Post-Translational Modification of p62: Roles and Regulations in Autophagy
Abstract
1. Introduction
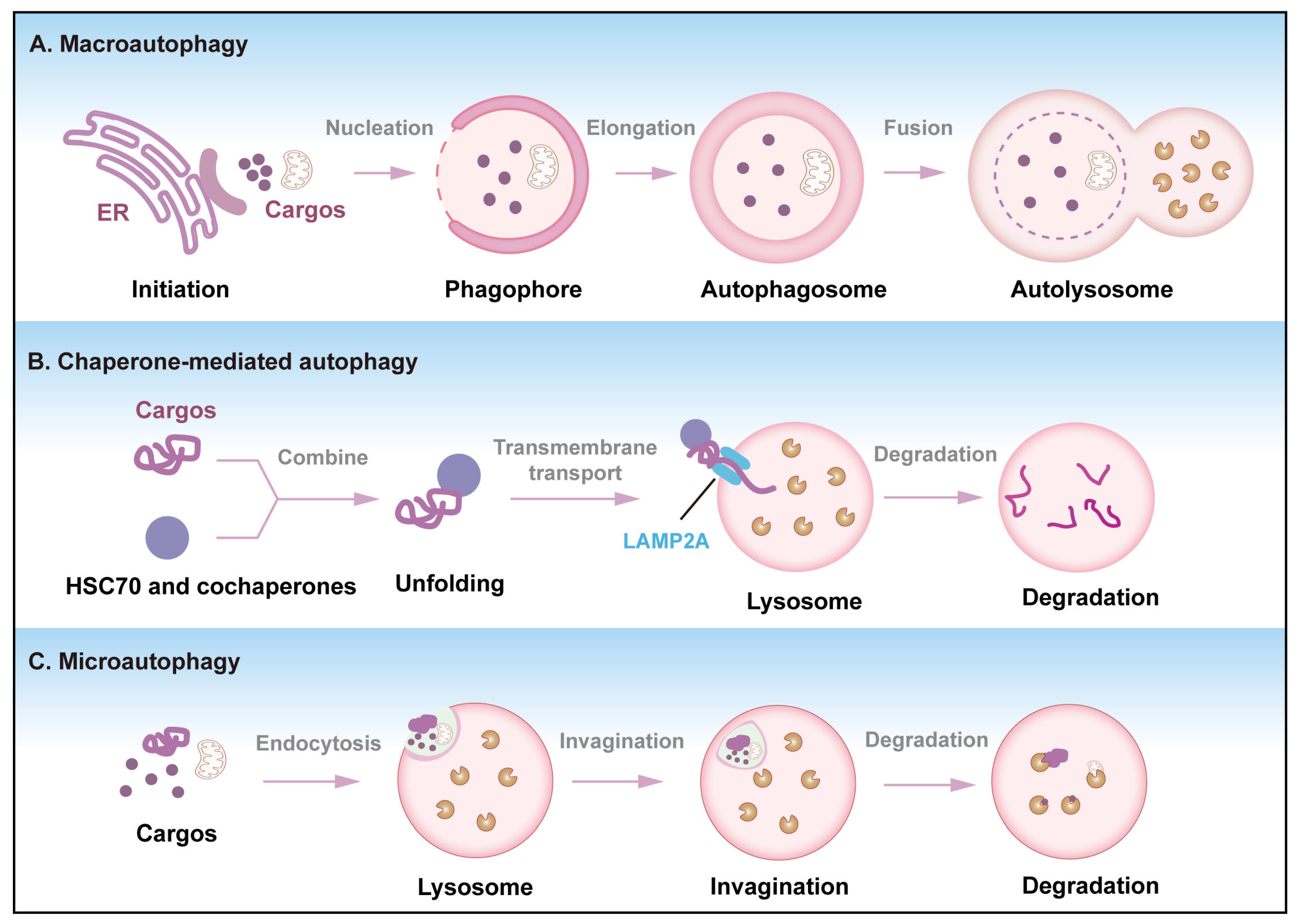
2. Overview of Autophagy
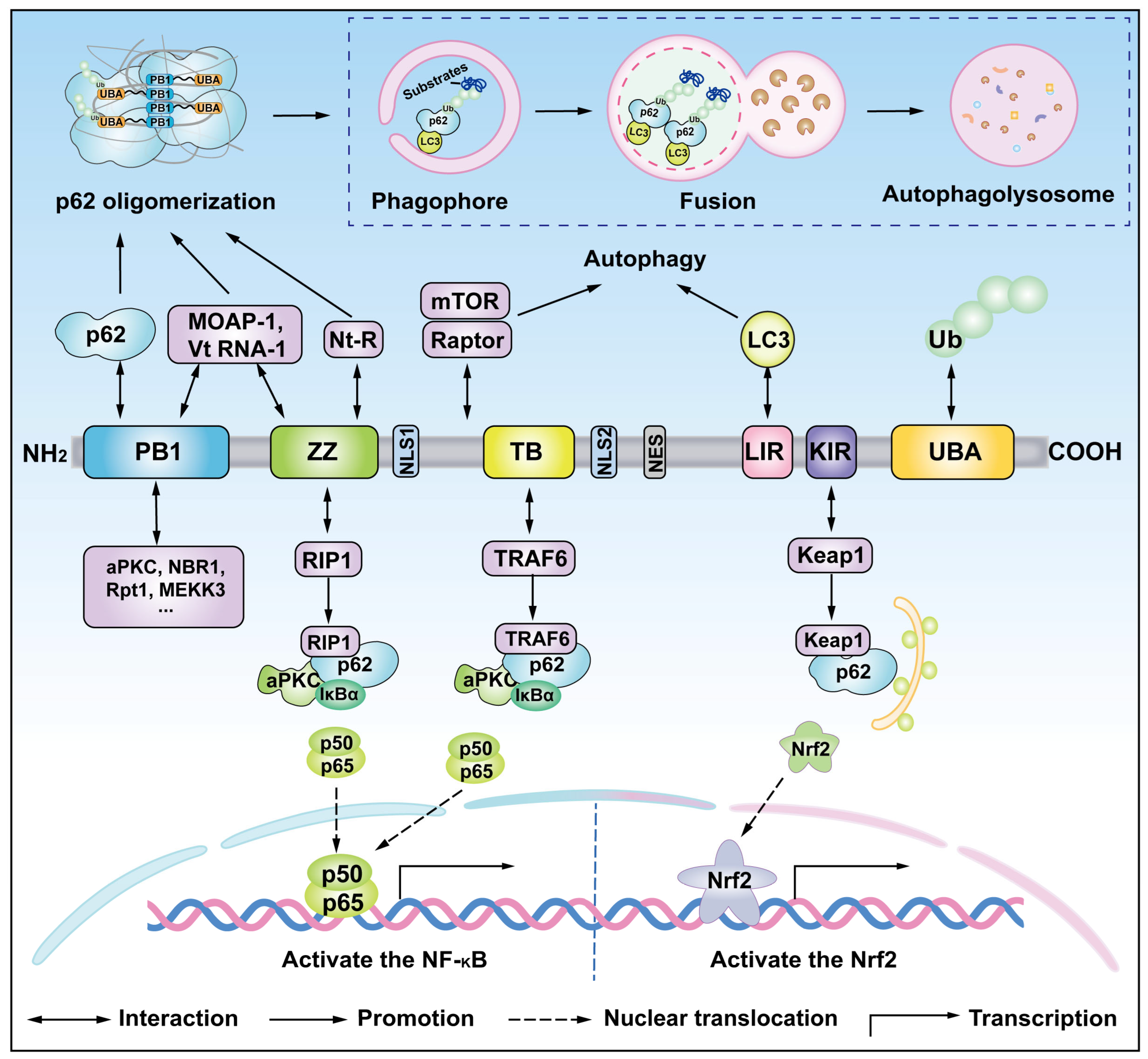
3. Structure and Function of p62
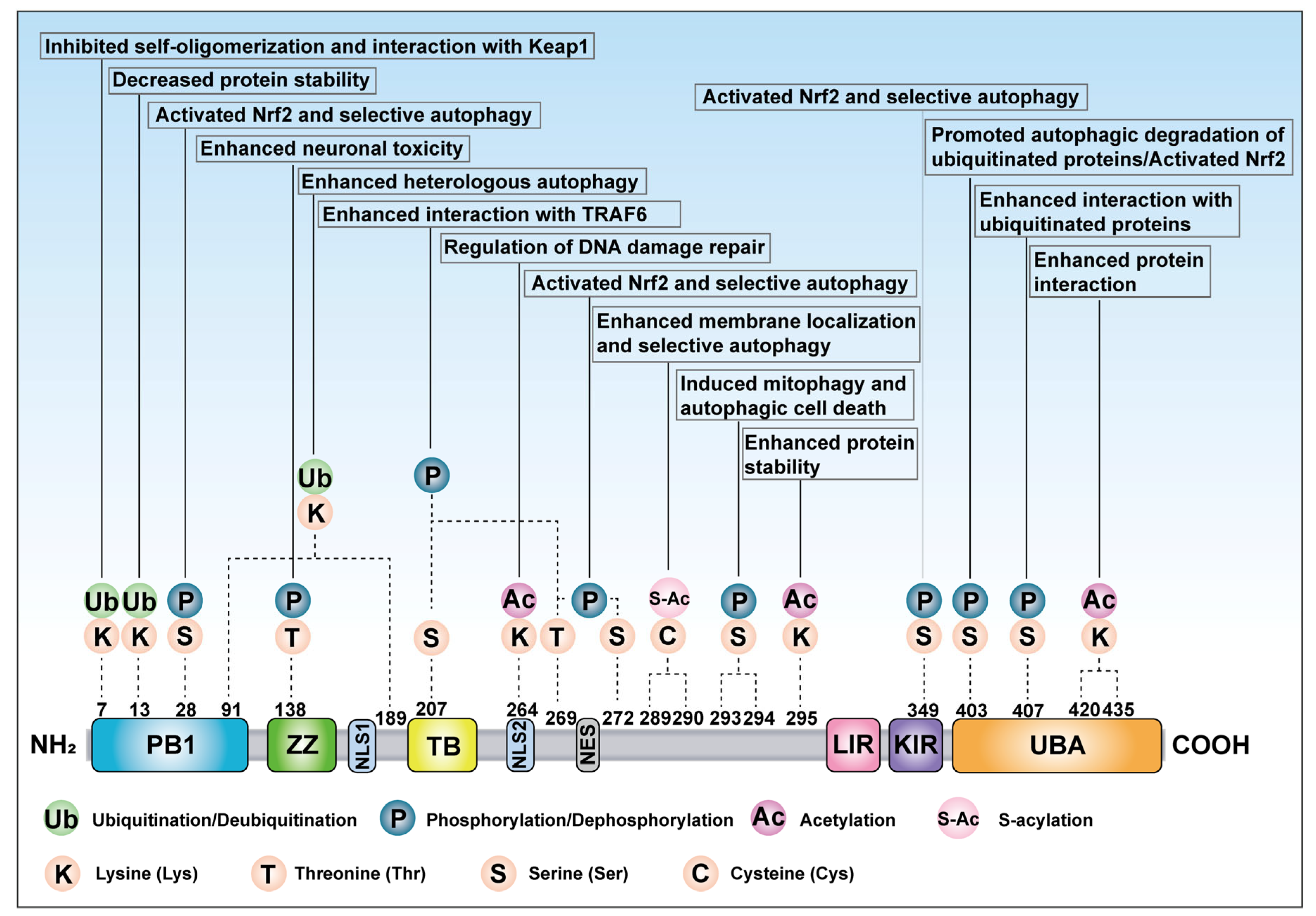
4. Post-Translational Modifications of p62
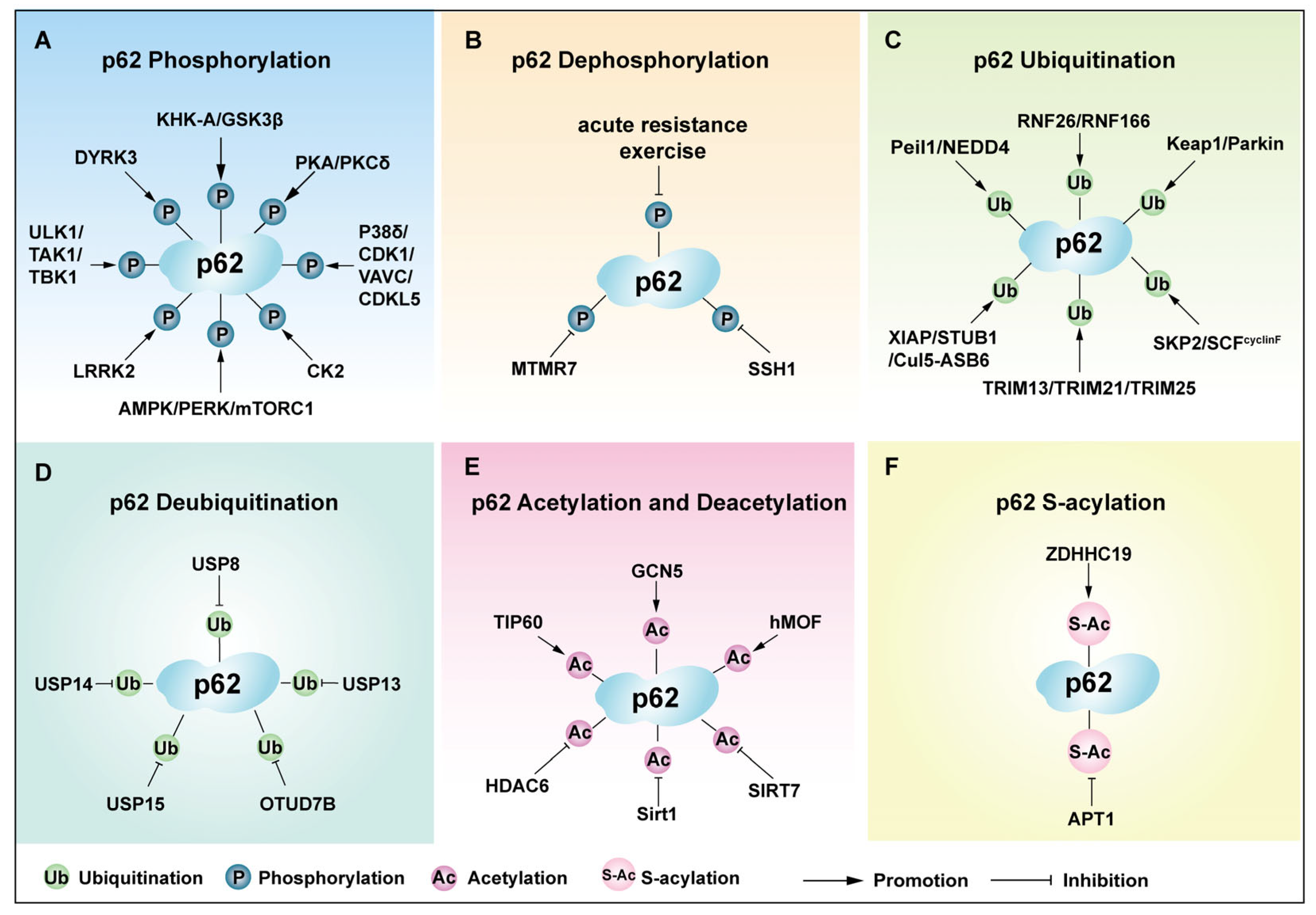
4.1. Phosphorylation of p62
4.1.1. p62 Phosphorylation by ULK1
4.1.2. p62 Phosphorylation by TBK1
4.1.3. P62 Phosphorylation by TAK1
4.1.4. P62 Phosphorylation by PKCδ
4.1.5. P62 Phosphorylation by Other Kinases
4.2. Dephosphorylation of p62
4.3. Ubiquitination of p62
4.4. Acetylation of p62
4.5. S-Acylation of p62
5. PTMs of p62 and Oligomerization
6. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef]
- Yamamoto, H.; Matsui, T. Molecular Mechanisms of Macroautophagy, Microautophagy, and Chaperone-Mediated Autophagy. J. Nippon. Med. Sch. 2024, 91, 2–9. [Google Scholar] [CrossRef]
- Griffey, C.J.; Yamamoto, A. Macroautophagy in CNS health and disease. Nat. Rev. Neurosci. 2022, 23, 411–427. [Google Scholar] [CrossRef] [PubMed]
- Kuchitsu, Y.; Taguchi, T. Lysosomal microautophagy: An emerging dimension in mammalian autophagy. Trends Cell Biol. 2024, 34, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Shen, J. Chaperone-mediated autophagy: Molecular mechanisms, biological functions, and diseases. MedComm (2020) 2023, 4, e347. [Google Scholar] [CrossRef]
- Kirkin, V.; Rogov, V.V. A Diversity of Selective Autophagy Receptors Determines the Specificity of the Autophagy Pathway. Mol. Cell 2019, 76, 268–285. [Google Scholar] [CrossRef] [PubMed]
- Mochida, K.; Nakatogawa, H. ER-phagy: Selective autophagy of the endoplasmic reticulum. EMBO Rep. 2022, 23, e55192. [Google Scholar] [CrossRef]
- Vargas, J.N.S.; Hamasaki, M.; Kawabata, T.; Youle, R.J.; Yoshimori, T. The mechanisms and roles of selective autophagy in mammals. Nat. Rev. Mol. Cell Biol. 2023, 24, 167–185. [Google Scholar] [CrossRef]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.A.; Outzen, H.; Overvatn, A.; Bjorkoy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef]
- Kirkin, V.; Lamark, T.; Sou, Y.S.; Bjorkoy, G.; Nunn, J.L.; Bruun, J.A.; Shvets, E.; McEwan, D.G.; Clausen, T.H.; Wild, P.; et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 2009, 33, 505–516. [Google Scholar] [CrossRef]
- Korac, J.; Schaeffer, V.; Kovacevic, I.; Clement, A.M.; Jungblut, B.; Behl, C.; Terzic, J.; Dikic, I. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J. Cell Sci. 2013, 126, 580–592. [Google Scholar] [CrossRef]
- Heo, J.M.; Ordureau, A.; Paulo, J.A.; Rinehart, J.; Harper, J.W. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol. Cell 2015, 60, 7–20. [Google Scholar] [CrossRef]
- Strappazzon, F.; Nazio, F.; Corrado, M.; Cianfanelli, V.; Romagnoli, A.; Fimia, G.M.; Campello, S.; Nardacci, R.; Piacentini, M.; Campanella, M.; et al. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015, 22, 419–432. [Google Scholar] [CrossRef]
- Di Rienzo, M.; Romagnoli, A.; Ciccosanti, F.; Refolo, G.; Consalvi, V.; Arena, G.; Valente, E.M.; Piacentini, M.; Fimia, G.M. AMBRA1 regulates mitophagy by interacting with ATAD3A and promoting PINK1 stability. Autophagy 2022, 18, 1752–1762. [Google Scholar] [CrossRef] [PubMed]
- Bhujabal, Z.; Birgisdottir, A.B.; Sjottem, E.; Brenne, H.B.; Overvatn, A.; Habisov, S.; Kirkin, V.; Lamark, T.; Johansen, T. FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 2017, 18, 947–961. [Google Scholar] [CrossRef]
- Wei, Y.; Chiang, W.C.; Sumpter, R., Jr.; Mishra, P.; Levine, B. Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell 2017, 168, 224–238.e210. [Google Scholar] [CrossRef]
- Zhang, Y.; Yao, Y.; Qiu, X.; Wang, G.; Hu, Z.; Chen, S.; Wu, Z.; Yuan, N.; Gao, H.; Wang, J.; et al. Listeria hijacks host mitophagy through a novel mitophagy receptor to evade killing. Nat. Immunol. 2019, 20, 433–446. [Google Scholar] [CrossRef]
- Chu, C.T.; Ji, J.; Dagda, R.K.; Jiang, J.F.; Tyurina, Y.Y.; Kapralov, A.A.; Tyurin, V.A.; Yanamala, N.; Shrivastava, I.H.; Mohammadyani, D.; et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 2013, 15, 1197–1205. [Google Scholar] [CrossRef] [PubMed]
- Sentelle, R.D.; Senkal, C.E.; Jiang, W.; Ponnusamy, S.; Gencer, S.; Selvam, S.P.; Ramshesh, V.K.; Peterson, Y.K.; Lemasters, J.J.; Szulc, Z.M.; et al. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat. Chem. Biol. 2012, 8, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, S.; Kumar, S.; Jain, A.; Ponpuak, M.; Mudd, M.H.; Kimura, T.; Choi, S.W.; Peters, R.; Mandell, M.; Bruun, J.A.; et al. TRIMs and Galectins Globally Cooperate and TRIM16 and Galectin-3 Co-direct Autophagy in Endomembrane Damage Homeostasis. Dev. Cell 2016, 39, 13–27. [Google Scholar] [CrossRef]
- Koerver, L.; Papadopoulos, C.; Liu, B.; Kravic, B.; Rota, G.; Brecht, L.; Veenendaal, T.; Polajnar, M.; Bluemke, A.; Ehrmann, M.; et al. The ubiquitin-conjugating enzyme UBE2QL1 coordinates lysophagy in response to endolysosomal damage. EMBO Rep. 2019, 20, e48014. [Google Scholar] [CrossRef] [PubMed]
- Deosaran, E.; Larsen, K.B.; Hua, R.; Sargent, G.; Wang, Y.; Kim, S.; Lamark, T.; Jauregui, M.; Law, K.; Lippincott-Schwartz, J.; et al. NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 2013, 126, 939–952. [Google Scholar] [CrossRef]
- Zhang, J.; Tripathi, D.N.; Jing, J.; Alexander, A.; Kim, J.; Powell, R.T.; Dere, R.; Tait-Mulder, J.; Lee, J.H.; Paull, T.T.; et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 2015, 17, 1259–1269. [Google Scholar] [CrossRef]
- Zheng, Y.T.; Shahnazari, S.; Brech, A.; Lamark, T.; Johansen, T.; Brumell, J.H. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 2009, 183, 5909–5916. [Google Scholar] [CrossRef] [PubMed]
- Thurston, T.L.; Ryzhakov, G.; Bloor, S.; von Muhlinen, N.; Randow, F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 2009, 10, 1215–1221. [Google Scholar] [CrossRef]
- Wild, P.; Farhan, H.; McEwan, D.G.; Wagner, S.; Rogov, V.V.; Brady, N.R.; Richter, B.; Korac, J.; Waidmann, O.; Choudhary, C.; et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011, 333, 228–233. [Google Scholar] [CrossRef]
- Tumbarello, D.A.; Manna, P.T.; Allen, M.; Bycroft, M.; Arden, S.D.; Kendrick-Jones, J.; Buss, F. The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy. PLoS Pathog. 2015, 11, e1005174. [Google Scholar] [CrossRef]
- Orvedahl, A.; MacPherson, S.; Sumpter, R., Jr.; Talloczy, Z.; Zou, Z.; Levine, B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 2010, 7, 115–127. [Google Scholar] [CrossRef]
- Mandell, M.A.; Kimura, T.; Jain, A.; Johansen, T.; Deretic, V. TRIM proteins regulate autophagy: TRIM5 is a selective autophagy receptor mediating HIV-1 restriction. Autophagy 2014, 10, 2387–2388. [Google Scholar] [CrossRef]
- Pohl, C.; Jentsch, S. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat. Cell Biol. 2009, 11, 65–70. [Google Scholar] [CrossRef]
- Isakson, P.; Lystad, A.H.; Breen, K.; Koster, G.; Stenmark, H.; Simonsen, A. TRAF6 mediates ubiquitination of KIF23/MKLP1 and is required for midbody ring degradation by selective autophagy. Autophagy 2013, 9, 1955–1964. [Google Scholar] [CrossRef] [PubMed]
- Mandell, M.A.; Jain, A.; Kumar, S.; Castleman, M.J.; Anwar, T.; Eskelinen, E.L.; Johansen, T.; Prekeris, R.; Deretic, V. TRIM17 contributes to autophagy of midbodies while actively sparing other targets from degradation. J. Cell Sci. 2016, 129, 3562–3573. [Google Scholar] [CrossRef]
- Gonzalez, A.; Covarrubias-Pinto, A.; Bhaskara, R.M.; Glogger, M.; Kuncha, S.K.; Xavier, A.; Seemann, E.; Misra, M.; Hoffmann, M.E.; Brauning, B.; et al. Ubiquitination regulates ER-phagy and remodelling of endoplasmic reticulum. Nature 2023, 618, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Grumati, P.; Morozzi, G.; Holper, S.; Mari, M.; Harwardt, M.I.E.; Yan, R.; Muller, S.; Reggiori, F.; Heilemann, M.; Dikic, I. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife 2017, 6, 25555. [Google Scholar] [CrossRef]
- Smith, M.D.; Harley, M.E.; Kemp, A.J.; Wills, J.; Lee, M.; Arends, M.; von Kriegsheim, A.; Behrends, C.; Wilkinson, S. CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev. Cell 2018, 44, 217–232.e211. [Google Scholar] [CrossRef]
- Chen, Q.; Xiao, Y.; Chai, P.; Zheng, P.; Teng, J.; Chen, J. ATL3 Is a Tubular ER-Phagy Receptor for GABARAP-Mediated Selective Autophagy. Curr. Biol. 2019, 29, 846–855.e846. [Google Scholar] [CrossRef]
- An, H.; Ordureau, A.; Paulo, J.A.; Shoemaker, C.J.; Denic, V.; Harper, J.W. TEX264 Is an Endoplasmic Reticulum-Resident ATG8-Interacting Protein Critical for ER Remodeling during Nutrient Stress. Mol. Cell 2019, 74, 891–908.e810. [Google Scholar] [CrossRef]
- Wyant, G.A.; Abu-Remaileh, M.; Frenkel, E.M.; Laqtom, N.N.; Dharamdasani, V.; Lewis, C.A.; Chan, S.H.; Heinze, I.; Ori, A.; Sabatini, D.M. NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 2018, 360, 751–758. [Google Scholar] [CrossRef]
- Dowdle, W.E.; Nyfeler, B.; Nagel, J.; Elling, R.A.; Liu, S.; Triantafellow, E.; Menon, S.; Wang, Z.; Honda, A.; Pardee, G.; et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat. Cell Biol. 2014, 16, 1069–1079. [Google Scholar] [CrossRef]
- Mancias, J.D.; Wang, X.; Gygi, S.P.; Harper, J.W.; Kimmelman, A.C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 2014, 509, 105–109. [Google Scholar] [CrossRef]
- Jiang, S.; Wells, C.D.; Roach, P.J. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem. Biophys. Res. Commun. 2011, 413, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Dou, Z.; Xu, C.; Donahue, G.; Shimi, T.; Pan, J.A.; Zhu, J.; Ivanov, A.; Capell, B.C.; Drake, A.M.; Shah, P.P.; et al. Autophagy mediates degradation of nuclear lamina. Nature 2015, 527, 105–109. [Google Scholar] [CrossRef]
- Lamark, T.; Kirkin, V.; Dikic, I.; Johansen, T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle 2009, 8, 1986–1990. [Google Scholar] [CrossRef] [PubMed]
- Katsuragi, Y.; Ichimura, Y.; Komatsu, M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015, 282, 4672–4678. [Google Scholar] [CrossRef]
- Bjorkoy, G.; Lamark, T.; Pankiv, S.; Overvatn, A.; Brech, A.; Johansen, T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzym. Enzymol. 2009, 452, 181–197. [Google Scholar] [CrossRef]
- Liang, X.; Yao, J.; Cui, D.; Zheng, W.; Liu, Y.; Lou, G.; Ye, B.; Shui, L.; Sun, Y.; Zhao, Y.; et al. The TRAF2-p62 axis promotes proliferation and survival of liver cancer by activating mTORC1 pathway. Cell Death Differ. 2023, 30, 1550–1562. [Google Scholar] [CrossRef]
- Li, M.D.; Chen, L.H.; Xiang, H.X.; Jiang, Y.L.; Lv, B.B.; Xu, D.X.; Zhao, H.; Fu, L. Benzo[a]pyrene evokes epithelial-mesenchymal transition and pulmonary fibrosis through AhR-mediated Nrf2-p62 signaling. J. Hazard. Mater. 2024, 473, 134560. [Google Scholar] [CrossRef]
- Nakamura, K.; Kimple, A.J.; Siderovski, D.P.; Johnson, G.L. PB1 domain interaction of p62/sequestosome 1 and MEKK3 regulates NF-kappaB activation. J. Biol. Chem. 2010, 285, 2077–2089. [Google Scholar] [CrossRef]
- Zhao, D.Y.; Bauerlein, F.J.B.; Saha, I.; Hartl, F.U.; Baumeister, W.; Wilfling, F. Autophagy preferentially degrades non-fibrillar polyQ aggregates. Mol. Cell 2024, 84, 1980–1994.e1988. [Google Scholar] [CrossRef]
- Yang, S.; Li, W.; Bai, X.; Di Nunzio, G.; Fan, L.; Zhao, Y.; Ren, L.; Zhao, R.; Bian, S.; Liu, M.; et al. Ginseng-derived nanoparticles alleviate inflammatory bowel disease via the TLR4/MAPK and p62/Nrf2/Keap1 pathways. J. Nanobiotechnol. 2024, 22, 48. [Google Scholar] [CrossRef]
- Yang, X.; Cao, X.; Zhu, Q. p62/SQSTM1 in cancer: Phenomena, mechanisms, and regulation in DNA damage repair. Cancer Metastasis Rev. 2025, 44, 33. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Sahu, M.; Srivastava, D.; Tiwari, S.; Ambasta, R.K.; Kumar, P. Post-translational modifications: Regulators of neurodegenerative proteinopathies. Ageing Res. Rev. 2021, 68, 101336. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jiang, X.; Li, B.; Zheng, J.; Guo, J.; Gao, L.; Du, M.; Weng, X.; Li, L.; Chen, S.; et al. A regulatory circuit comprising the CBP and SIRT7 regulates FAM134B-mediated ER-phagy. J. Cell Biol. 2023, 222, e202201068. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Kim, Y.H.; Lee, W.; Choi, H.Y.; Lee, J.; Kim, J.; Mai, D.N.; Jung, S.F.; Kwak, M.S.; Shin, J.S. USP13 deubiquitinates p62/SQSTM1 to induce autophagy and Nrf2 release for activating antioxidant response genes. Free Radic. Biol. Med. 2023, 208, 820–832. [Google Scholar] [CrossRef]
- Huang, X.; Yao, J.; Liu, L.; Chen, J.; Mei, L.; Huangfu, J.; Luo, D.; Wang, X.; Lin, C.; Chen, X.; et al. S-acylation of p62 promotes p62 droplet recruitment into autophagosomes in mammalian autophagy. Mol. Cell 2023, 83, 3485–3501.e3411. [Google Scholar] [CrossRef]
- Parzych, K.R.; Klionsky, D.J. An overview of autophagy: Morphology, mechanism, and regulation. Antioxid. Redox Signal 2014, 20, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Mehlkop, O.; Scharn, A.; Nolte, H.; Klemm, P.; Henschke, S.; Steuernagel, L.; Sotelo-Hitschfeld, T.; Kaya, E.; Wunderlich, C.M.; et al. Nutrient-sensing AgRP neurons relay control of liver autophagy during energy deprivation. Cell Metab. 2023, 35, 786–806.e713. [Google Scholar] [CrossRef]
- Kunchithapautham, K.; Rohrer, B. Apoptosis and autophagy in photoreceptors exposed to oxidative stress. Autophagy 2007, 3, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T.; Tooze, S.A. Emerging roles of ATG proteins and membrane lipids in autophagosome formation. Cell Discov. 2020, 6, 32. [Google Scholar] [CrossRef]
- Liu, L.; Tang, Y.; Zhou, Z.; Huang, Y.; Zhang, R.; Lyu, H.; Xiao, S.; Guo, D.; Ali, D.W.; Michalak, M.; et al. Membrane Curvature: The Inseparable Companion of Autophagy. Cells 2023, 12, 1132. [Google Scholar] [CrossRef]
- Lorincz, P.; Juhasz, G. Autophagosome-Lysosome Fusion. J. Mol. Biol. 2020, 432, 2462–2482. [Google Scholar] [CrossRef]
- Zhao, Y.G.; Codogno, P.; Zhang, H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat. Rev. Mol. Cell Biol. 2021, 22, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Torraca, V.; Lyu, H.; Xiao, S.; Guo, D.; Zhou, C.; Tang, J. RUNDC1 negatively mediates the fusion of autophagosomes with lysosomes via regulating SNARE complex assembly. Autophagy 2024, 20, 454–456. [Google Scholar] [CrossRef]
- Mizushima, N. The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 2020, 63, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Melia, T.J.; Lystad, A.H.; Simonsen, A. Autophagosome biogenesis: From membrane growth to closure. J. Cell Biol. 2020, 219, e202002085. [Google Scholar] [CrossRef]
- Li, J.; Chen, X.; Kang, R.; Zeh, H.; Klionsky, D.J.; Tang, D. Regulation and function of autophagy in pancreatic cancer. Autophagy 2021, 17, 3275–3296. [Google Scholar] [CrossRef]
- Zhou, C.; Qian, X.; Hu, M.; Zhang, R.; Liu, N.; Huang, Y.; Yang, J.; Zhang, J.; Bai, H.; Yang, Y.; et al. STYK1 promotes autophagy through enhancing the assembly of autophagy-specific class III phosphatidylinositol 3-kinase complex I. Autophagy 2020, 16, 1786–1806. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132-U171. [Google Scholar] [CrossRef]
- Yuan, W.; Fang, W.; Zhang, R.; Lyu, H.; Xiao, S.; Guo, D.; Ali, D.W.; Michalak, M.; Chen, X.Z.; Zhou, C.; et al. Therapeutic strategies targeting AMPK-dependent autophagy in cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2023, 1870, 119537. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Y.; Wang, D.; Xu, Y.; Dong, R.; Yang, Y.; Lv, Q.; Chen, X.; Zhang, Z. The Upstream Pathway of mTOR-Mediated Autophagy in Liver Diseases. Cells 2019, 8, 1597. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, H. Regulation of Autophagy by mTOR Signaling Pathway. Adv. Exp. Med. Biol. 2019, 1206, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Evans, T.D.; Sergin, I.; Zhang, X.; Razani, B. Target acquired: Selective autophagy in cardiometabolic disease. Sci. Signal 2017, 10, eaag2298. [Google Scholar] [CrossRef]
- Liu, J.; Wu, Y.; Meng, S.; Xu, P.; Li, S.; Li, Y.; Hu, X.; Ouyang, L.; Wang, G. Selective autophagy in cancer: Mechanisms, therapeutic implications, and future perspectives. Mol. Cancer 2024, 23, 22. [Google Scholar] [CrossRef] [PubMed]
- Shaid, S.; Brandts, C.H.; Serve, H.; Dikic, I. Ubiquitination and selective autophagy. Cell Death Differ. 2013, 20, 21–30. [Google Scholar] [CrossRef]
- Tao, M.; Liu, T.; You, Q.; Jiang, Z. p62 as a therapeutic target for tumor. Eur. J. Med. Chem. 2020, 193, 112231. [Google Scholar] [CrossRef] [PubMed]
- Pankiv, S.; Lamark, T.; Bruun, J.A.; Overvatn, A.; Bjorkoy, G.; Johansen, T. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J. Biol. Chem. 2010, 285, 5941–5953. [Google Scholar] [CrossRef]
- Noda, Y.; Kohjima, M.; Izaki, T.; Ota, K.; Yoshinaga, S.; Inagaki, F.; Ito, T.; Sumimoto, H. Molecular Recognition in Dimerization between PB1 Domains. J. Biol. Chem. 2003, 278, 43516–43524. [Google Scholar] [CrossRef]
- Yang, P.; Gao, S.; Shen, J.; Liu, T.; Lu, K.; Han, X.; Wang, J.; Ni, H.M.; Ding, W.X.; Li, H.; et al. TRIM21-mediated ubiquitination of SQSTM1/p62 abolishes its Ser403 phosphorylation and enhances palmitic acid cytotoxicity. Autophagy 2025, 21, 178–190. [Google Scholar] [CrossRef]
- Lee, S.J.; Pfluger, P.T.; Kim, J.Y.; Nogueiras, R.; Duran, A.; Pages, G.; Pouyssegur, J.; Tschop, M.H.; Diaz-Meco, M.T.; Moscat, J. A functional role for the p62-ERK1 axis in the control of energy homeostasis and adipogenesis. EMBO Rep. 2010, 11, 226–232. [Google Scholar] [CrossRef]
- Liu, Y.; Trnka, M.J.; He, L.; Burlingame, A.L.; Correia, M.A. In-Cell Chemical Crosslinking Identifies Hotspots for SQSTM-1/p62-IkappaBalpha Interaction That Underscore a Critical Role of p62 in Limiting NF-kappaB Activation Through IkappaBalpha Stabilization. Mol. Cell Proteom. 2023, 22, 100495. [Google Scholar] [CrossRef]
- Sanz, L.; Sanchez, P.; Lallena, M.J.; Diaz-Meco, M.T.; Moscat, J. The interaction of p62 with RIP links the atypical PKCs to NF-kappaB activation. EMBO J. 1999, 18, 3044–3053. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Kim, H.Y.; Lee, M.J.; Kim, S.B.; Kwon, Y.T.; Ji, C.H. Characterization and chemical modulation of p62/SQSTM1/Sequestosome-1 as an autophagic N-recognin. Methods Enzym. Enzymol. 2023, 686, 235–265. [Google Scholar] [CrossRef]
- Alcober-Boquet, L.; Zang, T.; Pietsch, L.; Suess, E.; Hartmann, M.; Proschak, E.; Gross, L.Z.F.; Sacerdoti, M.; Zeuzem, S.; Rogov, V.V.; et al. The PB1 and the ZZ domain of the autophagy receptor p62/SQSTM1 regulate the interaction of p62/SQSTM1 with the autophagosome protein LC3B. Protein Sci. 2024, 33, e4840. [Google Scholar] [CrossRef]
- Xu, J.; Wu, H.F.; Ang, E.S.; Yip, K.; Woloszyn, M.; Zheng, M.H.; Tan, R.X. NF-kappaB modulators in osteolytic bone diseases. Cytokine Growth Factor. Rev. 2009, 20, 7–17. [Google Scholar] [CrossRef]
- Duran, A.; Amanchy, R.; Linares, J.F.; Joshi, J.; Abu-Baker, S.; Porollo, A.; Hansen, M.; Moscat, J.; Diaz-Meco, M.T. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol. Cell 2011, 44, 134–146. [Google Scholar] [CrossRef]
- Birgisdottir, A.B.; Lamark, T.; Johansen, T. The LIR motif-crucial for selective autophagy. J. Cell Sci. 2013, 126, 3237–3247. [Google Scholar] [CrossRef]
- Seibenhener, M.L.; Babu, J.R.; Geetha, T.; Wong, H.C.; Krishna, N.R.; Wooten, M.W. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell Biol. 2004, 24, 8055–8068. [Google Scholar] [CrossRef] [PubMed]
- Zotti, T.; Scudiero, I.; Settembre, P.; Ferravante, A.; Mazzone, P.; D’Andrea, L.; Reale, C.; Vito, P.; Stilo, R. TRAF6-mediated ubiquitination of NEMO requires p62/sequestosome-1. Mol. Immunol. 2014, 58, 27–31. [Google Scholar] [CrossRef]
- Gallagher, E.R.; Holzbaur, E.L.F. The selective autophagy adaptor p62/SQSTM1 forms phase condensates regulated by HSP27 that facilitate the clearance of damaged lysosomes via lysophagy. Cell Rep. 2023, 42, 112037. [Google Scholar] [CrossRef]
- Li, T.; Jiang, D.; Wu, K. p62 promotes bladder cancer cell growth by activating KEAP1/NRF2-dependent antioxidative response. Cancer Sci. 2020, 111, 1156–1164. [Google Scholar] [CrossRef]
- Singh, V.; Ram, M.; Kumar, R.; Prasad, R.; Roy, B.K.; Singh, K.K. Phosphorylation: Implications in Cancer. Protein J. 2017, 36, 1–6. [Google Scholar] [CrossRef]
- Lin, Z.P.; Gan, G.; Xu, X.; Wen, C.; Ding, X.; Chen, X.Y.; Zhang, K.; Guo, W.Y.; Lin, M.; Wang, Y.Y.; et al. Comprehensive PTM profiling with SCASP-PTM uncovers mechanisms of p62 degradation and ALDOA-mediated tumor progression. Cell Rep. 2025, 44, 115500. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Li, X.; Shao, F.; Lv, G.; Lv, H.; Lee, J.H.; Qian, X.; Wang, Z.; Xia, Y.; Du, L.; et al. The protein kinase activity of fructokinase A specifies the antioxidant responses of tumor cells by phosphorylating p62. Sci. Adv. 2019, 5, eaav4570. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.; Jeong, S.H.; Yi, K.; Chung, K.M.; Hong, C.J.; Kim, S.W.; Kim, E.K.; Yu, S.W. Phosphorylation of p62 by AMP-activated protein kinase mediates autophagic cell death in adult hippocampal neural stem cells. J. Biol. Chem. 2017, 292, 13795–13808. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, R.; Noshiro, D.; Morishita, H.; Takada, S.; Kageyama, S.; Fujioka, Y.; Funakoshi, T.; Komatsu-Hirota, S.; Arai, R.; Ryzhii, E.; et al. Phosphorylation of phase-separated p62 bodies by ULK1 activates a redox-independent stress response. EMBO J. 2023, 42, e113349. [Google Scholar] [CrossRef]
- Yang, C.; Lu, T.; Liu, M.; Yuan, X.; Li, D.; Zhang, J.; Zhou, L.; Xu, M. Tiliroside targets TBK1 to induce ferroptosis and sensitize hepatocellular carcinoma to sorafenib. Phytomedicine 2023, 111, 154668. [Google Scholar] [CrossRef]
- Wang, Y.; Lyu, L.; Vu, T.; McCarty, N. TRIM44 enhances autophagy via SQSTM1 oligomerization in response to oxidative stress. Sci. Rep. 2024, 14, 18974. [Google Scholar] [CrossRef]
- Xia, Q.; Li, Y.; Xu, W.; Wu, C.; Zheng, H.; Liu, L.; Dong, L. Enhanced liquidity of p62 droplets mediated by Smurf1 links Nrf2 activation and autophagy. Cell Biosci. 2023, 13, 37. [Google Scholar] [CrossRef]
- Jiang, X.; Bao, Y.; Liu, H.; Kou, X.; Zhang, Z.; Sun, F.; Qian, Z.; Lin, Z.; Li, X.; Liu, X.; et al. VPS34 stimulation of p62 phosphorylation for cancer progression. Oncogene 2017, 36, 6850–6862. [Google Scholar] [CrossRef]
- Lee, D.H.; Park, J.S.; Lee, Y.S.; Bae, S.H. PERK prevents hepatic lipotoxicity by activating the p62-ULK1 axis-mediated noncanonical KEAP1-Nrf2 pathway. Redox Biol. 2022, 50, 102235. [Google Scholar] [CrossRef]
- Park, S.; Han, S.; Choi, I.; Kim, B.; Park, S.P.; Joe, E.H.; Suh, Y.H. Interplay between Leucine-Rich Repeat Kinase 2 (LRRK2) and p62/SQSTM-1 in Selective Autophagy. PLoS ONE 2016, 11, e0163029. [Google Scholar] [CrossRef] [PubMed]
- Pilli, M.; Arko-Mensah, J.; Ponpuak, M.; Roberts, E.; Master, S.; Mandell, M.A.; Dupont, N.; Ornatowski, W.; Jiang, S.; Bradfute, S.B.; et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 2012, 37, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Ro, S.H.; Semple, I.A.; Park, H.; Park, H.; Park, H.W.; Kim, M.; Kim, J.S.; Lee, J.H. Sestrin2 promotes Unc-51-like kinase 1 mediated phosphorylation of p62/sequestosome-1. FEBS J. 2014, 281, 3816–3827. [Google Scholar] [CrossRef]
- Kehl, S.R.; Soos, B.A.; Saha, B.; Choi, S.W.; Herren, A.W.; Johansen, T.; Mandell, M.A. TAK1 converts Sequestosome 1/p62 from an autophagy receptor to a signaling platform. EMBO Rep. 2019, 20, e46238. [Google Scholar] [CrossRef]
- Lee, S.; Jo, M.; Kwon, Y.; Jeon, Y.M.; Kim, S.; Lee, K.J.; Kim, H.J. PTK2 regulates tau-induced neurotoxicity via phosphorylation of p62 at Ser403. J. Neurogenet. 2023, 37, 10–19. [Google Scholar] [CrossRef]
- Matsumoto, G.; Wada, K.; Okuno, M.; Kurosawa, M.; Nukina, N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell 2011, 44, 279–289. [Google Scholar] [CrossRef]
- Lim, J.; Lachenmayer, M.L.; Wu, S.; Liu, W.; Kundu, M.; Wang, R.; Komatsu, M.; Oh, Y.J.; Zhao, Y.; Yue, Z. Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS Genet. 2015, 11, e1004987. [Google Scholar] [CrossRef]
- Deng, Z.; Lim, J.; Wang, Q.; Purtell, K.; Wu, S.; Palomo, G.M.; Tan, H.; Manfredi, G.; Zhao, Y.; Peng, J.; et al. ALS-FTLD-linked mutations of SQSTM1/p62 disrupt selective autophagy and NFE2L2/NRF2 anti-oxidative stress pathway. Autophagy 2020, 16, 917–931. [Google Scholar] [CrossRef]
- Kalogeropulou, A.F.; Zhao, J.; Bolliger, M.F.; Memou, A.; Narasimha, S.; Molitor, T.P.; Wilson, W.H.; Rideout, H.J.; Nichols, R.J. P62/SQSTM1 is a novel leucine-rich repeat kinase 2 (LRRK2) substrate that enhances neuronal toxicity. Biochem. J. 2018, 475, 1271–1293. [Google Scholar] [CrossRef]
- Lee, Y.H.; Yoon, A.R.; Yun, C.O.; Chung, K.C. Dual-specificity kinase DYRK3 phosphorylates p62 at the Thr-269 residue and promotes melanoma progression. J. Biol. Chem. 2024, 300, 107206. [Google Scholar] [CrossRef]
- Zhang, C.; Gao, J.; Li, M.; Deng, Y.; Jiang, C. p38delta MAPK regulates aggresome biogenesis by phosphorylating SQSTM1 in response to proteasomal stress. J. Cell Sci. 2018, 131, 216671. [Google Scholar] [CrossRef]
- Linares, J.F.; Amanchy, R.; Greis, K.; Diaz-Meco, M.T.; Moscat, J. Phosphorylation of p62 by cdk1 controls the timely transit of cells through mitosis and tumor cell proliferation. Mol. Cell Biol. 2011, 31, 105–117. [Google Scholar] [CrossRef]
- Krause, M.; Samolej, J.; Yakimovich, A.; Kriston-Vizi, J.; Huttunen, M.; Lara-Reyna, S.; Frickel, E.M.; Mercer, J. Vaccinia virus subverts xenophagy through phosphorylation and nuclear targeting of p62. J. Cell Biol. 2024, 223, e202104129. [Google Scholar] [CrossRef]
- Thinwa, J.W.; Zou, Z.; Parks, E.; Sebti, S.; Hui, K.; Wei, Y.; Goodarzi, M.; Singh, V.; Urquhart, G.; Jewell, J.L.; et al. CDKL5 regulates p62-mediated selective autophagy and confers protection against neurotropic viruses. J. Clin. Invest. 2024, 134. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Yang, Y.; Zhao, W.; Wang, M.; Chen, Y.; Wang, J.; Yang, D.; Yang, Y. MTMR7 suppresses the phenotypic switching of vascular smooth muscle cell and vascular intimal hyperplasia after injury via regulating p62/mTORC1-mediated glucose metabolism. Atherosclerosis 2024, 390, 117470. [Google Scholar] [CrossRef]
- Cazzaro, S.; Zhao, X.; Zhao, V.K.; Kim, Y.K.; Woo, J.A. Slingshot homolog-1 amplifies mitochondrial abnormalities by distinctly impairing health and clearance of mitochondria. Hum. Mol. Genet. 2023, 32, 1660–1672. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Dai, Q.; Meng, H.; Sun, A.; Wei, J.; Peng, K.; Childress, C.; Chen, M.; Shao, G.; Yang, W. The HECT E3 ubiquitin ligase NEDD4 interacts with and ubiquitylates SQSTM1 for inclusion body autophagy. J. Cell Sci. 2017, 130, 3839–3850. [Google Scholar] [CrossRef]
- Pan, J.A.; Sun, Y.; Jiang, Y.P.; Bott, A.J.; Jaber, N.; Dou, Z.; Yang, B.; Chen, J.S.; Catanzaro, J.M.; Du, C.; et al. TRIM21 Ubiquitylates SQSTM1/p62 and Suppresses Protein Sequestration to Regulate Redox Homeostasis. Mol. Cell 2016, 61, 720–733. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Li, S.; Wu, H.; Gao, R.; Rao, G.; Wang, D.; Chen, Z.; Ma, B.; Wang, H.; Sui, N.; et al. Parkin promotes proteasomal degradation of p62: Implication of selective vulnerability of neuronal cells in the pathogenesis of Parkinson’s disease. Protein Cell 2016, 7, 114–129. [Google Scholar] [CrossRef]
- Heath, R.J.; Goel, G.; Baxt, L.A.; Rush, J.S.; Mohanan, V.; Paulus, G.L.C.; Jani, V.; Lassen, K.G.; Xavier, R.J. RNF166 Determines Recruitment of Adaptor Proteins during Antibacterial Autophagy. Cell Rep. 2016, 17, 2183–2194. [Google Scholar] [CrossRef]
- Davidson, J.M.; Wu, S.S.L.; Rayner, S.L.; Cheng, F.; Duncan, K.; Russo, C.; Newbery, M.; Ding, K.; Scherer, N.M.; Balez, R.; et al. The E3 Ubiquitin Ligase SCF Cyclin F Promotes Sequestosome-1/p62 Insolubility and Foci Formation and is Dysregulated in ALS and FTD Pathogenesis. Mol. Neurobiol. 2023, 60, 5034–5054. [Google Scholar] [CrossRef]
- Lee, Y.; Chou, T.F.; Pittman, S.K.; Keith, A.L.; Razani, B.; Weihl, C.C. Keap1/Cullin3 Modulates p62/SQSTM1 Activity via UBA Domain Ubiquitination. Cell Rep. 2017, 19, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Pei, Q.; Dong, H.; Wei, X.; Li, L.; Duan, H.; Zhang, G.; Zhang, A. Tripartite motif 25 inhibits protein aggregate degradation during PRRSV infection by suppressing p62-mediated autophagy. J. Virol. 2024, 98, e0143724. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Lu, X.; Li, B.; Xu, S.; Fu, B.; Sha, Z.; Tian, R.; Yao, R.; Li, Q.; Yan, J.; et al. Bacteroides Fragilis Transplantation Reverses Reproductive Senescence by Transporting Extracellular Vesicles Through the Gut-Ovary Axis. Adv. Sci. 2025, 12, e2409740. [Google Scholar] [CrossRef]
- Jongsma, M.L.; Berlin, I.; Wijdeven, R.H.; Janssen, L.; Janssen, G.M.; Garstka, M.A.; Janssen, H.; Mensink, M.; van Veelen, P.A.; Spaapen, R.M.; et al. An ER-Associated Pathway Defines Endosomal Architecture for Controlled Cargo Transport. Cell 2016, 166, 152–166. [Google Scholar] [CrossRef]
- Yu, B.; Zhou, Y.; He, J. TRIM13 inhibits cell proliferation and induces autophagy in lung adenocarcinoma by regulating KEAP1/NRF2 pathway. Cell Cycle 2023, 22, 1496–1513. [Google Scholar] [CrossRef]
- Wang, W.; Yang, Y.; Shi, Y.; Xiang, T.; Xie, J. E3 ubiquitin ligase STUB1 affects the mTORC1 pathway through p62 and participates in regulating the differentiation of follicular helper T cells in rheumatoid arthritis. Clin. Immunol. 2023, 255, 109736. [Google Scholar] [CrossRef]
- Huang, X.; Wang, X.N.; Yuan, X.D.; Wu, W.Y.; Lobie, P.E.; Wu, Z. XIAP facilitates breast and colon carcinoma growth via promotion of p62 depletion through ubiquitination-dependent proteasomal degradation. Oncogene 2019, 38, 1448–1460. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Wang, K.; Wang, M.; Hu, R.; Li, H.; Gao, D.; Lin, M. CUL5-ASB6 Complex Promotes p62/SQSTM1 Ubiquitination and Degradation to Regulate Cell Proliferation and Autophagy. Front. Cell Dev. Biol. 2021, 9, 684885. [Google Scholar] [CrossRef]
- Xie, W.; Tian, S.; Yang, J.; Cai, S.; Jin, S.; Zhou, T.; Wu, Y.; Chen, Z.; Ji, Y.; Cui, J. OTUD7B deubiquitinates SQSTM1/p62 and promotes IRF3 degradation to regulate antiviral immunity. Autophagy 2022, 18, 2288–2302. [Google Scholar] [CrossRef]
- Peng, H.; Yang, F.; Hu, Q.; Sun, J.; Peng, C.; Zhao, Y.; Huang, C. The ubiquitin-specific protease USP8 directly deubiquitinates SQSTM1/p62 to suppress its autophagic activity. Autophagy 2020, 16, 698–708. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Wang, Y.; Chen, X.; Yu, K.; Luo, Z.Q.; Liu, X.; Qiu, J. Phosphoribosyl-linked serine ubiquitination of USP14 by the SidE family effectors of Legionella excludes p62 from the bacterial phagosome. Cell Rep. 2023, 42, 112817. [Google Scholar] [CrossRef]
- Li, M.; Xiong, J.; Yang, L.; Huang, J.; Zhang, Y.; Liu, M.; Wang, L.; Ji, J.; Zhao, Y.; Zhu, W.G.; et al. Acetylation of p62 regulates base excision repair through interaction with APE1. Cell Rep. 2022, 40, 111116. [Google Scholar] [CrossRef]
- Feng, L.; Chen, M.; Li, Y.; Li, M.; Hu, S.; Zhou, B.; Zhu, L.; Yu, L.; Zhou, Q.; Tan, L.; et al. Sirt1 deacetylates and stabilizes p62 to promote hepato-carcinogenesis. Cell Death Dis. 2021, 12, 405. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Jiang, W.X.; Qin, L.Y.; Gong, Z.; Wan, W.; Li, J.; Wang, Y.; Zhang, H.; Peng, C.; Zhou, T.; et al. Requirement for p62 acetylation in the aggregation of ubiquitylated proteins under nutrient stress. Nat. Commun. 2019, 10, 5792. [Google Scholar] [CrossRef]
- Huang, X.; Liu, L.; Yao, J.; Lin, C.; Xiang, T.; Yang, A. S-acylation regulates SQSTM1/p62-mediated selective autophagy. Autophagy 2024, 20, 1467–1469. [Google Scholar] [CrossRef]
- Alam, M.; Hasan, G.M.; Hassan, M.I. A review on the role of TANK-binding kinase 1 signaling in cancer. Int. J. Biol. Macromol. 2021, 183, 2364–2375. [Google Scholar] [CrossRef]
- Tsuchiya, M.; Kong, W.; Hiraoka, Y.; Haraguchi, T.; Ogawa, H. TBK1 inhibitors enhance transfection efficiency by suppressing p62/SQSTM1 phosphorylation. Genes. Cells 2023, 28, 68–77. [Google Scholar] [CrossRef]
- Schlutermann, D.; Berleth, N.; Deitersen, J.; Wallot-Hieke, N.; Friesen, O.; Wu, W.; Stuhldreier, F.; Sun, Y.; Berning, L.; Friedrich, A.; et al. FIP200 controls the TBK1 activation threshold at SQSTM1/p62-positive condensates. Sci. Rep. 2021, 11, 13863. [Google Scholar] [CrossRef]
- Foster, A.D.; Downing, P.; Figredo, E.; Polain, N.; Stott, A.; Layfield, R.; Rea, S.L. ALS-associated TBK1 variant p.G175S is defective in phosphorylation of p62 and impacts TBK1-mediated signalling and TDP-43 autophagic degradation. Mol. Cell Neurosci. 2020, 108, 103539. [Google Scholar] [CrossRef]
- Cho, C.S.; Park, H.W.; Ho, A.; Semple, I.A.; Kim, B.; Jang, I.; Park, H.; Reilly, S.; Saltiel, A.R.; Lee, J.H. Lipotoxicity induces hepatic protein inclusions through TANK binding kinase 1-mediated p62/sequestosome 1 phosphorylation. Hepatology 2018, 68, 1331–1346. [Google Scholar] [CrossRef] [PubMed]
- Prabakaran, T.; Bodda, C.; Krapp, C.; Zhang, B.C.; Christensen, M.H.; Sun, C.; Reinert, L.; Cai, Y.; Jensen, S.B.; Skouboe, M.K.; et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018, 37, e201797858. [Google Scholar] [CrossRef]
- Sakurai, H. Targeting of TAK1 in inflammatory disorders and cancer. Trends Pharmacol. Sci. 2012, 33, 522–530. [Google Scholar] [CrossRef]
- Yamada, M.; Warabi, E.; Oishi, H.; Lira, V.A.; Okutsu, M. Muscle p62 stimulates the expression of antioxidant proteins alleviating cancer cachexia. FASEB J. 2023, 37, e23156. [Google Scholar] [CrossRef]
- Hashimoto, K.; Simmons, A.N.; Kajino-Sakamoto, R.; Tsuji, Y.; Ninomiya-Tsuji, J. TAK1 Regulates the Nrf2 Antioxidant System Through Modulating p62/SQSTM1. Antioxid. Redox Signal 2016, 25, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Black, J.D.; Affandi, T.; Black, A.R.; Reyland, M.E. PKCalpha and PKCdelta: Friends and Rivals. J. Biol. Chem. 2022, 298, 102194. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, R.; Minato, Y.; Maeda, S.; Yagi, H. Nrf2 phosphorylation contributes to acquisition of pericyte reprogramming via the PKCdelta pathway. Neurobiol. Dis. 2025, 206, 106824. [Google Scholar] [CrossRef]
- Affandi, T.; Ohm, A.M.; Speidel, J.T.; Caino, M.C.; Boulton, D.P.; Reyland, M.E. PKCdelta regulates DNA damage and cell death through a SIRT6/Nrf2-dependent antioxidant response. Mol. Cancer Res. 2025, OF1–OF13. [Google Scholar] [CrossRef]
- Su, X.; Li, T.; Liu, Z.; Huang, Q.; Liao, K.; Ren, R.; Lu, L.; Qi, X.; Wang, M.; Chen, J.; et al. Licochalcone A activates Keap1-Nrf2 signaling to suppress arthritis via phosphorylation of p62 at serine 349. Free Radic. Biol. Med. 2018, 115, 471–483. [Google Scholar] [CrossRef]
- Shan, C.; Wang, Y.; Wang, Y. The Crosstalk between Autophagy and Nrf2 Signaling in Cancer: From Biology to Clinical Applications. Int. J. Biol. Sci. 2024, 20, 6181–6206. [Google Scholar] [CrossRef]
- Liao, K.; Su, X.; Lei, K.; Liu, Z.; Lu, L.; Wu, Q.; Pan, H.; Huang, Q.; Zhao, Y.; Wang, M.; et al. Sinomenine protects bone from destruction to ameliorate arthritis via activating p62(Thr269/Ser272)-Keap1-Nrf2 feedback loop. Biomed. Pharmacother. 2021, 135, 111195. [Google Scholar] [CrossRef] [PubMed]
- Linares, J.F.; Duran, A.; Reina-Campos, M.; Aza-Blanc, P.; Campos, A.; Moscat, J.; Diaz-Meco, M.T. Amino Acid Activation of mTORC1 by a PB1-Domain-Driven Kinase Complex Cascade. Cell Rep. 2015, 12, 1339–1352. [Google Scholar] [CrossRef]
- Kramer, N.; Mato, U.G.; Krauter, S.; Buscher, N.; Afifi, A.; Herhaus, L.; Florin, L.; Plachter, B.; Zimmermann, C. The Autophagy Receptor SQSTM1/p62 Is a Restriction Factor of HCMV Infection. Viruses 2024, 16, 1440. [Google Scholar] [CrossRef] [PubMed]
- Zeng, N.; D’Souza, R.F.; Figueiredo, V.C.; Markworth, J.F.; Roberts, L.A.; Peake, J.M.; Mitchell, C.J.; Cameron-Smith, D. Acute resistance exercise induces Sestrin2 phosphorylation and p62 dephosphorylation in human skeletal muscle. Physiol. Rep. 2017, 5, 13526. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Chen, J.; Li, K.; Li, H.; Zhu, Y.; Zhai, Y.; Lu, B.; Fan, Y.; Liu, Z.; Chen, X.; et al. Ubiquitination and deubiquitination in cancer: From mechanisms to novel therapeutic approaches. Mol. Cancer 2024, 23, 148. [Google Scholar] [CrossRef]
- Tang, C.; Lai, Y.; Li, L.; Situ, M.Y.; Li, S.; Cheng, B.; Chen, Y.; Lei, Z.; Ren, Y.; Zhou, J.; et al. SERPINH1 modulates apoptosis by inhibiting P62 ubiquitination degradation to promote bone metastasis of prostate cancer. iScience 2024, 27, 110427. [Google Scholar] [CrossRef]
- Yang, J.; Tong, T.; Zhu, C.; Zhou, M.; Jiang, Y.; Chen, H.; Que, L.; Liu, L.; Zhu, G.; Ha, T.; et al. Peli1 contributes to myocardial ischemia/reperfusion injury by impairing autophagy flux via its E3 ligase mediated ubiquitination of P62. J. Mol. Cell Cardiol. 2022, 173, 30–46. [Google Scholar] [CrossRef]
- Lu, S.; Xu, J.; Xu, Y.; Liu, Y.; Shi, D.; Wang, J.; Qiu, F. Glycyrol attenuates colon injury via promotion of SQSTM1/p62 ubiquitination and autophagy by inhibiting the ubiquitin-specific protease USP8. J. Funct. Foods 2023, 103, 105492. [Google Scholar] [CrossRef]
- Xia, C.; Tao, Y.; Li, M.; Che, T.; Qu, J. Protein acetylation and deacetylation: An important regulatory modification in gene transcription (Review). Exp. Ther. Med. 2020, 20, 2923–2940. [Google Scholar] [CrossRef]
- Xu, Y.; Wan, W. Acetylation in the regulation of autophagy. Autophagy 2022, 19, 379–387. [Google Scholar] [CrossRef]
- Chamberlain, L.H.; Shipston, M.J. The physiology of protein S-acylation. Physiol. Rev. 2015, 95, 341–376. [Google Scholar] [CrossRef]
- Abrar, F.; Davies, M.C.; Alshehabi, Y.; Kumar, A.; Dang, A.; Nguyen, Y.T.N.; Collins, J.; Caron, N.S.; Choudhary, J.S.; Sanders, S.S.; et al. Reduced Palmitoylation of SQSTM1/p62 in Huntington Disease Is Associated With Impaired Autophagy. FASEB J. 2025, 39, e70549. [Google Scholar] [CrossRef] [PubMed]
- Gemeinhardt, T.M.; Regy, R.M.; Phan, T.M.; Pal, N.; Sharma, J.; Senkovich, O.; Mendiola, A.J.; Ledterman, H.J.; Henrickson, A.; Lopes, D.; et al. A disordered linker in the Polycomb protein Polyhomeotic tunes phase separation and oligomerization. Mol. Cell 2025, 85, 2128–2146.e2115. [Google Scholar] [CrossRef] [PubMed]
- Kumari, N.; Yadav, S. Modulation of protein oligomerization: An overview. Prog. Progress. Biophys. Mol. Biol. 2019, 149, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Wu, R.; Zheng, J.; Li, P.; Yu, L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018, 28, 405–415. [Google Scholar] [CrossRef]
- Komatsu, M. p62 bodies: Phase separation, NRF2 activation, and selective autophagic degradation. IUBMB Life 2022, 74, 1200–1208. [Google Scholar] [CrossRef]
- Wurzer, B.; Zaffagnini, G.; Fracchiolla, D.; Turco, E.; Abert, C.; Romanov, J.; Martens, S. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. eLife 2015, 4, e08941. [Google Scholar] [CrossRef]
- Zaffagnini, G.; Savova, A.; Danieli, A.; Romanov, J.; Tremel, S.; Ebner, M.; Peterbauer, T.; Sztacho, M.; Trapannone, R.; Tarafder, A.K.; et al. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 2018, 37, e201798308. [Google Scholar] [CrossRef]
- Noda, N.N.; Wang, Z.; Zhang, H. Liquid-liquid phase separation in autophagy. J. Cell Biol. 2020, 219, e202004062. [Google Scholar] [CrossRef]
- Ercan-Herbst, E.; Ehrig, J.; Schondorf, D.C.; Behrendt, A.; Klaus, B.; Gomez Ramos, B.; Prat Oriol, N.; Weber, C.; Ehrnhoefer, D.E. A post-translational modification signature defines changes in soluble tau correlating with oligomerization in early stage Alzheimer’s disease brain. Acta Neuropathol. Commun. 2019, 7, 192. [Google Scholar] [CrossRef]
- Lamark, T.; Svenning, S.; Johansen, T. Regulation of selective autophagy: The p62/SQSTM1 paradigm. Essays Biochem. 2017, 61, 609–624. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Ding, W.X. SQSTM1/p62 and Hepatic Mallory-Denk Body Formation in Alcohol-Associated Liver Disease. Am. J. Pathol. 2023, 193, 1415–1426. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Ichimura, Y.; Taguchi, K.; Suzuki, T.; Mizushima, T.; Takagi, K.; Hirose, Y.; Nagahashi, M.; Iso, T.; Fukutomi, T.; et al. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat. Commun. 2016, 7, 12030. [Google Scholar] [CrossRef]
- Rubino, E.; Boschi, S.; Roveta, F.; Marcinno, A.; Cermelli, A.; Borghese, C.; Vigliani, M.C.; Rainero, I. Investigating p62 Concentrations in Cerebrospinal Fluid of Patients with Dementia: A Potential Autophagy Biomarker In Vivo? Brain Sci. 2022, 12, 1414. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zuo, Q.; Li, X.; Liu, Y.; Gan, L.; Wang, L.; Rao, Y.; Pan, R.; Dong, J. p62 Binding to Protein Kinase C Regulates HIV-1 gp120 V3 Loop Induced Microglial Inflammation. Inflammation 2024. [Google Scholar] [CrossRef]
- Zitkute, V.; Jasinevicius, A.; Vaitiekaite, G.; Kukcinaviciute, E.; Aleksandraviciute, B.; Eidenaite, E.; Sudeikis, L.; Jonusiene, V.; Sasnauskiene, A. The role of p62 in cell death and survival of 5-fluorouracil and oxaliplatin-resistant colorectal cancer cells. J. Cell Biochem. 2023, 124, 1779–1791. [Google Scholar] [CrossRef]
- Asghari, N.; Saei, A.K.; Cordani, M.; Nayeri, Z.; Moosavi, M.A. Drug repositioning identifies potential autophagy inhibitors for the LIR motif p62/SQSTM1 protein. Comput. Biol. Chem. 2024, 113, 108235. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, H.; You, S.; Xu, Z.; Liu, X.; Chen, X.; Zhang, W.; Ye, J.; Li, P.; Zhou, X. NEDD4L inhibits migration, invasion, cisplatin resistance and promotes apoptosis of bladder cancer cells by inactivating the p62/Keap1/Nrf2 pathway. Environ. Environ. Toxicol. 2023, 38, 1678–1689. [Google Scholar] [CrossRef]
- Zhang, S.; Huang, J.; Lan, Z.; Xiao, Y.; Liao, Y.; Basnet, S.; Huang, P.; Li, Y.; Yan, J.; Sheng, Y.; et al. CPEB1 Controls NRF2 Proteostasis and Ferroptosis Susceptibility in Pancreatic Cancer. Int. J. Biol. Sci. 2024, 20, 3156–3172. [Google Scholar] [CrossRef]
- Zhu, Y.F.; Yu, R.H.; Zhou, S.; Tang, P.P.; Zhang, R.; Wu, Y.X.; Xu, R.; Wei, J.M.; Wang, Y.Y.; Zhang, J.L.; et al. TAX1BP1 and FIP200 orchestrate non-canonical autophagy of p62 aggregates for mouse neural stem cell maintenance. Zool. Res. 2024, 45, 937–950. [Google Scholar] [CrossRef]
- Kalid, O.; Gotliv, I.; Levy-Apter, E.; Beker, D.F.; Cherniavsky-Lev, M.; Rotem, E.; Miron, N. PTX80, a novel compound targeting the autophagy receptor p62/SQSTM1 for treatment of cancer. Chem. Biol. Drug Des. 2022, 100, 623–638. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Garbutt, C.; Ma, H.; Gao, P.; Hornicek, F.J.; Kan, Q.; Shi, H.; Duan, Z. Expression and role of autophagy-associated p62 (SQSTM1) in multidrug resistant ovarian cancer. Gynecol. Oncol. 2018, 150, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Hennig, P.; Fenini, G.; Di Filippo, M.; Karakaya, T.; Beer, H.D. The Pathways Underlying the Multiple Roles of p62 in Inflammation and Cancer. Biomedicines 2021, 9, 707. [Google Scholar] [CrossRef]
- Adams, O.; Janser, F.A.; Dislich, B.; Berezowska, S.; Humbert, M.; Seiler, C.A.; Kroell, D.; Slotta-Huspenina, J.; Feith, M.; Ott, K.; et al. A specific expression profile of LC3B and p62 is associated with nonresponse to neoadjuvant chemotherapy in esophageal adenocarcinomas. PLoS ONE 2018, 13, e0197610. [Google Scholar] [CrossRef] [PubMed]


| Selective Autophagy | Substance | Mammalian Autophagy Receptors | References |
|---|---|---|---|
| Aggrephagy | Protein aggregate | p62, NBR1, OPTN | [9,10,11] |
| Mitophagy | Mitochondria | p62, NDP52, OPTN, NIX, TAX1BP1, NBR1, AMBRA1, BNIP3, FUNDC1, Bcl2L13, FKBP8, PHB2, NLRX1, cardiolipin, ceramide | [12,13,14,15,16,17,18,19] |
| Lysophagy | Lysosome | p62, TRIM16 | [20,21] |
| Pexophagy | Peroxisome | p62, NBR1 | [22,23] |
| Xenophagy | Bacteria, Viral | p62, NDP52, OPTN, TAX1BP1, TRIM5α | [24,25,26,27,28,29] |
| Midbody autophagy | Midbody rings | p62, NBR1, TRIM17 | [30,31,32] |
| ER-phagy | ER | FAM134B, RTN3, CCPG1, ATL3, TEX264, SEC62 | [33,34,35,36,37,38] |
| Ferritinophagy | Ferritin | NCO4A | [39,40] |
| Glycophagy | Glycogen | Stbd1 | [41] |
| Nuclear lamina autophagy | Nuclear lamina | Lamin B1 | [42] |
| Ribophagy | Ribosomes | NUFIP1 | [38] |
| Site | Location | Type of PTM | Regulators |
|---|---|---|---|
| Ser28 | PB1 | Phosphorylation | GSK3β [92], KHK-A [93] |
| Ser293, Ser294 | Linker | Phosphorylation | AMPK [94] |
| Ser349 | KIR | Phosphorylation | ULK1 [95], TBK1 [96], PKA [97], mTORC1 [98], PKCδ [99], PERK [100], LRRK2 [101] |
| Ser403 | UBA | Phosphorylation | TBK1 [102], ULK1 [103], TAK1 [104], PTK2 [105], LRRK2 [101], CK2 [106] |
| Ser407 | UBA | Phosphorylation | ULK1 [107], TBK1 [108] |
| Thr138 | ZZ | Phosphorylation | LRRK2 [109] |
| Ser207, Thr269 | TB, Linker | Phosphorylation | DYRK3 [110] |
| Thr269, Ser272 | Linker | Phosphorylation | P38δ [111], CDK1 [112], VAVC [113], CDKL5 [114] |
| Thr269, Ser272 | Linker | Dephosphorylation | MTMR7 [115] |
| Ser403 | UBA | Dephosphorylation | SSH1 [116] |
| K7 | PB1 | Ubiquitination | NEDD4 [117], TRIM21 [118] |
| K13 | PB1 | Ubiquitination | Parkin [119] |
| K91, K189 | PB1, Linker | Ubiquitination | RNF166 [120] |
| K281 | Linker | Ubiquitination | SCFcyclinF [121] |
| K420 | UBA | Ubiquitination | Keap1 [122] |
| - | - | Ubiquitination | TRIM25 [123], SKP2 [124], RNF26 [125], TRIM13 [126], STUB1 [127], XIAP [128], Cul5-ASB6 [129] |
| K7 | PB1 | Deubiquitination | USP13 [54], OTUD7B [130] |
| K420 | UBA | Deubiquitination | USP8 [131] |
| - | - | Deubiquitination | USP14 [132], USP15 [125] |
| K264 | NLS2 | Acetylation | hMOF [133] |
| K295 | Linker | Acetylation | GCN5 [134] |
| K420, K435 | UBA | Acetylation | TIP60 [135] |
| K264 | NLS2 | Deacetylation | SIRT7 [133] |
| K295 | Linker | Deacetylation | Sirt1 [134] |
| K420, K435 | UBA | Deacetylation | HDAC6 [135] |
| Cys289, Cys290 | Linker | S-acylation | ZDHHC19 [55,136] |
| Cys289, Cys290 | Linker | Deacylation | APT1 [55,136] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, S.; Yu, Y.; Liao, M.; Song, D.; Xu, X.; Tian, L.; Zhang, R.; Lyu, H.; Guo, D.; Zhang, Q.; et al. Post-Translational Modification of p62: Roles and Regulations in Autophagy. Cells 2025, 14, 1016. https://doi.org/10.3390/cells14131016
Xiao S, Yu Y, Liao M, Song D, Xu X, Tian L, Zhang R, Lyu H, Guo D, Zhang Q, et al. Post-Translational Modification of p62: Roles and Regulations in Autophagy. Cells. 2025; 14(13):1016. https://doi.org/10.3390/cells14131016
Chicago/Turabian StyleXiao, Shuai, Yeping Yu, Meng Liao, Dandan Song, Xiaozhen Xu, Lingli Tian, Rui Zhang, Hao Lyu, Dong Guo, Qi Zhang, and et al. 2025. "Post-Translational Modification of p62: Roles and Regulations in Autophagy" Cells 14, no. 13: 1016. https://doi.org/10.3390/cells14131016
APA StyleXiao, S., Yu, Y., Liao, M., Song, D., Xu, X., Tian, L., Zhang, R., Lyu, H., Guo, D., Zhang, Q., Chen, X.-Z., Zhou, C., & Tang, J. (2025). Post-Translational Modification of p62: Roles and Regulations in Autophagy. Cells, 14(13), 1016. https://doi.org/10.3390/cells14131016








