TIMP-1 Modulation Correlates with KRAS Dependency and EMT Induction in NSCLC
Abstract
1. Introduction
2. Methods
2.1. Cell Lines and Culture Conditions
2.2. Reverse Transcription Quantitative Real-Time PCR (RT-qPCR)
2.3. Western Blotting
2.4. RAS Activity Assay
2.5. siRNA Transfection
2.6. A549 KD Cells with Rescue Plasmid
- (1)
- CAAGCTTAAGCGGCCGCTAGTTATTAATAGTAATCAATTACG;
- (2)
- GTCGGAATTGCAaAAcGCaGTCTGTGGGTG;
- (3)
- CACCCACAGACtGCgTTtTGCAATTCCGAC;
- (4)
- GTCCCTCGACGAATTCCTGGGACCGAACCCCGCGT.
2.7. TUNEL Assay
2.8. Anchorage-Independent Soft Agar Assay
2.9. Wound Assay
2.10. Spheroid Formation Assay
2.11. Immunofluorescence Assay
2.12. PDK1 Inhibitor Treatment
2.13. Bioinformatic Analysis
2.14. Statistical Analysis
3. Results
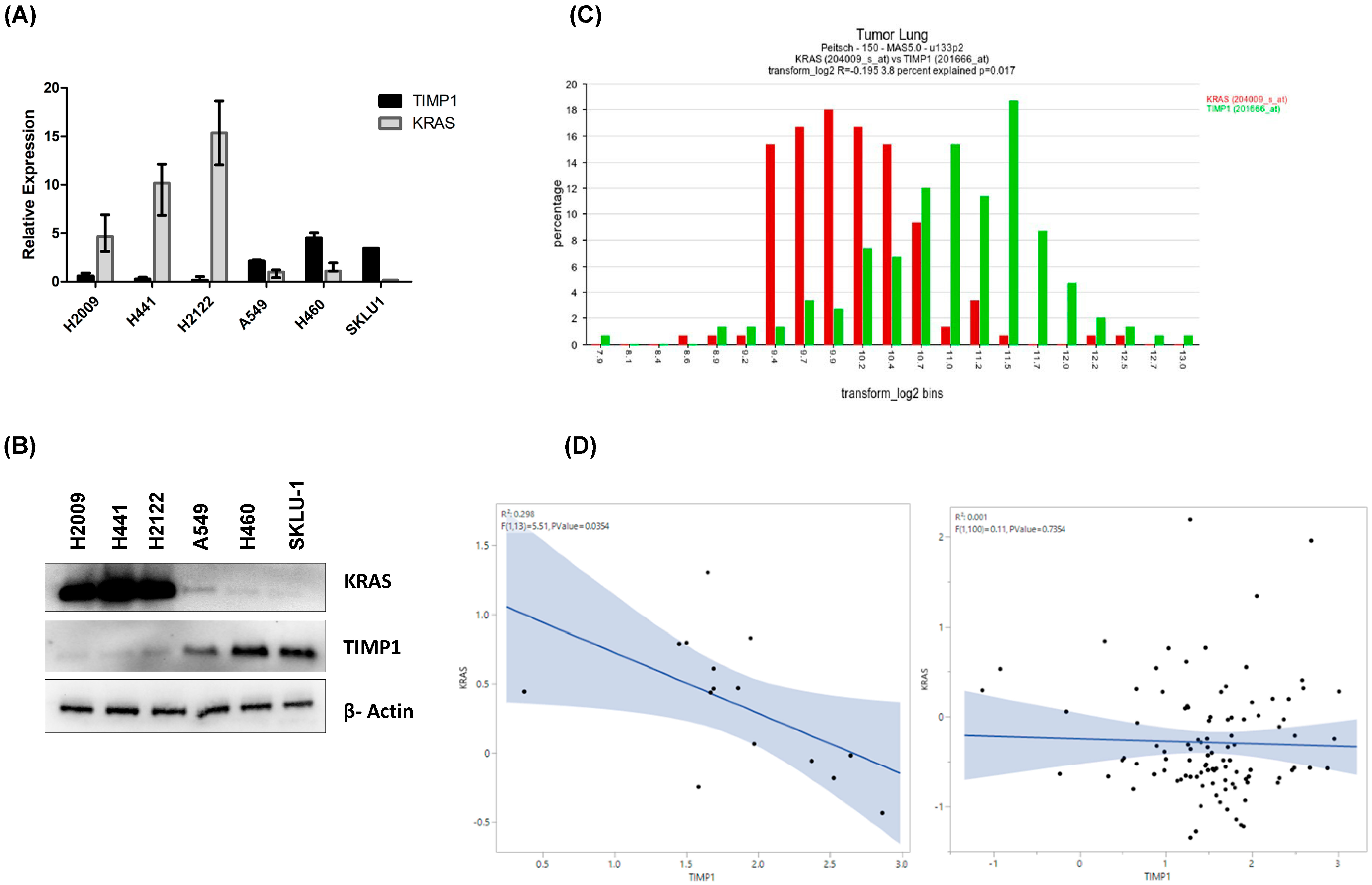
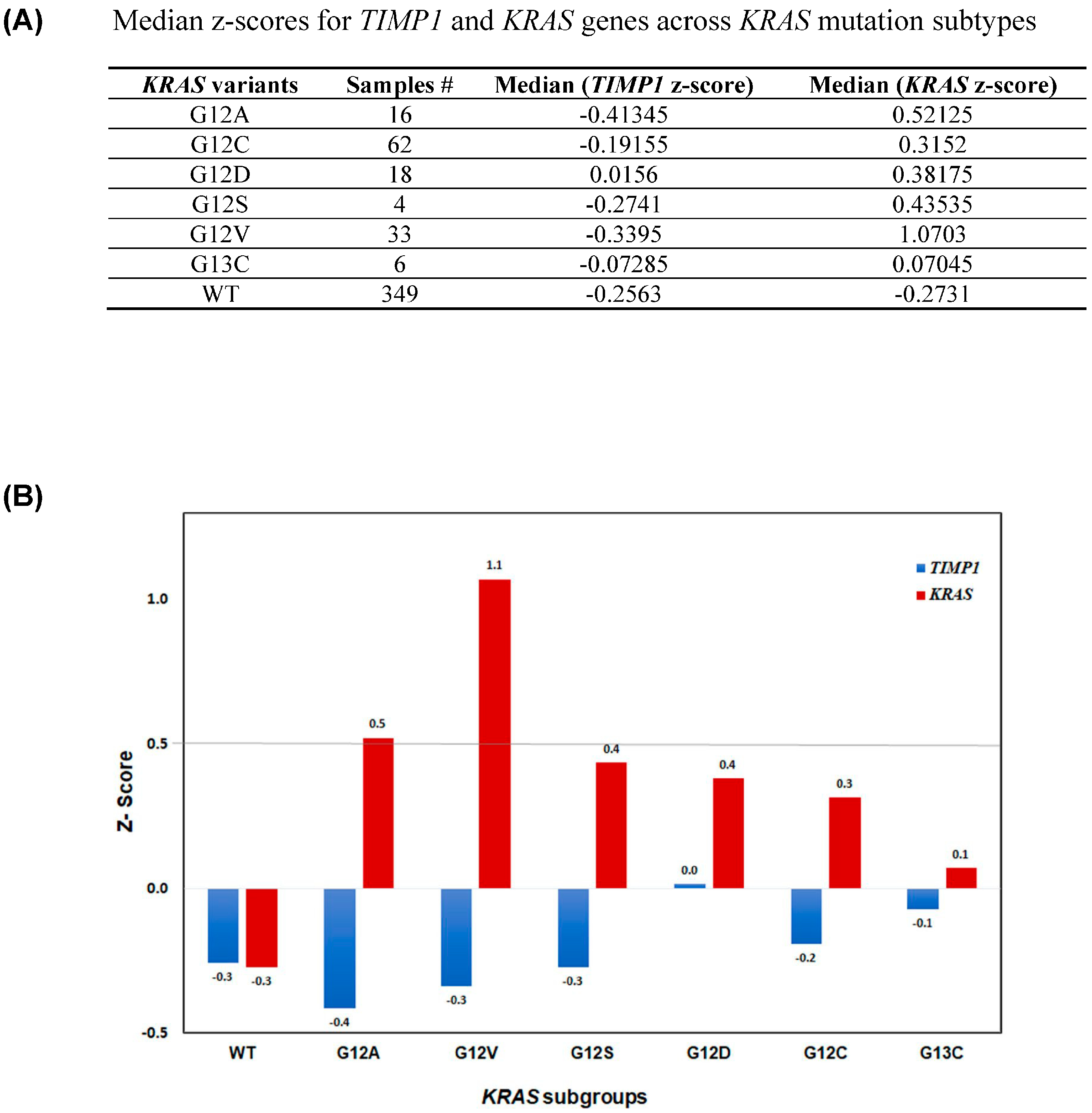
3.1. TIMP-1 Levels Correlate with KRAS Dependency
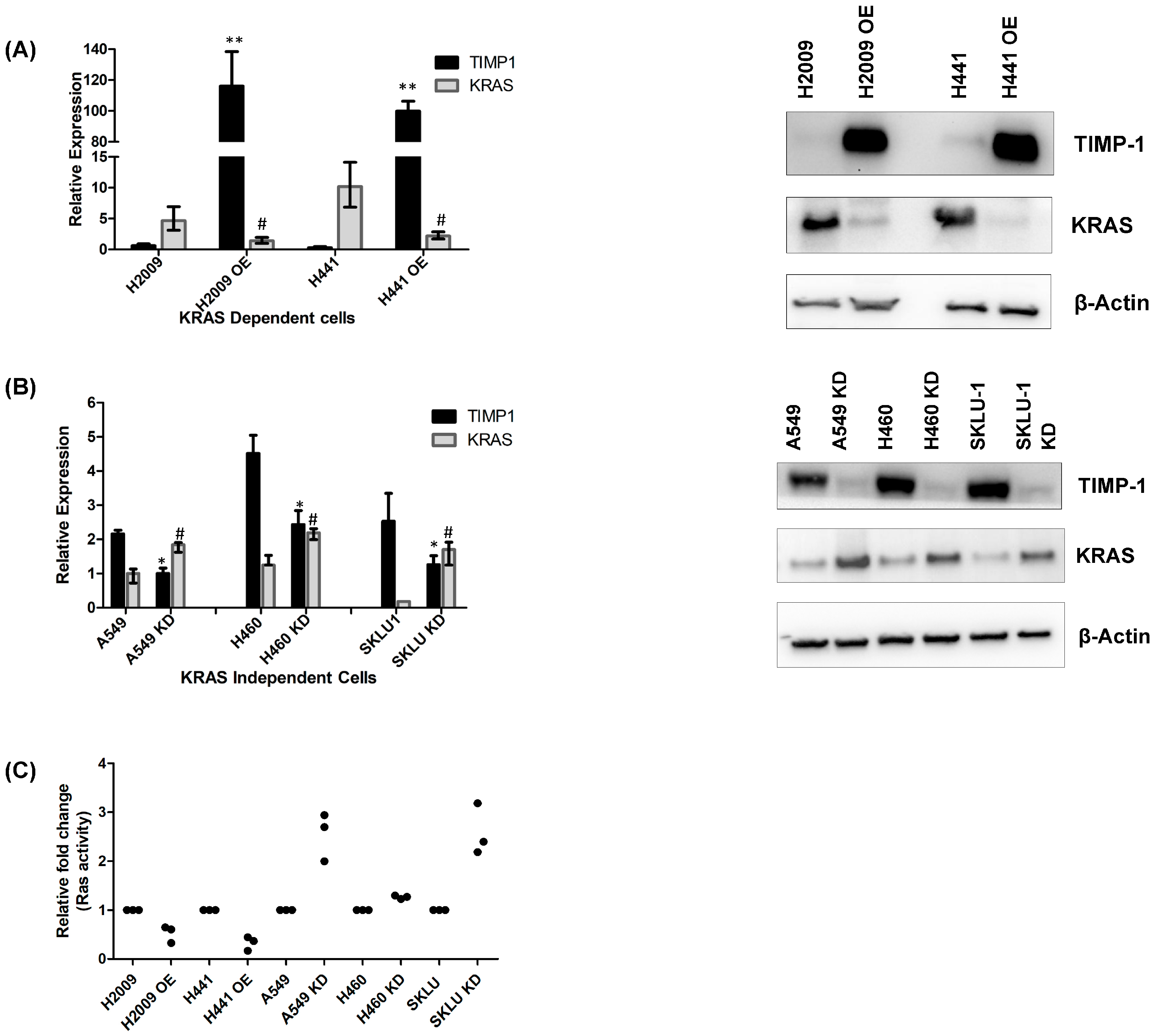
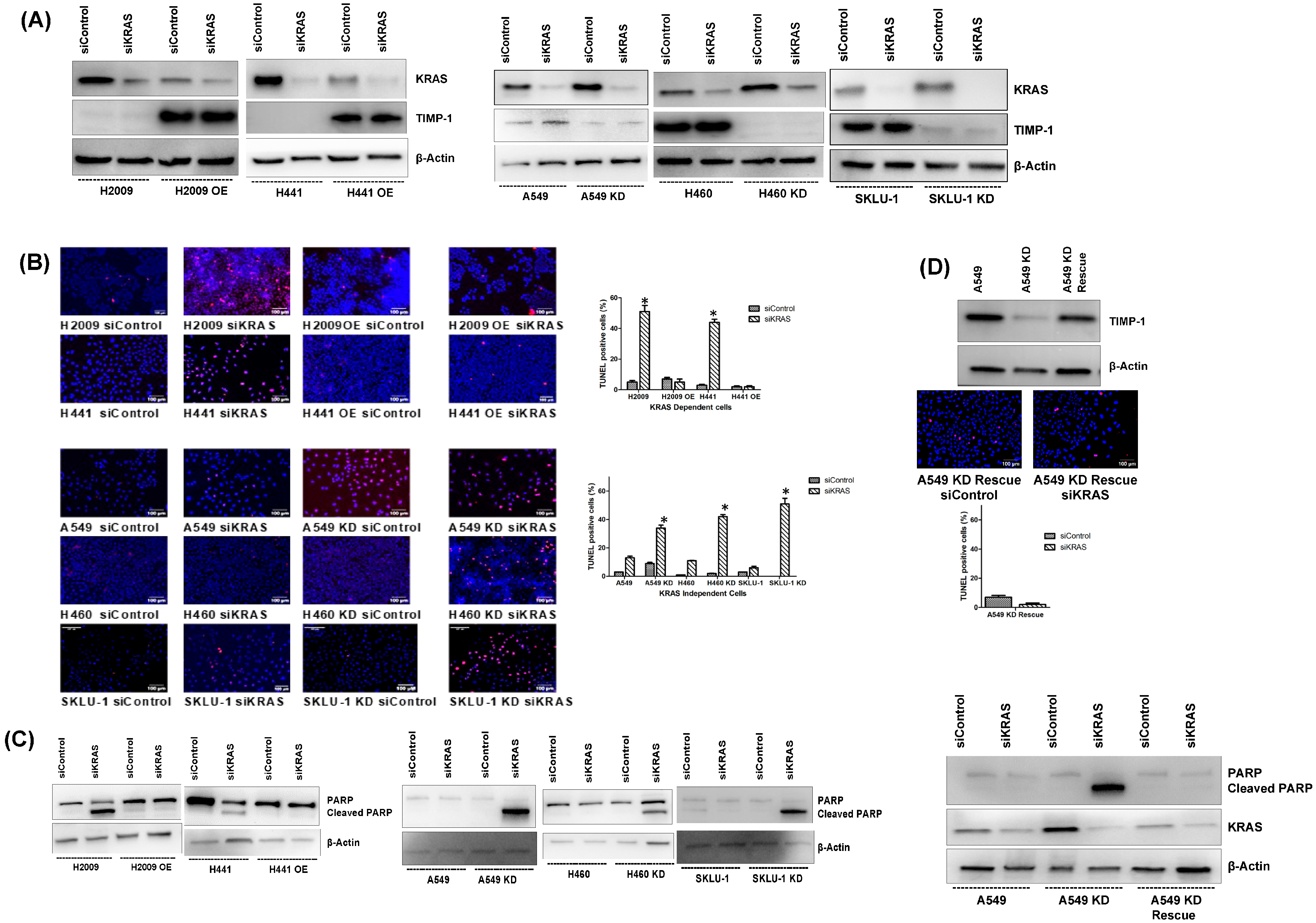
3.2. Modulating TIMP-1 Alters KRAS Levels and Affects KRAS Dependency
3.3. Modulating TIMP-1 Levels Alters Apoptosis upon KRAS Ablation
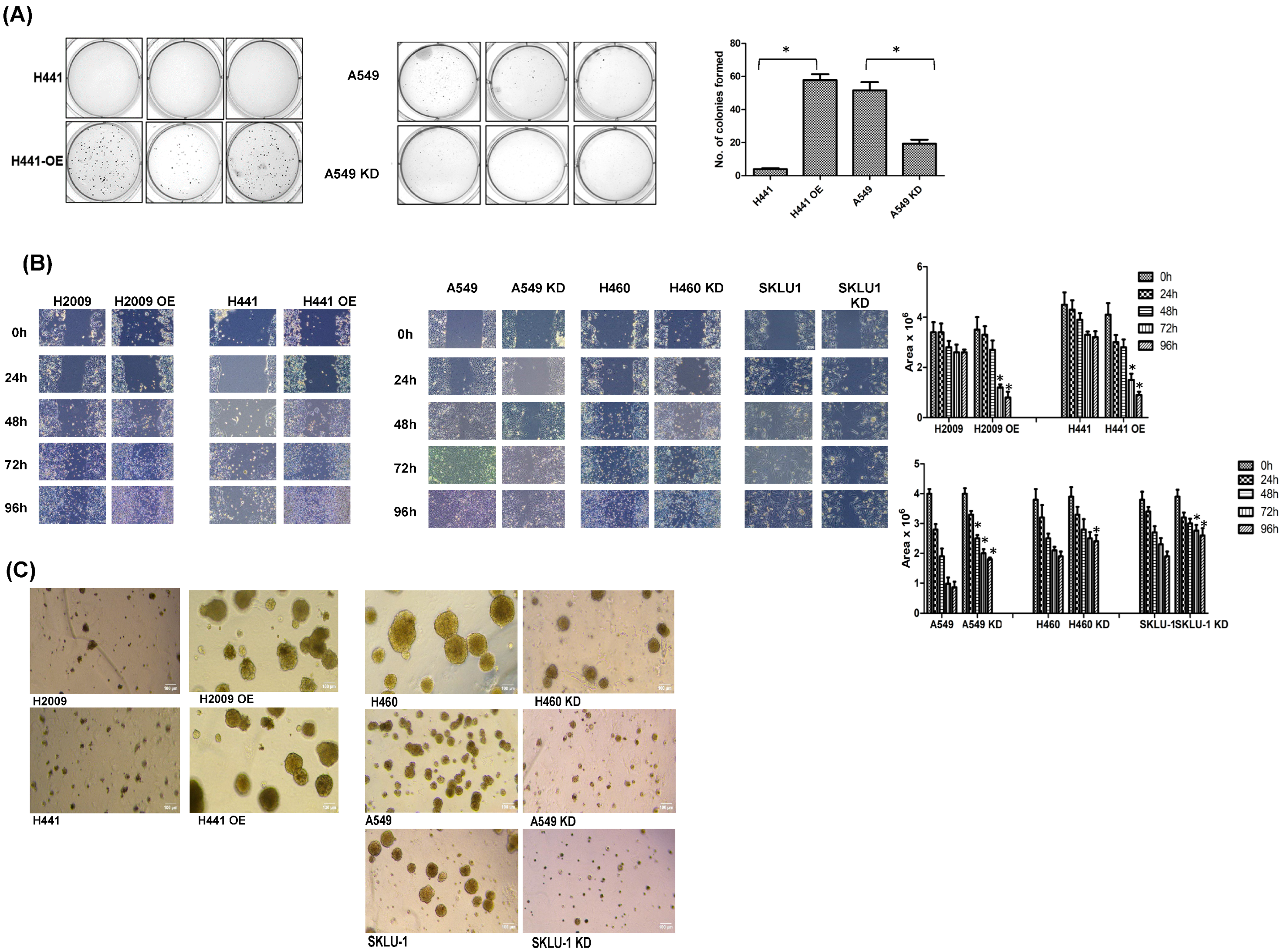
3.4. Modulating TIMP-1 Levels Alters In Vitro Tumorigenic Profile of NSCLC Cells
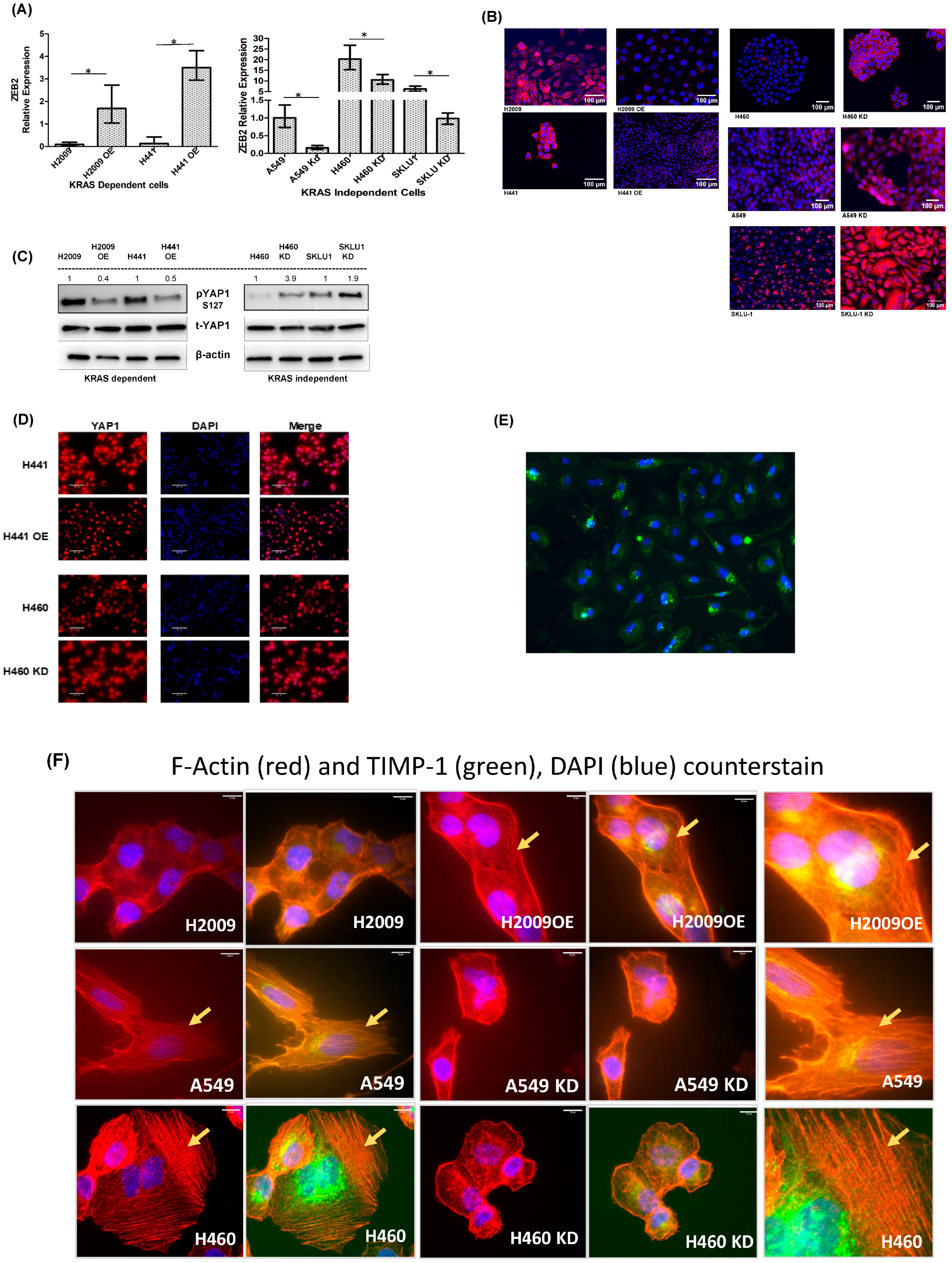
3.5. Differential EMT-Related Marker Expression Is Observed with TIMP-1 Modulation in KRAS-Dependent and -Independent Cells
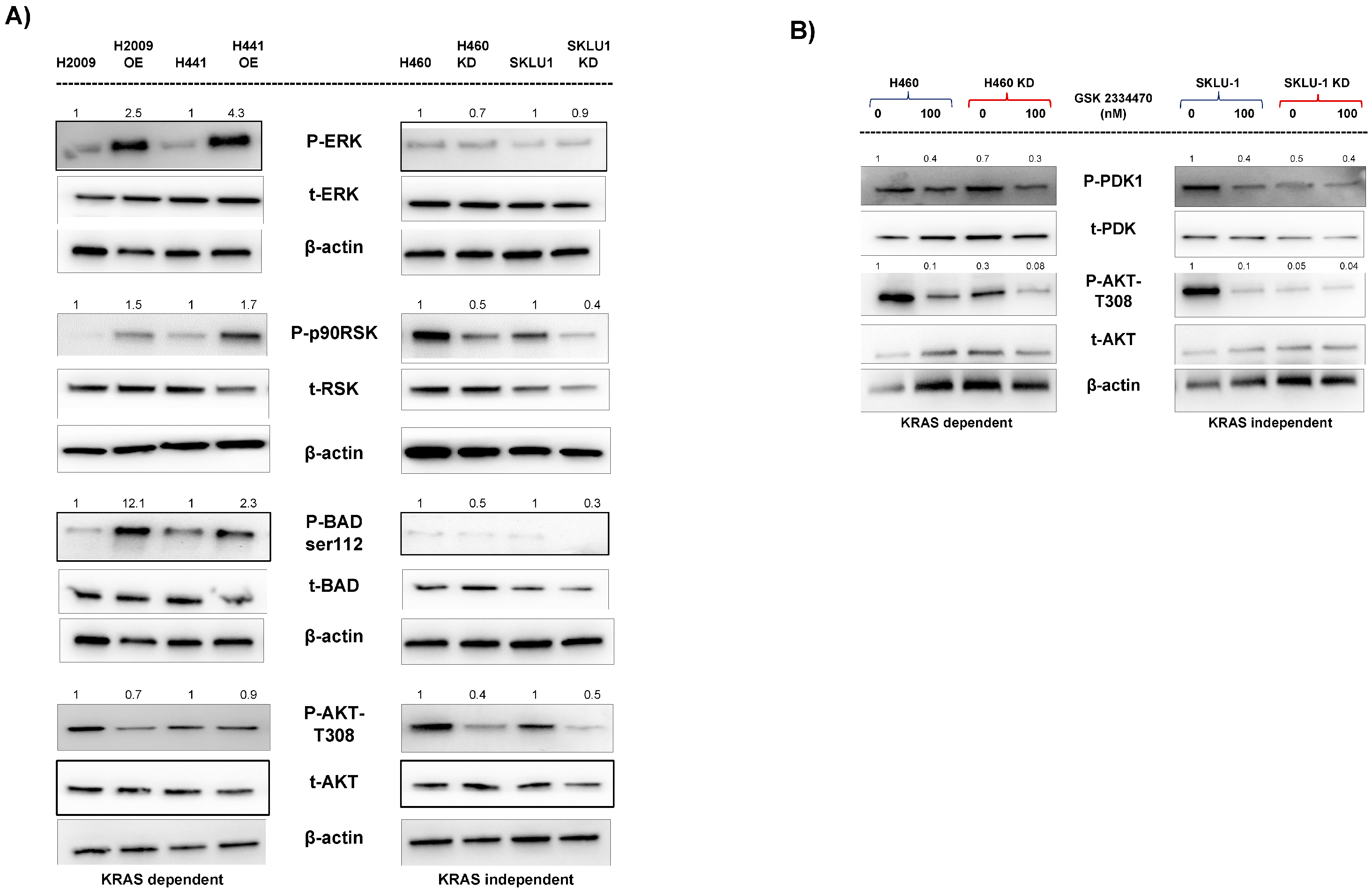
3.6. Different Signaling Pathways Utilized by KRAS-Dependent and -Independent Cell Lines
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AKT | Ak strain transforming |
| BAD | Bcl-2-associated death promoter |
| DMEM | Dulbecco’s Modified Eagle Medium |
| EGFR | Epidermal Growth Factor Receptor gene |
| EMT | Epithelial–Mesenchymal Transition |
| ERK | Extracellular signal-Regulated Kinase |
| KRAS | Kirsten rat sarcoma viral oncogene homolog |
| LUAD | Lung Adenocarcinoma |
| MDCK | Madin-Darby Canine Kidney cells |
| MMP | Matrix metalloproteinase |
| NSCLC | Non-Small Cell Lung Carcinoma |
| PARP | Poly ADP-ribose polymerase |
| PDAC | Pancreatic Ductal Adenocarcinoma |
| PMSF | Phenylmethylsulfonyl fluoride |
| PFA | Para Formaldehyde |
| RSK | Ribosomal S6 Kinase |
| SD | Standard Deviation |
| TIMP-1 | Tissue Inhibitor of Metalloproteinase-1 |
| TIMP-1 KD | TIMP-1 Knockdown |
| TIMP-1 OE | TIMP-1 Over-expressing |
| TUNEL | Terminal deoxynucleotidyl transferase dUTP nick end labeling |
| TWIST-1 | Twist-associated protein -1 |
| YAP1 | yes-associated protein 1 |
| ZEB | Zinc Finger E-Box Binding Homeobox 1 or 2 |
References
- Suda, K.; Tomizawa, K.; Mitsudomi, T. Biological and clinical significance of KRAS mutations in lung cancer: An oncogenic driver that contrasts with EGFR mutation. Cancer Metastasis Rev. 2010, 29, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Getz, G.; Wheeler, D.A.; Mardis, E.R.; McLellan, M.D.; Cibulskis, K.; Sougnez, C.; Greulich, H.; Muzny, D.M.; Morgan, M.B.; et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008, 455, 1069–1075. [Google Scholar] [CrossRef]
- Tomasini, P.; Walia, P.; Labbe, C.; Jao, K.; Leighl, N.B. Targeting the KRAS Pathway in Non-Small Cell Lung Cancer. Oncologist 2016, 21, 1450–1460. [Google Scholar] [CrossRef] [PubMed]
- McCormick, F. Cancer therapy based on oncogene addiction. J. Surg. Oncol. 2011, 103, 464–467. [Google Scholar] [CrossRef]
- Salgia, R.; Pharaon, R.; Mambetsariev, I.; Nam, A.; Sattler, M. The improbable targeted therapy: KRAS as an emerging target in non-small cell lung cancer (NSCLC). Cell Rep. Med. 2021, 2, 100186. [Google Scholar] [CrossRef]
- Riely, G.J.; Kris, M.G.; Rosenbaum, D.; Marks, J.; Li, A.; Chitale, D.A.; Nafa, K.; Riedel, E.R.; Hsu, M.; Pao, W.; et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin. Cancer Res. 2008, 14, 5731–5734. [Google Scholar] [CrossRef]
- Yuan, T.L.; Amzallag, A.; Bagni, R.; Yi, M.; Afghani, S.; Burgan, W.; Fer, N.; Strathern, L.A.; Powell, K.; Smith, B.; et al. Differential Effector Engagement by Oncogenic KRAS. Cell Rep. 2018, 22, 1889–1902. [Google Scholar] [CrossRef]
- Weinstein, I.B. Cancer. Addiction to oncogenes—The Achilles heal of cancer. Science 2002, 297, 63–64. [Google Scholar] [CrossRef]
- Singh, A.; Greninger, P.; Rhodes, D.; Koopman, L.; Violette, S.; Bardeesy, N.; Settleman, J. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 2009, 15, 489–500. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Brew, K.; Nagase, H. The tissue inhibitors of metalloproteinases (TIMPs): An ancient family with structural and functional diversity. Biochim. Biophys. Acta 2010, 1803, 55–71. [Google Scholar] [CrossRef]
- Jackson, H.W.; Defamie, V.; Waterhouse, P.; Khokha, R. TIMPs: Versatile extracellular regulators in cancer. Nat. Rev. Cancer 2017, 17, 38–53. [Google Scholar] [CrossRef]
- Nalluri, S.; Ghoshal-Gupta, S.; Kutiyanawalla, A.; Gayatri, S.; Lee, B.R.; Jiwani, S.; Rojiani, A.M.; Rojiani, M.V. TIMP-1 Inhibits Apoptosis in Lung Adenocarcinoma Cells via Interaction with Bcl-2. PLoS ONE 2015, 10, e0137673. [Google Scholar] [CrossRef]
- Toricelli, M.; Melo, F.H.M.; Hunger, A.; Zanatta, D.; Strauss, B.E.; Jasiulionis, M.G. Timp1 Promotes Cell Survival by Activating the PDK1 Signaling Pathway in Melanoma. Cancers 2017, 9, 37. [Google Scholar] [CrossRef]
- Liu, X.W.; Taube, M.E.; Jung, K.K.; Dong, Z.; Lee, Y.J.; Roshy, S.; Sloane, B.F.; Fridman, R.; Kim, H.R. Tissue inhibitor of metalloproteinase-1 protects human breast epithelial cells from extrinsic cell death: A potential oncogenic activity of tissue inhibitor of metalloproteinase-1. Cancer Res. 2005, 65, 898–906. [Google Scholar] [CrossRef]
- Lambert, E.; Bridoux, L.; Devy, J.; Dasse, E.; Sowa, M.L.; Duca, L.; Hornebeck, W.; Martiny, L.; Petitfrere-Charpentier, E. TIMP-1 binding to proMMP-9/CD44 complex localized at the cell surface promotes erythroid cell survival. Int. J. Biochem. Cell Biol. 2009, 41, 1102–1115. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, L.; Howard, J.; Kolhe, R.; Rojiani, A.M.; Rojiani, M.V. TIMP-1-Mediated Chemoresistance via Induction of IL-6 in NSCLC. Cancers 2019, 11, 1184. [Google Scholar] [CrossRef]
- Fu, Z.Y.; Lv, J.H.; Ma, C.Y.; Yang, D.P.; Wang, T. Tissue inhibitor of metalloproteinase-1 decreased chemosensitivity of MDA-435 breast cancer cells to chemotherapeutic drugs through the PI3K/AKT/NF-small ka, CyrillicB pathway. Biomed. Pharmacother. 2011, 65, 163–167. [Google Scholar] [CrossRef]
- D’Costa, Z.; Jones, K.; Azad, A.; van Stiphout, R.; Lim, S.Y.; Gomes, A.L.; Kinchesh, P.; Smart, S.C.; Gillies McKenna, W.; Buffa, F.M.; et al. Gemcitabine-Induced TIMP1 Attenuates Therapy Response and Promotes Tumor Growth and Liver Metastasis in Pancreatic Cancer. Cancer Res. 2017, 77, 5952–5962. [Google Scholar] [CrossRef]
- Sa, Y.; Li, C.; Li, H.; Guo, H. TIMP-1 Induces alpha-Smooth Muscle Actin in Fibroblasts to Promote Urethral Scar Formation. Cell Physiol. Biochem. 2015, 35, 2233–2243. [Google Scholar] [CrossRef]
- Jung, Y.S.; Liu, X.W.; Chirco, R.; Warner, R.B.; Fridman, R.; Kim, H.R. TIMP-1 induces an EMT-like phenotypic conversion in MDCK cells independent of its MMP-inhibitory domain. PLoS ONE 2012, 7, e38773. [Google Scholar] [CrossRef]
- D’Angelo, R.C.; Liu, X.W.; Najy, A.J.; Jung, Y.S.; Won, J.; Chai, K.X.; Fridman, R.; Kim, H.R. TIMP-1 via TWIST1 induces EMT phenotypes in human breast epithelial cells. Mol. Cancer Res. 2014, 12, 1324–1333. [Google Scholar] [CrossRef]
- Mathias, R.A.; Wang, B.; Ji, H.; Kapp, E.A.; Moritz, R.L.; Zhu, H.J.; Simpson, R.J. Secretome-based proteomic profiling of Ras-transformed MDCK cells reveals extracellular modulators of epithelial-mesenchymal transition. J. Proteome Res. 2009, 8, 2827–2837. [Google Scholar] [CrossRef]
- Ghoshal-Gupta, S.; Kutiyanawalla, A.; Lee, B.R.; Ojha, J.; Nurani, A.; Mondal, A.K.; Kolhe, R.; Rojiani, A.M.; Rojiani, M.V. TIMP-1 downregulation modulates miR-125a-5p expression and triggers the apoptotic pathway. Oncotarget 2018, 9, 8941–8956. [Google Scholar] [CrossRef] [PubMed]
- Rojiani, M.V.; Ghoshal-Gupta, S.; Kutiyanawalla, A.; Mathur, S.; Rojiani, A.M. TIMP-1 overexpression in lung carcinoma enhances tumor kinetics and angiogenesis in brain metastasis. J. Neuropathol. Exp. Neurol. 2015, 74, 293–304. [Google Scholar] [CrossRef]
- Xiao, W.; Ahluwalia, P.; Wang, L.; Howard, J.; Kolhe, R.; Rojiani, A.M.; Rojiani, M.V. TIMP-1 Dependent Modulation of Metabolic Profiles Impacts Chemoresistance in NSCLC. Cells 2022, 11, 3036. [Google Scholar] [CrossRef]
- Tomida, S.; Takeuchi, T.; Shimada, Y.; Arima, C.; Matsuo, K.; Mitsudomi, T.; Yatabe, Y.; Takahashi, T. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J. Clin. Oncol. 2009, 27, 2793–2799. [Google Scholar] [CrossRef]
- Tarca, A.L.; Bhatti, G.; Romero, R. A comparison of gene set analysis methods in terms of sensitivity, prioritization and specificity. PLoS ONE 2013, 8, e79217. [Google Scholar] [CrossRef]
- Pettazzoni, P.; Viale, A.; Shah, P.; Carugo, A.; Ying, H.; Wang, H.; Genovese, G.; Seth, S.; Minelli, R.; Green, T.; et al. Genetic events that limit the efficacy of MEK and RTK inhibitor therapies in a mouse model of KRAS-driven pancreatic cancer. Cancer Res. 2015, 75, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Adachi, Y.; Kimura, R.; Hirade, K.; Ebi, H. Escaping KRAS: Gaining Autonomy and Resistance to KRAS Inhibition in KRAS Mutant Cancers. Cancers 2021, 13, 5081. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Yao, W.; Ying, H.; Hua, S.; Liewen, A.; Wang, Q.; Zhong, Y.; Wu, C.J.; Sadanandam, A.; Hu, B.; et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 2014, 158, 185–197. [Google Scholar] [CrossRef]
- Yu, M.; Chen, Y.; Li, X.; Yang, R.; Zhang, L.; Huangfu, L.; Zheng, N.; Zhao, X.; Lv, L.; Hong, Y.; et al. YAP1 contributes to NSCLC invasion and migration by promoting Slug transcription via the transcription co-factor TEAD. Cell Death Dis. 2018, 9, 464. [Google Scholar] [CrossRef]
- Chen, N.; Golczer, G.; Ghose, S.; Lin, B.; Langenbucher, A.; Webb, J.; Bhanot, H.; Abt, N.B.; Lin, D.; Varvares, M.; et al. YAP1 maintains active chromatin state in head and neck squamous cell carcinomas that promotes tumorigenesis through cooperation with BRD4. Cell Rep. 2022, 39, 110970. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zhou, K.; Wang, Q.; Huang, Y.; Ji, J.; Peng, Y.; Zhang, X.; Zheng, T.; Zhang, Z.; Chong, D.; et al. The transcription factor RUNX2 fuels YAP1 signaling and gastric cancer tumorigenesis. Cancer Sci. 2021, 112, 3533–3544. [Google Scholar] [CrossRef]
- Shao, D.D.; Xue, W.; Krall, E.B.; Bhutkar, A.; Piccioni, F.; Wang, X.; Schinzel, A.C.; Sood, S.; Rosenbluh, J.; Kim, J.W.; et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 2014, 158, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Schneider, D.; Baronsky, T.; Pietuch, A.; Rother, J.; Oelkers, M.; Fichtner, D.; Wedlich, D.; Janshoff, A. Tension monitoring during epithelial-to-mesenchymal transition links the switch of phenotype to expression of moesin and cadherins in NMuMG cells. PLoS ONE 2013, 8, e80068. [Google Scholar] [CrossRef]
- Symonds, J.M.; Ohm, A.M.; Tan, A.C.; Reyland, M.E. PKCdelta regulates integrin alphaVbeta3 expression and transformed growth of K-ras dependent lung cancer cells. Oncotarget 2016, 7, 17905–17919. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, M.C.; Er, E.E.; Blenis, J. The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends Biochem. Sci. 2011, 36, 320–328. [Google Scholar] [CrossRef]
- Klomp, J.E.; Diehl, J.N.; Klomp, J.A.; Edwards, A.C.; Yang, R.; Morales, A.J.; Taylor, K.E.; Drizyte-Miller, K.; Bryant, K.L.; Schaefer, A.; et al. Determining the ERK-regulated phosphoproteome driving KRAS-mutant cancer. Science 2024, 384, eadk0850. [Google Scholar] [CrossRef]
- Tyc, K.M.; Kazi, A.; Ranjan, A.; Wang, R.; Sebti, S.M. Novel mutant KRAS addiction signature predicts response to the combination of ERBB and MEK inhibitors in lung and pancreatic cancers. iScience 2023, 26, 106082. [Google Scholar] [CrossRef]
- Wang, C.A.; Hou, Y.C.; Hong, Y.K.; Tai, Y.J.; Shen, C.; Hou, P.C.; Fu, J.L.; Wu, C.L.; Cheng, S.M.; Hwang, D.Y.; et al. Intercellular TIMP-1-CD63 signaling directs the evolution of immune escape and metastasis in KRAS-mutated pancreatic cancer cells. Mol. Cancer 2025, 24, 25. [Google Scholar] [CrossRef]
- Wang, G.M.; Wong, H.Y.; Konishi, H.; Blair, B.G.; Abukhdeir, A.M.; Gustin, J.P.; Rosen, D.M.; Denmeade, S.R.; Rasheed, Z.; Matsui, W.; et al. Single copies of mutant KRAS and mutant PIK3CA cooperate in immortalized human epithelial cells to induce tumor formation. Cancer Res. 2013, 73, 3248–3261. [Google Scholar] [CrossRef]
- Eser, S.; Reiff, N.; Messer, M.; Seidler, B.; Gottschalk, K.; Dobler, M.; Hieber, M.; Arbeiter, A.; Klein, S.; Kong, B.; et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell 2013, 23, 406–420. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Goldberg, I.D.; Shi, Y.E. Complex roles of tissue inhibitors of metalloproteinases in cancer. Oncogene 2002, 21, 2245–2252. [Google Scholar] [CrossRef] [PubMed]
- Akahane, T.; Akahane, M.; Shah, A.; Thorgeirsson, U.P. TIMP-1 stimulates proliferation of human aortic smooth muscle cells and Ras effector pathways. Biochem. Biophys Res. Commun. 2004, 324, 440–445. [Google Scholar] [CrossRef]
- Vitos-Faleato, J.; Real, S.M.; Gutierrez-Prat, N.; Villanueva, A.; Llonch, E.; Drosten, M.; Barbacid, M.; Nebreda, A.R. Requirement for epithelial p38alpha in KRAS-driven lung tumor progression. Proc. Natl. Acad. Sci. USA 2020, 117, 2588–2596. [Google Scholar] [CrossRef]
- Tarpgaard, L.S.; Orum-Madsen, M.S.; Christensen, I.J.; Nordgaard, C.; Noer, J.; Guren, T.K.; Glimelius, B.; Sorbye, H.; Ikdahl, T.; Kure, E.H.; et al. TIMP-1 is under regulation of the EGF signaling axis and promotes an aggressive phenotype in KRAS-mutated colorectal cancer cells: A potential novel approach to the treatment of metastatic colorectal cancer. Oncotarget 2016, 7, 59441–59457. [Google Scholar] [CrossRef]
- Chirco, R.; Liu, X.W.; Jung, K.K.; Kim, H.R. Novel functions of TIMPs in cell signaling. Cancer Metastasis Rev. 2006, 25, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Ueda, H.; Okamoto, K.; Bando, M.; Fujimoto, S.; Okada, Y.; Kawaguchi, T.; Wada, H.; Miyamoto, H.; Shimada, M.; et al. TIMP1 promotes cell proliferation and invasion capability of right-sided colon cancers via the FAK/Akt signaling pathway. Cancer Sci. 2022, 113, 4244–4257. [Google Scholar] [CrossRef]
- Collisson, E.A.; Sadanandam, A.; Olson, P.; Gibb, W.J.; Truitt, M.; Gu, S.; Cooc, J.; Weinkle, J.; Kim, G.E.; Jakkula, L.; et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011, 17, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Rosivatz, E.; Becker, I.; Specht, K.; Fricke, E.; Luber, B.; Busch, R.; Hofler, H.; Becker, K.F. Differential expression of the epithelial-mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. Am. J. Pathol. 2002, 161, 1881–1891. [Google Scholar] [CrossRef]
- Zhu, X.; Wei, L.; Bai, Y.; Wu, S.; Han, S. FoxC1 promotes epithelial-mesenchymal transition through PBX1 dependent transactivation of ZEB2 in esophageal cancer. Am. J. Cancer Res. 2017, 7, 1642–1653. [Google Scholar]
- Shi, Z.M.; Wang, L.; Shen, H.; Jiang, C.F.; Ge, X.; Li, D.M.; Wen, Y.Y.; Sun, H.R.; Pan, M.H.; Li, W.; et al. Downregulation of miR-218 contributes to epithelial-mesenchymal transition and tumor metastasis in lung cancer by targeting Slug/ZEB2 signaling. Oncogene 2017, 36, 2577–2588. [Google Scholar] [CrossRef]
- You, J.; Li, Y.; Fang, N.; Liu, B.; Zu, L.; Chang, R.; Li, X.; Zhou, Q. MiR-132 suppresses the migration and invasion of lung cancer cells via targeting the EMT regulator ZEB2. PLoS ONE 2014, 9, e91827. [Google Scholar] [CrossRef] [PubMed]
- Vandewalle, C.; Comijn, J.; De Craene, B.; Vermassen, P.; Bruyneel, E.; Andersen, H.; Tulchinsky, E.; Van Roy, F.; Berx, G. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005, 33, 6566–6578. [Google Scholar] [CrossRef] [PubMed]
- Comijn, J.; Berx, G.; Vermassen, P.; Verschueren, K.; van Grunsven, L.; Bruyneel, E.; Mareel, M.; Huylebroeck, D.; van Roy, F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell 2001, 7, 1267–1278. [Google Scholar] [CrossRef]
- Wu, D.M.; Zhang, T.; Liu, Y.B.; Deng, S.H.; Han, R.; Liu, T.; Li, J.; Xu, Y. The PAX6-ZEB2 axis promotes metastasis and cisplatin resistance in non-small cell lung cancer through PI3K/AKT signaling. Cell Death Dis. 2019, 10, 349. [Google Scholar] [CrossRef]
- Tu, B.; Yao, J.; Ferri-Borgogno, S.; Zhao, J.; Chen, S.; Wang, Q.; Yan, L.; Zhou, X.; Zhu, C.; Bang, S.; et al. YAP1 oncogene is a context-specific driver for pancreatic ductal adenocarcinoma. JCI Insight 2019, 4, e130811. [Google Scholar] [CrossRef] [PubMed]
- Ando, T.; Charindra, D.; Shrestha, M.; Umehara, H.; Ogawa, I.; Miyauchi, M.; Takata, T. Tissue inhibitor of metalloproteinase-1 promotes cell proliferation through YAP/TAZ activation in cancer. Oncogene 2018, 37, 263–270. [Google Scholar] [CrossRef]
- Shrestha, M.; Ando, T.; Chea, C.; Sakamoto, S.; Nishisaka, T.; Ogawa, I.; Miyauchi, M.; Takata, T. The transition of tissue inhibitor of metalloproteinases from −4 to −1 induces aggressive behavior and poor patient survival in dedifferentiated liposarcoma via YAP/TAZ activation. Carcinogenesis 2019, 40, 1288–1297. [Google Scholar] [CrossRef]
- Cheng, H.; Zhang, Z.; Rodriguez-Barrueco, R.; Borczuk, A.; Liu, H.; Yu, J.; Silva, J.M.; Cheng, S.K.; Perez-Soler, R.; Halmos, B. Functional genomics screen identifies YAP1 as a key determinant to enhance treatment sensitivity in lung cancer cells. Oncotarget 2016, 7, 28976–28988. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, W.; Han, X.; Li, F.; Wang, X.; Wang, R.; Fang, Z.; Tong, X.; Yao, S.; Li, F.; et al. YAP inhibits squamous transdifferentiation of Lkb1-deficient lung adenocarcinoma through ZEB2-dependent DNp63 repression. Nat. Commun. 2014, 5, 4629. [Google Scholar] [CrossRef] [PubMed]
- Ries, C. Cytokine functions of TIMP-1. Cell. Mol. Life Sci. 2014, 71, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Guo, Z.; Wang, F.; Fu, L. KRAS mutation: From undruggable to druggable in cancer. Signal Transduct. Target. Ther. 2021, 6, 386. [Google Scholar] [CrossRef] [PubMed]
- Ihle, N.T.; Byers, L.A.; Kim, E.S.; Saintigny, P.; Lee, J.J.; Blumenschein, G.R.; Tsao, A.; Liu, S.; Larsen, J.E.; Wang, J.; et al. Effect of KRAS oncogene substitutions on protein behavior: Implications for signaling and clinical outcome. J. Natl. Cancer Inst. 2012, 104, 228–239. [Google Scholar] [CrossRef]
- Tripathi, P.; Kumari, R.; Pathak, R. Drugging the undruggable: Advances in targeting KRAS signaling in solid tumors. Int. Rev. Cell Mol. Biol. 2024, 385, 1–39. [Google Scholar] [CrossRef]
- Davidsen, M.L.; Wurtz, S.O.; Romer, M.U.; Sorensen, N.M.; Johansen, S.K.; Christensen, I.J.; Larsen, J.K.; Offenberg, H.; Brunner, N.; Lademann, U. TIMP-1 gene deficiency increases tumour cell sensitivity to chemotherapy-induced apoptosis. Br. J. Cancer 2006, 95, 1114–1120. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
M-Thirusenthilarasan, I.; Ahluwalia, P.; Thorenoor, N.; Ghoshal-Gupta, S.; Lee, B.R.; Siddiqui, B.; Kolhe, R.; Rojiani, A.M.; Rojiani, M.V. TIMP-1 Modulation Correlates with KRAS Dependency and EMT Induction in NSCLC. Cells 2025, 14, 1413. https://doi.org/10.3390/cells14181413
M-Thirusenthilarasan I, Ahluwalia P, Thorenoor N, Ghoshal-Gupta S, Lee BR, Siddiqui B, Kolhe R, Rojiani AM, Rojiani MV. TIMP-1 Modulation Correlates with KRAS Dependency and EMT Induction in NSCLC. Cells. 2025; 14(18):1413. https://doi.org/10.3390/cells14181413
Chicago/Turabian StyleM-Thirusenthilarasan, Ilamathi, Pankaj Ahluwalia, Nithyananda Thorenoor, Sampa Ghoshal-Gupta, Byung Rho Lee, Bilal Siddiqui, Ravindra Kolhe, Amyn M. Rojiani, and Mumtaz V. Rojiani. 2025. "TIMP-1 Modulation Correlates with KRAS Dependency and EMT Induction in NSCLC" Cells 14, no. 18: 1413. https://doi.org/10.3390/cells14181413
APA StyleM-Thirusenthilarasan, I., Ahluwalia, P., Thorenoor, N., Ghoshal-Gupta, S., Lee, B. R., Siddiqui, B., Kolhe, R., Rojiani, A. M., & Rojiani, M. V. (2025). TIMP-1 Modulation Correlates with KRAS Dependency and EMT Induction in NSCLC. Cells, 14(18), 1413. https://doi.org/10.3390/cells14181413







