Comparative Analysis of Acute Kidney Injury Models and Related Fibrogenic Responses: Convergence on Methylation Patterns Regulated by Cold Shock Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Microarray Data
2.2. Animals
2.3. Genotyping
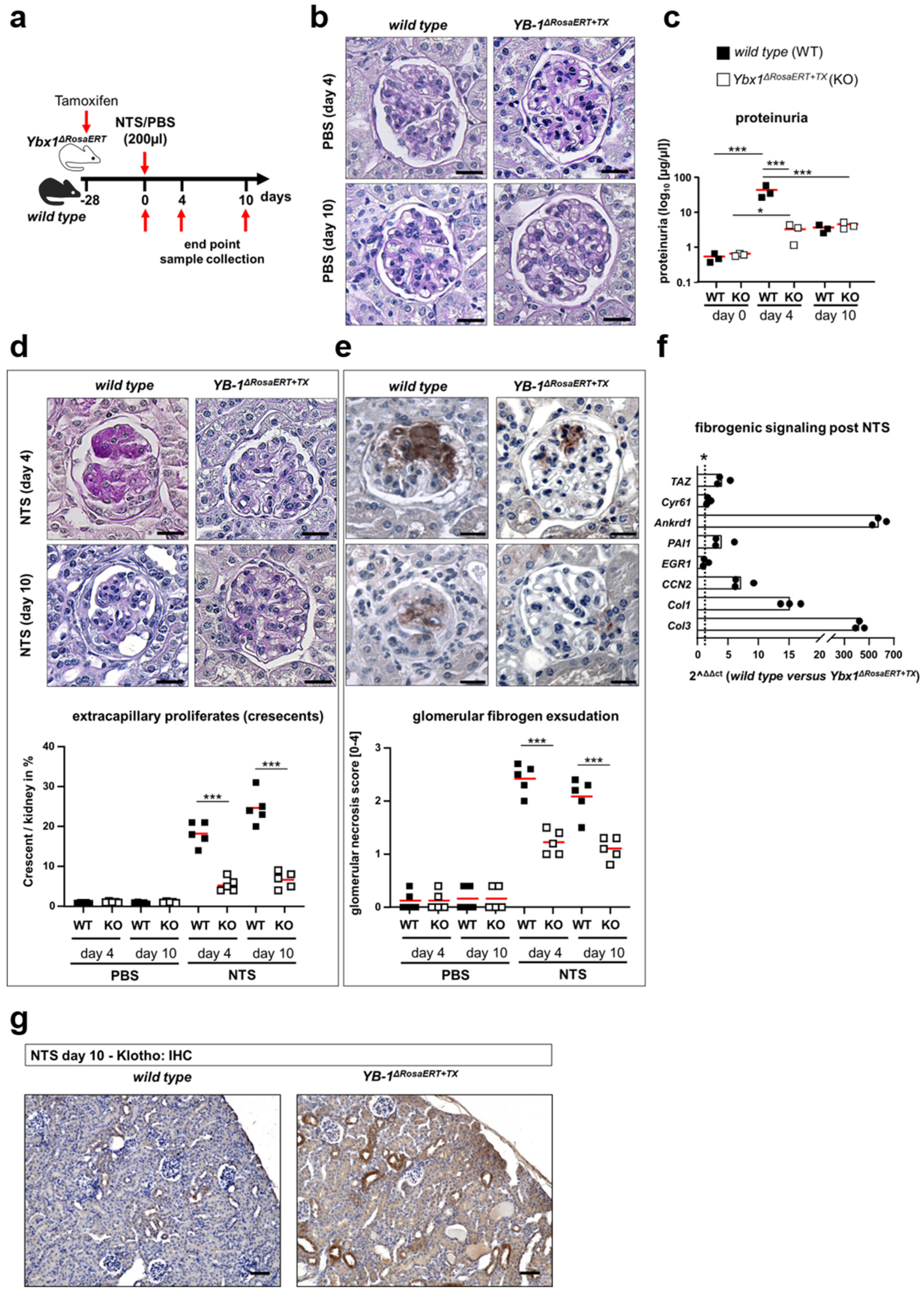
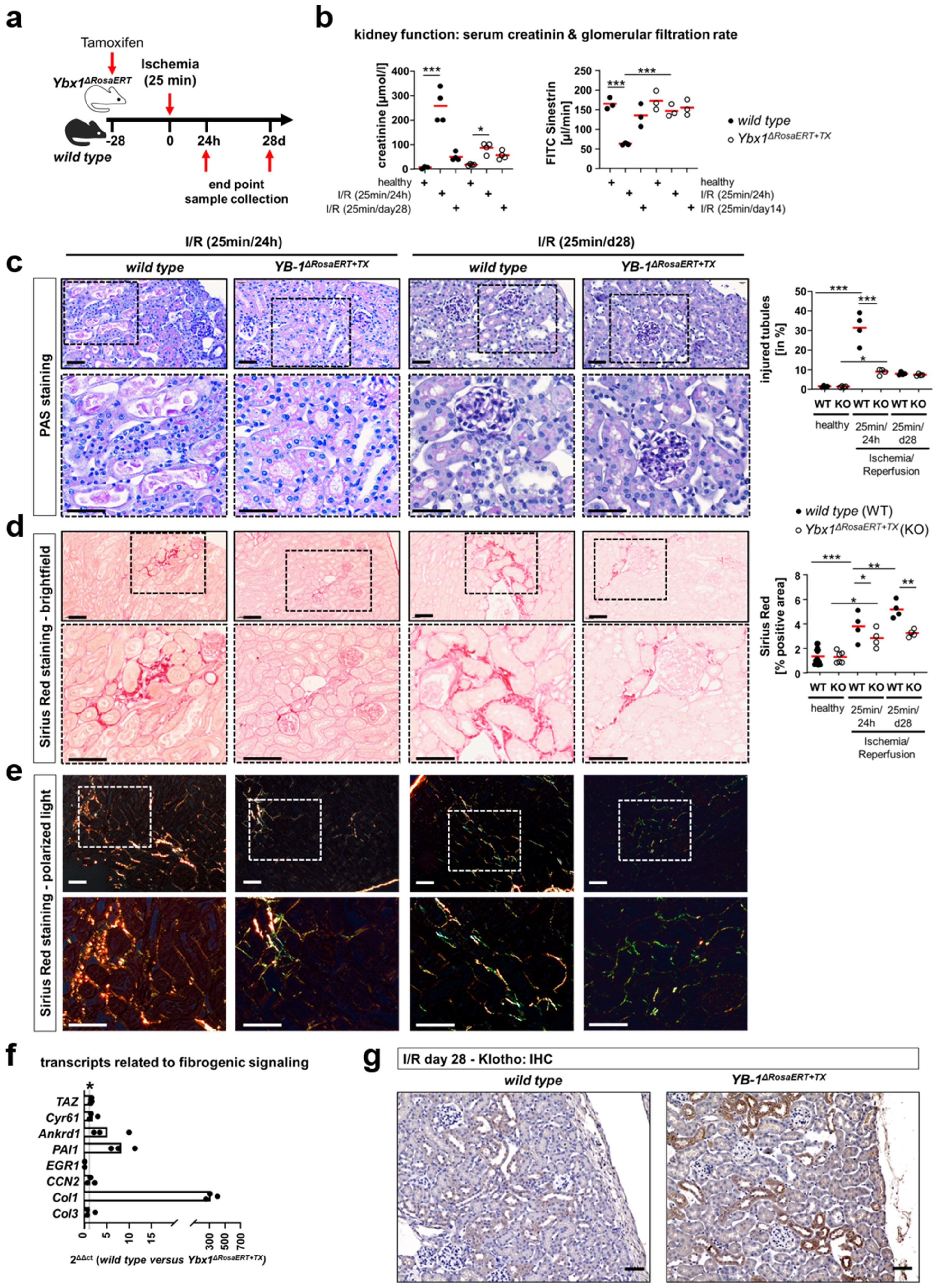
2.4. Experimental Kidney Disease
2.5. Analysis of Kidney Function
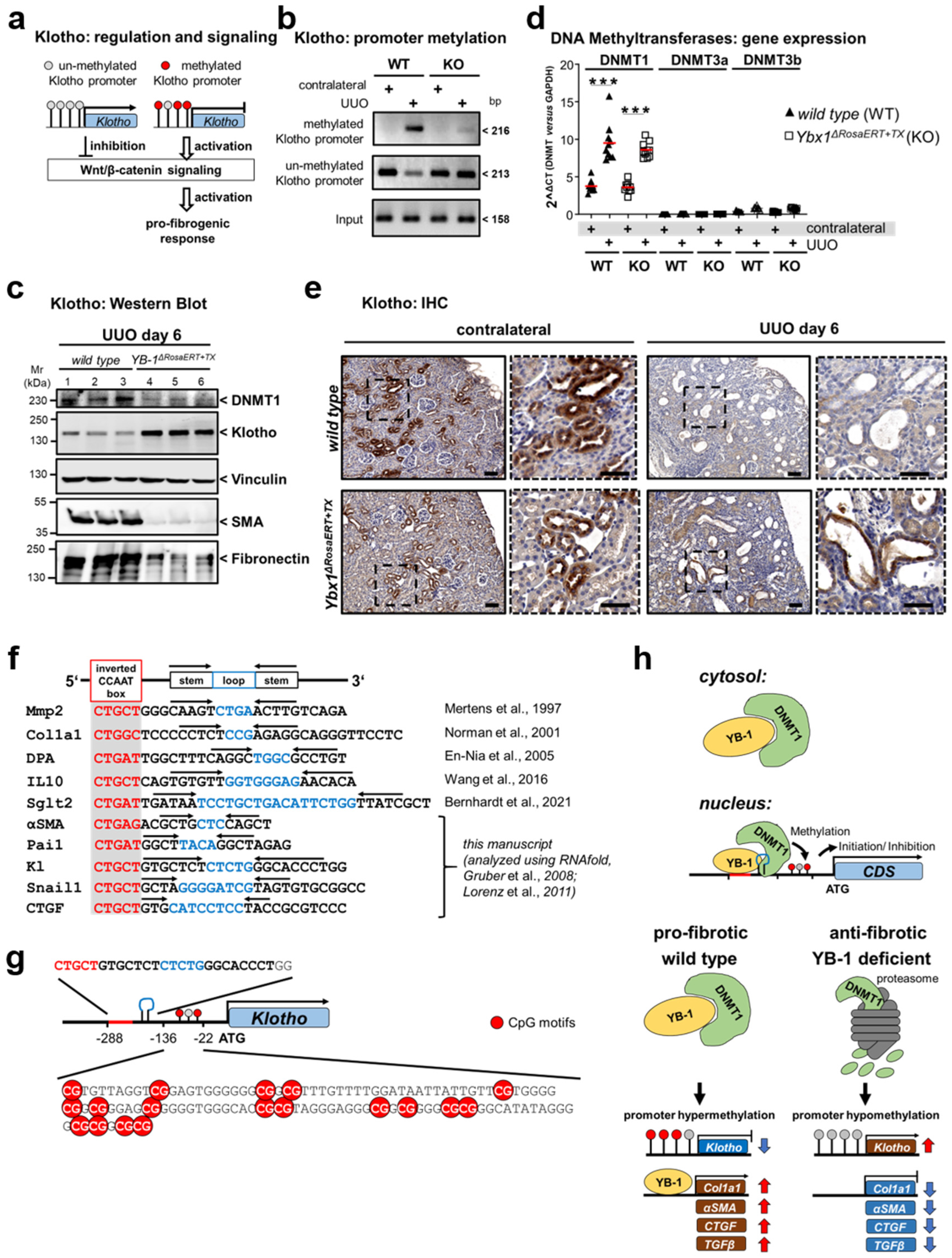
2.6. Methylation Analysis
2.7. Isolation of Total RNA
2.8. Quantitative Real-Time PCR
2.9. Gene Array
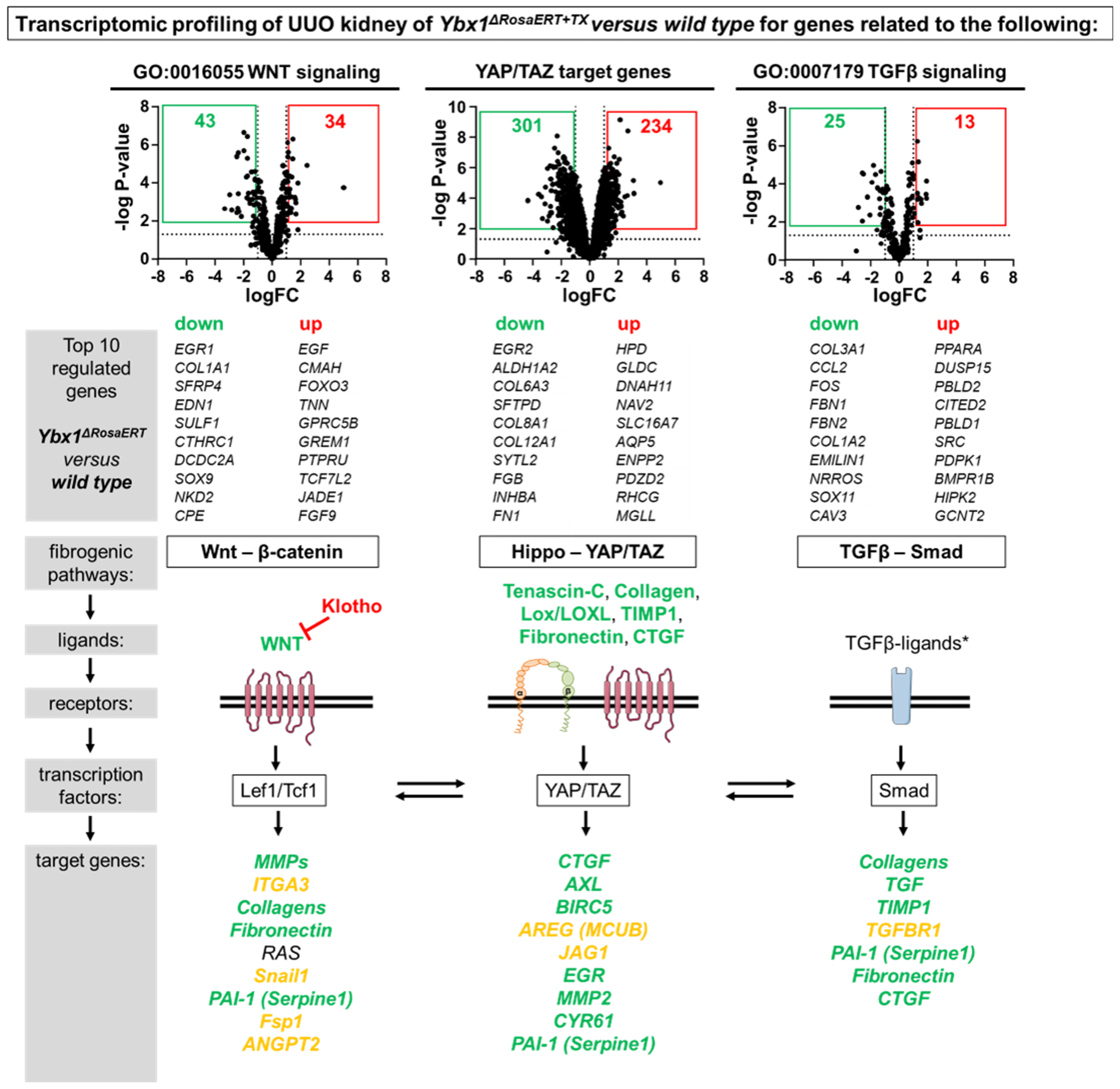
2.10. Transcriptome Analysis Using RNA-Seq
2.11. Tissue Lysate Preparation and Western Blot Analysis
2.12. Gelatin Zymography
2.13. Immunohistochemistry
2.14. Periodic Acid–Schiff (PAS) Staining
2.15. Sirius Red Staining
2.16. Bioinformatic Analysis
2.17. Statistical Analysis
3. Results
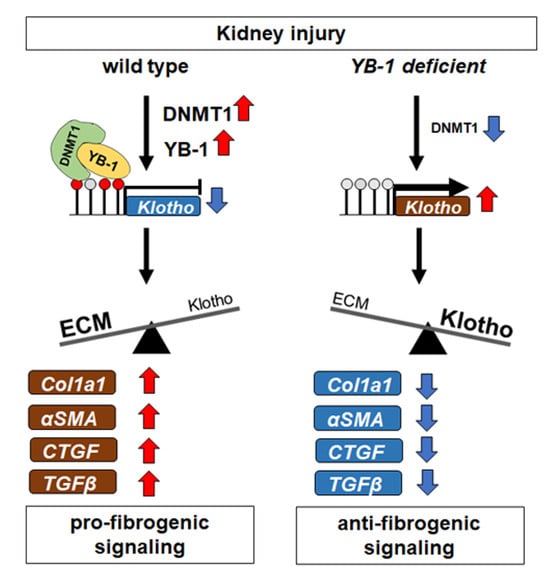
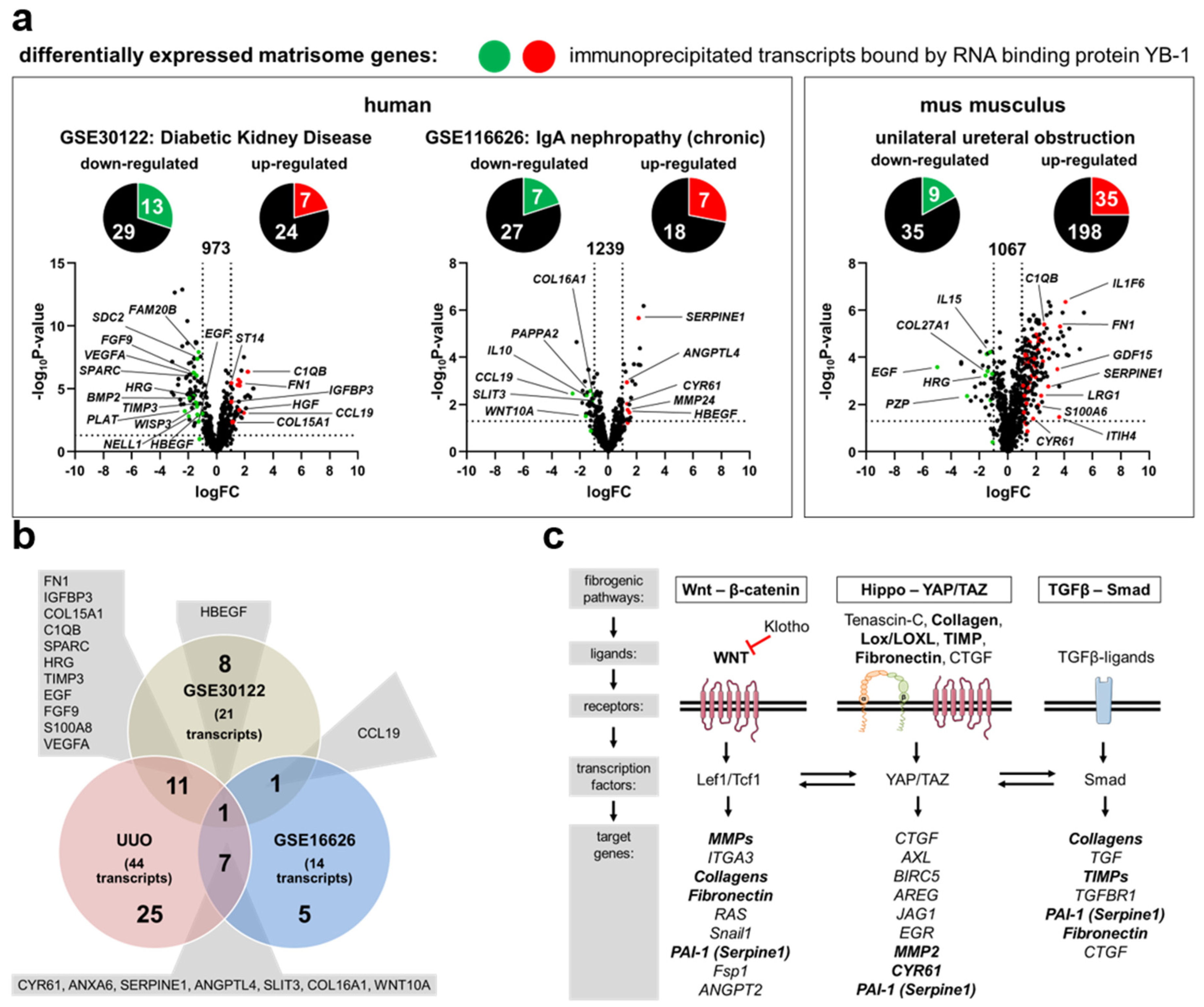
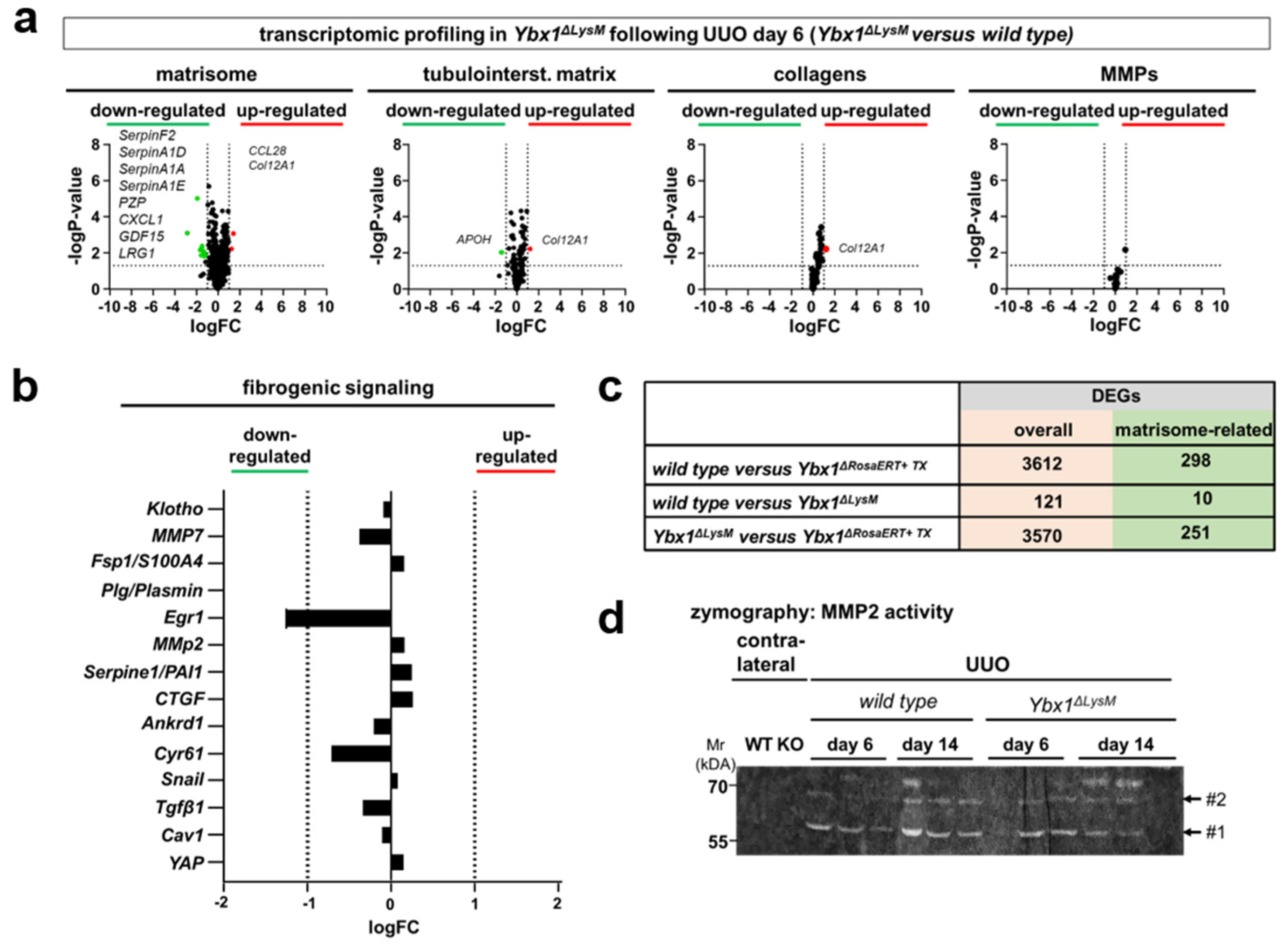
3.1. Transcriptional Programs for Kidney Fibrosis in Chronic Kidney Disease
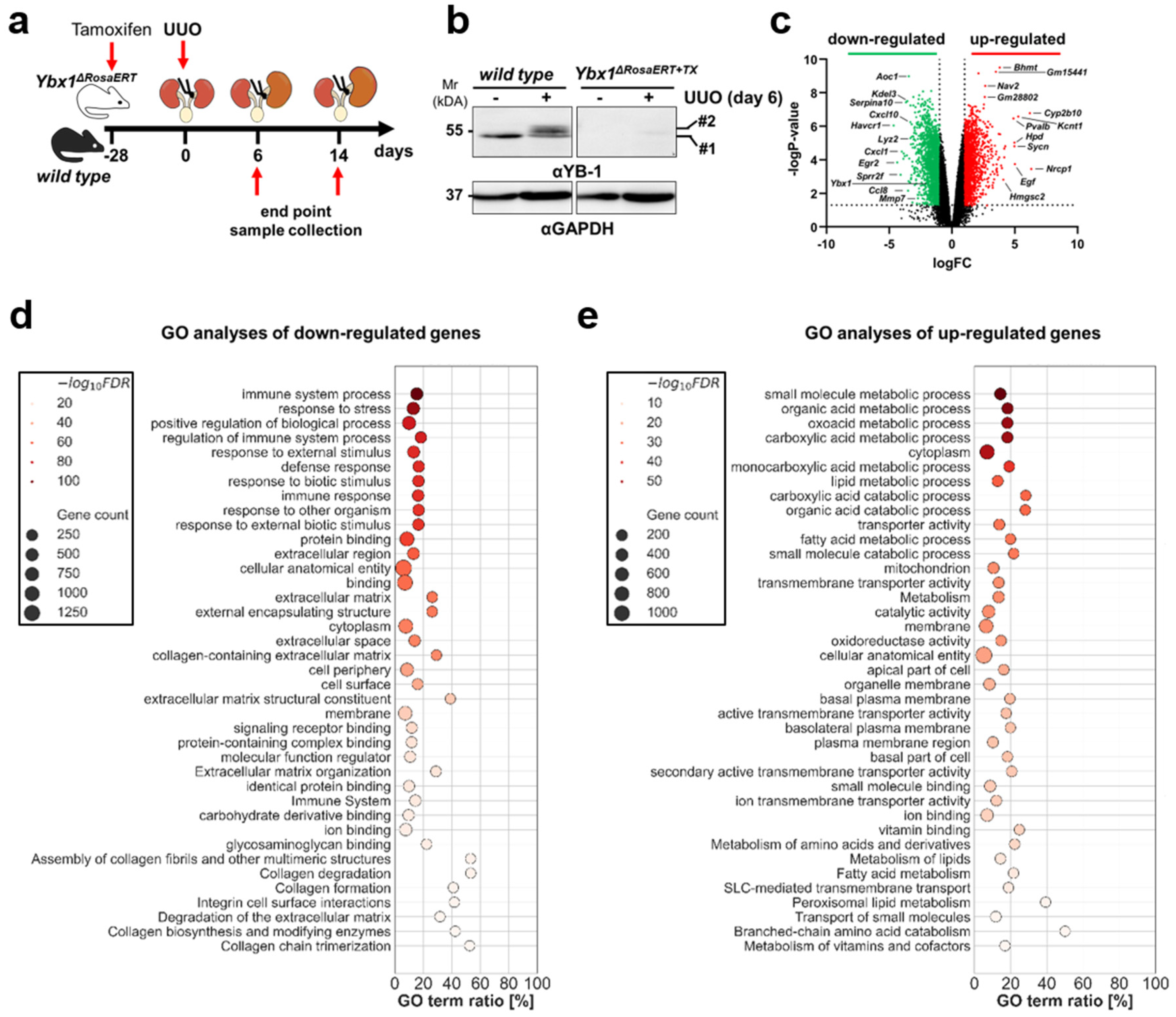
3.2. YB-1 Confers a Profibrogenic Response
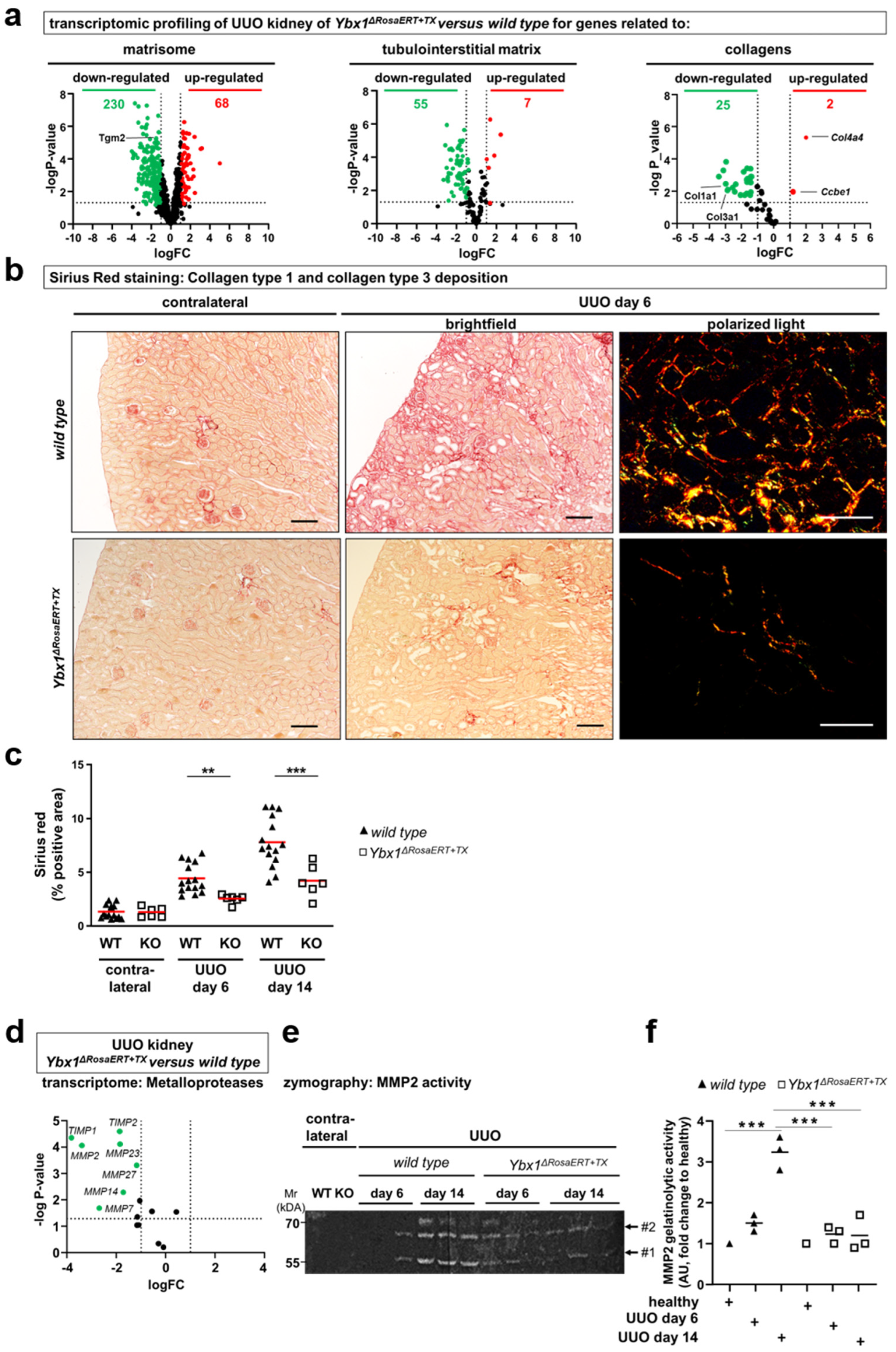
3.3. MMP2 Activity following UUO Is Dependent on YB-1 Expression
3.4. Key Fibrogenic Mechanisms Are Deregulated in YB-1-Deficient Kidneys
3.5. Similar Fibrogenic Pathways Are Regulated by YB-1 in a Glomerular Injury Model as Well as Following Ischemia–Reperfusion Injury
3.6. ECM Accumulation Is Orchestrated by YB-1-Expressing Tissue-Resident but Not Infiltrating Myeloid-Derived Innate Immune Cells
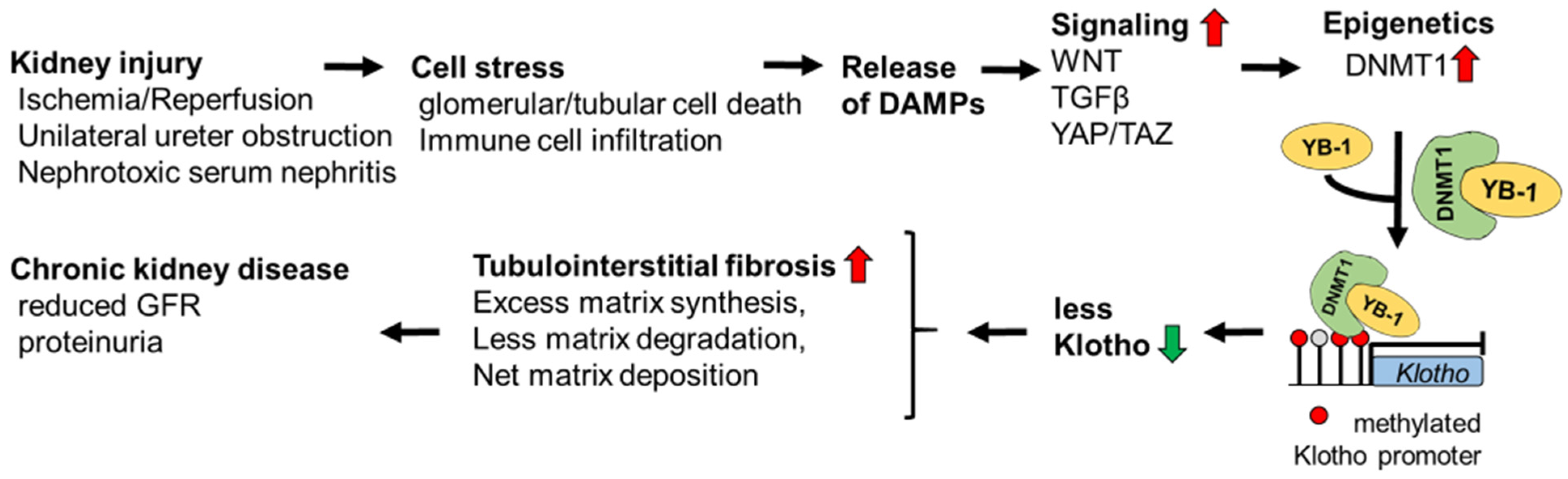
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Webster, A.C.; Nagler, E.V.; Morton, R.L.; Masson, P. Chronic kidney disease. Lancet 2017, 389, 1238–1252. [Google Scholar] [CrossRef] [PubMed]
- Bikbov, B.; Purcell, C.A.; Levey, A.S.; Smith, M.; Abdoli, A.; Abebe, M.; Adebayo, O.M.; Afarideh, M.; Agarwal, S.M.; Agudelo-Botero, M.; et al. Global, regional, and national burden of chronic kidney disease, 1990–2017: A systematic analysis for the global burden of disease study 2017. Lancet 2020, 395, 709–733. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.K.; Knicely, D.H.; Grams, M.E. Chronic kidney disease diagnosis and management: A review. J. Am. Med. Assoc. 2019, 322, 1294–1304. [Google Scholar] [CrossRef] [PubMed]
- Brauer, E.; Lippens, E.; Klein, O.; Nebrich, G.; Schreivogel, S.; Korus, G.; Duda, G.N.; Petersen, A. Collagen fibrils mechanically contribute to tissue contraction in an in vitro wound healing scenario. Adv. Sci. 2019, 6, 1801780. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A. Fibrotic disease and the t(h)1/t(h)2 paradigm. Nat. Rev. Immunol. 2004, 4, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Fu, H.; Liu, Y. The fibrogenic niche in kidney fibrosis: Components and mechanisms. Nat. Rev. Nephrol. 2022, 18, 545–557. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef]
- Yue, B. Biology of the extracellular matrix: An overview. J. Glaucoma 2014, 23, S20–S23. [Google Scholar] [CrossRef]
- Vogel, V. Unraveling the mechanobiology of extracellular matrix. Annu. Rev. Physiol. 2018, 80, 353–387. [Google Scholar] [CrossRef]
- Buravkova, L.; Larina, I.; Andreeva, E.; Grigoriev, A. Microgravity effects on the matrisome. Cells 2021, 10, 2226. [Google Scholar] [CrossRef]
- Naba, A.; Clauser, K.R.; Ding, H.; Whittaker, C.A.; Carr, S.A.; Hynes, R.O. The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol. J. Int. Soc. Matrix Biol. 2016, 49, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Lennon, R.; Byron, A.; Humphries, J.D.; Randles, M.J.; Carisey, A.; Murphy, S.; Knight, D.; Brenchley, P.E.; Zent, R.; Humphries, M.J. Global analysis reveals the complexity of the human glomerular extracellular matrix. J. Am. Soc. Nephrol. JASN 2014, 25, 939–951. [Google Scholar] [CrossRef]
- Hobeika, L.; Barati, M.T.; Caster, D.J.; McLeish, K.R.; Merchant, M.L. Characterization of glomerular extracellular matrix by proteomic analysis of laser-captured microdissected glomeruli. Kidney Int. 2017, 91, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Randles, M.; Lausecker, F.; Kong, Q.; Suleiman, H.; Reid, G.; Kolatsi-Joannou, M.; Tian, P.; Falcone, S.; Davenport, B.; Potter, P.; et al. Identification of an altered matrix signature in kidney aging and disease. J. Am. Soc. Nephrol. JASN 2021, 32, 1713. [Google Scholar] [CrossRef] [PubMed]
- Sobreiro-Almeida, R.; Quinteira, R.; Neves, N.M. Renal regeneration: The role of extracellular matrix and current ecm-based tissue engineered strategies. Adv. Healthc. Mater. 2021, 10, e2100160. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.; Dobie, R.; Wilson-Kanamori, J.R.; Dora, E.F.; Henderson, B.E.P.; Luu, N.T.; Portman, J.R.; Matchett, K.P.; Brice, M.; Marwick, J.A.; et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 2019, 575, 512–518. [Google Scholar] [CrossRef]
- Morrison, J.K.; DeRossi, C.; Alter, I.L.; Nayar, S.; Giri, M.; Zhang, C.; Cho, J.H.; Chu, J. Single-cell transcriptomics reveals conserved cell identities and fibrogenic phenotypes in zebrafish and human liver. Hepatol. Commun. 2022, 6, 1711–1724. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Tian, Y.; Zhou, L.; Zhou, D.; Tan, R.J.; Stolz, D.B.; Liu, Y. Tenascin-c is a major component of the fibrogenic niche in kidney fibrosis. J. Am. Soc. Nephrol. JASN 2017, 28, 785–801. [Google Scholar] [CrossRef]
- Arai, H.; Yanagita, M. Janus-faced: Molecular mechanisms and versatile nature of renal fibrosis. Kidney360 2020, 1, 697–704. [Google Scholar] [CrossRef]
- Zhu, H.; Liao, J.; Zhou, X.; Hong, X.; Song, D.; Hou, F.F.; Liu, Y.; Fu, H. Tenascin-c promotes acute kidney injury to chronic kidney disease progression by impairing tubular integrity via alphavbeta6 integrin signaling. Kidney Int. 2020, 97, 1017–1031. [Google Scholar] [CrossRef]
- Djudjaj, S.; Boor, P. Cellular and molecular mechanisms of kidney fibrosis. Mol. Asp. Med. 2019, 65, 16–36. [Google Scholar] [CrossRef] [PubMed]
- Lindquist, J.A.; Mertens, P.R. Cold shock proteins: From cellular mechanisms to pathophysiology and disease. Cell Commun. Signal. CCS 2018, 16, 63. [Google Scholar] [CrossRef]
- Jenkins, R.H.; Bennagi, R.; Martin, J.; Phillips, A.O.; Redman, J.E.; Fraser, D.J. A conserved stem loop motif in the 5’untranslated region regulates transforming growth factor-beta(1) translation. PLoS ONE 2010, 5, e12283. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stefanovic, B.; Lindquist, J.; Brenner, D.A. The 5’ stem-loop regulates expression of collagen alpha1(i) mrna in mouse fibroblasts cultured in a three-dimensional matrix. Nucleic Acids Res. 2000, 28, 641–647. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Stefanovic, B. Larp6 meets collagen mrna: Specific regulation of type i collagen expression. Int. J. Mol. Sci. 2016, 17, 419. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, U.; Roske, Y. Cold-shock domains-abundance, structure, properties, and nucleic-acid binding. Cancers 2021, 13, 190. [Google Scholar] [CrossRef] [PubMed]
- Brandt, S.; Raffetseder, U.; Djudjaj, S.; Schreiter, A.; Kadereit, B.; Michele, M.; Pabst, M.; Zhu, C.; Mertens, P.R. Cold shock y-box protein-1 participates in signaling circuits with auto-regulatory activities. Eur. J. Cell Biol. 2012, 91, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Lyabin, D.N.; Eliseeva, I.A.; Ovchinnikov, L.P. Yb-1 protein: Functions and regulation. Wiley Interdiscip. Rev. RNA 2014, 5, 95–110. [Google Scholar] [CrossRef]
- Wang, J.; Gibbert, L.; Djudjaj, S.; Alidousty, C.; Rauen, T.; Kunter, U.; Rembiak, A.; Enders, D.; Jankowski, V.; Braun, G.S.; et al. Therapeutic nuclear shuttling of yb-1 reduces renal damage and fibrosis. Kidney Int. 2016, 90, 1226–1237. [Google Scholar] [CrossRef]
- Brandt, S.; Mertens, P.R. A remedy for kidney disease successfully alters the cold shock protein response during inflammation. Kidney Int. 2016, 90, 1148–1150. [Google Scholar] [CrossRef]
- Dong, W.; Wang, H.; Shahzad, K.; Bock, F.; Al-Dabet, M.M.; Ranjan, S.; Wolter, J.; Kohli, S.; Hoffmann, J.; Dhople, V.M.; et al. Activated protein c ameliorates renal ischemia-reperfusion injury by restricting y-box binding protein-1 ubiquitination. J. Am. Soc. Nephrol. JASN 2015, 26, 2789–2799. [Google Scholar] [CrossRef]
- Dooley, S.; Said, H.M.; Gressner, A.M.; Floege, J.; En-Nia, A.; Mertens, P.R. Y-box protein-1 is the crucial mediator of antifibrotic interferon-gamma effects. J. Biol. Chem. 2006, 281, 1784–1795. [Google Scholar] [CrossRef] [PubMed]
- Higashi, K.; Tomigahara, Y.; Shiraki, H.; Miyata, K.; Mikami, T.; Kimura, T.; Moro, T.; Inagaki, Y.; Kaneko, H. A novel small compound that promotes nuclear translocation of yb-1 ameliorates experimental hepatic fibrosis in mice. J. Biol. Chem. 2011, 286, 4485–4492. [Google Scholar] [CrossRef]
- Clough, E.; Barrett, T. The gene expression omnibus database. Methods Mol. Biol. 2016, 1418, 93–110. [Google Scholar] [PubMed]
- Woroniecka, K.I.; Park, A.S.; Mohtat, D.; Thomas, D.B.; Pullman, J.M.; Susztak, K. Transcriptome analysis of human diabetic kidney disease. Diabetes 2011, 60, 2354–2369. [Google Scholar] [CrossRef] [PubMed]
- Cox, S.N.; Chiurlia, S.; Divella, C.; Rossini, M.; Serino, G.; Bonomini, M.; Sirolli, V.; Aiello, F.B.; Zaza, G.; Squarzoni, I.; et al. Formalin-fixed paraffin-embedded renal biopsy tissues: An underexploited biospecimen resource for gene expression profiling in iga nephropathy. Sci. Rep. 2020, 10, 15164. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, A.; Fehr, A.; Brandt, S.; Jerchel, S.; Ballhause, T.M.; Philipsen, L.; Stolze, S.; Geffers, R.; Weng, H.; Fischer, K.D.; et al. Inflammatory cell infiltration and resolution of kidney inflammation is orchestrated by the cold-shock protein y-box binding protein-1. Kidney Int. 2017, 92, 1157–1177. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, A.; Haberer, S.; Xu, J.; Damerau, H.; Steffen, J.; Reichardt, C.; Wolters, K.; Steffen, H.; Isermann, B.; Borucki, K.; et al. High salt diet-induced proximal tubular phenotypic changes and sodium-glucose cotransporter-2 expression are coordinated by cold shock y-box binding protein-1. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2021, 35, e21912. [Google Scholar] [CrossRef] [PubMed]
- Schock-Kusch, D.; Sadick, M.; Henninger, N.; Kraenzlin, B.; Claus, G.; Kloetzer, H.M.; Weiss, C.; Pill, J.; Gretz, N. Transcutaneous measurement of glomerular filtration rate using fitc-sinistrin in rats. Nephrol. Dial. Transplant. Off. Publ. Eur. Dial. Transpl. Assoc. Eur. Ren. Assoc. 2009, 24, 2997–3001. [Google Scholar] [CrossRef]
- Al-Dabet, M.M.; Shahzad, K.; Elwakiel, A.; Sulaj, A.; Kopf, S.; Bock, F.; Gadi, I.; Zimmermann, S.; Rana, R.; Krishnan, S.; et al. Reversal of the renal hyperglycemic memory in diabetic kidney disease by targeting sustained tubular p21 expression. Nat. Commun. 2022, 13, 5062. [Google Scholar] [CrossRef]
- Shahzad, K.; Gadi, I.; Nazir, S.; Al-Dabet, M.M.; Kohli, S.; Bock, F.; Breitenstein, L.; Ranjan, S.; Fuchs, T.; Halloul, Z.; et al. Activated protein c reverses epigenetically sustained p66(shc) expression in plaque-associated macrophages in diabetes. Commun. Biol. 2018, 1, 104. [Google Scholar] [CrossRef]
- Yin, S.; Zhang, Q.; Yang, J.; Lin, W.; Li, Y.; Chen, F.; Cao, W. Tgfbeta-incurred epigenetic aberrations of mirna and DNA methyltransferase suppress klotho and potentiate renal fibrosis. Biochim. Biophys. Acta. Mol. Cell Res. 2017, 1864, 1207–1216. [Google Scholar] [CrossRef]
- Brandt, S.; Ballhause, T.M.; Bernhardt, A.; Becker, A.; Salaru, D.; Le-Deffge, H.M.; Fehr, A.; Fu, Y.; Philipsen, L.; Djudjaj, S.; et al. Fibrosis and immune cell infiltration are separate events regulated by cell-specific receptor notch3 expression. J. Am. Soc. Nephrol. JASN 2020, 31, 2589–2608. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, M.R.; Astanehe, A.; To, K.; Fotovati, A.; Davies, A.H.; Zhao, Y.; Jiang, H.; Stratford, A.L.; Shadeo, A.; Boccaccio, C.; et al. Profiling yb-1 target genes uncovers a new mechanism for met receptor regulation in normal and malignant human mammary cells. Oncogene 2009, 28, 1421–1431. [Google Scholar] [CrossRef]
- Wu, H.; Kirita, Y.; Donnelly, E.L.; Humphreys, B.D. Advantages of single-nucleus over single-cell rna sequencing of adult kidney: Rare cell types and novel cell states revealed in fibrosis. J. Am. Soc. Nephrol. JASN 2019, 30, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Piersma, B.; Bank, R.A.; Boersema, M. Signaling in fibrosis: Tgf-beta, wnt, and yap/taz converge. Front. Med. 2015, 2, 59. [Google Scholar] [CrossRef]
- Kwon, E.; Todorova, K.; Wang, J.; Horos, R.; Lee, K.K.; Neel, V.A.; Negri, G.L.; Sorensen, P.H.; Lee, S.W.; Hentze, M.W.; et al. The rna-binding protein ybx1 regulates epidermal progenitors at a posttranscriptional level. Nat. Commun. 2018, 9, 1734. [Google Scholar] [CrossRef]
- Mertens, P.R.; Harendza, S.; Pollock, A.S.; Lovett, D.H. Glomerular mesangial cell-specific transactivation of matrix metalloproteinase 2 transcription is mediated by yb-1. J. Biol. Chem. 1997, 272, 22905–22912. [Google Scholar] [CrossRef] [PubMed]
- Bulow, R.D.; Boor, P. Extracellular matrix in kidney fibrosis: More than just a scaffold. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2019, 67, 643–661. [Google Scholar] [CrossRef]
- Mertens, P.R.; Alfonso-Jaume, M.A.; Steinmann, K.; Lovett, D.H. Yb-1 regulation of the human and rat gelatinase a genes via similar enhancer elements. J. Am. Soc. Nephrol. JASN 1999, 10, 2480–2487. [Google Scholar] [CrossRef]
- Turck, J.; Pollock, A.S.; Lee, L.K.; Marti, H.P.; Lovett, D.H. Matrix metalloproteinase 2 (gelatinase a) regulates glomerular mesangial cell proliferation and differentiation. J. Biol. Chem. 1996, 271, 15074–15083. [Google Scholar] [CrossRef]
- Kale, A.; Sankrityayan, H.; Anders, H.J.; Gaikwad, A.B. Epigenetic and non-epigenetic regulation of klotho in kidney disease. Life Sci. 2021, 264, 118644. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wen, D.; Ho, C.; Yu, L.; Zheng, D.; O’Reilly, S.; Gao, Y.; Li, Q.; Zhang, Y. Epigenetics as a versatile regulator of fibrosis. J. Transl. Med. 2023, 21, 164. [Google Scholar] [CrossRef] [PubMed]
- Norman, J.T.; Lindahl, G.E.; Shakib, K.; En-Nia, A.; Yilmaz, E.; Mertens, P.R. The y-box binding protein yb-1 suppresses collagen alpha 1(i) gene transcription via an evolutionarily conserved regulatory element in the proximal promoter. J. Biol. Chem. 2001, 276, 29880–29890. [Google Scholar] [CrossRef] [PubMed]
- En-Nia, A.; Yilmaz, E.; Klinge, U.; Lovett, D.H.; Stefanidis, I.; Mertens, P.R. Transcription factor yb-1 mediates DNA polymerase alpha gene expression. The Journal of biological chemistry 2005, 280, 7702–7711. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neubock, R.; Hofacker, I.L. The vienna rna websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Honer Zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. Viennarna package 2.0. Algorithms Mol. Biol. AMB 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Pratt, H.; Gao, M.; Wei, F.; Weng, Z.; Struhl, K. Yap and taz are transcriptional co-activators of ap-1 proteins and stat3 during breast cellular transformation. eLife 2021, 10, e67312. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.J.; Goldstein, L.; Chen, Y.J.; Lee, J.; Webster, J.D.; Roose-Girma, M.; Paudyal, S.C.; Modrusan, Z.; Dey, A.; Shaw, A.S. Single-cell transcriptome profiling of the kidney glomerulus identifies key cell types and reactions to injury. J. Am. Soc. Nephrol. JASN 2020, 31, 2341–2354. [Google Scholar] [CrossRef]
- Clausen, B.E.; Burkhardt, C.; Reith, W.; Renkawitz, R.; Forster, I. Conditional gene targeting in macrophages and granulocytes using lysmcre mice. Transgenic Res. 1999, 8, 265–277. [Google Scholar] [CrossRef]
- Wei, W.; Li, T.; Chen, J.; Fan, Z.; Gao, F.; Yu, Z.; Jiang, Y. Sirt3/6: An amazing challenge and opportunity in the fight against fibrosis and aging. Cell. Mol. Life Sci. 2024, 81, 69. [Google Scholar] [CrossRef]
- Henderson, N.C.; Rieder, F.; Wynn, T.A. Fibrosis: From mechanisms to medicines. Nature 2020, 587, 555–566. [Google Scholar] [CrossRef]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef]
- Gibbert, L.; Hermert, D.; Wang, J.; Breitkopf, D.M.; Alidousty, C.; Neusser, M.; Cohen, C.D.; Grone, E.; Macheleidt, I.; Rauen, T.; et al. Yb-1 increases glomerular, but decreases interstitial fibrosis in cni-induced nephropathy. Clin. Immunol. 2018, 194, 67–74. [Google Scholar] [CrossRef]
- Seufert, L.; Benzing, T.; Ignarski, M.; Muller, R.U. Rna-binding proteins and their role in kidney disease. Nat. Rev. Nephrol. 2022, 18, 153–170. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Qiu, X.; Liu, H.; Gan, C.; Tan, Z.; Xie, Y.; Wang, Y.; Ye, T. Epigenetic regulation in fibrosis progress. Pharmacol. Res. 2021, 173, 105910. [Google Scholar] [CrossRef]
- Zhang, Q.; Yin, S.; Liu, L.; Liu, Z.; Cao, W. Rhein reversal of DNA hypermethylation-associated klotho suppression ameliorates renal fibrosis in mice. Sci. Rep. 2016, 6, 34597. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Sheng, M.J.; Sun, Z.Y.; Liang, Y.; Yu, L.X.; Liu, Q.F. Upstream and downstream regulators of klotho expression in chronic kidney disease. Metab. Clin. Exp. 2023, 142, 155530. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Fu, P.; Ma, L. Kidney fibrosis: From mechanisms to therapeutic medicines. Signal Transduct. Target. Ther. 2023, 8, 129. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Cao, W. Epigenetic modifications of klotho expression in kidney diseases. J. Mol. Med. 2021, 99, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Gifford, C.C.; Lian, F.; Tang, J.; Costello, A.; Goldschmeding, R.; Samarakoon, R.; Higgins, P.J. Pai-1 induction during kidney injury promotes fibrotic epithelial dysfunction via deregulation of klotho, p53, and tgf-beta1-receptor signaling. FASEB J. 2021, 35, e21725. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Reddy, M.A.; Yuan, H.; Lanting, L.; Kato, M.; Natarajan, R. Epigenetic histone methylation modulates fibrotic gene expression. J. Am. Soc. Nephrol. JASN 2010, 21, 2069–2080. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Doke, T.; Guo, D.; Sheng, X.; Ma, Z.; Park, J.; Vy, H.M.T.; Nadkarni, G.N.; Abedini, A.; Miao, Z.; et al. Epigenomic and transcriptomic analyses define core cell types, genes and targetable mechanisms for kidney disease. Nat. Genet. 2022, 54, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Martin-Carro, B.; Martin-Virgala, J.; Fernandez-Villabrille, S.; Fernandez-Fernandez, A.; Perez-Basterrechea, M.; Navarro-Gonzalez, J.F.; Donate-Correa, J.; Mora-Fernandez, C.; Dusso, A.S.; Carrillo-Lopez, N.; et al. Role of klotho and age/rage-wnt/beta-catenin signalling pathway on the development of cardiac and renal fibrosis in diabetes. Int. J. Mol. Sci. 2023, 24, 5241. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Sun, Q.; Hua, M.R.; Suo, P.; Chen, J.R.; Yu, X.Y.; Zhao, Y.Y. Targeting the wnt/beta-catenin signaling pathway as a potential therapeutic strategy in renal tubulointerstitial fibrosis. Front. Pharmacol. 2021, 12, 719880. [Google Scholar] [CrossRef]
- Kuppe, C.; Ibrahim, M.M.; Kranz, J.; Zhang, X.; Ziegler, S.; Perales-Paton, J.; Jansen, J.; Reimer, K.C.; Smith, J.R.; Dobie, R.; et al. Decoding myofibroblast origins in human kidney fibrosis. Nature 2021, 589, 281–286. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, E.D.; Mylonas, K.J.; Xin, C.; Baird, D.P.; Carvalho, C.; Docherty, M.H.; Campbell, R.; Matchett, K.P.; Waddell, S.H.; Walker, A.D.; et al. Indian hedgehog release from tnf-activated renal epithelia drives local and remote organ fibrosis. Sci. Transl. Med. 2023, 15, eabn0736. [Google Scholar] [CrossRef]
- Zhou, J.; Abedini, A.; Balzer, M.; Shrestra, R.; Dhillon, P.; Liu, H.; Hu, H.; Susztak, K. Unified mouse and human kidney single-cell expression atlas reveal commonalities and differences in disease states. J. Am. Soc. Nephrol. JASN 2023, 34, 1843–1862. [Google Scholar] [CrossRef]
- Lake, B.B.; Menon, R.; Winfree, S.; Hu, Q.; Ferreira, R.M.; Kalhor, K.; Barwinska, D.; Otto, E.A.; Ferkowicz, M.; Diep, D.; et al. An atlas of healthy and injured cell states and niches in the human kidney. Nature 2023, 619, 585–594. [Google Scholar] [CrossRef]
| Name | Sequence (5′ > 3′) | Sequence (3′ > 5′) |
|---|---|---|
| Rosa26Cre | GCCTGCATTACCGGTCGATGCAACGA | GTGGCAGATGGCGCGGCAACACCATT |
| RosaWT | CTGTGGACAGAGGAGCCATAACTGC | CCACCACTGGCTGGCTAAACTCT |
| LysM | CTTGGGCTGCCAGAATTTCTC | TTACAGTCGGCCAGGCTGAC |
| CRE8 | CCCAGAAATGCCAGATTACG | |
| YB-1 | GCCTAAGGATAGTGAAGTTTCTGG | CCTAGCACACCTTAATCTACAGCC |
| Name | Sequence (5′ > 3′) | Sequence (3′ > 5′) |
|---|---|---|
| mKL/M | GGTATCGCGGGTATTTTTAATC | CGACATAATCCCTAAAATAATCGAC |
| mKL/unM | TTAATGGTATTGTGGGTATTTTTAATTG | CAACATAATCCCTAAAATAATCAAC |
| mIn | TAGTTTTAGGAAGGTAAAGGGAGTG | AAATCCCAAAAAAAACACAACAAA |
| mKL/M | GGTATCGCGGGTATTTTTAATC | CGACATAATCCCTAAAATAATCGAC |
| Target (Mouse) | Sequence (5′ > 3′) | Sequence (3′ > 5′) |
|---|---|---|
| Col1a1 | GAGAACCAGCAGAGCCA | GAACAAGGTGACAGAGGCATA |
| Col3a1 | GAAAGGATGGAGAGTCAGGAA | GAAAGGATGGAGAGTCAGGAA |
| Ccn2 | CCCTAGCTGCCTACCGACT | GGTAACTCGGGTGGAGATGC |
| Cyr61 | GCTCAGTCAGAAGGCAGACC | GTTCTTGGGGACACAGAGGA |
| Gapdh | AGGTCGGTGTGAACGGATTTG | GGGGTCGTTGATGGCAACAA |
| PAI-1 | GGC CAT TAC TAC GAC ATC CTG | GGT CAT GTT GCC TTT CCA GT |
| Egr1 | CAGTCCCAGCTCATCAAAC | TCTGCTTTCTTGTCCTTCTG |
| DNMT1 | CAGAGACTCCCGAGGACAGA | TTTACGTGTCGTTTTTCGTCTC |
| DNMT3a | CGTGAGTCCGGTGTGTCA | CTCCAACCACACACACAAGG |
| DNMT3b | TTCAGTGACCAGTCCTCAGACACGAA | TCAGAAGGCTGGAGACCTCCCTCTT |
| Immunoprecipitated Transcripts Bound by YB-1 | |
|---|---|
| Human Data Sets | |
| GSE30122 | down-regulated: NELL, SPARC, WISP3, SDC2, FAM20B, HRG, PLAT, TIMP3, EGF, FGF9, HBEGF, S100A8, VEGFA up-regulated: FN1, IGFBP3, COL15A1, C1QB, ST14, CCL19, HGF |
| GSE116626 | down-regulated: SLIT3, COL16A1, CLEC4F, PAPPA2, CCL19, IL10, WNT10A up-regulated: CYR61, ANXA6, MMP24, SERPINA6, SERPINE1, ANGPTL4, HBEGF |
| mouse data sets | |
| UUO | down-regulated: ADIPOQ, IGFBP3, COL27A1, HRG, PZP, TIMP3, EGF, FGF9, VEGFA up-regulated: CYR61, FGA, FN1, IGFBP6, LRG1, MFAP2, MFAP5, MGP, SLIT3, SPARC, WISP1, COL15A1, COL16A1, COL5A1, COL6A2, ANXA5, ANXA6, C1QB, CLEC4D, LGALS3, MUC4, ADAMTS1, ADAMTS2, ITIH4, LOXL2, SERPINE1, ANGPTL4, CSF1, GDF15, HBEGF, IL1F6, S100A6, S100A8, WNT10A, WNT4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brandt, S.; Bernhardt, A.; Häberer, S.; Wolters, K.; Gehringer, F.; Reichardt, C.; Krause, A.; Geffers, R.; Kahlfuß, S.; Jeron, A.; et al. Comparative Analysis of Acute Kidney Injury Models and Related Fibrogenic Responses: Convergence on Methylation Patterns Regulated by Cold Shock Protein. Cells 2024, 13, 367. https://doi.org/10.3390/cells13050367
Brandt S, Bernhardt A, Häberer S, Wolters K, Gehringer F, Reichardt C, Krause A, Geffers R, Kahlfuß S, Jeron A, et al. Comparative Analysis of Acute Kidney Injury Models and Related Fibrogenic Responses: Convergence on Methylation Patterns Regulated by Cold Shock Protein. Cells. 2024; 13(5):367. https://doi.org/10.3390/cells13050367
Chicago/Turabian StyleBrandt, Sabine, Anja Bernhardt, Saskia Häberer, Katharina Wolters, Fabian Gehringer, Charlotte Reichardt, Anna Krause, Robert Geffers, Sascha Kahlfuß, Andreas Jeron, and et al. 2024. "Comparative Analysis of Acute Kidney Injury Models and Related Fibrogenic Responses: Convergence on Methylation Patterns Regulated by Cold Shock Protein" Cells 13, no. 5: 367. https://doi.org/10.3390/cells13050367
APA StyleBrandt, S., Bernhardt, A., Häberer, S., Wolters, K., Gehringer, F., Reichardt, C., Krause, A., Geffers, R., Kahlfuß, S., Jeron, A., Bruder, D., Lindquist, J. A., Isermann, B., & Mertens, P. R. (2024). Comparative Analysis of Acute Kidney Injury Models and Related Fibrogenic Responses: Convergence on Methylation Patterns Regulated by Cold Shock Protein. Cells, 13(5), 367. https://doi.org/10.3390/cells13050367


.png)