Human Differentiated Adipocytes as Surrogate Mature Adipocytes for Adipocyte-Derived Extracellular Vesicle Analysis
Abstract
1. Introduction
2. Materials and Methods
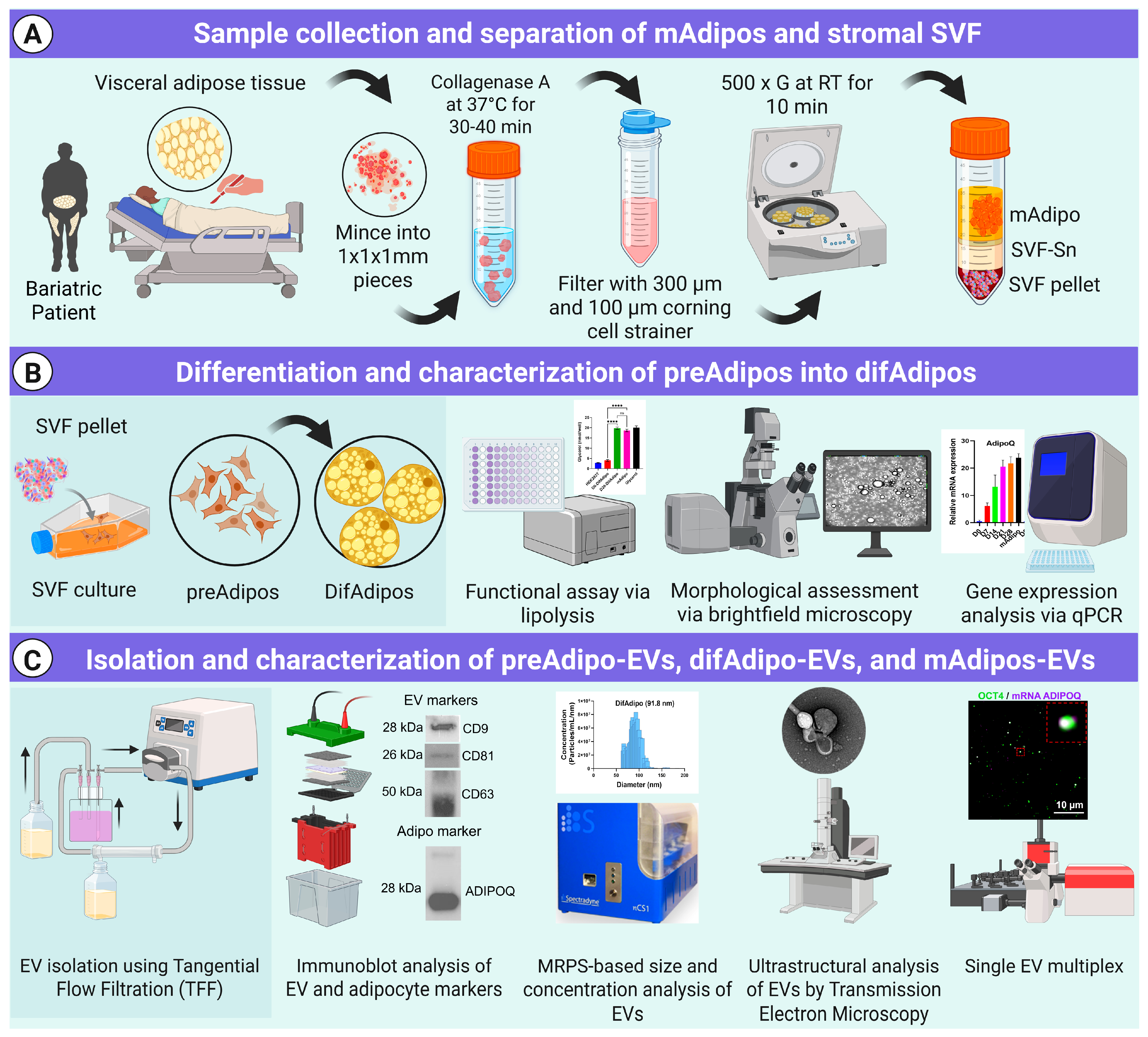
2.1. Study Participants
2.2. Adipose Tissue Dissociation and Mature Adipocyte Culture for EV Isolation
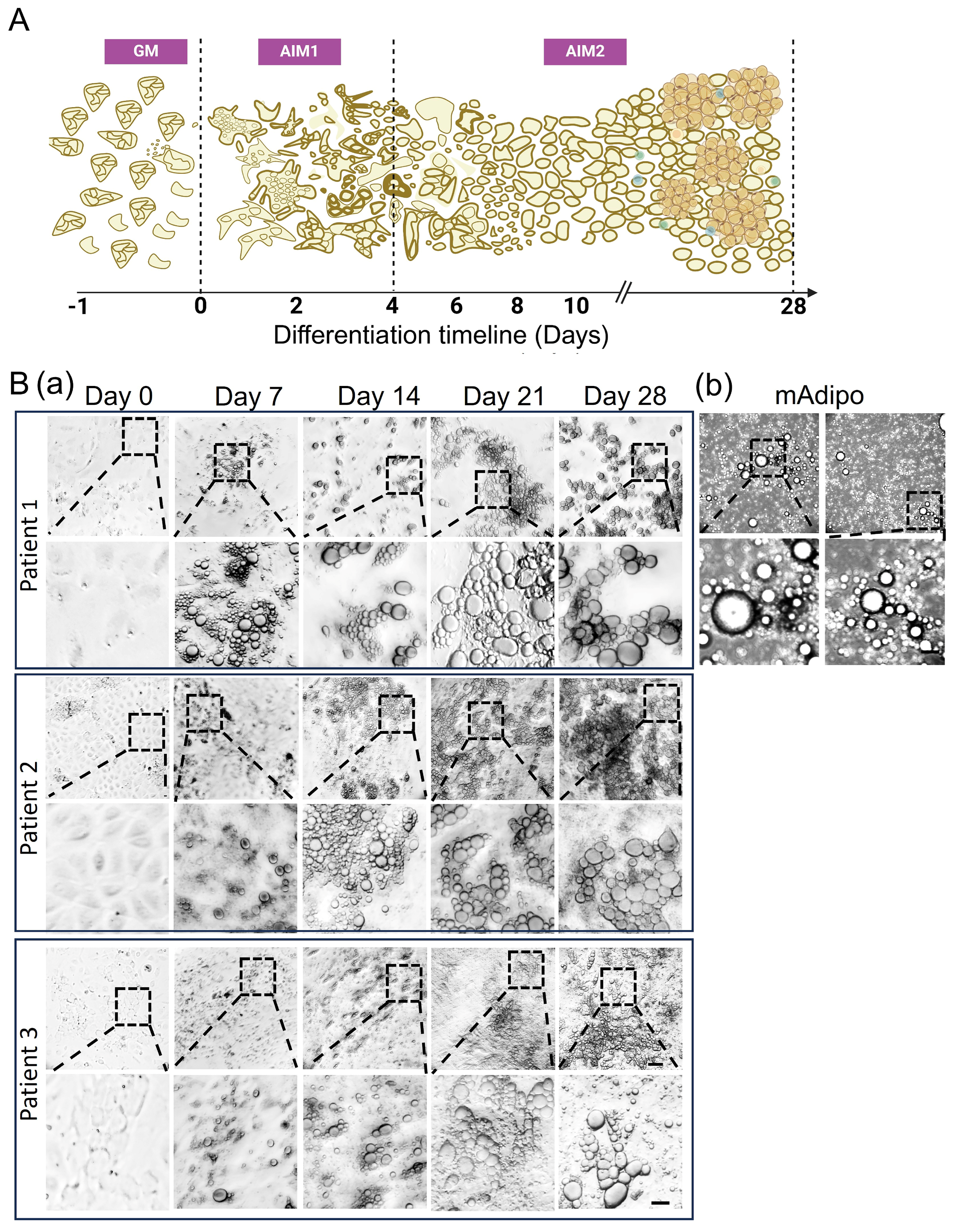
2.3. Preadipocyte (preAdipo) Isolation and Differentiation into Differentiated Mature Adipocytes (difAdipos)
2.4. Cell Line and Culture
2.5. ADEV Isolation via Tangential Flow Filtration (TFF)
2.6. Microfluidic Resistive Pulse Sensing (MRPS)
2.7. Western Blotting
2.8. Quantification of Real-Time Polymerase Chain Reaction (qRT-PCR)
2.9. Lipolysis Assay
2.10. Transmission Electron Microscopy (TEM)
2.11. Single EV Analysis Using Total Internal Reflection Fluorescence Microscopy (TIRF)
- Antibody functionalization of the gold-coated biochip:
- Immobilization of biotinylated capture antibodies:
- Hybridization of molecular beacons (MBs):
- EV capture on the biochip surface:
- Detection of surface proteins using fluorophore-conjugated antibodies:
2.12. EV Labeling and Uptake Assay
2.13. Statistical Analysis
3. Results
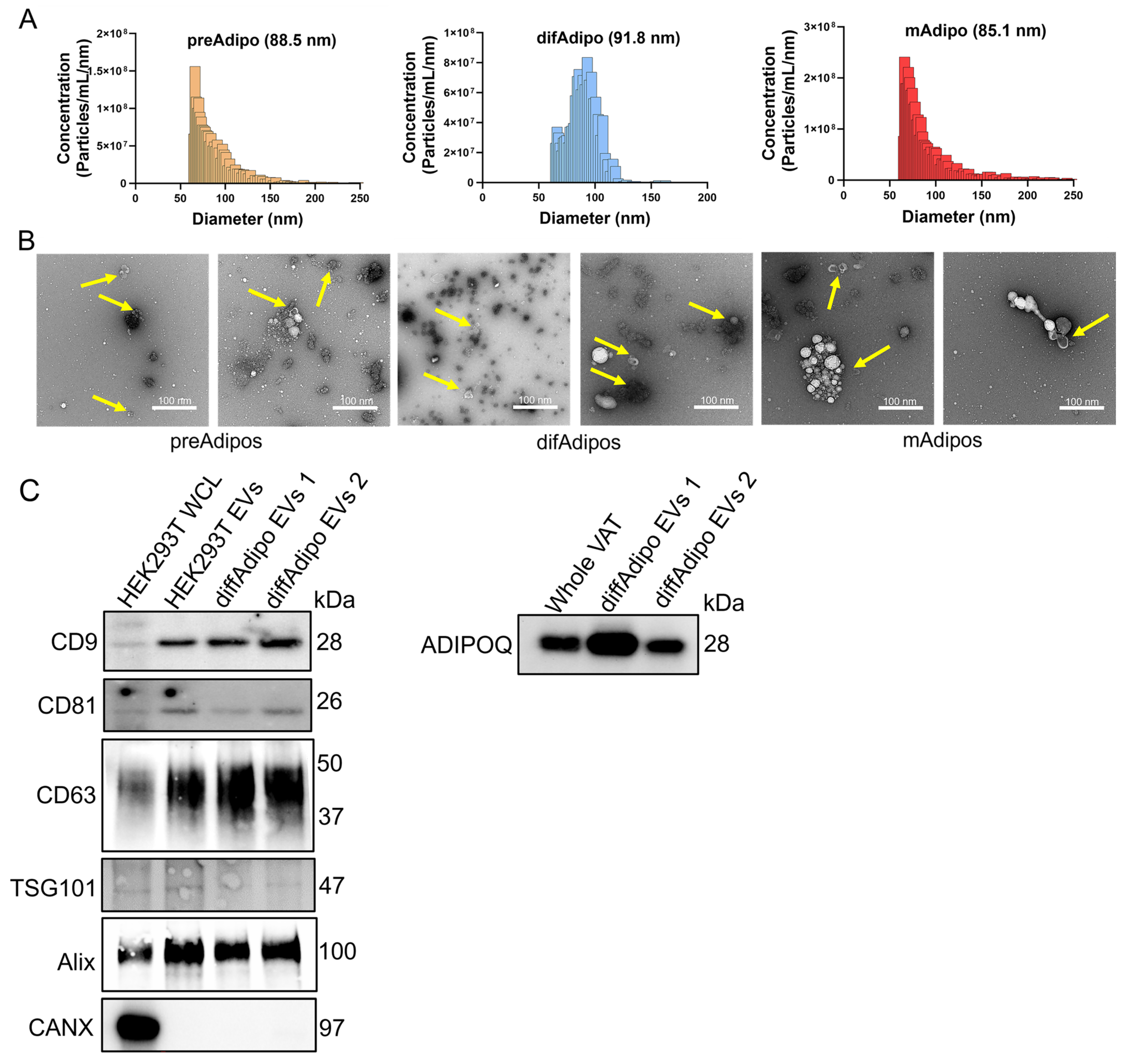
3.1. Multiparametric Isolation and Characterization of ADEVs
3.2. Visceral Adipose-Derived Preadipocytes Serve as a Surrogate Source for Mature Adipocytes
3.3. Differential Expression of Preadipocyte and Mature Adipocyte Markers During Adipocyte Differentiation
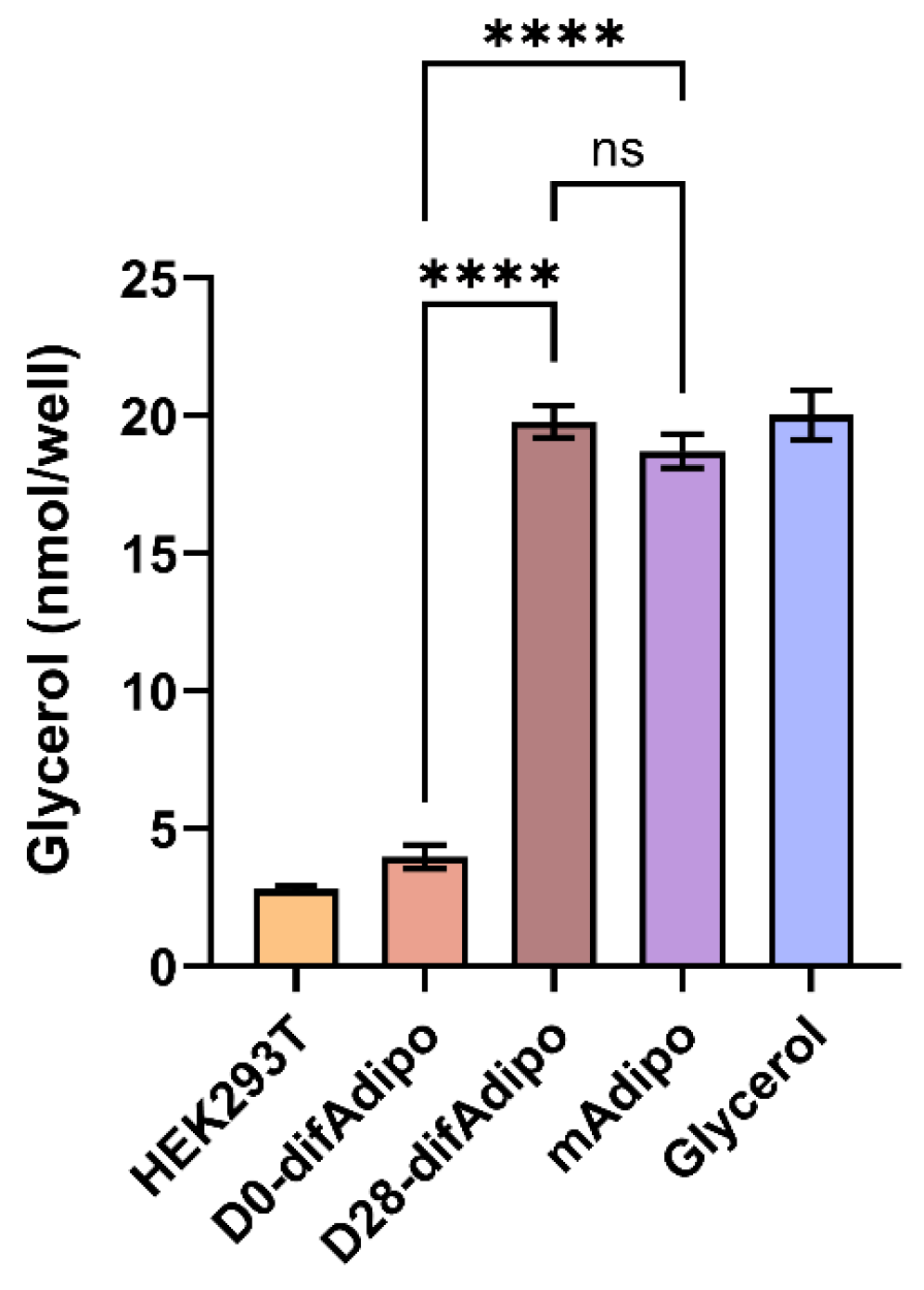
3.4. Differentiated Adipocytes Exhibit Lipolytic Function Comparable to Mature Adipocytes
3.5. Multiparametric Characterization of EVs from Differentiated Adipocytes
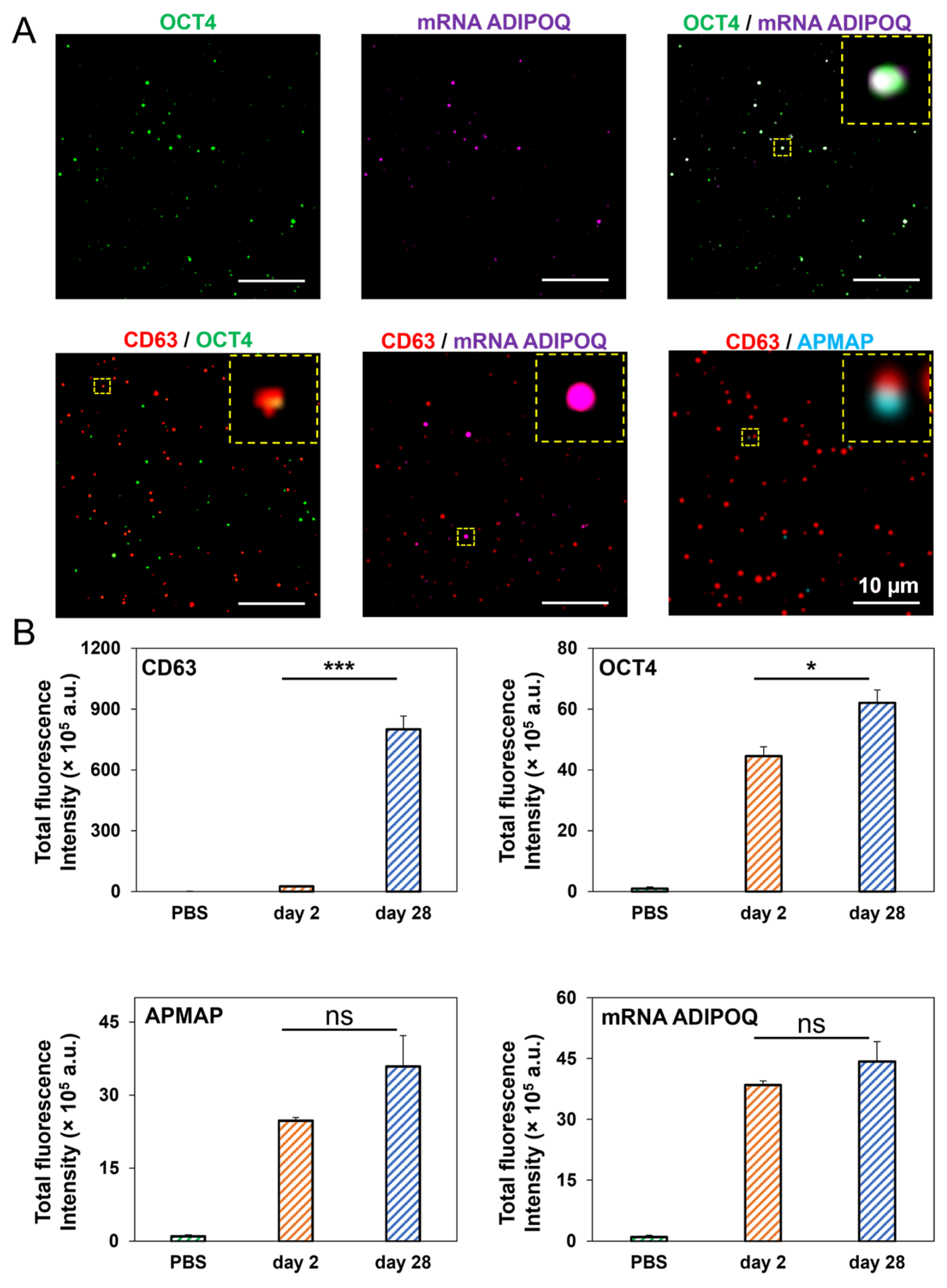
3.6. High-Resolution, Simultaneous Detection of Human Adipose-Derived EV Proteins and RNA Using Total Internal Reflection Fluorescence (TIRF) Microscopy
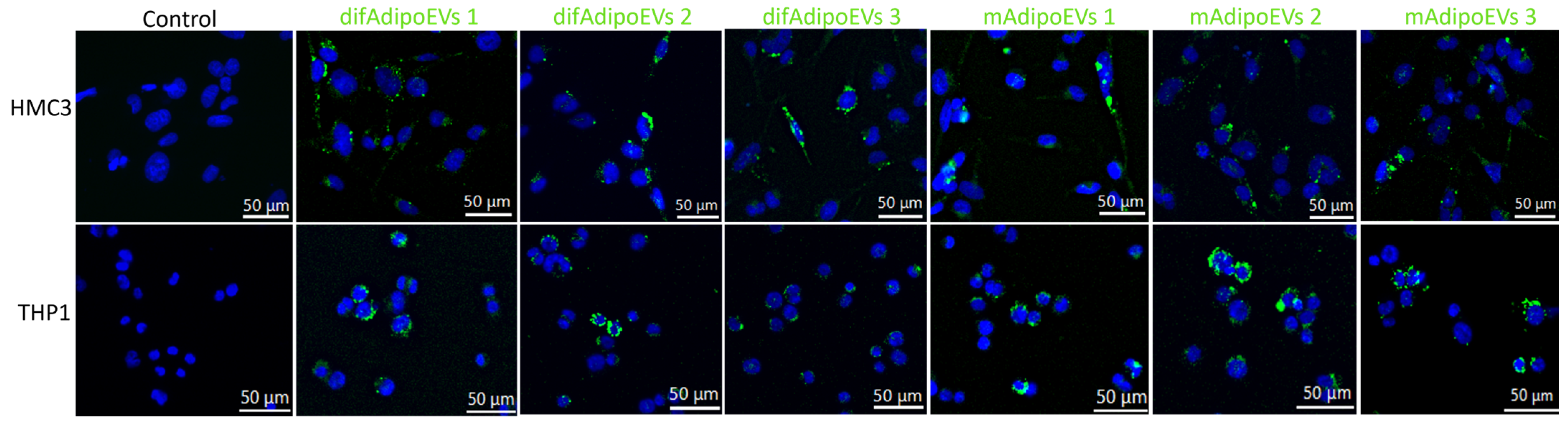
3.7. Efficient Internalization of DifAdipo- and MAdipo-Derived EVs by HMC3 Microglia and THP-1 Macrophages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blüher, M. Obesity: Global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 2019, 15, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Seidell, J.C.; Halberstadt, J. The global burden of obesity and the challenges of prevention. Ann. Nutr. Metab. 2015, 66 (Suppl. S2), 7–12. [Google Scholar] [CrossRef]
- Chong, B.; Jayabaskaran, J.; Kong, G.; Chan, Y.H.; Chin, Y.H.; Goh, R.; Kannan, S.; Ng, C.H.; Loong, S.; Kueh, M.T.W.; et al. Trends and predictions of malnutrition and obesity in 204 countries and territories: An analysis of the Global Burden of Disease Study 2019. eClinicalMedicine 2023, 57, 101850. [Google Scholar] [CrossRef]
- Powell-Wiley, T.M.; Poirier, P.; Burke, L.E.; Despres, J.P.; Gordon-Larsen, P.; Lavie, C.J.; Lear, S.A.; Ndumele, C.E.; Neeland, I.J.; Sanders, P.; et al. Obesity and Cardiovascular Disease: A Scientific Statement From the American Heart Association. Circulation 2021, 143, e984–e1010. [Google Scholar] [CrossRef]
- Avgerinos, K.I.; Spyrou, N.; Mantzoros, C.S.; Dalamaga, M. Obesity and cancer risk: Emerging biological mechanisms and perspectives. Metabolism 2019, 92, 121–135. [Google Scholar] [CrossRef]
- Sarwar, R.; Pierce, N.; Koppe, S. Obesity and nonalcoholic fatty liver disease: Current perspectives. Diabetes Metab. Syndr. Obes. 2018, 11, 533–542. [Google Scholar] [CrossRef]
- Picone, P.; Di Carlo, M.; Nuzzo, D. Obesity and Alzheimer’s disease: Molecular bases. Eur. J. Neurosci. 2020, 52, 3944–3950. [Google Scholar] [CrossRef]
- Shaikh, S.R.; Beck, M.A.; Alwarawrah, Y.; MacIver, N.J. Emerging mechanisms of obesity-associated immune dysfunction. Nat. Rev. Endocrinol. 2024, 20, 136–148. [Google Scholar] [CrossRef]
- Kawai, T.; Autieri, M.V.; Scalia, R. Adipose tissue inflammation and metabolic dysfunction in obesity. Am. J. Physiol. Cell Physiol. 2021, 320, C375–C391. [Google Scholar] [CrossRef]
- Kershaw, E.E.; Flier, J.S. Adipose tissue as an endocrine organ. J. Clin. Endocrinol. Metab. 2004, 89, 2548–2556. [Google Scholar] [CrossRef]
- Booth, A.; Magnuson, A.; Fouts, J.; Foster, M.T. Adipose tissue: An endocrine organ playing a role in metabolic regulation. Horm. Mol. Biol. Clin. Investig. 2016, 26, 25–42. [Google Scholar] [CrossRef]
- Ouchi, N.; Parker, J.L.; Lugus, J.J.; Walsh, K. Adipokines in inflammation and metabolic disease. Nat. Rev. Immunol. 2011, 11, 85–97. [Google Scholar] [CrossRef]
- Longo, M.; Zatterale, F.; Naderi, J.; Parrillo, L.; Formisano, P.; Raciti, G.A.; Beguinot, F.; Miele, C. Adipose Tissue Dysfunction as Determinant of Obesity-Associated Metabolic Complications. Int. J. Mol. Sci. 2019, 20, 2358. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Hade, M.D.; Suire, C.N.; Suo, Z. Mesenchymal Stem Cell-Derived Exosomes: Applications in Regenerative Medicine. Cells 2021, 10, 1959. [Google Scholar] [CrossRef]
- Li, M.-X.; Hu, S.; Lei, H.-H.; Yuan, M.; Li, X.; Hou, W.-K.; Huang, X.-J.; Xiao, B.-W.; Yu, T.-X.; Zhang, X.-H.; et al. Tumor-derived miR-9-5p-loaded EVs regulate cholesterol homeostasis to promote breast cancer liver metastasis in mice. Nat. Commun. 2024, 15, 10539. [Google Scholar] [CrossRef]
- Tkach, M.; Thery, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef]
- Chen, S.; Datta-Chaudhuri, A.; Deme, P.; Dickens, A.; Dastgheyb, R.; Bhargava, P.; Bi, H.; Haughey, N.J. Lipidomic characterization of extracellular vesicles in human serum. J. Circ. Biomark. 2019, 8, 1849454419879848. [Google Scholar] [CrossRef]
- Connolly, K.D.; Rees, D.A.; James, P.E. Role of adipocyte-derived extracellular vesicles in vascular inflammation. Free Radic. Biol. Med. 2021, 172, 58–64. [Google Scholar] [CrossRef]
- Isaac, R.; Reis, F.C.G.; Ying, W.; Olefsky, J.M. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab. 2021, 33, 1744–1762. [Google Scholar] [CrossRef]
- Zhou, C.; Huang, Y.-Q.; Da, M.-X.; Jin, W.-L.; Zhou, F.-H. Adipocyte-derived extracellular vesicles: Bridging the communications between obesity and tumor microenvironment. Discov. Oncol. 2023, 14, 92. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kiran, S.; Kumar, S.; Singh, U.P. Extracellular vesicles in obesity and its associated inflammation. Int. Rev. Immunol. 2022, 41, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Crewe, C. The challenges of interrogating adipose tissue extracellular vesicle functions in physiology. Commun. Biol. 2022, 5, 581. [Google Scholar] [CrossRef]
- Crewe, C.; Joffin, N.; Rutkowski, J.M.; Kim, M.; Zhang, F.; Towler, D.A.; Gordillo, R.; Scherer, P.E. An Endothelial-to-Adipocyte Extracellular Vesicle Axis Governed by Metabolic State. Cell 2018, 175, 695–708.e613. [Google Scholar] [CrossRef]
- Akbar, N.; Pinnick, K.E.; Paget, D.; Choudhury, R.P. Isolation and Characterization of Human Adipocyte-Derived Extracellular Vesicles using Filtration and Ultracentrifugation. J. Vis. Exp. 2021, 170, e61979. [Google Scholar] [CrossRef]
- Borgeson, E.; Boucher, J.; Hagberg, C.E. Of mice and men: Pinpointing species differences in adipose tissue biology. Front. Cell Dev. Biol. 2022, 10, 1003118. [Google Scholar] [CrossRef]
- Palacio, P.L.; Greenwald, J.; Nguyen, K.T.; Shantaram, D.; Butsch, B.L.; Kim, Y.; Dattu, M.H.; Noria, S.; Brethauer, S.A.; Needleman, B.J.; et al. Novel multiparametric bulk and single EV pipeline for adipose cell-specific biomarker discovery in paired human biospecimens. bioRxiv 2024. [Google Scholar] [CrossRef]
- Nguyen, K.T.; Xia, J.; Chang, C.; Hade, M.D.; Rima, X.Y.; An, J.Y.; Min, B.H.; Nagaraj, C.K.; Wang, L.; Huang, T.J.; et al. Integrated Techniques for Extracellular Particle Separation and Single-Particle Multiparametric Characterization to Track Cancer Biomarkers from Tissue to Biofluids. bioRxiv 2025. [Google Scholar] [CrossRef]
- Rima, X.Y.; Zhang, J.; Nguyen, L.T.H.; Rajasuriyar, A.; Yoon, M.J.; Chiang, C.L.; Walters, N.; Kwak, K.J.; Lee, L.J.; Reátegui, E. Microfluidic harvesting of breast cancer tumor spheroid-derived extracellular vesicles from immobilized microgels for single-vesicle analysis. Lab. Chip 2022, 22, 2502–2518. [Google Scholar] [CrossRef]
- Nguyen, L.T.H.; Zhang, J.; Rima, X.Y.; Wang, X.; Kwak, K.J.; Okimoto, T.; Amann, J.; Yoon, M.J.; Shukuya, T.; Chiang, C.L.; et al. An immunogold single extracellular vesicular RNA and protein ((Au) SERP) biochip to predict responses to immunotherapy in non-small cell lung cancer patients. J. Extracell. Vesicles 2022, 11, e12258. [Google Scholar] [CrossRef]
- Vella, L.J.; Scicluna, B.J.; Cheng, L.; Bawden, E.G.; Masters, C.L.; Ang, C.S.; Willamson, N.; McLean, C.; Barnham, K.J.; Hill, A.F. A rigorous method to enrich for exosomes from brain tissue. J. Extracell. Vesicles 2017, 6, 1348885. [Google Scholar] [CrossRef] [PubMed]
- Crewe, C.; Scherer, P.E. Intercellular and interorgan crosstalk through adipocyte extracellular vesicles. Rev. Endocr. Metab. Disord. 2022, 23, 61–69. [Google Scholar] [CrossRef]
- Kranendonk, M.E.; Visseren, F.L.; van Balkom, B.W.; Nolte-’t Hoen, E.N.; van Herwaarden, J.A.; de Jager, W.; Schipper, H.S.; Brenkman, A.B.; Verhaar, M.C.; Wauben, M.H.; et al. Human adipocyte extracellular vesicles in reciprocal signaling between adipocytes and macrophages. Obesity 2014, 22, 1296–1308. [Google Scholar] [CrossRef]
- Lazar, I.; Clement, E.; Dauvillier, S.; Milhas, D.; Ducoux-Petit, M.; LeGonidec, S.; Moro, C.; Soldan, V.; Dalle, S.; Balor, S.; et al. Adipocyte Exosomes Promote Melanoma Aggressiveness through Fatty Acid Oxidation: A Novel Mechanism Linking Obesity and Cancer. Cancer Res. 2016, 76, 4051–4057. [Google Scholar] [CrossRef]
- Clement, E.; Lazar, I.; Attané, C.; Carrié, L.; Dauvillier, S.; Ducoux-Petit, M.; Esteve, D.; Menneteau, T.; Moutahir, M.; Le Gonidec, S.; et al. Adipocyte extracellular vesicles carry enzymes and fatty acids that stimulate mitochondrial metabolism and remodeling in tumor cells. Embo J. 2020, 39, e102525. [Google Scholar] [CrossRef]
- Matilainen, J.; Berg, V.; Vaittinen, M.; Impola, U.; Mustonen, A.M.; Männistö, V.; Malinen, M.; Luukkonen, V.; Rosso, N.; Turunen, T.; et al. Increased secretion of adipocyte-derived extracellular vesicles is associated with adipose tissue inflammation and the mobilization of excess lipid in human obesity. J. Transl. Med. 2024, 22, 623. [Google Scholar] [CrossRef]
- Kulaj, K.; Harger, A.; Bauer, M.; Caliskan, Ö.S.; Gupta, T.K.; Chiang, D.M.; Milbank, E.; Reber, J.; Karlas, A.; Kotzbeck, P.; et al. Adipocyte-derived extracellular vesicles increase insulin secretion through transport of insulinotropic protein cargo. Nat. Commun. 2023, 14, 709. [Google Scholar] [CrossRef]
- Tong, Q.; Dalgin, G.; Xu, H.; Ting, C.-N.; Leiden, J.M.; Hotamisligil, G.S. Function of GATA Transcription Factors in Preadipocyte-Adipocyte Transition. Science 2000, 290, 134–138. [Google Scholar] [CrossRef]
- da Silva, C.; Durandt, C.; Kallmeyer, K.; Ambele, M.A.; Pepper, M.S. The Role of Pref-1 during Adipogenic Differentiation: An Overview of Suggested Mechanisms. Int. J. Mol. Sci. 2020, 21, 4104. [Google Scholar] [CrossRef]
- Rim, J.S.; Lewis, P.; Dave, A.; Rim, J.; Szuszka, K.; Acosta, C.; Kim, J.; Kim, J.-D.; Staszkiewicz, J.; Gao, R.; et al. Direct Conversion of Human Preadipocytes into Hematopoietic, Neuronal, and Pancreatic α Cells by Oct4 and Klf4 Overexpression. BioResearch Open Access 2015, 4, 389–397. [Google Scholar] [CrossRef]
- Dang, T.N.; Tiongco, R.P.; Brown, L.M.; Taylor, J.L.; Lyons, J.M.; Lau, F.H.; Floyd, Z.E. Expression of the preadipocyte marker ZFP423 is dysregulated between well-differentiated and dedifferentiated liposarcoma. BMC Cancer 2022, 22, 300. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, Y.; Murakami, M.; Murata, M.; Kano, F. Microscopic image-based classification of adipocyte differentiation by machine learning. Histochem. Cell Biol. 2023, 159, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Audano, M.; Pedretti, S.; Caruso, D.; Crestani, M.; De Fabiani, E.; Mitro, N. Regulatory mechanisms of the early phase of white adipocyte differentiation: An overview. Cell. Mol. Life Sci. 2022, 79, 139. [Google Scholar] [CrossRef] [PubMed]
- Al-Mansoori, L.; Al-Jaber, H.; Madani, A.Y.; Mazloum, N.A.; Agouni, A.; Ramanjaneya, M.; Abou-Samra, A.-B.; Elrayess, M.A. Suppression of GATA-3 increases adipogenesis, reduces inflammation and improves insulin sensitivity in 3T3L-1 preadipocytes. Cell. Signal. 2020, 75, 109735. [Google Scholar] [CrossRef]
- Itabe, H.; Yamaguchi, T.; Nimura, S.; Sasabe, N. Perilipins: A diversity of intracellular lipid droplet proteins. Lipids Health Dis. 2017, 16, 83. [Google Scholar] [CrossRef]
- Sztalryd, C.; Brasaemle, D.L. The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 1221–1232. [Google Scholar] [CrossRef]
- Fu, Y.; Luo, N.; Klein, R.L.; Garvey, W.T. Adiponectin promotes adipocyte differentiation, insulin sensitivity, and lipid accumulation. J. Lipid Res. 2005, 46, 1369–1379. [Google Scholar] [CrossRef]
- Rosen, E.D.; Spiegelman, B.M. Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol. 2000, 16, 145–171. [Google Scholar] [CrossRef]
- Farmer, S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006, 4, 263–273. [Google Scholar] [CrossRef]
- Gregoire, F.M. Adipocyte differentiation: From fibroblast to endocrine cell. Exp. Biol. Med. 2001, 226, 997–1002. [Google Scholar] [CrossRef]
- Green, H.; Kehinde, O. Spontaneous heritable changes leading to increased adipose conversion in 3T3 cells. Cell 1976, 7, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Buschmann, D.; Kirchner, B.; Hermann, S.; Märte, M.; Wurmser, C.; Brandes, F.; Kotschote, S.; Bonin, M.; Steinlein, O.K.; Pfaffl, M.W.; et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell. Vesicles 2018, 7, 1481321. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, K.S.; Harris, K.; Arian, K.A.; Ma, L.; Schueng Zancanela, B.; Church, K.A.; McAndrews, K.M.; Kalluri, R. High throughput and rapid isolation of extracellular vesicles and exosomes with purity using size exclusion liquid chromatography. Bioact. Mater. 2024, 40, 683–695. [Google Scholar] [CrossRef]
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Crewe, C.; Funcke, J.B.; Li, S.; Joffin, N.; Gliniak, C.M.; Ghaben, A.L.; An, Y.A.; Sadek, H.A.; Gordillo, R.; Akgul, Y.; et al. Extracellular vesicle-based interorgan transport of mitochondria from energetically stressed adipocytes. Cell Metab. 2021, 33, 1853–1868.e1811. [Google Scholar] [CrossRef]
- Ferrante Jr, A.W. Obesity-induced inflammation: A metabolic dialogue in the language of inflammation. J. Intern. Med. 2007, 262, 408–414. [Google Scholar] [CrossRef]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384.e312. [Google Scholar] [CrossRef]
- Muniyappa, R.; Chen, H.; Muzumdar, R.H.; Einstein, F.H.; Yan, X.; Yue, L.Q.; Barzilai, N.; Quon, M.J. Comparison between surrogate indexes of insulin sensitivity/resistance and hyperinsulinemic euglycemic clamp estimates in rats. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E1023–E1029. [Google Scholar] [CrossRef]
- Gómez-Serrano, M.; Ponath, V.; Preußer, C.; Pogge von Strandmann, E. Beyond the Extracellular Vesicles: Technical Hurdles, Achieved Goals and Current Challenges When Working on Adipose Cells. Int. J. Mol. Sci. 2021, 22, 3362. [Google Scholar] [CrossRef]
- Sarango, G.; Richetta, C.; Pereira, M.; Kumari, A.; Ghosh, M.; Bertrand, L.; Pionneau, C.; Le Gall, M.; Grégoire, S.; Jeger-Madiot, R.; et al. The Autophagy Receptor TAX1BP1 (T6BP) improves antigen presentation by MHC-II molecules. EMBO Rep. 2022, 23, e55470. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, E.; Betti, M.J.; Sheng, Q.; Lin, P.; Emont, M.P.; Li, G.; Amancherla, K.; Limpitikul, W.B.; Whittaker, O.R.; Luong, K.; et al. The extracellular vesicle transcriptome provides tissue-specific functional genomic annotation relevant to disease susceptibility in obesity. medRxiv 2024. [Google Scholar] [CrossRef]
- Sethi, J.K.; Vidal-Puig, A.J. Thematic review series: Adipocyte Biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J. Lipid Res. 2007, 48, 1253–1262. [Google Scholar] [CrossRef]
- Connolly, K.D.; Wadey, R.M.; Mathew, D.; Johnson, E.; Rees, D.A.; James, P.E. Evidence for Adipocyte-Derived Extracellular Vesicles in the Human Circulation. Endocrinology 2018, 159, 3259–3267. [Google Scholar] [CrossRef]
- Lötvall, J.; Hill, A.F.; Hochberg, F.; Buzás, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef]
- Adnan, S.-s.; Benjamin, G.T.; David, R.W. Advances in extracellular vesicle isolation methods: A path towards cell-type specific EV isolation. Extracell. Vesicles Circ. Nucleic Acids 2023, 4, 447–460. [Google Scholar] [CrossRef]
- Beck, B.R.; Shin, B.; Choi, Y.; Park, S.; Kang, K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020, 18, 784–790. [Google Scholar] [CrossRef]
- Li, Y.; He, X.; Li, Q.; Lai, H.; Zhang, H.; Hu, Z.; Li, Y.; Huang, S. EV-origin: Enumerating the tissue-cellular origin of circulating extracellular vesicles using exLR profile. Comput. Struct. Biotechnol. J. 2020, 18, 2851–2859. [Google Scholar] [CrossRef]
- Connolly, K.D.; Guschina, I.A.; Yeung, V.; Clayton, A.; Draman, M.S.; Von Ruhland, C.; Ludgate, M.; James, P.E.; Rees, D.A. Characterisation of adipocyte-derived extracellular vesicles released pre- and post-adipogenesis. J. Extracell. Vesicles 2015, 4, 29159. [Google Scholar] [CrossRef]
- Ferguson, S.; Yang, K.S.; Zelga, P.; Liss, A.S.; Carlson, J.C.T.; Del Castillo, C.F.; Weissleder, R. Single-EV analysis (sEVA) of mutated proteins allows detection of stage 1 pancreatic cancer. Sci. Adv. 2022, 8, eabm3453. [Google Scholar] [CrossRef]
- Li, Z.; Guo, K.; Gao, Z.; Chen, J.; Ye, Z.; Cao, M.; Wang, S.E.; Yin, Y.; Zhong, W. Colocalization of protein and microRNA markers reveals unique extracellular vesicle subpopulations for early cancer detection. Sci. Adv. 2024, 10, eadh8689. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hade, M.D.; Butsch, B.L.; Loreto Palacio, P.; Nguyen, K.T.; Shantaram, D.; Noria, S.F.; Brethauer, S.A.; Needleman, B.J.; Hsueh, W.; Reátegui, E.; et al. Human Differentiated Adipocytes as Surrogate Mature Adipocytes for Adipocyte-Derived Extracellular Vesicle Analysis. Cells 2025, 14, 757. https://doi.org/10.3390/cells14110757
Hade MD, Butsch BL, Loreto Palacio P, Nguyen KT, Shantaram D, Noria SF, Brethauer SA, Needleman BJ, Hsueh W, Reátegui E, et al. Human Differentiated Adipocytes as Surrogate Mature Adipocytes for Adipocyte-Derived Extracellular Vesicle Analysis. Cells. 2025; 14(11):757. https://doi.org/10.3390/cells14110757
Chicago/Turabian StyleHade, Mangesh Dattu, Bradley L. Butsch, Paola Loreto Palacio, Kim Truc Nguyen, Dharti Shantaram, Sabrena F. Noria, Stacy A. Brethauer, Bradley J. Needleman, Willa Hsueh, Eduardo Reátegui, and et al. 2025. "Human Differentiated Adipocytes as Surrogate Mature Adipocytes for Adipocyte-Derived Extracellular Vesicle Analysis" Cells 14, no. 11: 757. https://doi.org/10.3390/cells14110757
APA StyleHade, M. D., Butsch, B. L., Loreto Palacio, P., Nguyen, K. T., Shantaram, D., Noria, S. F., Brethauer, S. A., Needleman, B. J., Hsueh, W., Reátegui, E., & Magaña, S. M. (2025). Human Differentiated Adipocytes as Surrogate Mature Adipocytes for Adipocyte-Derived Extracellular Vesicle Analysis. Cells, 14(11), 757. https://doi.org/10.3390/cells14110757





