Examining Stromal Cell Interactions in an In Vitro Blood–Brain Barrier Model with Human Umbilical Vein Endothelial Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol Overview
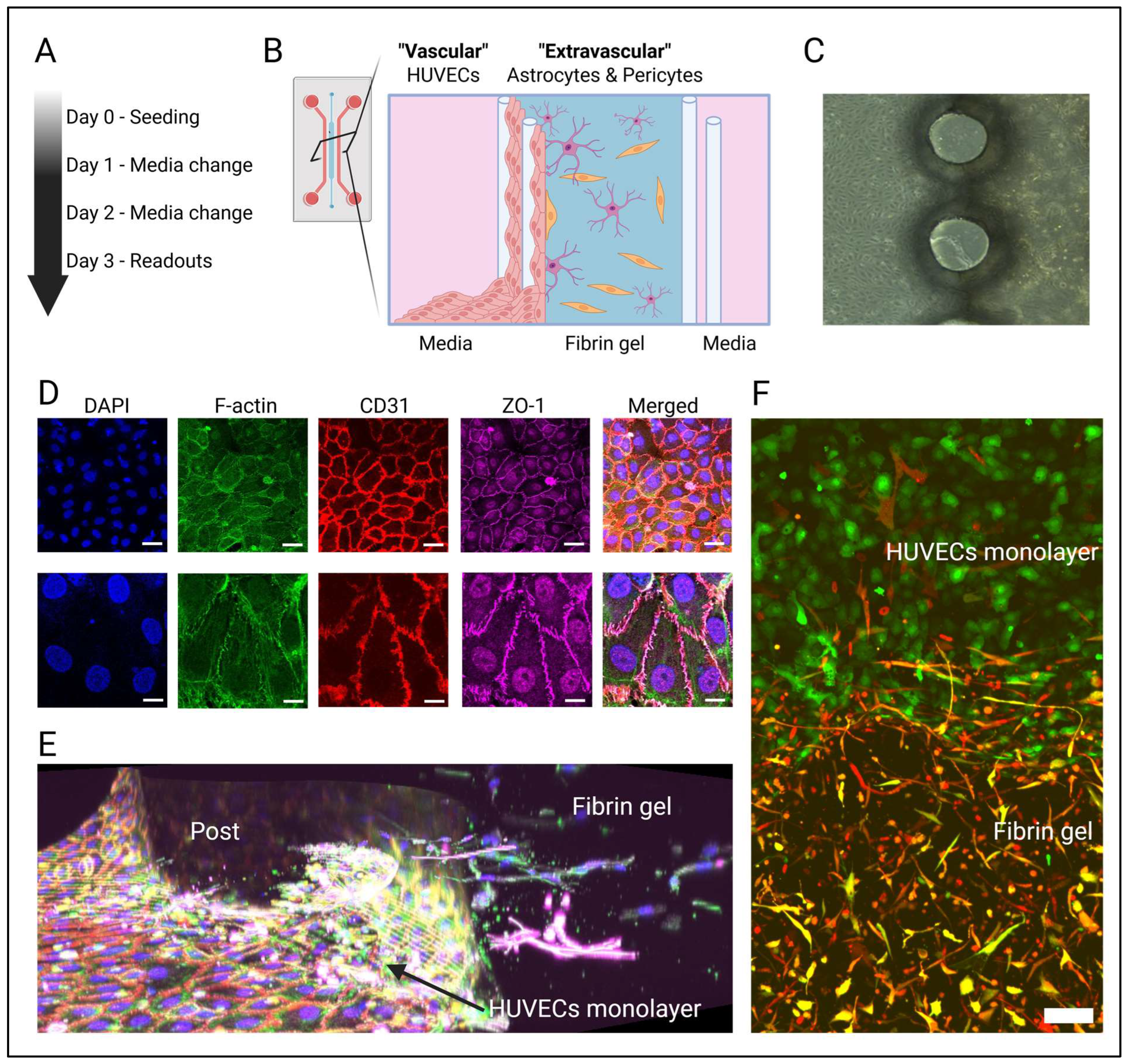
2.2. Model Design
2.3. Microfluidic Device Fabrication
2.4. Cell Culture
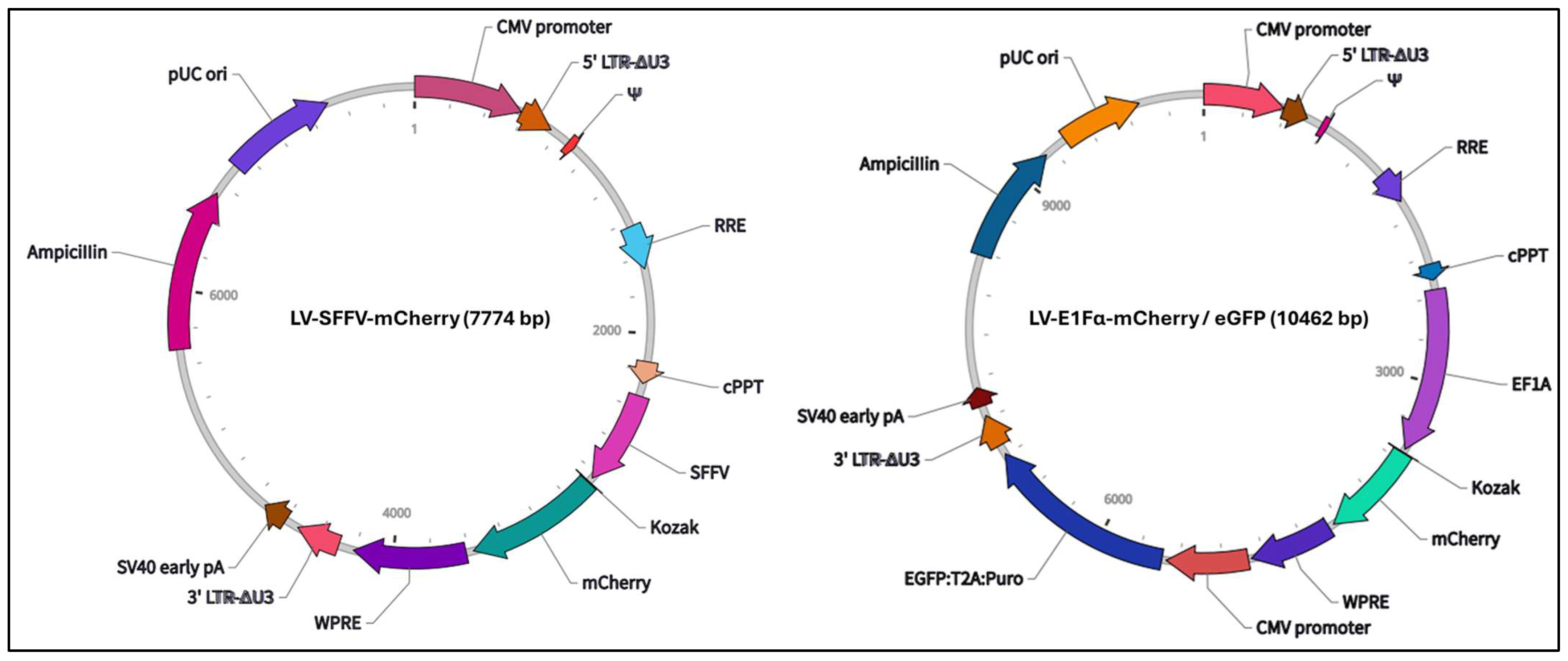
2.5. Lentiviral Transduction
2.6. Three-Dimensional Cell Culture in the Microfluidic Platform
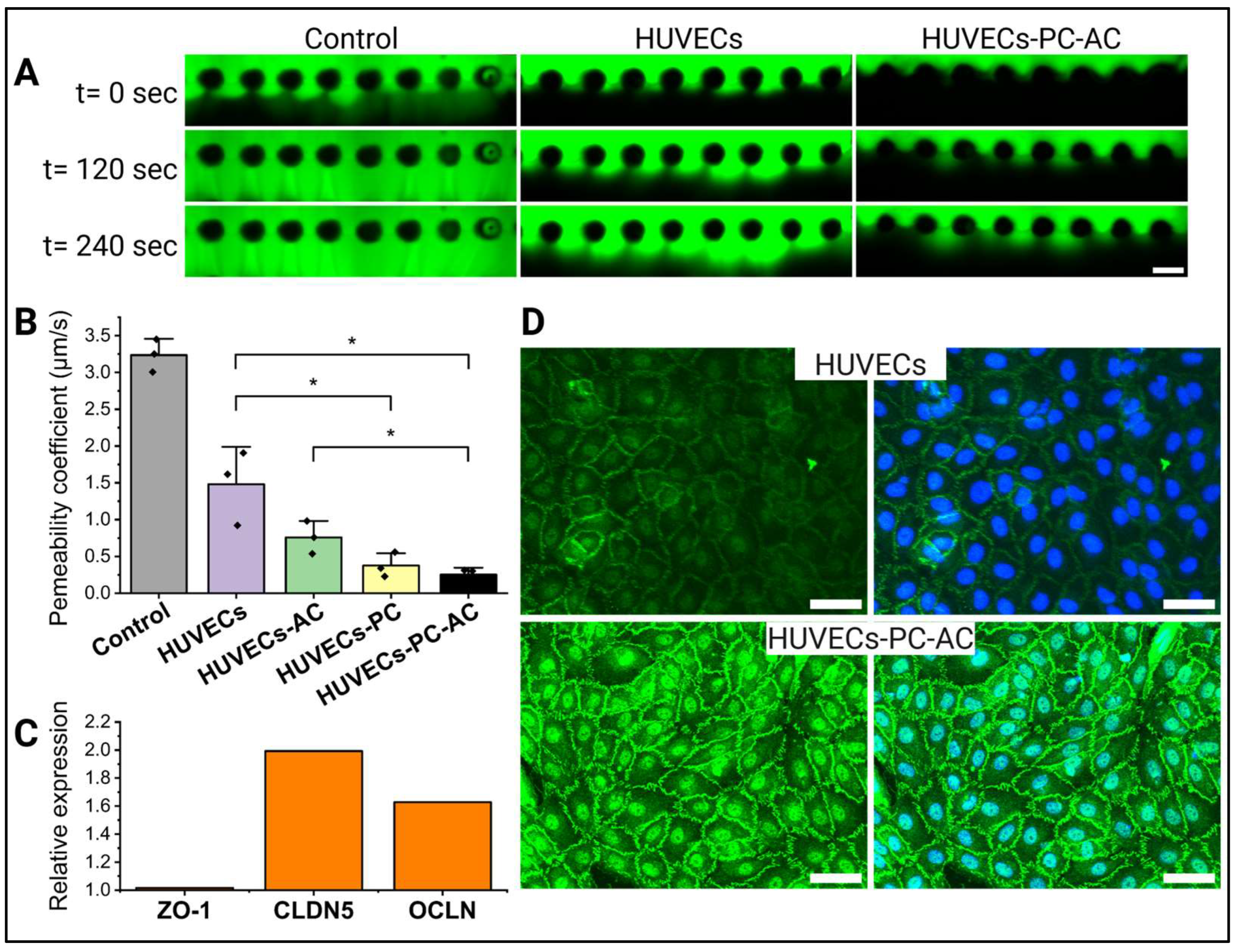
2.7. Permeability Measurements
2.8. RNA Isolation and Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
2.9. Immunofluorescent Staining and Image Acquisition and Analysis
2.10. Statistics
3. Results
3.1. An In Vitro Model of the Human Blood–Brain Barrier Consisting of HUVECs, Astrocytes, and Pericytes
3.2. Impact of Astrocytes and Pericytes on HUVEC Barrier Formation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BBB | Blood–brain barrier |
| HUVECs | Human umbilical vein endothelial cells |
| ZO-1 | Zona occludens-1 |
| CLDN5 | Claudin-5 |
| OCLN | Occludin |
| PECAM | Platelet endothelial cell adhesion molecule-1 |
| 2D | Two-dimensional |
| 3D | Three-dimensional |
| PDMS | Polydimethylsiloxane |
Appendix A

| Target Gene | Forward Primer Sequence 5′–3′ | Reverse Primer Sequence 5′–3′ |
|---|---|---|
| PECAM | GAAAGCTGTCCCTGATGCCG | GGAGCAGGGCAGGTTCATAA |
| ZO-1 | TGGACAACCAGATGTGGATTTACC | TCCCGTCTTCATGAGCTGAATT |
| CLDN5 | CTCTGCTGGTTCGCCAACAT | CAGCTCGTACTTCTGCGACA |
| OCLN | ACAAGCGGT TTT ATC CAG AGTC | GTCATCCACAGGCGAAGTTAAT |
References
- Serlin, Y.; Shelef, I.; Knyazer, B.; Friedman, A. Anatomy and physiology of the blood-brain barrier. Semin. Cell Dev. Biol. 2015, 38, 2–6. [Google Scholar] [CrossRef]
- Daneman, R.; Prat, A. The blood-brain barrier. Cold Spring Harb. Perspect. Biol. 2015, 7, a020412. [Google Scholar] [CrossRef]
- Pachter, J.S.; de Vries, H.E.; Fabry, Z. The blood-brain barrier and its role in immune privilege in the central nervous system. J. Neuropathol. Exp. Neurol. 2003, 62, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Pandit, R.; Chen, L.; Götz, J. The blood-brain barrier: Physiology and strategies for drug delivery. Adv. Drug Deliv. Rev. 2020, 165–166, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Chen, Q.; Chen, X.; Han, F.; Chen, Z.; Wang, Y. The blood–brain barrier: Structure, regulation and drug delivery. Signal Transduct. Target. Ther. 2023, 8, 217. [Google Scholar] [CrossRef] [PubMed]
- Nehra, G.; Bauer, B.; Hartz, A.M.S. Blood-brain barrier leakage in Alzheimer’s disease: From discovery to clinical relevance. Pharmacol. Ther. 2022, 234, 108119. [Google Scholar] [CrossRef]
- Kealy, J.; Greene, C.; Campbell, M. Blood-brain barrier regulation in psychiatric disorders. Neurosci. Lett. 2020, 726, 133664. [Google Scholar] [CrossRef]
- Bronger, H.; König, J.; Kopplow, K.; Steiner, H.-H.; Ahmadi, R.; Herold-Mende, C.; Keppler, D.; Nies, A.T. ABCC Drug Efflux Pumps and Organic Anion Uptake Transporters in Human Gliomas and the Blood-Tumor Barrier. Cancer Res. 2005, 65, 11419–11428. [Google Scholar] [CrossRef]
- Desai, B.S.; Monahan, A.J.; Carvey, P.M.; Hendey, B. Blood–Brain Barrier Pathology in Alzheimer’s and Parkinson’s Disease: Implications for Drug Therapy. Cell Transplant. 2007, 16, 285–299. [Google Scholar] [CrossRef]
- Kawakita, S.; Mandal, K.; Mou, L.; Mecwan, M.M.; Zhu, Y.; Li, S.; Sharma, S.; Hernandez, A.L.; Nguyen, H.T.; Maity, S.; et al. Organ-On-A-Chip Models of the Blood–Brain Barrier: Recent Advances and Future Prospects. Small 2022, 18, 2201401. [Google Scholar] [CrossRef]
- Oddo, A.; Peng, B.; Tong, Z.; Wei, Y.; Tong, W.Y.; Thissen, H.; Voelcker, N.H. Advances in Microfluidic Blood–Brain Barrier (BBB) Models. Trends Biotechnol. 2019, 37, 1295–1314. [Google Scholar] [CrossRef] [PubMed]
- Williams-Medina, A.; Deblock, M.; Janigro, D. In vitro Models of the Blood-Brain Barrier: Tools in Translational Medicine. Front. Med. Technol. 2021, 2, 623950. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhu, M.; Huang, R.; Wang, K.; Zeng, Z.; Xiao, L.; Lin, Y.; Liu, D. Blood–brain barrier microfluidic chips and their applications. Organs--A-Chip 2023, 5, 100027. [Google Scholar] [CrossRef]
- Stebbins, M.J.; Gastfriend, B.D.; Canfield, S.G.; Lee, M.S.; Richards, D.; Faubion, M.G.; Li, W.J.; Daneman, R.; Palecek, S.P.; Shusta, E.V. Human pluripotent stem cell-derived brain pericyte-like cells induce blood-brain barrier properties. Sci. Adv. 2019, 5, eaau7375. [Google Scholar] [CrossRef]
- Cucullo, L.; Hossain, M.; Puvenna, V.; Marchi, N.; Janigro, D. The role of shear stress in Blood-Brain Barrier endothelial physiology. BMC Neurosci. 2011, 12, 40. [Google Scholar] [CrossRef]
- Choublier, N.; Taghi, M.; Menet, M.C.; Le Gall, M.; Bruce, J.; Chafey, P.; Guillonneau, F.; Moreau, A.; Denizot, C.; Parmentier, Y.; et al. Exposure of human cerebral microvascular endothelial cells hCMEC/D3 to laminar shear stress induces vascular protective responses. Fluids Barriers CNS 2022, 19, 41. [Google Scholar] [CrossRef] [PubMed]
- Javanmardi, Y.; Agrawal, A.; Malandrino, A.; Lasli, S.; Chen, M.; Shahreza, S.; Serwinski, B.; Cammoun, L.; Li, R.; Jorfi, M.; et al. Endothelium and Subendothelial Matrix Mechanics Modulate Cancer Cell Transendothelial Migration. Adv. Sci. 2023, 10, 2206554. [Google Scholar] [CrossRef]
- Campisi, M.; Shin, Y.; Osaki, T.; Hajal, C.; Chiono, V.; Kamm, R.D. 3D self-organized microvascular model of the human blood-brain barrier with endothelial cells, pericytes and astrocytes. Biomaterials 2018, 180, 117–129. [Google Scholar] [CrossRef]
- Hajal, C.; Offeddu, G.S.; Shin, Y.; Zhang, S.; Morozova, O.; Hickman, D.; Knutson, C.G.; Kamm, R.D. Engineered human blood–brain barrier microfluidic model for vascular permeability analyses. Nat. Protoc. 2022, 17, 95–128. [Google Scholar] [CrossRef]
- Kim, J.A.; Kim, H.N.; Im, S.-K.; Chung, S.; Kang, J.Y.; Choi, N. Collagen-based brain microvasculature model in vitro using three-dimensional printed template. Biomicrofluidics 2015, 9, 024115. [Google Scholar] [CrossRef]
- Lee, S.W.L.; Campisi, M.; Osaki, T.; Possenti, L.; Mattu, C.; Adriani, G.; Kamm, R.D.; Chiono, V. Modeling Nanocarrier Transport across a 3D In Vitro Human Blood-Brain–Barrier Microvasculature. Adv. Healthc. Mater. 2020, 9, 1901486. [Google Scholar] [CrossRef] [PubMed]
- Drolez, A.; Vandenhaute, E.; Julien, S.; Gosselet, F.; Burchell, J.; Cecchelli, R.; Delannoy, P.; Dehouck, M.-P.; Mysiorek, C. Selection of a Relevant In Vitro Blood-Brain Barrier Model to Investigate Pro-Metastatic Features of Human Breast Cancer Cell Lines. PLoS ONE 2016, 11, e0151155. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.I.; Abaci, H.E.; Shuler, M.L. Microfluidic blood–brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol. Bioeng. 2017, 114, 184–194. [Google Scholar] [CrossRef]
- Whisler, J.; Shahreza, S.; Schlegelmilch, K.; Ege, N.; Javanmardi, Y.; Malandrino, A.; Agrawal, A.; Fantin, A.; Serwinski, B.; Azizgolshani, H.; et al. Emergent mechanical control of vascular morphogenesis. Sci. Adv. 2023, 9, eadg9781. [Google Scholar] [CrossRef]
- Yuan, W.; Lv, Y.; Zeng, M.; Fu, B.M. Non-invasive measurement of solute permeability in cerebral microvessels of the rat. Microvasc. Res. 2009, 77, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Zervantonakis, I.K.; Hughes-Alford, S.K.; Charest, J.L.; Condeelis, J.S.; Gertler, F.B.; Kamm, R.D. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl. Acad. Sci. USA 2012, 109, 13515–13520. [Google Scholar] [CrossRef]
- Ho, Y.T.; Adriani, G.; Beyer, S.; Nhan, P.T.; Kamm, R.D.; Kah, J.C.Y. A Facile Method to Probe the Vascular Permeability of Nanoparticles in Nanomedicine Applications. Sci. Rep. 2017, 7, 707. [Google Scholar] [CrossRef]
- Man, S.; Ubogu, E.E.; Williams, K.A.; Tucky, B.; Callahan, M.K.; Ransohoff, R.M. Human Brain Microvascular Endothelial Cells and Umbilical Vein Endothelial Cells Differentially Facilitate Leukocyte Recruitment and Utilize Chemokines for T Cell Migration. J. Immunol. Res. 2008, 2008, 384982. [Google Scholar] [CrossRef]
- Minagar, A.; Long, A.; Ma, T.; Jackson, T.H.; Kelley, R.E.; Ostanin, D.V.; Sasaki, M.; Warren, A.C.; Jawahar, A.; Cappell, B.; et al. Interferon (IFN)-ß1a and IFN-ß1b Block IFN-?-Induced Disintegration of Endothelial Junction Integrity and Barrier. Endothelium 2003, 10, 299–307. [Google Scholar] [CrossRef]
- Akiyama, H.; Kondoh, T.; Kokunai, T.; Nagashima, T.; Saito, N.; Tamaki, N. Blood-brain barrier formation of grafted human umbilical vein endothelial cells in athymic mouse brain. Brain Res. 2000, 858, 172–176. [Google Scholar] [CrossRef]
- Mathiisen, T.M.; Lehre, K.P.; Danbolt, N.C.; Ottersen, O.P. The perivascular astroglial sheath provides a complete covering of the brain microvessels: An electron microscopic 3D reconstruction. Glia 2010, 58, 1094–1103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wan, Z.; Pavlou, G.; Zhong, A.X.; Xu, L.; Kamm, R.D. Interstitial Flow Promotes the Formation of Functional Microvascular Networks In Vitro through Upregulation of Matrix Metalloproteinase-2. Adv. Funct. Mater. 2022, 32, 2206767. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Margari, A.; Konig, S.; Jayarajan, V.; Rizzato, S.; Maruccio, G.; Moeendarbary, E. Examining Stromal Cell Interactions in an In Vitro Blood–Brain Barrier Model with Human Umbilical Vein Endothelial Cells. Cells 2025, 14, 759. https://doi.org/10.3390/cells14110759
Margari A, Konig S, Jayarajan V, Rizzato S, Maruccio G, Moeendarbary E. Examining Stromal Cell Interactions in an In Vitro Blood–Brain Barrier Model with Human Umbilical Vein Endothelial Cells. Cells. 2025; 14(11):759. https://doi.org/10.3390/cells14110759
Chicago/Turabian StyleMargari, Andrea, Simon Konig, Vignesh Jayarajan, Silvia Rizzato, Giuseppe Maruccio, and Emad Moeendarbary. 2025. "Examining Stromal Cell Interactions in an In Vitro Blood–Brain Barrier Model with Human Umbilical Vein Endothelial Cells" Cells 14, no. 11: 759. https://doi.org/10.3390/cells14110759
APA StyleMargari, A., Konig, S., Jayarajan, V., Rizzato, S., Maruccio, G., & Moeendarbary, E. (2025). Examining Stromal Cell Interactions in an In Vitro Blood–Brain Barrier Model with Human Umbilical Vein Endothelial Cells. Cells, 14(11), 759. https://doi.org/10.3390/cells14110759









