gut Microbiota Regulates Blood-cerebrospinal Fluid Barrier Function and Aβ Pathology. EMBO J. 2023, 42, e111515. [Google Scholar] [CrossRef] [PubMed]
- Al, K.F.; Craven, L.J.; Gibbons, S.; Parvathy, S.N.; Wing, A.C.; Graf, C.; Parham, K.A.; Kerfoot, S.M.; Wilcox, H.; Burton, J.P.; et al. Fecal Microbiota Transplantation Is Safe and Tolerable in Patients with Multiple Sclerosis: A Pilot Randomized Controlled Trial. Mult. Scler. J.-Exp. Transl. Clin. 2022, 8, 20552173221086662. [Google Scholar] [CrossRef] [PubMed]
- Scheperjans, F.; Levo, R.; Bosch, B.; Lääperi, M.; Pereira, P.A.B.; Smolander, O.-P.; Aho, V.T.E.; Vetkas, N.; Toivio, L.; Kainulainen, V.; et al. Fecal Microbiota Transplantation for Treatment of Parkinson Disease: A Randomized Clinical Trial. JAMA Neurol. 2024, 81, 925–938. [Google Scholar] [CrossRef]
- Feng, R.; Zhu, Q.; Wang, A.; Wang, H.; Wang, J.; Chen, P.; Zhang, R.; Liang, D.; Teng, J.; Ma, M.; et al. Effect of Fecal Microbiota Transplantation on Patients with Sporadic Amyotrophic Lateral Sclerosis: A Randomized, Double-Blind, Placebo-Controlled Trial. BMC Med. 2024, 22, 566. [Google Scholar] [CrossRef]
- Mandrioli, J.; Amedei, A.; Cammarota, G.; Niccolai, E.; Zucchi, E.; D’Amico, R.; Ricci, F.; Quaranta, G.; Spanu, T.; Masucci, L. FETR-ALS Study Protocol: A Randomized Clinical Trial of Fecal Microbiota Transplantation in Amyotrophic Lateral Sclerosis. Front. Neurol. 2019, 10, 1021. [Google Scholar] [CrossRef]
- Kim, H.S.; Son, J.; Lee, D.; Tsai, J.; Wang, D.; Chocron, E.S.; Jeong, S.; Kittrell, P.; Murchison, C.F.; Kennedy, R.E.; et al. Gut- and Oral-Dysbiosis Differentially Impact Spinal- and Bulbar-Onset ALS, Predicting ALS Severity and Potentially Determining the Location of Disease Onset. BMC Neurol. 2022, 22, 62. [Google Scholar] [CrossRef]
- Spector, R.; Robert Snodgrass, S.; Johanson, C.E. A Balanced View of the Cerebrospinal Fluid Composition and Functions: Focus on Adult Humans. Exp. Neurol. 2015, 273, 57–68. [Google Scholar] [CrossRef]
- Buishas, J.; Gould, I.G.; Linninger, A.A. A Computational Model of Cerebrospinal Fluid Production and Reabsorption Driven by Starling Forces. Croat. Med. J. 2014, 55, 481–497. [Google Scholar] [CrossRef]
- Shen, D.; Artru, A.; Adkison, K. Principles and Applicability of CSF Sampling for the Assessment of CNS Drug Delivery and Pharmacodynamics. Adv. Drug Deliv. Rev. 2004, 56, 1825–1857. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Liu, J.; Liang, C.; Yang, L.; Wang, G. Aquaporin-4 in Glymphatic System, and Its Implication for Central Nervous System Disorders. Neurobiol. Dis. 2023, 179, 106035. [Google Scholar] [CrossRef] [PubMed]
- Aspelund, A.; Antila, S.; Proulx, S.T.; Karlsen, T.V.; Karaman, S.; Detmar, M.; Wiig, H.; Alitalo, K. A Dural Lymphatic Vascular System That Drains Brain Interstitial Fluid and Macromolecules. J. Exp. Med. 2015, 212, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Louveau, A.; Smirnov, I.; Keyes, T.J.; Eccles, J.D.; Rouhani, S.J.; Peske, J.D.; Derecki, N.C.; Castle, D.; Mandell, J.W.; Lee, K.S.; et al. Structural and Functional Features of Central Nervous System Lymphatic Vessels. Nature 2015, 523, 337–341. [Google Scholar] [CrossRef]
- Yankova, G.; Bogomyakova, O.; Tulupov, A. The Glymphatic System and Meningeal Lymphatics of the Brain: New Understanding of Brain Clearance. Rev. Neurosci. 2021, 32, 693–705. [Google Scholar] [CrossRef]
- Absinta, M.; Ha, S.-K.; Nair, G.; Sati, P.; Luciano, N.J.; Palisoc, M.; Louveau, A.; Zaghloul, K.A.; Pittaluga, S.; Kipnis, J.; et al. Human and Nonhuman Primate Meninges Harbor Lymphatic Vessels That Can Be Visualized Noninvasively by MRI. eLife 2017, 6, e29738. [Google Scholar] [CrossRef]
- Al-Diwani, A.; Provine, N.M.; Murchison, A.; Laban, R.; Swann, O.J.; Koychev, I.; Sheerin, F.; Da Mesquita, S.; Heslegrave, A.; Zetterberg, H.; et al. Neurodegenerative Fluid Biomarkers Are Enriched in Human Cervical Lymph Nodes. Brain 2025, 148, 394–400. [Google Scholar] [CrossRef]
- Abu-Rumeileh, S.; Scholle, L.; Mensch, A.; Großkopf, H.; Ratti, A.; Kölsch, A.; Stoltenburg-Didinger, G.; Conrad, J.; De Gobbi, A.; Barba, L.; et al. Phosphorylated Tau 181 and 217 Are Elevated in Serum and Muscle of Patients with Amyotrophic Lateral Sclerosis. Nat. Commun. 2025, 16, 2019. [Google Scholar] [CrossRef]
- Sturmey, E.; Malaspina, A. Blood Biomarkers in ALS : Challenges, Applications and Novel Frontiers. Acta Neuro. Scand. 2022, 146, 375–388. [Google Scholar] [CrossRef]
- Behzadi, A.; Pujol-Calderón, F.; Tjust, A.E.; Wuolikainen, A.; Höglund, K.; Forsberg, K.; Portelius, E.; Blennow, K.; Zetterberg, H.; Andersen, P.M. Neurofilaments Can Differentiate ALS Subgroups and ALS from Common Diagnostic Mimics. Sci. Rep. 2021, 11, 22128. [Google Scholar] [CrossRef]
- Steinacker, P.; Verde, F.; Fang, L.; Feneberg, E.; Oeckl, P.; Roeber, S.; Anderl-Straub, S.; Danek, A.; Diehl-Schmid, J.; Fassbender, K.; et al. Chitotriosidase (CHIT1) Is Increased in Microglia and Macrophages in Spinal Cord of Amyotrophic Lateral Sclerosis and Cerebrospinal Fluid Levels Correlate with Disease Severity and Progression. J. Neurol. Neurosurg. Psychiatry 2018, 89, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Agnello, L.; Colletti, T.; Lo Sasso, B.; Vidali, M.; Spataro, R.; Gambino, C.M.; Giglio, R.V.; Piccoli, T.; Bivona, G.; La Bella, V.; et al. Tau Protein as a Diagnostic and Prognostic Biomarker in Amyotrophic Lateral Sclerosis. Euro J. Neurol. 2021, 28, 1868–1875. [Google Scholar] [CrossRef] [PubMed]
- Irwin, K.E.; Jasin, P.; Braunstein, K.E.; Sinha, I.R.; Garret, M.A.; Bowden, K.D.; Chang, K.; Troncoso, J.C.; Moghekar, A.; Oh, E.S.; et al. A Fluid Biomarker Reveals Loss of TDP-43 Splicing Repression in Presymptomatic ALS–FTD. Nat. Med. 2024, 30, 382–393. [Google Scholar] [CrossRef]
- Teunissen, C.E.; Petzold, A.; Bennett, J.L.; Berven, F.S.; Brundin, L.; Comabella, M.; Franciotta, D.; Frederiksen, J.L.; Fleming, J.O.; Furlan, R.; et al. A Consensus Protocol for the Standardization of Cerebrospinal Fluid Collection and Biobanking. Neurology 2009, 73, 1914–1922. [Google Scholar] [CrossRef]
- Alkire, K.; Collingwood, J. Physiology of Blood and Bone Marrow. Semin. Oncol. Nurs. 1990, 6, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Krebs, H.A. Chemical Composition of Blood Plasma and Serum. Annu. Rev. Biochem. 1950, 19, 409–430. [Google Scholar] [CrossRef]
- Weiss, C.; Jelkmann, W. Functions of the Blood. In Human Physiology; Schmidt, R.F., Thews, G., Eds.; Springer: Berlin/Heidelberg, Germany, 1989; pp. 402–438. ISBN 978-3-642-73833-3. [Google Scholar]
- Pinho, S.; Frenette, P.S. Haematopoietic Stem Cell Activity and Interactions with the Niche. Nat. Rev. Mol. Cell Biol. 2019, 20, 303–320. [Google Scholar] [CrossRef]
- Benjamin, R.J.; McLaughlin, L.S. Plasma Components: Properties, Differences, and Uses. Transfusion 2012, 52, 9S–19S. [Google Scholar] [CrossRef]
- Donini, L.; Tanel, R.; Zuccarino, R.; Basso, M. Protein Biomarkers for the Diagnosis and Prognosis of Amyotrophic Lateral Sclerosis. Neurosci. Res. 2023, 197, 31–41. [Google Scholar] [CrossRef]
- Mondesert, E.; Delaby, C.; De La Cruz, E.; Kuhle, J.; Benkert, P.; Pradeilles, N.; Duchiron, M.; Morchikh, M.; Camu, W.; Cristol, J.-P.; et al. Comparative Performances of 4 Serum NfL Assays, pTau181, and GFAP in Patients With Amyotrophic Lateral Sclerosis. Neurology 2025, 104, e213400. [Google Scholar] [CrossRef]
- Verde, F.; Milone, I.; Maranzano, A.; Colombo, E.; Torre, S.; Solca, F.; Doretti, A.; Gentile, F.; Manini, A.; Bonetti, R.; et al. Serum Levels of Glial Fibrillary Acidic Protein in Patients with Amyotrophic Lateral Sclerosis. Ann. Clin. Transl. Neurol. 2023, 10, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Kläppe, U.; Chamoun, S.; Shen, Q.; Finn, A.; Evertsson, B.; Zetterberg, H.; Blennow, K.; Press, R.; Samuelsson, K.; Månberg, A.; et al. Cardiac Troponin T Is Elevated and Increases Longitudinally in ALS Patients. Amyotroph. Lateral Scler. Front. Degener. 2022, 23, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.T.; Bowser, R. Fluid-Based Biomarkers for Amyotrophic Lateral Sclerosis. Neurotherapeutics 2017, 14, 119–134. [Google Scholar] [CrossRef]
- Feneberg, E.; Gray, E.; Ansorge, O.; Talbot, K.; Turner, M.R. Towards a TDP-43-Based Biomarker for ALS and FTLD. Mol. Neurobiol. 2018, 55, 7789–7801. [Google Scholar] [CrossRef] [PubMed]
- Kasai, T.; Kojima, Y.; Ohmichi, T.; Tatebe, H.; Tsuji, Y.; Noto, Y.; Kitani-Morii, F.; Shinomoto, M.; Allsop, D.; Mizuno, T.; et al. Combined Use of CSF NfL and CSF TDP-43 Improves Diagnostic Performance in ALS. Ann. Clin. Transl. Neurol. 2019, 6, 2489–2502. [Google Scholar] [CrossRef]
- Matsuura, S.; Tatebe, H.; Higuchi, M.; Tokuda, T. Validation of a Newly Developed Immunoassay for TDP-43 in Human Plasma. Heliyon 2024, 10, e24672. [Google Scholar] [CrossRef]
- Casiraghi, V.; Milone, I.; Brusati, A.; Peverelli, S.; Doretti, A.; Poletti, B.; Maderna, L.; Morelli, C.; Ticozzi, N.; Silani, V.; et al. Quantification of Serum TDP-43 and Neurofilament Light Chain in Patients with Amyotrophic Lateral Sclerosis Stratified by UNC13A Genotype. J. Neurol. Sci. 2024, 466, 123210. [Google Scholar] [CrossRef]
- Chatterjee, M.; Özdemir, S.; Fritz, C.; Möbius, W.; Kleineidam, L.; Mandelkow, E.; Biernat, J.; Doğdu, C.; Peters, O.; Cosma, N.C.; et al. Plasma Extracellular Vesicle Tau and TDP-43 as Diagnostic Biomarkers in FTD and ALS. Nat. Med. 2024, 30, 1771–1783. [Google Scholar] [CrossRef]
- Ogobuiro, I.; Tuma, F. Physiology, Renal. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Hopsort, G.; Latapie, L.; Groenen Serrano, K.; Loubière, K.; Tzedakis, T. Deciphering the Human Urine Matrix: A New Approach to Simultaneously Quantify the Main Ions and Organic Compounds by Ion Chromatography/Mass Spectrometry (IC-MS). Anal. Bioanal. Chem. 2023, 415, 5337–5352. [Google Scholar] [CrossRef]
- Karagiannidis, A.G.; Theodorakopoulou, M.P.; Pella, E.; Sarafidis, P.A.; Ortiz, A. Uromodulin Biology. Nephrol. Dial. Transplant. 2024, 39, 1073–1087. [Google Scholar] [CrossRef]
- Julian, B.A.; Suzuki, H.; Suzuki, Y.; Tomino, Y.; Spasovski, G.; Novak, J. Sources of Urinary Proteins and Their Analysis by Urinary Proteomics for the Detection of Biomarkers of Disease. Proteom. Clin. Apps 2009, 3, 1029–1043. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Zhao, M.; Chen, X.; Sun, H.; Yang, Y.; Xiao, X.; Guo, Z.; Liu, X.; Lv, Y.; Chen, X.; et al. Comprehensive Analysis of Individual Variation in the Urinary Proteome Revealed Significant Gender Differences. Mol. Cell. Proteom. 2019, 18, 1110–1122. [Google Scholar] [CrossRef]
- Nagaraj, N.; Mann, M. Quantitative Analysis of the Intra- and Inter-Individual Variability of the Normal Urinary Proteome. J. Proteome Res. 2011, 10, 637–645. [Google Scholar] [CrossRef]
- Parikh, P.C.; Souza, S.D.; Obeid, W. Changes in the Composition of Urine over Six Hours Using Urine Dipstick Analysis and Automated Microscopy. BMC Nephrol. 2025, 26, 11. [Google Scholar] [CrossRef]
- Shepheard, S.R.; Wuu, J.; Cardoso, M.; Wiklendt, L.; Dinning, P.G.; Chataway, T.; Schultz, D.; Benatar, M.; Rogers, M.-L. Urinary P75ECD: A Prognostic, Disease Progression, and Pharmacodynamic Biomarker in ALS. Neurology 2017, 88, 1137–1143. [Google Scholar] [CrossRef] [PubMed]
- Rogers, M.-L.; Schultz, D.W.; Karnaros, V.; Shepheard, S.R. Urinary Biomarkers for Amyotrophic Lateral Sclerosis: Candidates, Opportunities and Considerations. Brain Commun. 2023, 5, fcad287. [Google Scholar] [CrossRef]
- Alhajj, M.; Babos, M. Physiology, Salivation. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Carpenter, G.H. The Secretion, Components, and Properties of Saliva. Annu. Rev. Food Sci. Technol. 2013, 4, 267–276. [Google Scholar] [CrossRef]
- Veerman, E.C.I.; Van Den Keybus, P.A.M.; Vissink, A.; Amerongen, A.V.N. Human Glandular Salivas: Their Separate Collection and Analysis. Eur. J. Oral. Sci. 1996, 104, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, S.P.; Williamson, R.T. A Review of Saliva: Normal Composition, Flow, and Function. J. Prosthet. Dent. 2001, 85, 162–169. [Google Scholar] [CrossRef]
- Scott, B.J.; Hassanwalia, R.; Linden, R.W. The Masticatory–Parotid Salivary Reflex in Edentulous Subjects. J. Oral. Rehabil. 1998, 25, 28–33. [Google Scholar] [CrossRef]
- Carlomagno, C.; Banfi, P.I.; Gualerzi, A.; Picciolini, S.; Volpato, E.; Meloni, M.; Lax, A.; Colombo, E.; Ticozzi, N.; Verde, F.; et al. Human Salivary Raman Fingerprint as Biomarker for the Diagnosis of Amyotrophic Lateral Sclerosis. Sci. Rep. 2020, 10, 10175. [Google Scholar] [CrossRef] [PubMed]
- Sjoqvist, S.; Otake, K. Saliva and Saliva Extracellular Vesicles for Biomarker Candidate Identification-Assay Development and Pilot Study in Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2023, 24, 5237. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Wu, Y.; Liu, G.; Jiang, Y.; Wang, X.; Wang, Z.; Feng, T. α-Synuclein in Salivary Extracellular Vesicles as a Potential Biomarker of Parkinson’s Disease. Neurosci. Lett. 2019, 696, 114–120. [Google Scholar] [CrossRef]
- Zhou, L.; Zhao, S.Z.; Koh, S.K.; Chen, L.; Vaz, C.; Tanavde, V.; Li, X.R.; Beuerman, R.W. In-Depth Analysis of the Human Tear Proteome. J. Proteom. 2012, 75, 3877–3885. [Google Scholar] [CrossRef]
- Von Thun Und Hohenstein-Blaul, N.; Funke, S.; Grus, F.H. Tears as a Source of Biomarkers for Ocular and Systemic Diseases. Exp. Eye Res. 2013, 117, 126–137. [Google Scholar] [CrossRef]
- Gachon, A.-M.; Richard, J.; Dastugue, B. Human Tears: Normal Protein Pattern and Individual Protein Determinations in Adults. Curr. Eye Res. 1982, 2, 301–308. [Google Scholar] [CrossRef]
- Gachon, A.M.; Verrelle, P.; Betail, G.; Dastugue, B. Immunological and Electrophoretic Studies of Human Tear Proteins. Exp. Eye Res. 1979, 29, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Tiffany, J.M. Tears in Health and Disease. Eye 2003, 17, 923–926. [Google Scholar] [CrossRef]
- Pieczyński, J.; Szulc, U.; Harazna, J.; Szulc, A.; Kiewisz, J. Tear Fluid Collection Methods: Review of Current Techniques. Eur. J. Ophthalmol. 2021, 31, 2245–2251. [Google Scholar] [CrossRef]
- Hagan, S.; Martin, E.; Enríquez-de-Salamanca, A. Tear Fluid Biomarkers in Ocular and Systemic Disease: Potential Use for Predictive, Preventive and Personalised Medicine. EPMA J. 2016, 7, 15. [Google Scholar] [CrossRef]
- Scholl, L.-S.; Demleitner, A.F.; Riedel, J.; Adachi, S.; Neuenroth, L.; Meijs, C.; Tzeplaeff, L.; Gomes, L.C.; Galhoz, A.; Cordts, I.; et al. Identification and Validation of a Tear Fluid-Derived Protein Biomarker Signature in Patients with Amyotrophic Lateral Sclerosis. Res. Sq. 2025. [Google Scholar] [CrossRef]
- Khanna, R.K.; Catanese, S.; Mortemousque, G.; Dupuy, C.; Lefevre, A.; Emond, P.; Beltran, S.; Gissot, V.; Pisella, P.-J.; Blasco, H.; et al. Metabolomics of Basal Tears in Amyotrophic Lateral Sclerosis: A Cross-Sectional Study. Ocul. Surf. 2024, 34, 363–369. [Google Scholar] [CrossRef]
- Gagliardi, D.; Rizzuti, M.; Masrori, P.; Saccomanno, D.; Del Bo, R.; Sali, L.; Meneri, M.; Scarcella, S.; Milone, I.; Hersmus, N.; et al. Exploiting the Role of CSF NfL, CHIT1, and miR-181b as Potential Diagnostic and Prognostic Biomarkers for ALS. J. Neurol. 2024, 271, 7557–7571. [Google Scholar] [CrossRef]
- Varghese, A.M.; Ghosh, M.; Bhagat, S.K.; Vijayalakshmi, K.; Preethish-Kumar, V.; Vengalil, S.; Chevula, P.-C.-R.; Nashi, S.; Polavarapu, K.; Sharma, M.; et al. Chitotriosidase, a Biomarker of Amyotrophic Lateral Sclerosis, Accentuates Neurodegeneration in Spinal Motor Neurons through Neuroinflammation. J. Neuroinflam. 2020, 17, 232. [Google Scholar] [CrossRef]
- Rosén, C.; Mitre, B.; Nellgård, B.; Axelsson, M.; Constantinescu, R.; Andersen, P.M.; Dalla, K.; Blennow, K.; Nilsson, G.; Zetterberg, H.; et al. High Levels of Neurofilament Light and YKL-40 in Cerebrospinal Fluid Are Related to Poor Outcome in ALS. J. Neurol. Sci. 2024, 463, 123112. [Google Scholar] [CrossRef]
- Basso, M.; Bonetto, V. Extracellular Vesicles and a Novel Form of Communication in the Brain. Front. Neurosci. 2016, 10, 127. [Google Scholar] [CrossRef]
- Bravo-Miana, R.D.C.; Arizaga-Echebarria, J.K.; Otaegui, D. Central Nervous System-Derived Extracellular Vesicles: The next Generation of Neural Circulating Biomarkers? Transl. Neurodegener. 2024, 13, 32. [Google Scholar] [CrossRef] [PubMed]
- Gomes, D.E.; Witwer, K.W. L1CAM-associated Extracellular Vesicles: A Systematic Review of Nomenclature, Sources, Separation, and Characterization. J. Extracell. Biol. 2022, 1, e35. [Google Scholar] [CrossRef]
- Janas, A.M.; Sapoń, K.; Janas, T.; Stowell, M.H.B.; Janas, T. Exosomes and Other Extracellular Vesicles in Neural Cells and Neurodegenerative Diseases. Biochim. Biophys. Acta-Biomembr. 2016, 1858, 1139–1151. [Google Scholar] [CrossRef]
- Ter-Ovanesyan, D.; Whiteman, S.; Gilboa, T.; Kowal, E.J.; Trieu, W.; Iyer, S.; Budnik, B.; Babila, C.M.; Heimberg, G.; Burgess, M.W.; et al. Identification of Markers for the Isolation of Neuron-Specific Extracellular Vesicles. bioRxiv 2024. [Google Scholar] [CrossRef]
- Tian, C.; Stewart, T.; Hong, Z.; Guo, Z.; Aro, P.; Soltys, D.; Pan, C.; Peskind, E.R.; Zabetian, C.P.; Shaw, L.M.; et al. Blood Extracellular Vesicles Carrying Synaptic Function- and Brain-related Proteins as Potential Biomarkers for Alzheimer’s Disease. Alzheimer’s Dement. 2023, 19, 909–923. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Zhang, Z.; Sultana, N.; Ericsson, M.; Martens, Y.A.; Sun, M.; Kanekiyo, T.; Ikezu, S.; Shaffer, S.A.; Ikezu, T. ATP1A3 as a Target for Isolating Neuron-Specific Extracellular Vesicles from Human Brain and Biofluids. Sci. Adv. 2023, 9, eadi3647. [Google Scholar] [CrossRef]
- Ramos-Zaldívar, H.M.; Polakovicova, I.; Salas-Huenuleo, E.; Corvalán, A.H.; Kogan, M.J.; Yefi, C.P.; Andia, M.E. Extracellular Vesicles through the Blood–Brain Barrier: A Review. Fluids Barriers CNS 2022, 19, 60. [Google Scholar] [CrossRef]
- Torrini, F.; Gil-Garcia, M.; Cardellini, J.; Frigerio, R.; Basso, M.; Gori, A.; Arosio, P. Monitoring Neurodegeneration through Brain-Derived Extracellular Vesicles in Biofluids. Trends Pharmacol. Sci. 2025, 46, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Counil, H.; Silva, R.O.; Rabanel, J.; Zaouter, C.; Haddad, M.; Ben Khedher, M.R.; Brambilla, D.; Fülöp, T.; Patten, S.A.; Ramassamy, C. Brain Penetration of Peripheral Extracellular Vesicles from Alzheimer’s Patients and Induction of Microglia Activation. J. Extracell. Biol. 2025, 4, e70027. [Google Scholar] [CrossRef]
- Chen, C.C.; Liu, L.; Ma, F.; Wong, C.W.; Guo, X.E.; Chacko, J.V.; Farhoodi, H.P.; Zhang, S.X.; Zimak, J.; Ségaliny, A.; et al. Elucidation of Exosome Migration Across the Blood–Brain Barrier Model In Vitro. Cel. Mol. Bioeng. 2016, 9, 509–529. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, J.; Stewart, T.; Sheng, L.; Li, N.; Bullock, K.; Song, N.; Shi, M.; Banks, W.A.; Zhang, J. Transmission of α-Synuclein-Containing Erythrocyte-Derived Extracellular Vesicles across the Blood-Brain Barrier via Adsorptive Mediated Transcytosis: Another Mechanism for Initiation and Progression of Parkinson’s Disease? Acta Neuropathol. Commun. 2017, 5, 71. [Google Scholar] [CrossRef]
- Morad, G.; Carman, C.V.; Hagedorn, E.J.; Perlin, J.R.; Zon, L.I.; Mustafaoglu, N.; Park, T.-E.; Ingber, D.E.; Daisy, C.C.; Moses, M.A. Tumor-Derived Extracellular Vesicles Breach the Intact Blood–Brain Barrier via Transcytosis. ACS Nano 2019, 13, 13853–13865. [Google Scholar] [CrossRef]
- Banks, W.A.; Sharma, P.; Bullock, K.M.; Hansen, K.M.; Ludwig, N.; Whiteside, T.L. Transport of Extracellular Vesicles across the Blood-Brain Barrier: Brain Pharmacokinetics and Effects of Inflammation. Int. J. Mol. Sci. 2020, 21, 4407. [Google Scholar] [CrossRef]
- Kur, I.-M.; Prouvot, P.-H.; Fu, T.; Fan, W.; Müller-Braun, F.; Das, A.; Das, S.; Deller, T.; Roeper, J.; Stroh, A.; et al. Neuronal Activity Triggers Uptake of Hematopoietic Extracellular Vesicles In Vivo. PLoS Biol. 2020, 18, e3000643. [Google Scholar] [CrossRef]
- Perets, N.; Betzer, O.; Shapira, R.; Brenstein, S.; Angel, A.; Sadan, T.; Ashery, U.; Popovtzer, R.; Offen, D. Golden Exosomes Selectively Target Brain Pathologies in Neurodegenerative and Neurodevelopmental Disorders. Nano Lett. 2019, 19, 3422–3431. [Google Scholar] [CrossRef]
- Rufino-Ramos, D.; Lule, S.; Mahjoum, S.; Ughetto, S.; Cristopher Bragg, D.; Pereira De Almeida, L.; Breakefield, X.O.; Breyne, K. Using Genetically Modified Extracellular Vesicles as a Non-Invasive Strategy to Evaluate Brain-Specific Cargo. Biomaterials 2022, 281, 121366. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Park, J.H.; Jo, A.; Lim, C.-W.; Park, J.-M.; Hwang, J.W.; Lee, K.S.; Kim, Y.-S.; Lee, H.; Moon, J. Blood-Derived APLP1+ Extracellular Vesicles Are Potential Biomarkers for the Early Diagnosis of Brain Diseases. Sci. Adv. 2025, 11, eado6894. [Google Scholar] [CrossRef]
- Katsu, M.; Hama, Y.; Utsumi, J.; Takashina, K.; Yasumatsu, H.; Mori, F.; Wakabayashi, K.; Shoji, M.; Sasaki, H. MicroRNA Expression Profiles of Neuron-Derived Extracellular Vesicles in Plasma from Patients with Amyotrophic Lateral Sclerosis. Neurosci. Lett. 2019, 708, 134176. [Google Scholar] [CrossRef]
- Banack, S.A.; Dunlop, R.A.; Cox, P.A. An miRNA Fingerprint Using Neural-Enriched Extracellular Vesicles from Blood Plasma: Towards a Biomarker for Amyotrophic Lateral Sclerosis/Motor Neuron Disease. Open Biol. 2020, 10, 200116. [Google Scholar] [CrossRef]
- Dunlop, R.A.; Banack, S.A.; Cox, P.A. L1CAM Immunocapture Generates a Unique Extracellular Vesicle Population with a Reproducible miRNA Fingerprint. RNA Biol. 2023, 20, 140–148. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Nuclear Acids In Human Blood Plasma. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Kustanovich, A.; Schwartz, R.; Peretz, T.; Grinshpun, A. Life and Death of Circulating Cell-Free DNA. Cancer Biol. Ther. 2019, 20, 1057–1067. [Google Scholar] [CrossRef]
- Giacona, M.B.; Ruben, G.C.; Iczkowski, K.A.; Roos, T.B.; Porter, D.M.; Sorenson, G.D. Cell-Free DNA in Human Blood Plasma: Length Measurements in Patients with Pancreatic Cancer and Healthy Controls. Pancreas 1998, 17, 89–97. [Google Scholar] [CrossRef]
- Rumore, P.; Muralidhar, B.; Lin, M.; Lai, C.; Steinman, C.R. Haemodialysis as a Model for Studying Endogenous Plasma DNA: Oligonucleosome-like Structure and Clearance. Clin. Exp. Immunol. 2008, 90, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Snyder, M.W.; Kircher, M.; Hill, A.J.; Daza, R.M.; Shendure, J. Cell-Free DNA Comprises an In Vivo Nucleosome Footprint That Informs Its Tissues-Of-Origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef]
- Moss, J.; Magenheim, J.; Neiman, D.; Zemmour, H.; Loyfer, N.; Korach, A.; Samet, Y.; Maoz, M.; Druid, H.; Arner, P.; et al. Comprehensive Human Cell-Type Methylation Atlas Reveals Origins of Circulating Cell-Free DNA in Health and Disease. Nat. Commun. 2018, 9, 5068. [Google Scholar] [CrossRef]
- Gaitsch, H.; Franklin, R.J.M.; Reich, D.S. Cell-Free DNA-Based Liquid Biopsies in Neurology. Brain 2023, 146, 1758–1774. [Google Scholar] [CrossRef]
- Lehmann-Werman, R.; Neiman, D.; Zemmour, H.; Moss, J.; Magenheim, J.; Vaknin-Dembinsky, A.; Rubertsson, S.; Nellgård, B.; Blennow, K.; Zetterberg, H.; et al. Identification of Tissue-Specific Cell Death Using Methylation Patterns of Circulating DNA. Proc. Natl. Acad. Sci. USA 2016, 113, E1826–E1834. [Google Scholar] [CrossRef]
- Vasioukhin, V.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Stroun, M. Point Mutations of the N-ras Gene in the Blood Plasma DNA of Patients with Myelodysplastic Syndrome or Acute Myelogenous Leukaemia. Br. J. Haematol. 1994, 86, 774–779. [Google Scholar] [CrossRef]
- Stroun, M.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Beljanski, M. Neoplastic Characteristics of the DNA Found in the Plasma of Cancer Patients. Oncology 1989, 46, 318–322. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Liu, A.P.Y.; Smith, K.S.; Kumar, R.; Paul, L.; Bihannic, L.; Lin, T.; Maass, K.K.; Pajtler, K.W.; Chintagumpala, M.; Su, J.M.; et al. Serial Assessment of Measurable Residual Disease in Medulloblastoma Liquid Biopsies. Cancer Cell 2021, 39, 1519–1530.E4. [Google Scholar] [CrossRef]
- Wang, Y.; Springer, S.; Zhang, M.; McMahon, K.W.; Kinde, I.; Dobbyn, L.; Ptak, J.; Brem, H.; Chaichana, K.; Gallia, G.L.; et al. Detection of Tumor-Derived DNA in Cerebrospinal Fluid of Patients with Primary Tumors of the Brain and Spinal Cord. Proc. Natl. Acad. Sci. USA 2015, 112, 9704–9709. [Google Scholar] [CrossRef]
- Iser, F.; Hinz, F.; Hoffmann, D.C.; Grassl, N.; Güngör, C.; Meyer, J.; Dörner, L.; Hofmann, L.; Kelbch, V.; Göbel, K.; et al. Cerebrospinal Fluid cfDNA Sequencing for Classification of Central Nervous System Glioma. Clin. Cancer Res. 2024, 30, 2974–2985. [Google Scholar] [CrossRef]
- Miller, A.M.; Shah, R.H.; Pentsova, E.I.; Pourmaleki, M.; Briggs, S.; Distefano, N.; Zheng, Y.; Skakodub, A.; Mehta, S.A.; Campos, C.; et al. Tracking Tumour Evolution in Glioma through Liquid Biopsies of Cerebrospinal Fluid. Nature 2019, 565, 654–658. [Google Scholar] [CrossRef]
- Robichaud, P.-P.; Arseneault, M.; O’Connell, C.; Ouellette, R.J.; Morin, P.J. Circulating Cell-Free DNA as Potential Diagnostic Tools for Amyotrophic Lateral Sclerosis. Neurosci. Lett. 2021, 750, 135813. [Google Scholar] [CrossRef]
- Caggiano, C.; Celona, B.; Garton, F.; Mefford, J.; Black, B.L.; Henderson, R.; Lomen-Hoerth, C.; Dahl, A.; Zaitlen, N. Comprehensive Cell Type Decomposition of Circulating Cell-Free DNA with CelFiE. Nat. Commun. 2021, 12, 2717. [Google Scholar] [CrossRef]
- Pollard, C.; Aston, K.; Emery, B.R.; Hill, J.; Jenkins, T. Detection of Neuron-Derived cfDNA in Blood Plasma: A New Diagnostic Approach for Neurodegenerative Conditions. Front. Neurol. 2023, 14, 1272960. [Google Scholar] [CrossRef]
- Trumpff, C.; Michelson, J.; Lagranha, C.J.; Taleon, V.; Karan, K.R.; Sturm, G.; Lindqvist, D.; Fernström, J.; Moser, D.; Kaufman, B.A.; et al. Stress and Circulating Cell-Free Mitochondrial DNA: A Systematic Review of Human Studies, Physiological Considerations, and Technical Recommendations. Mitochondrion 2021, 59, 225–245. [Google Scholar] [CrossRef]
- Wojtkowska, M.; Karczewska, N.; Pacewicz, K.; Pacak, A.; Kopeć, P.; Florczak-Wyspiańska, J.; Popławska-Domaszewicz, K.; Małkiewicz, T.; Sokół, B. Quantification of Circulating Cell-Free DNA in Idiopathic Parkinson’s Disease Patients. Int. J. Mol. Sci. 2024, 25, 2818. [Google Scholar] [CrossRef]
- Takousis, P.; Devonshire, A.S.; Redshaw, N.; Von Baumgarten, L.; Whale, A.S.; Jones, G.M.; Fernandez-Gonzalez, A.; Martin, J.; Foy, C.A.; Alexopoulos, P.; et al. A Standardised Methodology for the Extraction and Quantification of Cell-Free DNA in Cerebrospinal Fluid and Application to Evaluation of Alzheimer’s Disease and Brain Cancers. New Biotechnol. 2022, 72, 97–106. [Google Scholar] [CrossRef]
- Li, J.; Gao, C.; Wang, Q.; Liu, J.; Xie, Z.; Zhao, Y.; Yu, M.; Zheng, Y.; Lv, H.; Zhang, W.; et al. Elevated Serum Circulating Cell-free Mitochondrial DNA in Amyotrophic Lateral Sclerosis. Eur. J. Neurol. 2024, 31, e16493. [Google Scholar] [CrossRef]
- Mugoni, V.; Ciani, Y.; Quaini, O.; Tomasini, S.; Notarangelo, M.; Vannuccini, F.; Marinelli, A.; Leonardi, E.; Pontalti, S.; Martinelli, A.; et al. Integrating Extracellular Vesicle and Circulating Cell-free DNA Analysis Using a Single Plasma Aliquot Improves the Detection of HER2 Positivity in Breast Cancer Patients. J. Extracell. Biol. 2023, 2, e108. [Google Scholar] [CrossRef]
- Gentile, J.E.; Heiss, C.; Corridon, T.L.; Mortberg, M.A.; Fruhwürth, S.; Guzman, K.; Grötschel, L.; Chan, K.; Herring, N.C.; Janicki, T.; et al. Evidence That Minocycline Treatment Confounds the Interpretation of Neurofilament as a Biomarker. Brain Commun. 2025, 7, fcaf175. [Google Scholar] [CrossRef]
- A Virata, M.C.; Catahay, J.A.; Lippi, G.; Henry, B.M. Neurofilament Light Chain: A Biomarker at the Crossroads of Clarity and Confusion for Gene-Directed Therapies. Neurodegener. Dis. Manag. 2024, 14, 227–239. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pisoni, L.; Donini, L.; Gagni, P.; Pennuto, M.; Ratti, A.; Verde, F.; Ticozzi, N.; Mandrioli, J.; Calvo, A.; Basso, M. Barriers in the Nervous System: Challenges and Opportunities for Novel Biomarkers in Amyotrophic Lateral Sclerosis. Cells 2025, 14, 848. https://doi.org/10.3390/cells14110848
Pisoni L, Donini L, Gagni P, Pennuto M, Ratti A, Verde F, Ticozzi N, Mandrioli J, Calvo A, Basso M. Barriers in the Nervous System: Challenges and Opportunities for Novel Biomarkers in Amyotrophic Lateral Sclerosis. Cells. 2025; 14(11):848. https://doi.org/10.3390/cells14110848
Chicago/Turabian StylePisoni, Lorena, Luisa Donini, Paola Gagni, Maria Pennuto, Antonia Ratti, Federico Verde, Nicola Ticozzi, Jessica Mandrioli, Andrea Calvo, and Manuela Basso. 2025. "Barriers in the Nervous System: Challenges and Opportunities for Novel Biomarkers in Amyotrophic Lateral Sclerosis" Cells 14, no. 11: 848. https://doi.org/10.3390/cells14110848
APA StylePisoni, L., Donini, L., Gagni, P., Pennuto, M., Ratti, A., Verde, F., Ticozzi, N., Mandrioli, J., Calvo, A., & Basso, M. (2025). Barriers in the Nervous System: Challenges and Opportunities for Novel Biomarkers in Amyotrophic Lateral Sclerosis. Cells, 14(11), 848. https://doi.org/10.3390/cells14110848









