Functional Selectivity of Cannabinoid Type 1 G Protein-Coupled Receptor Agonists in Transactivating Glycosylated Receptors on Cancer Cells to Induce Epithelial–Mesenchymal Transition Metastatic Phenotype
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Reagents
2.3. CB1 Agonist Treatment Protocol (Time and Dosage)
2.4. Antibodies
2.5. Sialidase Assay
2.6. NF-kB Dependent Secreted Embryonic Alkaline Phosphatase (SEAP) Assay
2.7. Co-Localization
2.8. Immunofluorescence Staining
2.9. Statistics
3. Results
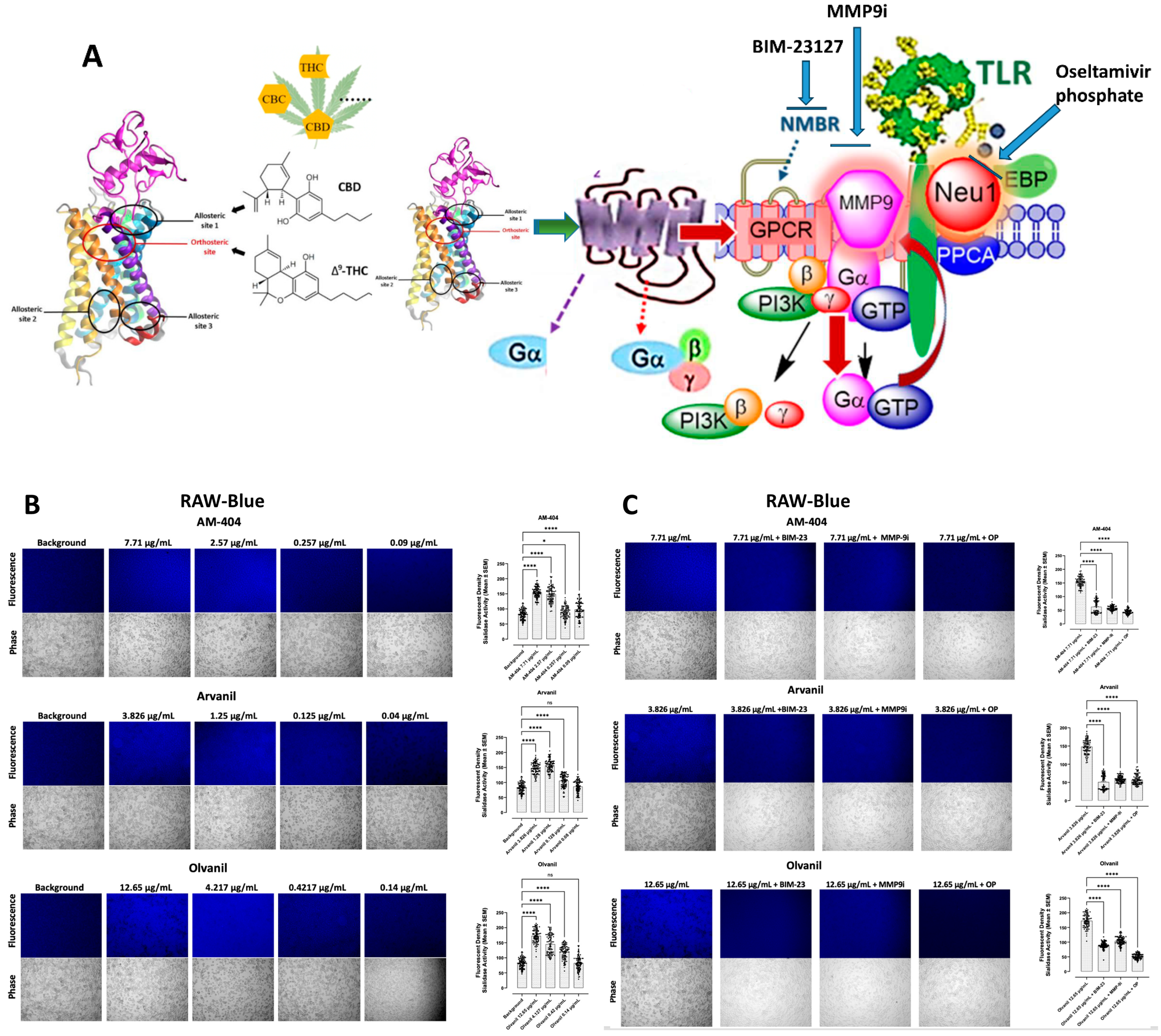
3.1. CB1 G Protein-Coupled Receptor Agonists Dose-Dependently Induce Neu1 Sialidase Activity in RAW-Blue Macrophage Cells
3.2. CB1 G Protein-Coupled Receptor Agonists Dose-Dependently Induce Neu1 Sialidase Activity in Pancreatic PANC-1 and Colorectal SW-620 Cancer Cell Lines
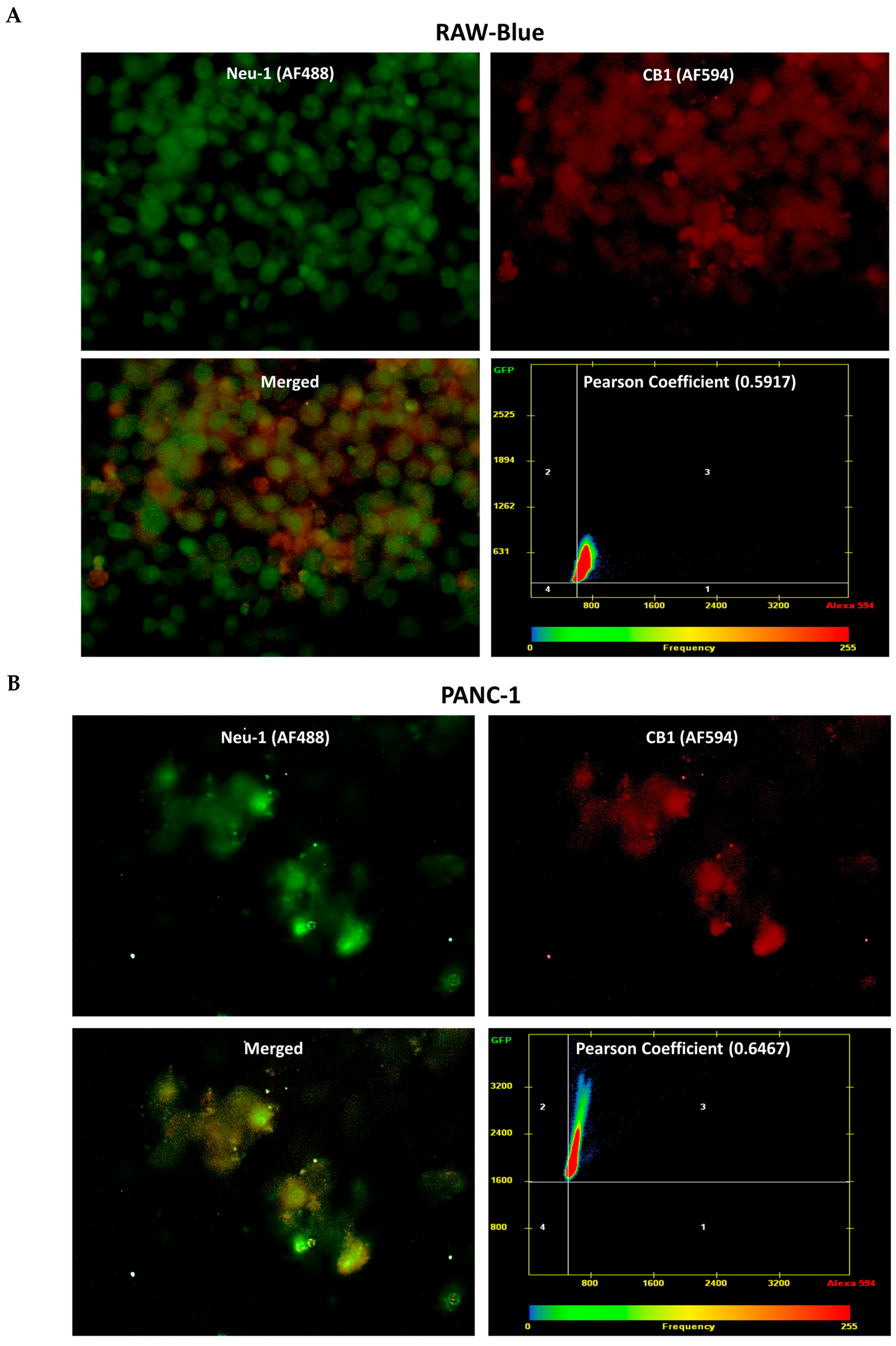
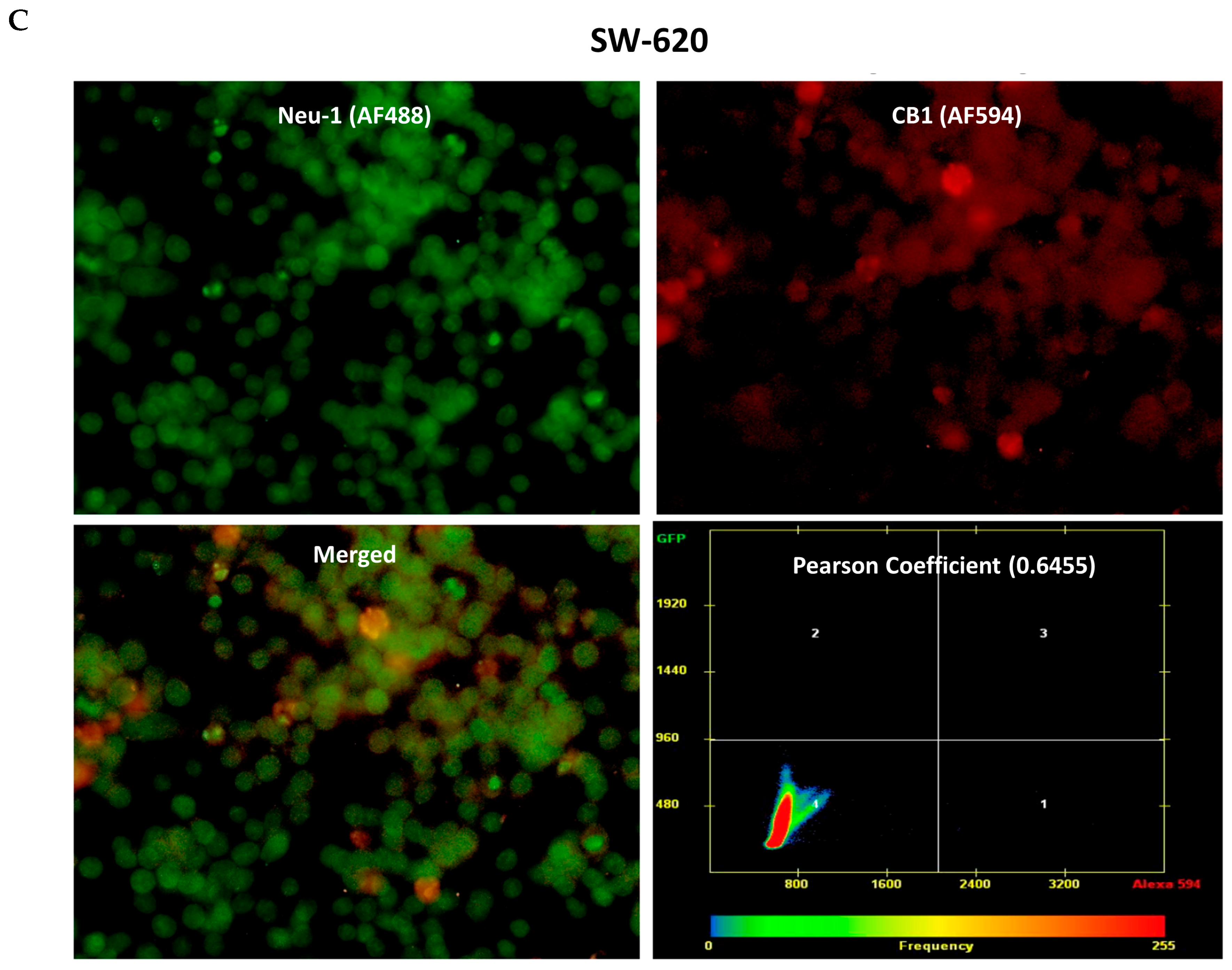
3.3. CB1 Receptor Co-Localizes with Neu1 on the Cell Surface of Naïve Unstimulated RAW-Blue Macrophages, PANC-1, and SW-620 Cells
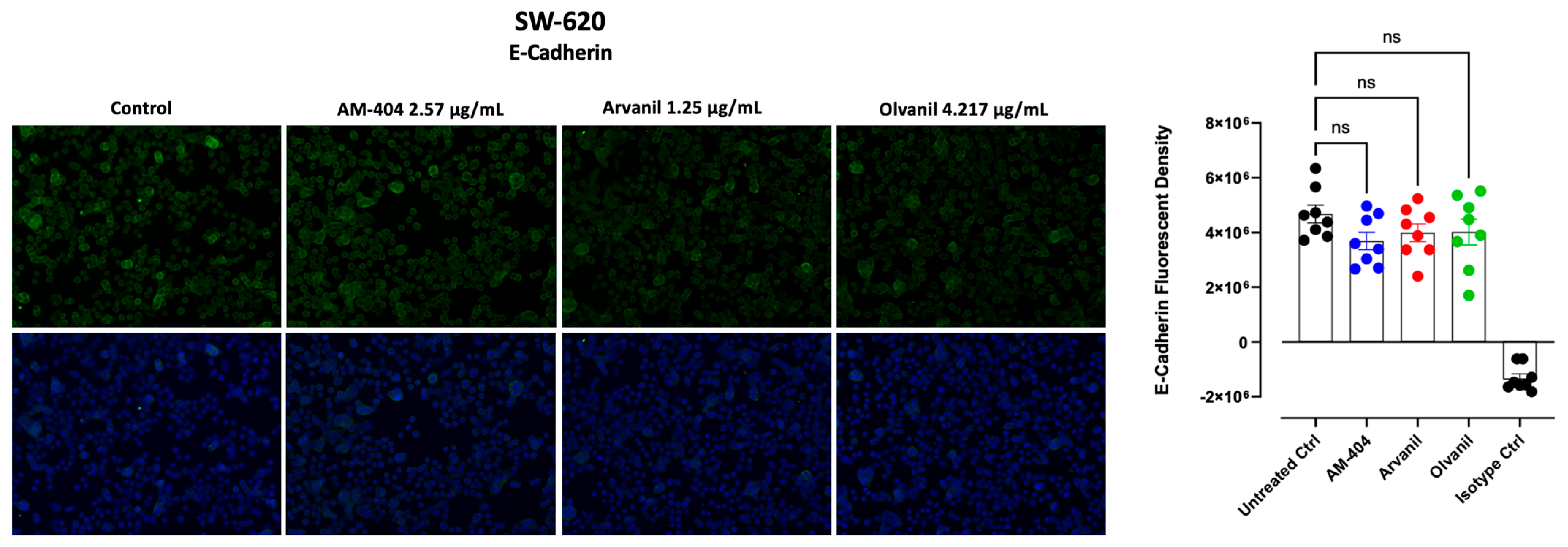
3.4. Synthetic CB1 Cannabinoids AM-404, Aravnil, and Olvanil Marginally Reduce the Expression of E-Cadherin in SW-620 Colorectal Cancer Cells
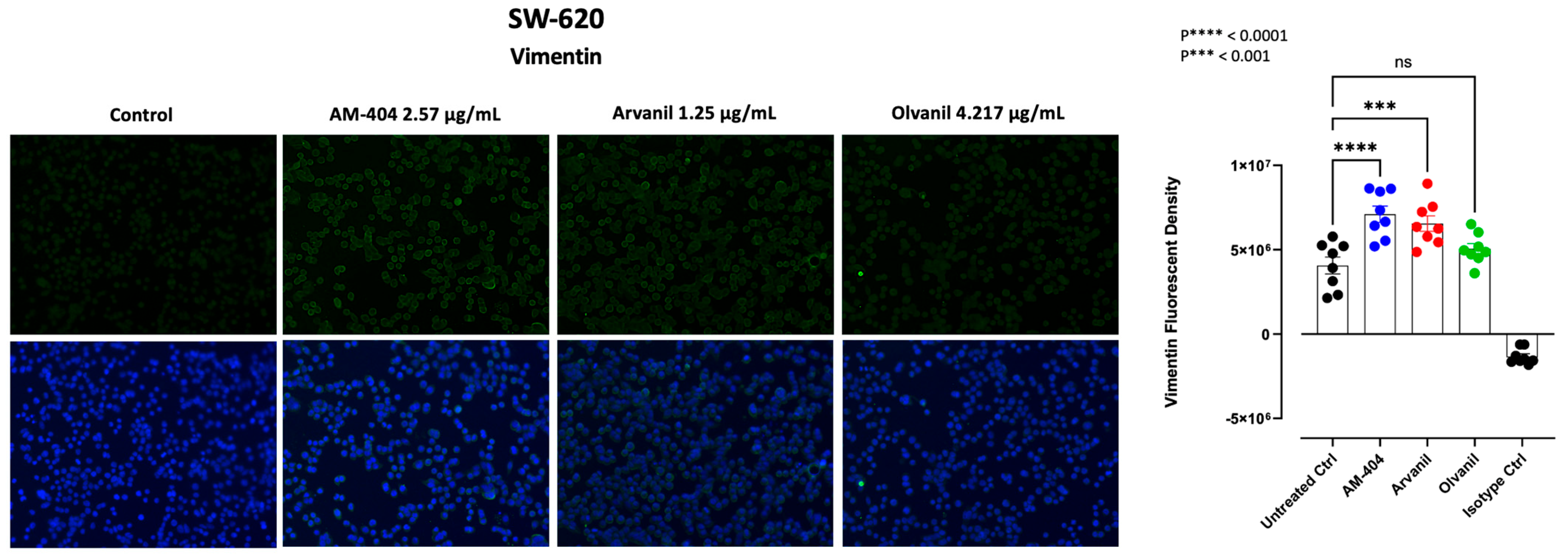
3.5. Synthetic CB1 Cannabinoids AM-404, Arvanil, and Olvanil Significantly Upregulate the Expression of Vimentin in SW-620 Cells
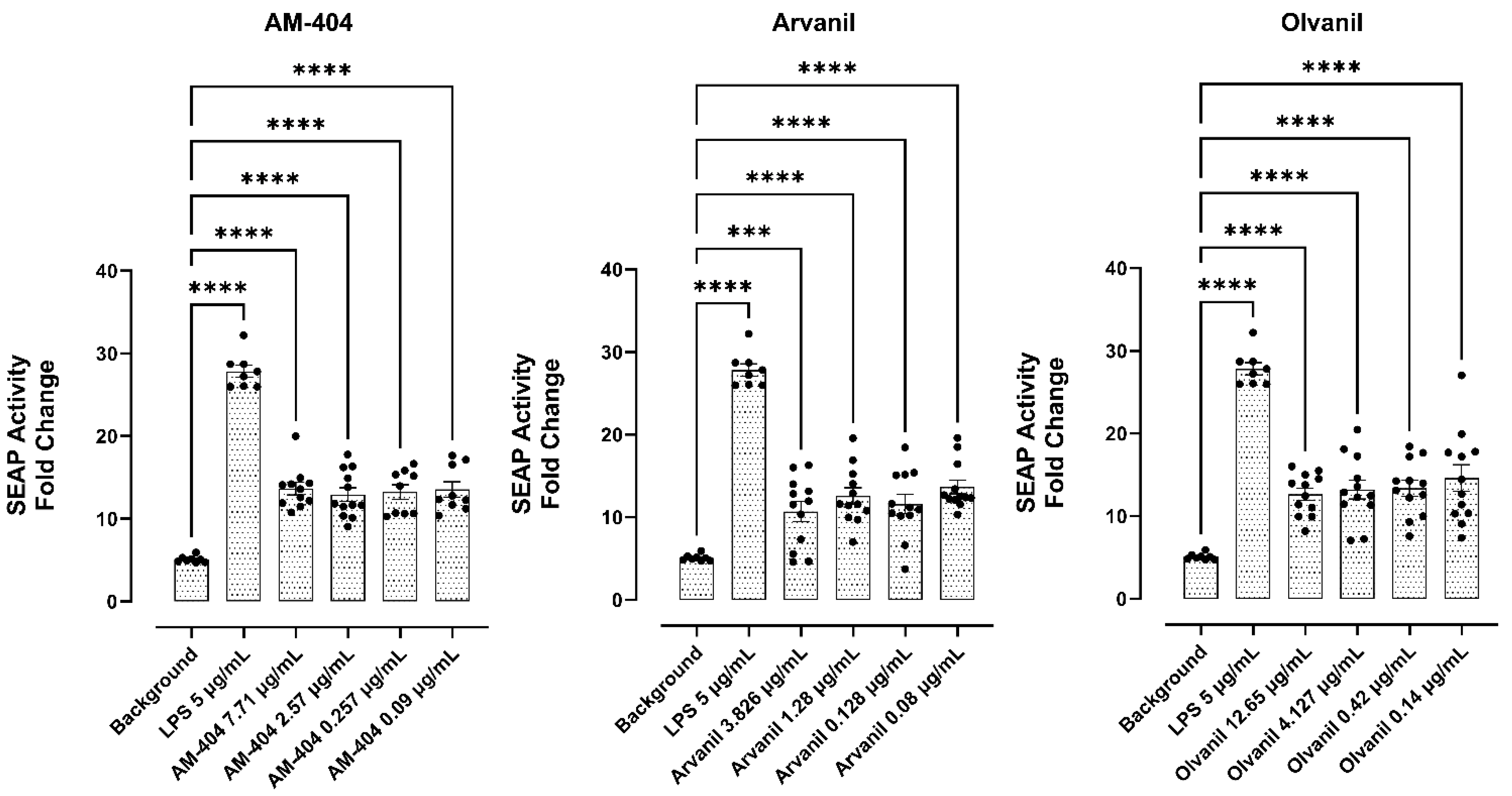
3.6. CB1 Agonists, AM-404, Arvanil, and Olvanil Induce Upregulation of NF-kB
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Rosenbaum, D.M.; Rasmussen, S.G.; Kobilka, B.K. The structure and function of G-protein-coupled receptors. Nature 2009, 459, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Rask-Andersen, M.; Almen, M.S.; Schioth, H.B. Trends in the exploitation of novel drug targets. Nat. Rev. Drug Discov. 2011, 10, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Wisler, J.W.; Xiao, K.; Thomsen, A.R.; Lefkowitz, R.J. Recent developments in biased agonism. Curr. Opin. Cell Biol. 2014, 27, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Shah, B.H.; Catt, K.J. GPCR-mediated transactivation of RTKs in the CNS: Mechanisms and consequences. Trends Neurosci. 2004, 27, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Delcourt, N.; Bockaert, J.; Marin, P. GPCR-jacking: From a new route in RTK signalling to a new concept in GPCR activation. Trends Pharmacol. Sci. 2007, 28, 602–607. [Google Scholar] [CrossRef]
- Jayanth, P.; Amith, S.R.; Gee, K.; Szewczuk, M.R. Neu1 sialidase and matrix metalloproteinase-9 cross-talk is essential for neurotrophin activation of Trk receptors and cellular signaling. Cell. Signal. 2010, 22, 1193–1205. [Google Scholar] [CrossRef]
- Alghamdi, F.; Guo, M.; Abdulkhalek, S.; Crawford, N.; Amith, S.R.; Szewczuk, M.R. A novel insulin receptor-signaling platform and its link to insulin resistance and type 2 diabetes. Cell. Signal. 2014, 26, 1355–1368. [Google Scholar] [CrossRef]
- Haxho, F.; Haq, S.; Szewczuk, M.R. Biased G protein-coupled receptor agonism mediates Neu1 sialidase and matrix metalloproteinase-9 crosstalk to induce transactivation of insulin receptor signaling. Cell. Signal. 2018, 43, 71–84. [Google Scholar] [CrossRef]
- Haxho, F.; Neufeld, R.J.; Szewczuk, M.R. Neuraminidase-1: A novel therapeutic target in multistage tumorigenesis. Oncotarget 2016, 7, 40860–40881. [Google Scholar] [CrossRef]
- Haxho, F.; Allison, S.; Alghamdi, F.; Brodhagen, L.; Kuta, V.E.L.; Abdulkhalek, S.; Neufeld, R.J.; Szewczuk, M.R. Oseltamivir phosphate monotherapy ablates tumor neovascularization, growth, and metastasis in mouse model of human triple-negative breast adenocarcinoma. Breast Cancer Targets Ther. 2014, 6, 191. [Google Scholar]
- Gilmour, A.M.; Abdulkhalek, S.; Cheng, T.S.; Alghamdi, F.; Jayanth, P.; O’Shea, L.K.; Geen, O.; Arvizu, L.A.; Szewczuk, M.R. A novel epidermal growth factor receptor-signaling platform and its targeted translation in pancreatic cancer. Cell. Signal. 2013, 25, 2587–2603. [Google Scholar] [CrossRef]
- Abdulkhalek, S.; Szewczuk, M.R. Neu1 sialidase and matrix metalloproteinase-9 cross-talk regulates nucleic acid-induced endosomal TOLL-like receptor-7 and-9 activation, cellular signaling and pro-inflammatory responses. Cell. Signal. 2013, 25, 2093–2105. [Google Scholar] [CrossRef] [PubMed]
- Abdulkhalek, S.; Amith, S.R.; Franchuk, S.L.; Jayanth, P.; Guo, M.; Finlay, T.; Gilmour, A.; Guzzo, C.; Gee, K.; Beyaert, R. Neu1 sialidase and matrix metalloproteinase-9 cross-talk is essential for Toll-like receptor activation and cellular signaling. J. Biol. Chem. 2011, 286, 36532–36549. [Google Scholar] [CrossRef] [PubMed]
- Abdulkhalek, S.; Guo, M.; Amith, S.R.; Jayanth, P.; Szewczuk, M.R. G-protein coupled receptor agonists mediate Neu1 sialidase and matrix metalloproteinase-9 cross-talk to induce transactivation of TOLL-like receptors and cellular signaling. Cell. Signal. 2012, 24, 2035–2042. [Google Scholar] [CrossRef] [PubMed]
- Rozenfeld, R.; Gupta, A.; Gagnidze, K.; Lim, M.P.; Gomes, I.; Lee-Ramos, D.; Nieto, N.; Devi, L.A. AT1R-CB₁R heteromerization reveals a new mechanism for the pathogenic properties of angiotensin II. Embo J. 2011, 30, 2350–2363. [Google Scholar] [CrossRef] [PubMed]
- Blasco-Benito, S.; Moreno, E.; Seijo-Vila, M.; Tundidor, I.; Andradas, C.; Caffarel, M.M.; Caro-Villalobos, M.; Urigüen, L.; Diez-Alarcia, R.; Moreno-Bueno, G.; et al. Therapeutic targeting of HER2-CB(2)R heteromers in HER2-positive breast cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 3863–3872. [Google Scholar] [CrossRef] [PubMed]
- Wager-Miller, J.; Westenbroek, R.; Mackie, K. Dimerization of G protein-coupled receptors: CB1 cannabinoid receptors as an example. Chem. Phys. Lipids 2002, 121, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Kearn, C.; Mackie, K.; Glass, M. Physical interactions of CB1 cannabinoid and D2 receptors. In Proceedings of the 2004 Symposium of the Cannabinoids, Paestum, Italy, 22–27 June 2004; p. 16. [Google Scholar]
- Glass, M.; Felder, C.C. Concurrent stimulation of cannabinoid CB1 and dopamine D2 receptors augments cAMP accumulation in striatal neurons: Evidence for a Gs linkage to the CB1 receptor. J. Neurosci. 1997, 17, 5327–5333. [Google Scholar] [CrossRef]
- Jarrahian, A.; Watts, V.J.; Barker, E.L. D2 dopamine receptors modulate Gα-subunit coupling of the CB1 cannabinoid receptor. J. Pharmacol. Exp. Ther. 2004, 308, 880–886. [Google Scholar] [CrossRef]
- Hojo, M.; Sudo, Y.; Ando, Y.; Minami, K.; Takada, M.; Matsubara, T.; Kanaide, M.; Taniyama, K.; Sumikawa, K.; Uezono, Y. μ-Opioid receptor forms a functional heterodimer with cannabinoid CB1 receptor: Electrophysiological and FRET assay analysis. J. Pharmacol. Sci. 2008, 108, 308–319. [Google Scholar] [CrossRef]
- Carriba, P.; Ortiz, O.; Patkar, K.; Justinova, Z.; Stroik, J.; Themann, A.; Müller, C.; Woods, A.S.; Hope, B.T.; Ciruela, F. Striatal adenosine A2A and cannabinoid CB1 receptors form functional heteromeric complexes that mediate the motor effects of cannabinoids. Neuropsychopharmacology 2007, 32, 2249–2259. [Google Scholar] [CrossRef] [PubMed]
- Hudson, B.D.; Hébert, T.E.; Kelly, M.E. Physical and functional interaction between CB1 cannabinoid receptors and beta2-adrenoceptors. Br. J. Pharmacol. 2010, 160, 627–642. [Google Scholar] [CrossRef] [PubMed]
- Glass, M.; Northup, J.K. Agonist selective regulation of G proteins by cannabinoid CB1 and CB2 receptors. Mol. Pharmacol. 1999, 56, 1362–1369. [Google Scholar] [CrossRef] [PubMed]
- Villaseca, S.; Romero, G.; Ruiz, M.J.; Pérez, C.; Leal, J.I.; Tovar, L.M.; Torrejón, M. Gαi protein subunit: A step toward understanding its non-canonical mechanisms. Front. Cell Dev. Biol. 2022, 10, 941870. [Google Scholar] [CrossRef]
- Hudson, B.D.; Hebert, T.E.; Kelly, M.E. Ligand- and heterodimer-directed signaling of the CB(1) cannabinoid receptor. Mol. Pharmacol. 2010, 77, 1–9. [Google Scholar] [CrossRef]
- Carriba, P.; Navarro, G.; Ciruela, F.; Ferré, S.; Casadó, V.; Agnati, L.; Cortés, A.; Mallol, J.; Fuxe, K.; Canela, E.I. Detection of heteromerization of more than two proteins by sequential BRET-FRET. Nat. Methods 2008, 5, 727–733. [Google Scholar] [CrossRef]
- Peeri, H.; Koltai, H. Cannabis Biomolecule Effects on Cancer Cells and Cancer Stem Cells: Cytotoxic, Anti-Proliferative, and Anti-Migratory Activities. Biomolecules 2022, 12, 491. [Google Scholar] [CrossRef]
- Caffarel, M.M.; Sarrió, D.; Palacios, J.; Guzmán, M.; Sánchez, C. Delta9-tetrahydrocannabinol inhibits cell cycle progression in human breast cancer cells through Cdc2 regulation. Cancer Res. 2006, 66, 6615–6621. [Google Scholar] [CrossRef] [PubMed]
- Qamri, Z.; Preet, A.; Nasser, M.W.; Bass, C.E.; Leone, G.; Barsky, S.H.; Ganju, R.K. Synthetic cannabinoid receptor agonists inhibit tumor growth and metastasis of breast cancer. Mol. Cancer Ther. 2009, 8, 3117–3129. [Google Scholar] [CrossRef] [PubMed]
- Preet, A.; Ganju, R.K.; Groopman, J.E. Delta9-Tetrahydrocannabinol inhibits epithelial growth factor-induced lung cancer cell migration in vitro as well as its growth and metastasis in vivo. Oncogene 2008, 27, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, C.; Salazar, M.; Carracedo, A.; Lorente, M.; Egia, A.; González-Feria, L.; Haro, A.; Velasco, G.; Guzmán, M. Cannabinoids inhibit glioma cell invasion by down-regulating matrix metalloproteinase-2 expression. Cancer Res. 2008, 68, 1945–1952. [Google Scholar] [CrossRef]
- Moon, K.-Y.; Hahn, B.-S.; Lee, J.; Kim, Y.S. A Cell-Based Assay System for Monitoring NF-κB Activity in Human HaCaT Transfectant Cells. Anal. Biochem. 2001, 292, 17–21. [Google Scholar] [CrossRef]
- Hillard, C.J.; Huang, H.; Vogt, C.D.; Rodrigues, B.E.; Neumann, T.S.; Sem, D.S.; Schroeder, F.; Cunningham, C.W. Endocannabinoid Transport Proteins: Discovery of Tools to Study Sterol Carrier Protein-2. Methods Enzym. 2017, 593, 99–121. [Google Scholar] [CrossRef]
- Marzęda, P.; Wróblewska-Łuczka, P.; Florek-Łuszczki, M.; Drozd, M.; Góralczyk, A.; Łuszczki, J.J. Comparison of the Anticancer Effects of Arvanil and Olvanil When Combined with Cisplatin and Mitoxantrone in Various Melanoma Cell Lines-An Isobolographic Analysis. Int. J. Mol. Sci. 2022, 23, 4192. [Google Scholar] [CrossRef] [PubMed]
- De Petrocellis, L.; Bisogno, T.; Davis, J.B.; Pertwee, R.G.; Di Marzo, V. Overlap between the ligand recognition properties of the anandamide transporter and the VR1 vanilloid receptor: Inhibitors of anandamide uptake with negligible capsaicin-like activity. FEBS Lett. 2000, 483, 52–56. [Google Scholar] [CrossRef]
- Fegley, D.; Kathuria, S.; Mercier, R.; Li, C.; Goutopoulos, A.; Makriyannis, A.; Piomelli, D. Anandamide transport is independent of fatty-acid amide hydrolase activity and is blocked by the hydrolysis-resistant inhibitor AM1172. Proc. Natl. Acad. Sci. USA 2004, 101, 8756–8761. [Google Scholar] [CrossRef] [PubMed]
- Ralevic, V.; Kendall, D.A.; Jerman, J.C.; Middlemiss, D.N.; Smart, D. Cannabinoid activation of recombinant and endogenous vanilloid receptors. Eur. J. Pharmacol. 2001, 424, 211–219. [Google Scholar] [CrossRef]
- López-Rodríguez, M.L.; Viso, A.; Ortega-Gutiérrez, S.; Lastres-Becker, I.; González, S.; Fernández-Ruiz, J.; Ramos, J.A. Design, synthesis and biological evaluation of novel arachidonic acid derivatives as highly potent and selective endocannabinoid transporter inhibitors. J. Med. Chem. 2001, 44, 4505–4508. [Google Scholar] [CrossRef]
- Zygmunt, P.M.; Chuang, H.-h.; Movahed, P.; Julius, D.; Högestätt, E.D. The anandamide transport inhibitor AM404 activates vanilloid receptors. Eur. J. Pharmacol. 2000, 396, 39–42. [Google Scholar] [CrossRef]
- Glaser, S.T.; Abumrad, N.A.; Fatade, F.; Kaczocha, M.; Studholme, K.M.; Deutsch, D.G. Evidence against the presence of an anandamide transporter. Proc. Natl. Acad. Sci. USA 2003, 100, 4269–4274. [Google Scholar] [CrossRef]
- Melck, D.; Bisogno, T.; De Petrocellis, L.; Chuang, H.-h.; Julius, D.; Bifulco, M.; Di Marzo, V. Unsaturated long-chain N-acyl-vanillyl-amides (N-AVAMs): Vanilloid receptor ligands that inhibit anandamide-facilitated transport and bind to CB1 cannabinoid receptors. Biochem. Biophys. Res. Commun. 1999, 262, 275–284. [Google Scholar] [CrossRef]
- Sancho, R.; De La Vega, L.; Appendino, G.; Di Marzo, V.; Macho, A.; Muñoz, E. The CB1/VR1 agonist arvanil induces apoptosis through an FADD/caspase-8-dependent pathway. Br. J. Pharmacol. 2003, 140, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Di Marzo, V.; Bisogno, T.; Melck, D.; Ross, R.; Brockie, H.; Stevenson, L.; Pertwee, R.; De Petrocellis, L. Interactions between synthetic vanilloids and the endogenous cannabinoid system. FEBS Lett. 1998, 436, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Beltramo, M.; Piomelli, D. Anandamide transport inhibition by the vanilloid agonist olvanil. Eur. J. Pharmacol. 1999, 364, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Leelawat, S.; Leelawat, K.; Narong, S.; Matangkasombut, O. The dual effects of Δ9-tetrahydrocannabinol on cholangiocarcinoma cells: Anti-invasion activity at low concentration and apoptosis induction at high concentration. Cancer Investig. 2010, 28, 357–363. [Google Scholar] [CrossRef]
- Carracedo, A.; Gironella, M.; Lorente, M.; Garcia, S.; Guzmán, M.; Velasco, G.; Iovanna, J.L. Cannabinoids induce apoptosis of pancreatic tumor cells via endoplasmic reticulum stress–related genes. Cancer Res. 2006, 66, 6748–6755. [Google Scholar] [CrossRef] [PubMed]
- Preet, A.; Qamri, Z.; Nasser, M.W.; Prasad, A.; Shilo, K.; Zou, X.; Groopman, J.E.; Ganju, R.K. Cannabinoid receptors, CB1 and CB2, as novel targets for inhibition of non-small cell lung cancer growth and metastasis. Cancer Prev. Res. 2011, 4, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Au-Amith, S.R.; Au-Jayanth, P.; Au-Finlay, T.; Au-Franchuk, S.; Au-Gilmour, A.; Au-Abdulkhalek, S.; Au-Szewczuk, M.R. Detection of Neu1 Sialidase Activity in Regulating Toll-like Receptor Activation. JoVE 2010, 43, e2142. [Google Scholar] [CrossRef]
- Amith, S.R.; Jayanth, P.; Franchuk, S.; Siddiqui, S.; Seyrantepe, V.; Gee, K.; Basta, S.; Beyaert, R.; Pshezhetsky, A.V.; Szewczuk, M.R. Dependence of pathogen molecule-induced Toll-like receptor activation and cell function on Neu1 sialidase. Glycoconj. J. 2009, 26, 1197. [Google Scholar] [CrossRef]
- O’Shea, L.K.; Abdulkhalek, S.; Allison, S.; Neufeld, R.J.; Szewczuk, M.R. Therapeutic targeting of Neu1 sialidase with oseltamivir phosphate (Tamiflu®) disables cancer cell survival in human pancreatic cancer with acquired chemoresistance. Onco Targets Ther. 2014, 7, 117–134. [Google Scholar] [CrossRef]
- Bunsick, D.A.; Matsukubo, J.; Szewczuk, M.R. Cannabinoids Transmogrify Cancer Metabolic Phenotype via Epigenetic Reprogramming and a Novel CBD Biased G Protein-Coupled Receptor Signaling Platform. Cancers 2023, 15, 1030. [Google Scholar] [CrossRef]
- Herold, C.L.; Behm, D.J.; Buckley, P.T.; Foley, J.J.; Wixted, W.E.; Sarau, H.M.; Douglas, S.A. The neuromedin B receptor antagonist, BIM-23127, is a potent antagonist at human and rat urotensin-II receptors. Br. J. Pharmacol. 2003, 139, 203–207. [Google Scholar] [CrossRef]
- Yang, H.; Wang, Z.; Capó-Aponte, J.E.; Zhang, F.; Pan, Z.; Reinach, P.S. Epidermal growth factor receptor transactivation by the cannabinoid receptor (CB1) and transient receptor potential vanilloid 1 (TRPV1) induces differential responses in corneal epithelial cells. Exp. Eye Res. 2010, 91, 462–471. [Google Scholar] [CrossRef]
- Melck, D.; De Petrocellis, L.; Orlando, P.; Bisogno, T.; Laezza, C.; Bifulco, M.; Di Marzo, V. Suppression of nerve growth factor Trk receptors and prolactin receptors by endocannabinoids leads to inhibition of human breast and prostate cancer cell proliferation. Endocrinology 2000, 141, 118–126. [Google Scholar] [CrossRef]
- Duncan, M.; Galic, M.A.; Wang, A.; Chambers, A.P.; McCafferty, D.M.; McKay, D.M.; Sharkey, K.A.; Pittman, Q.J. Cannabinoid 1 receptors are critical for the innate immune response to TLR4 stimulation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 305, R224–R231. [Google Scholar] [CrossRef]
- Khajehali, E.; Malone, D.T.; Glass, M.; Sexton, P.M.; Christopoulos, A.; Leach, K. Biased Agonism and Biased Allosteric Modulation at the CB1 Cannabinoid Receptor. Mol. Pharmacol. 2015, 88, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Al-Zoubi, R.; Morales, P.; Reggio, P.H. Structural Insights into CB1 Receptor Biased Signaling. Int. J. Mol. Sci. 2019, 20, 1837. [Google Scholar] [CrossRef] [PubMed]
- Van Roy, F.; Berx, G. The cell-cell adhesion molecule E-cadherin. Cell Mol. Life Sci. 2008, 65, 3756–3788. [Google Scholar] [CrossRef] [PubMed]
- Burandt, E.; Lübbersmeyer, F.; Gorbokon, N.; Büscheck, F.; Luebke, A.M.; Menz, A.; Kluth, M.; Hube-Magg, C.; Hinsch, A.; Höflmayer, D.; et al. E-Cadherin expression in human tumors: A tissue microarray study on 10,851 tumors. Biomark. Res. 2021, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Li, N.; Tu, T.; Tao, Y.; Bi, Y.; Yuan, D.; Zhang, N.; Yang, X.; Kong, D.; You, H.; et al. Hepatitis B virus core protein promotes the expression of neuraminidase 1 to facilitate hepatocarcinogenesis. Lab. Investig. 2020, 100, 1602–1617. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.Q.; Liu, K.; Shi, N.; Ma, G.; Wang, P.; Xie, H.M.; Jin, S.J.; Wei, T.T.; Yu, X.Y.; Wang, Y.; et al. Neuraminidase 1 promotes renal fibrosis development in male mice. Nat. Commun. 2023, 14, 1713. [Google Scholar] [CrossRef]
- Hollestelle, A.; Peeters, J.K.; Smid, M.; Timmermans, M.; Verhoog, L.C.; Westenend, P.J.; Heine, A.A.; Chan, A.; Sieuwerts, A.M.; Wiemer, E.A.; et al. Loss of E-cadherin is not a necessity for epithelial to mesenchymal transition in human breast cancer. Breast Cancer Res. Treat. 2013, 138, 47–57. [Google Scholar] [CrossRef]
- Chen, A.; Beetham, H.; Black, M.A.; Priya, R.; Telford, B.J.; Guest, J.; Wiggins, G.A.; Godwin, T.D.; Yap, A.S.; Guilford, P.J. E-cadherin loss alters cytoskeletal organization and adhesion in non-malignant breast cells but is insufficient to induce an epithelial-mesenchymal transition. BMC Cancer 2014, 14, 552. [Google Scholar] [CrossRef]
- Ridge, K.M.; Eriksson, J.E.; Pekny, M.; Goldman, R.D. Roles of vimentin in health and disease. Genes. Dev. 2022, 36, 391–407. [Google Scholar] [CrossRef]
- Berr, A.L.; Wiese, K.; Dos Santos, G.; Koch, C.M.; Anekalla, K.R.; Kidd, M.; Davis, J.M.; Cheng, Y.; Hu, Y.S.; Ridge, K.M. Vimentin is required for tumor progression and metastasis in a mouse model of non-small cell lung cancer. Oncogene 2023, 42, 2074–2087. [Google Scholar] [CrossRef]
- Hewitt, R.E.; McMarlin, A.; Kleiner, D.; Wersto, R.; Martin, P.; Tsokos, M.; Stamp, G.W.; Stetler-Stevenson, W.G. Validation of a model of colon cancer progression. J. Pathol. 2000, 192, 446–454. [Google Scholar] [CrossRef]
- Toussaint, K.; Appert-Collin, A.; Morjani, H.; Albrecht, C.; Sartelet, H.; Romier-Crouzet, B.; Maurice, P.; Duca, L.; Blaise, S.; Bennasroune, A. Neuraminidase-1: A Sialidase Involved in the Development of Cancers and Metabolic Diseases. Cancers 2022, 14, 4868. [Google Scholar] [CrossRef]
- Rodgers, J.; Sundararaj, K.; Bruner, E.; Wolf, B.; Nowling, T.K. The role of neuraminidase 1 (NEU1) in cytokine release by primary mouse mesangial cells and disease outcomes in murine lupus nephritis. Autoimmunity 2021, 54, 163–175. [Google Scholar] [CrossRef]
- Fougerat, A.; Pan, X.; Smutova, V.; Heveker, N.; Cairo, C.W.; Issad, T.; Larrivée, B.; Medin, J.A.; Pshezhetsky, A.V. Neuraminidase 1 activates insulin receptor and reverses insulin resistance in obese mice. Mol. Metab. 2018, 12, 76–88. [Google Scholar] [CrossRef]
- Mackie, K. Cannabinoid receptor homo-and heterodimerization. Life Sci. 2005, 77, 1667–1673. [Google Scholar] [CrossRef]
- Tóth, A.D.; Turu, G.; Hunyady, L.; Balla, A. Novel mechanisms of G-protein-coupled receptors functions: AT1 angiotensin receptor acts as a signaling hub and focal point of receptor cross-talk. Best Pract. Res. Clin. Endocrinol. Metab. 2018, 32, 69–82. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, J.-T.; Hang, L.; Liu, T. Mu opioid receptor heterodimers emerge as novel therapeutic targets: Recent progress and future perspective. Front. Pharmacol. 2020, 11, 1078. [Google Scholar] [CrossRef]
- De Petrocellis, L.; Marini, P.; Matias, I.; Moriello, A.S.; Starowicz, K.; Cristino, L.; Nigam, S.; Di Marzo, V. Mechanisms for the coupling of cannabinoid receptors to intracellular calcium mobilization in rat insulinoma β-cells. Exp. Cell Res. 2007, 313, 2993–3004. [Google Scholar] [CrossRef]
- Ohki-Hamazaki, H.; Wada, E.; Matsui, K.; Wada, K. Cloning and expression of the neuromedin B receptor and the third subtype of bombesin receptor genes in the mouse. Brain Res. 1997, 762, 165–172. [Google Scholar] [CrossRef]
- Ohki-Hamazaki, H.; Sakai, Y.; Kamata, K.; Ogura, H.; Okuyama, S.; Watase, K.; Yamada, K.; Wada, K. Functional properties of two bombesin-like peptide receptors revealed by the analysis of mice lacking neuromedin B receptor. J. Neurosci. 1999, 19, 948–954. [Google Scholar] [CrossRef][Green Version]
- Dalton, G.D.; Howlett, A.C. Cannabinoid CB1 receptors transactivate multiple receptor tyrosine kinases and regulate serine/threonine kinases to activate ERK in neuronal cells. Br. J. Pharmacol. 2012, 165, 2497–2511. [Google Scholar] [CrossRef]
- Leifer, C.A.; Medvedev, A.E. Molecular mechanisms of regulation of Toll-like receptor signaling. J. Leukoc. Biol. 2016, 100, 927–941. [Google Scholar] [CrossRef]
- Leo, L.M.; Abood, M.E. CB1 Cannabinoid Receptor Signaling and Biased Signaling. Molecules 2021, 26, 5413. [Google Scholar] [CrossRef]
- Huang, H. Matrix Metalloproteinase-9 (MMP-9) as a Cancer Biomarker and MMP-9 Biosensors: Recent Advances. Sensors 2018, 18, 3249. [Google Scholar] [CrossRef]
- Luo, C.K.; Chou, P.H.; Ng, S.K.; Lin, W.Y.; Wei, T.T. Cannabinoids orchestrate cross-talk between cancer cells and endothelial cells in colorectal cancer. Cancer Gene Ther. 2022, 29, 597–611. [Google Scholar] [CrossRef]
- Rosenthaler, S.; Pöhn, B.; Kolmanz, C.; Huu, C.N.; Krewenka, C.; Huber, A.; Kranner, B.; Rausch, W.D.; Moldzio, R. Differences in receptor binding affinity of several phytocannabinoids do not explain their effects on neural cell cultures. Neurotoxicol. Teratol. 2014, 46, 49–56. [Google Scholar] [CrossRef]
- Morales, P.; Goya, P.; Jagerovic, N. Emerging strategies targeting CB2 cannabinoid receptor: Biased agonism and allosterism. Biochem. Pharmacol. 2018, 157, 8–17. [Google Scholar] [CrossRef]
- Morales, P.; Reggio, P.H. An update on non-CB1, non-CB2 cannabinoid related G-protein-coupled receptors. Cannabis Cannabinoid Res. 2017, 2, 265–273. [Google Scholar] [CrossRef]
- Laprairie, R.B.; Bagher, A.M.; Kelly, M.E.; Denovan-Wright, E.M. Biased type 1 cannabinoid receptor signaling influences neuronal viability in a cell culture model of Huntington disease. Mol. Pharmacol. 2016, 89, 364–375. [Google Scholar] [CrossRef]
- Qorri, B.; Kalaydina, R.V.; Velickovic, A.; Kaplya, Y.; Decarlo, A.; Szewczuk, M.R. Agonist-Biased Signaling via Matrix Metalloproteinase-9 Promotes Extracellular Matrix Remodeling. Cells 2018, 7, 117. [Google Scholar] [CrossRef]
- Jakowiecki, J.; Abel, R.; Orzeł, U.; Pasznik, P.; Preissner, R.; Filipek, S. Allosteric Modulation of the CB1 Cannabinoid Receptor by Cannabidiol-A Molecular Modeling Study of the N-Terminal Domain and the Allosteric-Orthosteric Coupling. Molecules 2021, 26, 2456. [Google Scholar] [CrossRef]
- Reber, L.; Vermeulen, L.; Haegeman, G.; Frossard, N. Ser276 phosphorylation of NF-kB p65 by MSK1 controls SCF expression in inflammation. PLoS ONE 2009, 4, e4393. [Google Scholar] [CrossRef]
- Verstrepen, L.; Bekaert, T.; Chau, T.L.; Tavernier, J.; Chariot, A.; Beyaert, R. TLR-4, IL-1R and TNF-R signaling to NF-κB: Variations on a common theme. Cell. Mol. Life Sci. 2008, 65, 2964–2978. [Google Scholar] [CrossRef]
- Marquardt, J.U.; Factor, V.M.; Thorgeirsson, S.S. Epigenetic regulation of cancer stem cells in liver cancer: Current concepts and clinical implications. J. Hepatol. 2010, 53, 568–577. [Google Scholar] [CrossRef]
- Xia, Y.; Shen, S.; Verma, I.M. NF-κB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef]
- Seltzer, E.S.; Watters, A.K.; MacKenzie, D., Jr.; Granat, L.M.; Zhang, D. Cannabidiol (CBD) as a Promising Anti-Cancer Drug. Cancers 2020, 12, 3203. [Google Scholar] [CrossRef]
- Alaaeldin, R.; Ali, F.E.M.; Bekhit, A.A.; Zhao, Q.L.; Fathy, M. Inhibition of NF-kB/IL-6/JAK2/STAT3 Pathway and Epithelial-Mesenchymal Transition in Breast Cancer Cells by Azilsartan. Molecules 2022, 27, 7825. [Google Scholar] [CrossRef]
- Abdel-Latif, R.T.; Wadie, W.; Abdel-Mottaleb, Y.; Abdallah, D.M.; El-Maraghy, N.N.; El-Abhar, H.S. Reposition of the anti-inflammatory drug diacerein in an in-vivo colorectal cancer model. Saudi Pharm. J. 2022, 30, 72–90. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bunsick, D.A.; Matsukubo, J.; Aldbai, R.; Baghaie, L.; Szewczuk, M.R. Functional Selectivity of Cannabinoid Type 1 G Protein-Coupled Receptor Agonists in Transactivating Glycosylated Receptors on Cancer Cells to Induce Epithelial–Mesenchymal Transition Metastatic Phenotype. Cells 2024, 13, 480. https://doi.org/10.3390/cells13060480
Bunsick DA, Matsukubo J, Aldbai R, Baghaie L, Szewczuk MR. Functional Selectivity of Cannabinoid Type 1 G Protein-Coupled Receptor Agonists in Transactivating Glycosylated Receptors on Cancer Cells to Induce Epithelial–Mesenchymal Transition Metastatic Phenotype. Cells. 2024; 13(6):480. https://doi.org/10.3390/cells13060480
Chicago/Turabian StyleBunsick, David A., Jenna Matsukubo, Rashelle Aldbai, Leili Baghaie, and Myron R. Szewczuk. 2024. "Functional Selectivity of Cannabinoid Type 1 G Protein-Coupled Receptor Agonists in Transactivating Glycosylated Receptors on Cancer Cells to Induce Epithelial–Mesenchymal Transition Metastatic Phenotype" Cells 13, no. 6: 480. https://doi.org/10.3390/cells13060480
APA StyleBunsick, D. A., Matsukubo, J., Aldbai, R., Baghaie, L., & Szewczuk, M. R. (2024). Functional Selectivity of Cannabinoid Type 1 G Protein-Coupled Receptor Agonists in Transactivating Glycosylated Receptors on Cancer Cells to Induce Epithelial–Mesenchymal Transition Metastatic Phenotype. Cells, 13(6), 480. https://doi.org/10.3390/cells13060480






