Cell Death, by Any Other Name…
Abstract
1. Introduction: The Dead Cell
2. Cell–Cell Communication: Dying Alone or Together
2.1. Cell Death and Group Dynamics in Early Development
2.2. Cell Death versus Cell Communication
2.2.1. Extruding the Dead
2.2.2. Setting Fire
2.2.3. Trying to Fit In
2.2.4. Surviving Attacks
2.3. The “Bystander Effect”
3. Concluding Remarks
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vaux, D.L.; Korsmeyer, S.J. Cell death in development. Cell 1999, 96, 245–254. [Google Scholar] [CrossRef]
- Kerr, J.F. History of the events leading to the formulation of the apoptosis concept. Toxicology 2002, 181–182, 471–474. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Galluzzi, L.; Vandenabeele, P.; Abrams, J.; Alnemri, E.S.; Baehrecke, E.H.; Blagosklonny, M.V.; El-Deiry, W.S.; Golstein, P.; Green, D.R.; et al. Classification of cell death: Recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009, 16, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.L.; Yuen, K.L.; Tang, H.M.; Fung, M.C. Reversibility of apoptosis in cancer cells. Br. J. Cancer 2009, 100, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.L.; Tang, H.M.; Mak, K.H.; Hu, S.; Wang, S.S.; Wong, K.M.; Wong, C.S.; Wu, H.Y.; Law, H.T.; Liu, K.; et al. Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response. Mol. Biol. Cell 2012, 23, 2240–2252. [Google Scholar] [CrossRef] [PubMed]
- Zakharov, I.I.; Savitskaya, M.A.; Onishchenko, G.E. The Problem of Apoptotic Processes Reversibility. Biochemistry 2020, 85, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, R.N.; Khosravi, M.; Rahman, H.S.; Adili, A.; Kamali, N.; Soloshenkov, P.P.; Thangavelu, L.; Saeedi, H.; Shomali, N.; Tamjidifar, R.; et al. Anastasis: Cell recovery mechanisms and potential role in cancer. Cell Commun. Signal. 2022, 20, 81. [Google Scholar] [CrossRef] [PubMed]
- Zaitceva, V.; Kopeina, G.S.; Zhivotovsky, B. Anastasis: Return Journey from Cell Death. Cancers 2021, 13, 3671. [Google Scholar] [CrossRef]
- Chakraborty, S.; Mir, K.B.; Seligson, N.D.; Nayak, D.; Kumar, R.; Goswami, A. Integration of EMT and cellular survival instincts in reprogramming of programmed cell death to anastasis. Cancer Metastasis Rev. 2020, 39, 553–566. [Google Scholar] [CrossRef]
- Gong, Y.N.; Crawford, J.C.; Heckmann, B.L.; Green, D.R. To the edge of cell death and back. FEBS J. 2019, 286, 430–440. [Google Scholar] [CrossRef]
- Sun, G. Death and survival from executioner caspase activation. Semin. Cell Dev. Biol. 2024, 156, 66–73. [Google Scholar] [CrossRef]
- Lovric, M.M.; Hawkins, C.J. TRAIL treatment provokes mutations in surviving cells. Oncogene 2010, 29, 5048–5060. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; He, Y.; Li, F.; Huang, Q.; Kato, T.A.; Hall, R.P.; Li, C.Y. Caspase-3 promotes genetic instability and carcinogenesis. Mol. Cell 2015, 58, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Ichim, G.; Lopez, J.; Ahmed, S.U.; Muthalagu, N.; Giampazolias, E.; Delgado, M.E.; Haller, M.; Riley, J.S.; Mason, S.M.; Athineos, D.; et al. Limited mitochondrial permeabilization causes DNA damage and genomic instability in the absence of cell death. Mol. Cell 2015, 57, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Guzman, E.; Balasanyan, V.; Conner, C.M.; Wong, K.; Zhou, H.R.; Kosik, K.S.; Montell, D.J. A molecular signature for anastasis, recovery from the brink of apoptotic cell death. J. Cell Biol. 2017, 216, 3355–3368. [Google Scholar] [CrossRef] [PubMed]
- Nano, M.; Montell, D.J. Apoptotic signaling: Beyond cell death. Semin. Cell Dev. Biol. 2024, 156, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Boya, P.; Kroemer, G. Lysosomal membrane permeabilization in cell death. Oncogene 2008, 27, 6434–6451. [Google Scholar] [CrossRef] [PubMed]
- Aits, S.; Jaattela, M. Lysosomal cell death at a glance. J. Cell Sci. 2013, 126 Pt 9, 1905–1912. [Google Scholar] [CrossRef]
- Battistelli, M.; Falcieri, E. Apoptotic Bodies: Particular Extracellular Vesicles Involved in Intercellular Communication. Biology 2020, 9, 21. [Google Scholar] [CrossRef]
- Kakarla, R.; Hur, J.; Kim, Y.J.; Kim, J.; Chwae, Y.J. Apoptotic cell-derived exosomes: Messages from dying cells. Exp. Mol. Med. 2020, 52, 1–6. [Google Scholar] [CrossRef]
- Zhang, M.; Lin, Y.; Chen, R.; Yu, H.; Li, Y.; Chen, M.; Dou, C.; Yin, P.; Zhang, L.; Tang, P. Ghost messages: Cell death signals spread. Cell Commun. Signal. 2023, 21, 6. [Google Scholar] [CrossRef]
- Golstein, P.; Kroemer, G. Cell death by necrosis: Towards a molecular definition. Trends Biochem. Sci. 2007, 32, 37–43. [Google Scholar] [CrossRef]
- Festjens, N.; Vanden Berghe, T.; Vandenabeele, P. Necrosis, a well-orchestrated form of cell demise: Signalling cascades, important mediators and concomitant immune response. Biochim. Biophys. Acta 2006, 1757, 1371–1387. [Google Scholar] [CrossRef]
- Silva, M.T.; do Vale, A.; dos Santos, N.M. Secondary necrosis in multicellular animals: An outcome of apoptosis with pathogenic implications. Apoptosis 2008, 13, 463–482. [Google Scholar] [CrossRef]
- Wyllie, A.H.; Kerr, J.F.; Currie, A.R. Cell death: The significance of apoptosis. Int. Rev. Cytol. 1980, 68, 251–306. [Google Scholar] [CrossRef] [PubMed]
- Gaipl, U.S.; Franz, S.; Voll, R.E.; Sheriff, A.; Kalden, J.R.; Herrmann, M. Defects in the disposal of dying cells lead to autoimmunity. Curr. Rheumatol. Rep. 2004, 6, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Lauber, K.; Blumenthal, S.G.; Waibel, M.; Wesselborg, S. Clearance of apoptotic cells: Getting rid of the corpses. Mol. Cell 2004, 14, 277–287. [Google Scholar] [CrossRef]
- de Castro, M.A.; Bunt, G.; Wouters, F.S. Cathepsin B launches an apoptotic exit effort upon cell death-associated disruption of lysosomes. Cell Death Discov. 2016, 2, 16012. [Google Scholar] [CrossRef]
- Vercammen, D.; Beyaert, R.; Denecker, G.; Goossens, V.; Van Loo, G.; Declercq, W.; Grooten, J.; Fiers, W.; Vandenabeele, P. Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J. Exp. Med. 1998, 187, 1477–1485. [Google Scholar] [CrossRef] [PubMed]
- Holler, N.; Zaru, R.; Micheau, O.; Thome, M.; Attinger, A.; Valitutti, S.; Bodmer, J.L.; Schneider, P.; Seed, B.; Tschopp, J. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat. Immunol. 2000, 1, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.W.; Shao, J.; Lin, J.; Zhang, N.; Lu, B.J.; Lin, S.C.; Dong, M.Q.; Han, J. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 2009, 325, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, A.; Vandenabeele, P.; Krysko, D.V. Necroptosis: The release of damage-associated molecular patterns and its physiological relevance. Immunity 2013, 38, 209–223. [Google Scholar] [CrossRef] [PubMed]
- Newton, K.; Manning, G. Necroptosis and Inflammation. Annu. Rev. Biochem. 2016, 85, 743–763. [Google Scholar] [CrossRef] [PubMed]
- Chan, F.K.; Luz, N.F.; Moriwaki, K. Programmed necrosis in the cross talk of cell death and inflammation. Annu. Rev. Immunol. 2015, 33, 79–106. [Google Scholar] [CrossRef] [PubMed]
- Pasparakis, M.; Vandenabeele, P. Necroptosis and its role in inflammation. Nature 2015, 517, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Nailwal, H.; Chan, F.K. Necroptosis in anti-viral inflammation. Cell Death Differ. 2019, 26, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, S.B.; Miao, E.A. Gasdermins: Effectors of Pyroptosis. Trends Cell Biol. 2017, 27, 673–684. [Google Scholar] [CrossRef]
- Shi, J.; Gao, W.; Shao, F. Pyroptosis: Gasdermin-Mediated Programmed Necrotic Cell Death. Trends Biochem. Sci. 2017, 42, 245–254. [Google Scholar] [CrossRef]
- Cookson, B.T.; Brennan, M.A. Pro-inflammatory programmed cell death. Trends Microbiol. 2001, 9, 113–114. [Google Scholar] [CrossRef]
- Pilla, D.M.; Hagar, J.A.; Haldar, A.K.; Mason, A.K.; Degrandi, D.; Pfeffer, K.; Ernst, R.K.; Yamamoto, M.; Miao, E.A.; Coers, J. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc. Natl. Acad. Sci. USA 2014, 111, 6046–6051. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665. [Google Scholar] [CrossRef]
- Jorgensen, I.; Miao, E.A. Pyroptotic cell death defends against intracellular pathogens. Immunol. Rev. 2015, 265, 130–142. [Google Scholar] [CrossRef] [PubMed]
- Miao, E.A.; Leaf, I.A.; Treuting, P.M.; Mao, D.P.; Dors, M.; Sarkar, A.; Warren, S.E.; Wewers, M.D.; Aderem, A. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 2010, 11, 1136–1142. [Google Scholar] [CrossRef] [PubMed]
- Aachoui, Y.; Leaf, I.A.; Hagar, J.A.; Fontana, M.F.; Campos, C.G.; Zak, D.E.; Tan, M.H.; Cotter, P.A.; Vance, R.E.; Aderem, A.; et al. Caspase-11 protects against bacteria that escape the vacuole. Science 2013, 339, 975–978. [Google Scholar] [CrossRef] [PubMed]
- Fink, S.L.; Cookson, B.T. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell Microbiol. 2006, 8, 1812–1825. [Google Scholar] [CrossRef] [PubMed]
- Sborgi, L.; Ruhl, S.; Mulvihill, E.; Pipercevic, J.; Heilig, R.; Stahlberg, H.; Farady, C.J.; Muller, D.J.; Broz, P.; Hiller, S. GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J. 2016, 35, 1766–1778. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Wang, K.; Liu, W.; She, Y.; Sun, Q.; Shi, J.; Sun, H.; Wang, D.C.; Shao, F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 2016, 535, 111–116. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Z.; Ruan, J.; Pan, Y.; Magupalli, V.G.; Wu, H.; Lieberman, J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 2016, 535, 153–158. [Google Scholar] [CrossRef]
- Man, S.M.; Kanneganti, T.D. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat. Rev. Immunol. 2016, 16, 7–21. [Google Scholar] [CrossRef]
- Lockshin, R.A.; Williams, C.M. Programmed cell death. IV. The influence of drugs on the breakdown of the intersegmental muscles of silkmoths. J. Insect. Physiol. 1965, 11, 803–809. [Google Scholar] [CrossRef]
- Tata, J.R. Requirement for RNA and protein synthesis for induced regression of the tadpole tail in organ culture. Dev. Biol. 1966, 13, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.F.; Wyllie, A.H.; Currie, A.R. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Nossing, C.; Ryan, K.M. 50 years on and still very much alive: Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 2023, 128, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Horvitz, H.R. Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res. 1999, 59 (Suppl. S7), 1701s–1706s. [Google Scholar] [PubMed]
- Abrams, J.M.; White, K.; Fessler, L.I.; Steller, H. Programmed cell death during Drosophila embryogenesis. Development 1993, 117, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Woodfield, S.E.; Lee, T.V.; Fan, Y.; Antonio, C.; Bergmann, A. Genetic control of programmed cell death (apoptosis) in Drosophila. Fly 2009, 3, 78–90. [Google Scholar] [CrossRef]
- Ranger, A.M.; Malynn, B.A.; Korsmeyer, S.J. Mouse models of cell death. Nat. Genet. 2001, 28, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Joza, N.; Kroemer, G.; Penninger, J.M. Genetic analysis of the mammalian cell death machinery. Trends Genet. 2002, 18, 142–149. [Google Scholar] [CrossRef]
- Woo, M.; Hakem, R.; Mak, T.W. Executionary pathway for apoptosis: Lessons from mutant mice. Cell Res. 2000, 10, 267–278. [Google Scholar] [CrossRef]
- Fink, S.L.; Cookson, B.T. Apoptosis, pyroptosis, and necrosis: Mechanistic description of dead and dying eukaryotic cells. Infect. Immun. 2005, 73, 1907–1916. [Google Scholar] [CrossRef]
- Davidovich, P.; Kearney, C.J.; Martin, S.J. Inflammatory outcomes of apoptosis, necrosis and necroptosis. Biol. Chem. 2014, 395, 1163–1171. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S. Apoptosis and Clearance of Apoptotic Cells. Annu. Rev. Immunol. 2018, 36, 489–517. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed]
- Snyder, A.G.; Oberst, A. The Antisocial Network: Cross Talk Between Cell Death Programs in Host Defense. Annu. Rev. Immunol. 2021, 39, 77–101. [Google Scholar] [CrossRef] [PubMed]
- Bedoui, S.; Herold, M.J.; Strasser, A. Emerging connectivity of programmed cell death pathways and its physiological implications. Nat. Rev. Mol. Cell Biol. 2020, 21, 678–695. [Google Scholar] [CrossRef]
- Tang, D.; Kang, R.; Berghe, T.V.; Vandenabeele, P.; Kroemer, G. The molecular machinery of regulated cell death. Cell Res. 2019, 29, 347–364. [Google Scholar] [CrossRef]
- Mizushima, N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 2018, 20, 521–527. [Google Scholar] [CrossRef]
- Klionsky, D.J. Autophagy revisited: A conversation with Christian de Duve. Autophagy 2008, 4, 740–743. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef]
- Vargas, J.N.S.; Hamasaki, M.; Kawabata, T.; Youle, R.J.; Yoshimori, T. The mechanisms and roles of selective autophagy in mammals. Nat. Rev. Mol. Cell Biol. 2023, 24, 167–185. [Google Scholar] [CrossRef]
- Wang, L.; Klionsky, D.J.; Shen, H.M. The emerging mechanisms and functions of microautophagy. Nat. Rev. Mol. Cell Biol. 2023, 24, 186–203. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Lee, M.S. Autophagy—A key player in cellular and body metabolism. Nat. Rev. Endocrinol. 2014, 10, 322–337. [Google Scholar] [CrossRef] [PubMed]
- Maiuri, M.C.; Zalckvar, E.; Kimchi, A.; Kroemer, G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Ploumi, C.; Papandreou, M.E.; Tavernarakis, N. The complex interplay between autophagy and cell death pathways. Biochem. J. 2022, 479, 75–90. [Google Scholar] [CrossRef] [PubMed]
- Denton, D.; Kumar, S. Autophagy-dependent cell death. Cell Death Differ. 2019, 26, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Loos, B.; Engelbrecht, A.M.; Lockshin, R.A.; Klionsky, D.J.; Zakeri, Z. The variability of autophagy and cell death susceptibility: Unanswered questions. Autophagy 2013, 9, 1270–1285. [Google Scholar] [CrossRef] [PubMed]
- Mahapatra, K.K.; Mishra, S.R.; Behera, B.P.; Patil, S.; Gewirtz, D.A.; Bhutia, S.K. The lysosome as an imperative regulator of autophagy and cell death. Cell Mol. Life Sci. 2021, 78, 7435–7449. [Google Scholar] [CrossRef]
- Johansson, A.C.; Appelqvist, H.; Nilsson, C.; Kagedal, K.; Roberg, K.; Ollinger, K. Regulation of apoptosis-associated lysosomal membrane permeabilization. Apoptosis 2010, 15, 527–540. [Google Scholar] [CrossRef]
- Skowyra, M.L.; Schlesinger, P.H.; Naismith, T.V.; Hanson, P.I. Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 2018, 360, eaar5078. [Google Scholar] [CrossRef]
- Radulovic, M.; Schink, K.O.; Wenzel, E.M.; Nahse, V.; Bongiovanni, A.; Lafont, F.; Stenmark, H. ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J. 2018, 37, e99753. [Google Scholar] [CrossRef]
- Ono, K.; Kim, S.O.; Han, J. Susceptibility of lysosomes to rupture is a determinant for plasma membrane disruption in tumor necrosis factor alpha-induced cell death. Mol. Cell Biol. 2003, 23, 665–676. [Google Scholar] [CrossRef]
- Galluzzi, L.; Bravo-San Pedro, J.M.; Kroemer, G. Organelle-specific initiation of cell death. Nat. Cell Biol. 2014, 16, 728–736. [Google Scholar] [CrossRef] [PubMed]
- Joshi, G.N.; Knecht, D.A. Silica phagocytosis causes apoptosis and necrosis by different temporal and molecular pathways in alveolar macrophages. Apoptosis 2013, 18, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Gomez-Sintes, R.; Boya, P. Lysosomal membrane permeabilization and cell death. Traffic 2018, 19, 918–931. [Google Scholar] [CrossRef] [PubMed]
- Eaton, J.W.; Qian, M. Molecular bases of cellular iron toxicity. Free Radic. Biol. Med. 2002, 32, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Borner, C.; Monney, L. Apoptosis without caspases: An inefficient molecular guillotine? Cell Death Differ. 1999, 6, 497–507. [Google Scholar] [CrossRef]
- Mrschtik, M.; Ryan, K.M. Lysosomal proteins in cell death and autophagy. FEBS J. 2015, 282, 1858–1870. [Google Scholar] [CrossRef] [PubMed]
- Counis, M.F.; Torriglia, A. DNases and apoptosis. Biochem. Cell Biol. 2000, 78, 405–414. [Google Scholar] [CrossRef]
- Keyel, P.A. Dnases in health and disease. Dev. Biol. 2017, 429, 1–11. [Google Scholar] [CrossRef]
- Daugas, E.; Susin, S.A.; Zamzami, N.; Ferri, K.F.; Irinopoulou, T.; Larochette, N.; Prevost, M.C.; Leber, B.; Andrews, D.; Penninger, J.; et al. Mitochondrio-nuclear translocation of AIF in apoptosis and necrosis. FASEB J. 2000, 14, 729–739. [Google Scholar] [CrossRef]
- Li, L.Y.; Luo, X.; Wang, X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 2001, 412, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.J.; Green, D.R. Protease activation during apoptosis: Death by a thousand cuts? Cell 1995, 82, 349–352. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Lu, N.; Zhou, Z. Cellular and nuclear degradation during apoptosis. Curr. Opin. Cell Biol. 2009, 21, 900–912. [Google Scholar] [CrossRef]
- Taylor, R.C.; Cullen, S.P.; Martin, S.J. Apoptosis: Controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 2008, 9, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Adrain, C.; Brumatti, G.; Martin, S.J. Apoptosomes: Protease activation platforms to die from. Trends Biochem. Sci. 2006, 31, 243–247. [Google Scholar] [CrossRef]
- Dorstyn, L.; Akey, C.W.; Kumar, S. New insights into apoptosome structure and function. Cell Death Differ. 2018, 25, 1194–1208. [Google Scholar] [CrossRef]
- Williams, J.R.; Little, J.B.; Shipley, W.U. Association of mammalian cell death with a specific endonucleolytic degradation of DNA. Nature 1974, 252, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Luthi, A.U.; Martin, S.J. The CASBAH: A searchable database of caspase substrates. Cell Death Differ. 2007, 14, 641–650. [Google Scholar] [CrossRef]
- Monier, B.; Suzanne, M. Orchestration of Force Generation and Nuclear Collapse in Apoptotic Cells. Int. J. Mol. Sci. 2021, 22, 10257. [Google Scholar] [CrossRef]
- Stewart, C.L.; Roux, K.J.; Burke, B. Blurring the boundary: The nuclear envelope extends its reach. Science 2007, 318, 1408–1412. [Google Scholar] [CrossRef]
- Burke, B. Lamins and apoptosis: A two-way street? J. Cell Biol. 2001, 153, F5–F7. [Google Scholar] [CrossRef] [PubMed]
- Rao, L.; Perez, D.; White, E. Lamin proteolysis facilitates nuclear events during apoptosis. J. Cell Biol. 1996, 135 Pt 1, 1441–1455. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.; Ward, C.; Sheldrake, T.A.; Dransfield, I.; Rossi, A.G.; Pryde, J.G.; Haslett, C. Golgi fragmentation during Fas-mediated apoptosis is associated with the rapid loss of GM130. Biochem. Biophys. Res. Commun. 2004, 316, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Lane, J.D.; Lucocq, J.; Pryde, J.; Barr, F.A.; Woodman, P.G.; Allan, V.J.; Lowe, M. Caspase-mediated cleavage of the stacking protein GRASP65 is required for Golgi fragmentation during apoptosis. J. Cell Biol. 2002, 156, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.D.; Orrenius, S.; Zhivotovsky, B. Review: Nuclear events in apoptosis. J. Struct. Biol. 2000, 129, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Doonan, F.; Cotter, T.G. Morphological assessment of apoptosis. Methods 2008, 44, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Buendia, B.; Courvalin, J.C.; Collas, P. Dynamics of the nuclear envelope at mitosis and during apoptosis. Cell Mol. Life Sci. 2001, 58, 1781–1789. [Google Scholar] [CrossRef]
- Kutay, U.; Hetzer, M.W. Reorganization of the nuclear envelope during open mitosis. Curr. Opin. Cell Biol. 2008, 20, 669–677. [Google Scholar] [CrossRef]
- Ungricht, R.; Kutay, U. Mechanisms and functions of nuclear envelope remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 229–245. [Google Scholar] [CrossRef]
- Shahin, V. Strategic disruption of nuclear pores structure, integrity and barrier for nuclear apoptosis. Semin. Cell Dev. Biol. 2017, 68, 85–90. [Google Scholar] [CrossRef]
- Youle, R.J.; van der Bliek, A.M. Mitochondrial fission, fusion, and stress. Science 2012, 337, 1062–1065. [Google Scholar] [CrossRef] [PubMed]
- Hoppins, S.; Nunnari, J. Cell Biology. Mitochondrial dynamics and apoptosis--the ER connection. Science 2012, 337, 1052–1054. [Google Scholar] [CrossRef] [PubMed]
- Machamer, C.E. Golgi disassembly in apoptosis: Cause or effect? Trends Cell Biol. 2003, 13, 279–281. [Google Scholar] [CrossRef] [PubMed]
- Gonatas, N.K.; Stieber, A.; Gonatas, J.O. Fragmentation of the Golgi apparatus in neurodegenerative diseases and cell death. J. Neurol. Sci. 2006, 246, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.V.; Ellerby, H.M.; Bredesen, D.E. Coupling endoplasmic reticulum stress to the cell death program. Cell Death Differ. 2004, 11, 372–380. [Google Scholar] [CrossRef]
- Suen, D.F.; Norris, K.L.; Youle, R.J. Mitochondrial dynamics and apoptosis. Genes Dev. 2008, 22, 1577–1590. [Google Scholar] [CrossRef] [PubMed]
- Elgass, K.; Pakay, J.; Ryan, M.T.; Palmer, C.S. Recent advances into the understanding of mitochondrial fission. Biochim. Biophys. Acta 2013, 1833, 150–161. [Google Scholar] [CrossRef]
- Arnoult, D. Mitochondrial fragmentation in apoptosis. Trends Cell Biol. 2007, 17, 6–12. [Google Scholar] [CrossRef]
- Martinou, J.C.; Youle, R.J. Which came first, the cytochrome c release or the mitochondrial fission? Cell Death Differ. 2006, 13, 1291–1295. [Google Scholar] [CrossRef]
- Bock, F.J.; Tait, S.W.G. Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 2020, 21, 85–100. [Google Scholar] [CrossRef]
- Lane, J.D.; Allan, V.J.; Woodman, P.G. Active relocation of chromatin and endoplasmic reticulum into blebs in late apoptotic cells. J. Cell Sci. 2005, 118 Pt 17, 4059–4071. [Google Scholar] [CrossRef] [PubMed]
- Keller, H.; Rentsch, P.; Hagmann, J. Differences in cortical actin structure and dynamics document that different types of blebs are formed by distinct mechanisms. Exp. Cell Res. 2002, 277, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.C.; Stone, N.L.; Pittman, R.N. Extranuclear apoptosis. The role of the cytoplasm in the execution phase. J. Cell Biol. 1999, 146, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.C.; Stone, N.L.; Erhardt, J.; Pittman, R.N. Apoptotic membrane blebbing is regulated by myosin light chain phosphorylation. J. Cell Biol. 1998, 140, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Charras, G.T.; Yarrow, J.C.; Horton, M.A.; Mahadevan, L.; Mitchison, T.J. Non-equilibration of hydrostatic pressure in blebbing cells. Nature 2005, 435, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Croft, D.R.; Coleman, M.L.; Li, S.; Robertson, D.; Sullivan, T.; Stewart, C.L.; Olson, M.F. Actin-myosin-based contraction is responsible for apoptotic nuclear disintegration. J. Cell Biol. 2005, 168, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Moss, D.K.; Betin, V.M.; Malesinski, S.D.; Lane, J.D. A novel role for microtubules in apoptotic chromatin dynamics and cellular fragmentation. J. Cell Sci. 2006, 119 Pt 11, 2362–2374. [Google Scholar] [CrossRef] [PubMed]
- Bozzaro, S. The past, present and future of Dictyostelium as a model system. Int. J. Dev. Biol. 2019, 63, 321–331. [Google Scholar] [CrossRef]
- Kandouz, M. Dying to communicate: Apoptotic functions of Eph/Ephrin proteins. Apoptosis 2018, 23, 265–289. [Google Scholar] [CrossRef]
- Bates, R.C.; Buret, A.; van Helden, D.F.; Horton, M.A.; Burns, G.F. Apoptosis induced by inhibition of intercellular contact. J. Cell Biol. 1994, 125, 403–415. [Google Scholar] [CrossRef]
- Krysko, D.V.; Leybaert, L.; Vandenabeele, P.; D’Herde, K. Gap junctions and the propagation of cell survival and cell death signals. Apoptosis 2005, 10, 459–469. [Google Scholar] [CrossRef]
- Carette, D.; Gilleron, J.; Chevallier, D.; Segretain, D.; Pointis, G. Connexin a check-point component of cell apoptosis in normal and physiopathological conditions. Biochimie 2014, 101, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gilleron, J.; Carette, D.; Segretain, D.; Pointis, G. Multiple and complex influences of connexins and pannexins on cell death. Biochim. Biophys. Acta Biomembr. 2018, 1860, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Caruso, S.; Poon, I.K.H. Apoptotic Cell-Derived Extracellular Vesicles: More Than Just Debris. Front. Immunol. 2018, 9, 1486. [Google Scholar] [CrossRef] [PubMed]
- Sanwlani, R.; Gangoda, L. Role of Extracellular Vesicles in Cell Death and Inflammation. Cells 2021, 10, 2663. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gerdes, H.H. Transfer of mitochondria via tunneling nanotubes rescues apoptotic PC12 cells. Cell Death Differ. 2015, 22, 1181–1191. [Google Scholar] [CrossRef]
- Zheng, F.; Luo, Z.; Lin, X.; Wang, W.; Aschner, M.; Cai, P.; Wang, Y.L.; Shao, W.; Yu, G.; Guo, Z.; et al. Intercellular transfer of mitochondria via tunneling nanotubes protects against cobalt nanoparticle-induced neurotoxicity and mitochondrial damage. Nanotoxicology 2021, 15, 1358–1379. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Davis, D.; Fang, Y. Intercellular transfer of mitochondria rescues virus-induced cell death but facilitates cell-to-cell spreading of porcine reproductive and respiratory syndrome virus. Virology 2018, 517, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Luchetti, F.; Canonico, B.; Arcangeletti, M.; Guescini, M.; Cesarini, E.; Stocchi, V.; Degli Esposti, M.; Papa, S. Fas signalling promotes intercellular communication in T cells. PLoS ONE 2012, 7, e35766. [Google Scholar] [CrossRef]
- Bittins, M.; Wang, X. TNT-Induced Phagocytosis: Tunneling Nanotubes Mediate the Transfer of Pro-Phagocytic Signals From Apoptotic to Viable Cells. J. Cell. Physiol. 2017, 232, 2271–2279. [Google Scholar] [CrossRef]
- Lu, S.; Sun, Z.; Liu, L.; Li, P.; Li, B.; Li, W.; Wu, Z.; Zhao, M.; Liu, W.; Wang, Y.; et al. Tumor-Derived Exosomes Regulate Apoptosis of CD45(+)EpCAM(+) Cells in Lung Cancer. Front. Immunol. 2022, 13, 903882. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, Y.; Steller, H. Programmed cell death in animal development and disease. Cell 2011, 147, 742–758. [Google Scholar] [CrossRef] [PubMed]
- Voss, A.K.; Strasser, A. The essentials of developmental apoptosis. F1000Research 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Wong, F.K.; Marin, O. Developmental Cell Death in the Cerebral Cortex. Annu. Rev. Cell Dev. Biol. 2019, 35, 523–542. [Google Scholar] [CrossRef] [PubMed]
- Southwell, D.G.; Paredes, M.F.; Galvao, R.P.; Jones, D.L.; Froemke, R.C.; Sebe, J.Y.; Alfaro-Cervello, C.; Tang, Y.; Garcia-Verdugo, J.M.; Rubenstein, J.L.; et al. Intrinsically determined cell death of developing cortical interneurons. Nature 2012, 491, 109–113. [Google Scholar] [CrossRef]
- Doseff, A.I. Apoptosis: The sculptor of development. Stem Cells Dev. 2004, 13, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Boehm, I. Apoptosis in physiological and pathological skin: Implications for therapy. Curr. Mol. Med. 2006, 6, 375–394. [Google Scholar] [CrossRef] [PubMed]
- Anderton, H.; Alqudah, S. Cell death in skin function, inflammation, and disease. Biochem. J. 2022, 479, 1621–1651. [Google Scholar] [CrossRef]
- Tower, J. Programmed cell death in aging. Ageing Res. Rev. 2015, 23 Pt A, 90–100. [Google Scholar] [CrossRef]
- Hardy, K.; Handyside, A.H. Metabolism and cell allocation during parthenogenetic preimplantation mouse development. Mol. Reprod. Dev. 1996, 43, 313–322. [Google Scholar] [CrossRef]
- Fleming, T.P.; Sheth, B.; Fesenko, I. Cell adhesion in the preimplantation mammalian embryo and its role in trophectoderm differentiation and blastocyst morphogenesis. Front. Biosci. 2001, 6, D1000-7. [Google Scholar] [CrossRef]
- Bloor, D.J.; Metcalfe, A.D.; Rutherford, A.; Brison, D.R.; Kimber, S.J. Expression of cell adhesion molecules during human preimplantation embryo development. Mol. Hum. Reprod. 2002, 8, 237–245. [Google Scholar] [CrossRef]
- Brison, D.R.; Sturmey, R.G.; Leese, H.J. Metabolic heterogeneity during preimplantation development: The missing link? Hum. Reprod. Update 2014, 20, 632–640. [Google Scholar] [CrossRef] [PubMed]
- Venturas, M.; Shah, J.S.; Yang, X.; Sanchez, T.H.; Conway, W.; Sakkas, D.; Needleman, D.J. Metabolic state of human blastocysts measured by fluorescence lifetime imaging microscopy. Hum. Reprod. 2022, 37, 411–427. [Google Scholar] [CrossRef] [PubMed]
- Leese, H.J. Quiet please, do not disturb: A hypothesis of embryo metabolism and viability. Bioessays 2002, 24, 845–849. [Google Scholar] [CrossRef] [PubMed]
- Baumann, C.G.; Morris, D.G.; Sreenan, J.M.; Leese, H.J. The quiet embryo hypothesis: Molecular characteristics favoring viability. Mol. Reprod. Dev. 2007, 74, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Leese, H.J. Metabolism of the preimplantation embryo: 40 years on. Reproduction 2012, 143, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Byrne, A.T.; Southgate, J.; Brison, D.R.; Leese, H.J. Analysis of apoptosis in the preimplantation bovine embryo using TUNEL. J. Reprod. Fertil. 1999, 117, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Gjorret, J.O.; Knijn, H.M.; Dieleman, S.J.; Avery, B.; Larsson, L.I.; Maddox-Hyttel, P. Chronology of apoptosis in bovine embryos produced in vivo and in vitro. Biol. Reprod. 2003, 69, 1193–1200. [Google Scholar] [CrossRef]
- Brison, D.R.; Schultz, R.M. Apoptosis during mouse blastocyst formation: Evidence for a role for survival factors including transforming growth factor alpha. Biol. Reprod. 1997, 56, 1088–1096. [Google Scholar] [CrossRef]
- Hardy, K. Cell death in the mammalian blastocyst. Mol. Hum. Reprod. 1997, 3, 919–925. [Google Scholar] [CrossRef]
- Moura, M.T.; Latorraca, L.B.; Paula-Lopes, F.F. Contextualizing Autophagy during Gametogenesis and Preimplantation Embryonic Development. Int. J. Mol. Sci. 2021, 22, 6313. [Google Scholar] [CrossRef] [PubMed]
- Escobar, M.L.; Echeverria, O.M.; Ortiz, R.; Vazquez-Nin, G.H. Combined apoptosis and autophagy, the process that eliminates the oocytes of atretic follicles in immature rats. Apoptosis 2008, 13, 1253–1266. [Google Scholar] [CrossRef] [PubMed]
- Song, W.H.; Yi, Y.J.; Sutovsky, M.; Meyers, S.; Sutovsky, P. Autophagy and ubiquitin-proteasome system contribute to sperm mitophagy after mammalian fertilization. Proc. Natl. Acad. Sci. USA 2016, 113, E5261–E5270. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, S.; Kuma, A.; Murakami, M.; Kishi, C.; Yamamoto, A.; Mizushima, N. Autophagy is essential for preimplantation development of mouse embryos. Science 2008, 321, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, S.; Kuma, A.; Mizushima, N. The role of autophagy during the oocyte-to-embryo transition. Autophagy 2008, 4, 1076–1078. [Google Scholar] [CrossRef] [PubMed]
- Al Rawi, S.; Louvet-Vallee, S.; Djeddi, A.; Sachse, M.; Culetto, E.; Hajjar, C.; Boyd, L.; Legouis, R.; Galy, V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 2011, 334, 1144–1147. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Sato, K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 2011, 334, 1141–1144. [Google Scholar] [CrossRef] [PubMed]
- Politi, Y.; Gal, L.; Kalifa, Y.; Ravid, L.; Elazar, Z.; Arama, E. Paternal mitochondrial destruction after fertilization is mediated by a common endocytic and autophagic pathway in Drosophila. Dev. Cell 2014, 29, 305–320. [Google Scholar] [CrossRef]
- Allen, E.A.; Baehrecke, E.H. Autophagy in animal development. Cell Death Differ. 2020, 27, 903–918. [Google Scholar] [CrossRef]
- Qu, X.; Zou, Z.; Sun, Q.; Luby-Phelps, K.; Cheng, P.; Hogan, R.N.; Gilpin, C.; Levine, B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 2007, 128, 931–946. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Yu, J.; Bhagat, G.; Furuya, N.; Hibshoosh, H.; Troxel, A.; Rosen, J.; Eskelinen, E.L.; Mizushima, N.; Ohsumi, Y.; et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Investig. 2003, 112, 1809–1820. [Google Scholar] [CrossRef] [PubMed]
- Yue, Z.; Jin, S.; Yang, C.; Levine, A.J.; Heintz, N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA 2003, 100, 15077–15082. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H.; et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.C.; Wang, C.; Peng, X.; Gan, B.; Guan, J.L. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J. Biol. Chem. 2010, 285, 3499–3509. [Google Scholar] [CrossRef] [PubMed]
- Le Douarin, N.M.; Dupin, E. The “beginnings” of the neural crest. Dev. Biol. 2018, 444 (Suppl. S1), S3–S13. [Google Scholar] [CrossRef] [PubMed]
- Dupin, E.; Calloni, G.W.; Coelho-Aguiar, J.M.; Le Douarin, N.M. The issue of the multipotency of the neural crest cells. Dev. Biol. 2018, 444 (Suppl. S1), S47–S59. [Google Scholar] [CrossRef] [PubMed]
- Szabo, A.; Mayor, R. Mechanisms of Neural Crest Migration. Annu. Rev. Genet. 2018, 52, 43–63. [Google Scholar] [CrossRef]
- Teddy, J.M.; Kulesa, P.M. In vivo evidence for short- and long-range cell communication in cranial neural crest cells. Development 2004, 131, 6141–6151. [Google Scholar] [CrossRef]
- Huang, G.Y.; Cooper, E.S.; Waldo, K.; Kirby, M.L.; Gilula, N.B.; Lo, C.W. Gap junction-mediated cell-cell communication modulates mouse neural crest migration. J. Cell Biol. 1998, 143, 1725–1734. [Google Scholar] [CrossRef]
- Sullivan, R.; Huang, G.Y.; Meyer, R.A.; Wessels, A.; Linask, K.K.; Lo, C.W. Heart malformations in transgenic mice exhibiting dominant negative inhibition of gap junctional communication in neural crest cells. Dev. Biol. 1998, 204, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.W.; Waldo, K.L.; Kirby, M.L. Gap junction communication and the modulation of cardiac neural crest cells. Trends Cardiovasc. Med. 1999, 9, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.W.; Cohen, M.F.; Huang, G.Y.; Lazatin, B.O.; Patel, N.; Sullivan, R.; Pauken, C.; Park, S.M. Cx43 gap junction gene expression and gap junctional communication in mouse neural crest cells. Dev. Genet. 1997, 20, 119–132. [Google Scholar] [CrossRef]
- Gustafson, C.M.; Roffers-Agarwal, J.; Gammill, L.S. Chick cranial neural crest cells release extracellular vesicles that are critical for their migration. J. Cell Sci. 2022, 135, jcs260272. [Google Scholar] [CrossRef]
- Nakagawa, S.; Takeichi, M. Neural crest emigration from the neural tube depends on regulated cadherin expression. Development 1998, 125, 2963–2971. [Google Scholar] [CrossRef]
- Xu, X.; Li, W.E.; Huang, G.Y.; Meyer, R.; Chen, T.; Luo, Y.; Thomas, M.P.; Radice, G.L.; Lo, C.W. Modulation of mouse neural crest cell motility by N-cadherin and connexin 43 gap junctions. J. Cell Biol. 2001, 154, 217–230. [Google Scholar] [CrossRef]
- Fishwick, K.J.; Neiderer, T.E.; Jhingory, S.; Bronner, M.E.; Taneyhill, L.A. The tight junction protein claudin-1 influences cranial neural crest cell emigration. Mech. Dev. 2012, 129, 275–283. [Google Scholar] [CrossRef]
- Wu, C.Y.; Jhingory, S.; Taneyhill, L.A. The tight junction scaffolding protein cingulin regulates neural crest cell migration. Dev. Dyn. 2011, 240, 2309–2323. [Google Scholar] [CrossRef]
- Vacca, B.; Sanchez-Heras, E.; Steed, E.; Busson, S.L.; Balda, M.S.; Ohnuma, S.I.; Sasai, N.; Mayor, R.; Matter, K. Control of neural crest induction by MarvelD3-mediated attenuation of JNK signalling. Sci. Rep. 2018, 8, 1204. [Google Scholar] [CrossRef]
- Smith, A.; Robinson, V.; Patel, K.; Wilkinson, D.G. The EphA4 and EphB1 receptor tyrosine kinases and ephrin-B2 ligand regulate targeted migration of branchial neural crest cells. Curr. Biol. 1997, 7, 561–570. [Google Scholar] [CrossRef]
- Bannerman, P.; Nichols, W.; Puhalla, S.; Oliver, T.; Berman, M.; Pleasure, D. Early migratory rat neural crest cells express functional gap junctions: Evidence that neural crest cell survival requires gap junction function. J. Neurosci. Res. 2000, 61, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, D.; Khan, B.; Simoes-Costa, M. Neural crest metabolism: At the crossroads of development and disease. Dev. Biol. 2021, 475, 245–255. [Google Scholar] [CrossRef]
- Wang, G.; Chen, E.N.; Liang, C.; Liang, J.; Gao, L.R.; Chuai, M.; Munsterberg, A.; Bao, Y.; Cao, L.; Yang, X. Atg7-Mediated Autophagy Is Involved in the Neural Crest Cell Generation in Chick Embryo. Mol. Neurobiol. 2018, 55, 3523–3536. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Li, S.; Wang, G.; Ma, Z.L.; Chuai, M.; Cao, L.; Yang, X. High glucose environment inhibits cranial neural crest survival by activating excessive autophagy in the chick embryo. Sci. Rep. 2015, 5, 18321. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. The intestinal crypt, a prototype stem cell compartment. Cell 2013, 154, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H.; Batlle, E. SnapShot: The intestinal crypt. Cell 2013, 152, 1198–1198.e2. [Google Scholar] [CrossRef] [PubMed]
- Maloy, K.J.; Powrie, F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 2011, 474, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Eisenhoffer, G.T.; Loftus, P.D.; Yoshigi, M.; Otsuna, H.; Chien, C.B.; Morcos, P.A.; Rosenblatt, J. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature 2012, 484, 546–549. [Google Scholar] [CrossRef]
- Patterson, A.M.; Watson, A.J.M. Deciphering the Complex Signaling Systems That Regulate Intestinal Epithelial Cell Death Processes and Shedding. Front. Immunol. 2017, 8, 841. [Google Scholar] [CrossRef]
- Ngo, P.A.; Neurath, M.F.; Lopez-Posadas, R. Impact of Epithelial Cell Shedding on Intestinal Homeostasis. Int. J. Mol. Sci. 2022, 23, 4160. [Google Scholar] [CrossRef]
- Rosenblatt, J.; Raff, M.C.; Cramer, L.P. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Curr. Biol. 2001, 11, 1847–1857. [Google Scholar] [CrossRef]
- Marchiando, A.M.; Shen, L.; Graham, W.V.; Edelblum, K.L.; Duckworth, C.A.; Guan, Y.; Montrose, M.H.; Turner, J.R.; Watson, A.J. The epithelial barrier is maintained by in vivo tight junction expansion during pathologic intestinal epithelial shedding. Gastroenterology 2011, 140, 1208–1218. [Google Scholar] [CrossRef] [PubMed]
- Zihni, C.; Mills, C.; Matter, K.; Balda, M.S. Tight junctions: From simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 2016, 17, 564–580. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.A. The role of tight junctions in cancer metastasis. Semin Cell Dev. Biol. 2014, 36, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.A.; Jiang, W.G. Loss of tight junction barrier function and its role in cancer metastasis. Biochim. Biophys. Acta 2009, 1788, 872–891. [Google Scholar] [CrossRef] [PubMed]
- Nehme, Z.; Roehlen, N.; Dhawan, P.; Baumert, T.F. Tight Junction Protein Signaling and Cancer Biology. Cells 2023, 12, 243. [Google Scholar] [CrossRef] [PubMed]
- Kyuno, D.; Takasawa, A.; Kikuchi, S.; Takemasa, I.; Osanai, M.; Kojima, T. Role of tight junctions in the epithelial-to-mesenchymal transition of cancer cells. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183503. [Google Scholar] [CrossRef] [PubMed]
- Aijaz, S.; Balda, M.S.; Matter, K. Tight junctions: Molecular architecture and function. Int. Rev. Cytol. 2006, 248, 261–298. [Google Scholar] [CrossRef]
- Cummins, P.M. Occludin: One protein, many forms. Mol. Cell Biol. 2012, 32, 242–250. [Google Scholar] [CrossRef]
- Steed, E.; Balda, M.S.; Matter, K. Dynamics and functions of tight junctions. Trends Cell Biol. 2010, 20, 142–149. [Google Scholar] [CrossRef]
- Bhat, A.A.; Uppada, S.; Achkar, I.W.; Hashem, S.; Yadav, S.K.; Shanmugakonar, M.; Al-Naemi, H.A.; Haris, M.; Uddin, S. Tight Junction Proteins and Signaling Pathways in Cancer and Inflammation: A Functional Crosstalk. Front. Physiol. 2018, 9, 1942. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.A.; Nelson, W.J.; Chavez, N. Cell-Cell Junctions Organize Structural and Signaling Networks. Cold Spring Harb. Perspect. Biol. 2018, 10, a029181. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.B.; Uppada, S.B.; Dhawan, P. Claudin proteins, outside-in signaling, and carcinogenesis. Pflug. Arch. 2017, 469, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Franke, W.W. Discovering the molecular components of intercellular junctions—A historical view. Cold Spring Harb. Perspect. Biol. 2009, 1, a003061. [Google Scholar] [CrossRef] [PubMed]
- Rubsam, M.; Broussard, J.A.; Wickstrom, S.A.; Nekrasova, O.; Green, K.J.; Niessen, C.M. Adherens Junctions and Desmosomes Coordinate Mechanics and Signaling to Orchestrate Tissue Morphogenesis and Function: An Evolutionary Perspective. Cold Spring Harb. Perspect. Biol. 2018, 10, a029207. [Google Scholar] [CrossRef] [PubMed]
- Green, K.J.; Roth-Carter, Q.; Niessen, C.M.; Nichols, S.A. Tracing the Evolutionary Origin of Desmosomes. Curr. Biol. 2020, 30, R535–R543. [Google Scholar] [CrossRef] [PubMed]
- Madara, J.L. Maintenance of the macromolecular barrier at cell extrusion sites in intestinal epithelium: Physiological rearrangement of tight junctions. J. Membr. Biol. 1990, 116, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.R. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 2009, 9, 799–809. [Google Scholar] [CrossRef]
- Cliffe, L.J.; Humphreys, N.E.; Lane, T.E.; Potten, C.S.; Booth, C.; Grencis, R.K. Accelerated intestinal epithelial cell turnover: A new mechanism of parasite expulsion. Science 2005, 308, 1463–1465. [Google Scholar] [CrossRef]
- Nenci, A.; Becker, C.; Wullaert, A.; Gareus, R.; van Loo, G.; Danese, S.; Huth, M.; Nikolaev, A.; Neufert, C.; Madison, B.; et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature 2007, 446, 557–561. [Google Scholar] [CrossRef]
- Clayburgh, D.R.; Shen, L.; Turner, J.R. A porous defense: The leaky epithelial barrier in intestinal disease. Lab. Investig. 2004, 84, 282–291. [Google Scholar] [CrossRef]
- Bruewer, M.; Luegering, A.; Kucharzik, T.; Parkos, C.A.; Madara, J.L.; Hopkins, A.M.; Nusrat, A. Proinflammatory cytokines disrupt epithelial barrier function by apoptosis-independent mechanisms. J. Immunol. 2003, 171, 6164–6172. [Google Scholar] [CrossRef]
- Gunther, C.; Neumann, H.; Neurath, M.F.; Becker, C. Apoptosis, necrosis and necroptosis: Cell death regulation in the intestinal epithelium. Gut 2013, 62, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Patankar, J.V.; Becker, C. Cell death in the gut epithelium and implications for chronic inflammation. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 543–556. [Google Scholar] [CrossRef]
- Ayyar, K.K.; Moss, A.C. Exosomes in Intestinal Inflammation. Front. Pharmacol. 2021, 12, 658505. [Google Scholar] [CrossRef]
- van Niel, G.; Raposo, G.; Candalh, C.; Boussac, M.; Hershberg, R.; Cerf-Bensussan, N.; Heyman, M. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology 2001, 121, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Mallegol, J.; Van Niel, G.; Lebreton, C.; Lepelletier, Y.; Candalh, C.; Dugave, C.; Heath, J.K.; Raposo, G.; Cerf-Bensussan, N.; Heyman, M. T84-intestinal epithelial exosomes bear MHC class II/peptide complexes potentiating antigen presentation by dendritic cells. Gastroenterology 2007, 132, 1866–1876. [Google Scholar] [CrossRef] [PubMed]
- Canbay, A.; Feldstein, A.E.; Higuchi, H.; Werneburg, N.; Grambihler, A.; Bronk, S.F.; Gores, G.J. Kupffer cell engulfment of apoptotic bodies stimulates death ligand and cytokine expression. Hepatology 2003, 38, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhao, Y.; Shi, L.; Li, W.; Chen, K.; Li, M.; Chen, X.; Zhang, H.; Li, T.; Matsuzawa-Ishimoto, Y.; et al. Gut epithelial TSC1/mTOR controls RIPK3-dependent necroptosis in intestinal inflammation and cancer. J. Clin. Investig. 2020, 130, 2111–2128. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, S.; Jin, W.; Zhou, H.; Zhong, T.; Cheng, X.; Fu, Y.; Xiao, P.; Cheng, H.; Wang, D.; et al. Epithelial Gab1 calibrates RIPK3-dependent necroptosis to prevent intestinal inflammation. JCI Insight 2023, 8, e162701. [Google Scholar] [CrossRef] [PubMed]
- Churchill, M.J.; Mitchell, P.S.; Rauch, I. Epithelial Pyroptosis in Host Defense. J. Mol. Biol. 2022, 434, 167278. [Google Scholar] [CrossRef]
- Rauch, I.; Deets, K.A.; Ji, D.X.; von Moltke, J.; Tenthorey, J.L.; Lee, A.Y.; Philip, N.H.; Ayres, J.S.; Brodsky, I.E.; Gronert, K.; et al. NAIP-NLRC4 Inflammasomes Coordinate Intestinal Epithelial Cell Expulsion with Eicosanoid and IL-18 Release via Activation of Caspase-1 and -8. Immunity 2017, 46, 649–659. [Google Scholar] [CrossRef] [PubMed]
- He, W.T.; Wan, H.; Hu, L.; Chen, P.; Wang, X.; Huang, Z.; Yang, Z.H.; Zhong, C.Q.; Han, J. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res. 2015, 25, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.H.; Lin, H.Y.; Kuo, C.C.; Yang, L.T. N-terminal functional domain of Gasdermin A3 regulates mitochondrial homeostasis via mitochondrial targeting. J. Biomed. Sci. 2015, 22, 44. [Google Scholar] [CrossRef]
- Shi, P.; Tang, A.; Xian, L.; Hou, S.; Zou, D.; Lv, Y.; Huang, Z.; Wang, Q.; Song, A.; Lin, Z.; et al. Loss of conserved Gsdma3 self-regulation causes autophagy and cell death. Biochem. J. 2015, 468, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Tamura, M.; Shiroishi, T. GSDM family genes meet autophagy. Biochem. J. 2015, 469, e5–e7. [Google Scholar] [CrossRef]
- Privitera, G.; Rana, N.; Armuzzi, A.; Pizarro, T.T. The gasdermin protein family: Emerging roles in gastrointestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 366–387. [Google Scholar] [CrossRef] [PubMed]
- Randall-Demllo, S.; Chieppa, M.; Eri, R. Intestinal epithelium and autophagy: Partners in gut homeostasis. Front. Immunol. 2013, 4, 301. [Google Scholar] [CrossRef]
- Benjamin, J.L.; Sumpter, R., Jr.; Levine, B.; Hooper, L.V. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe 2013, 13, 723–734. [Google Scholar] [CrossRef]
- Telpaz, S.; Bel, S. Autophagy in intestinal epithelial cells prevents gut inflammation. Trends Cell Biol. 2023, 33, 817–819. [Google Scholar] [CrossRef]
- Chen, S.L.; Li, C.M.; Li, W.; Liu, Q.S.; Hu, S.Y.; Zhao, M.Y.; Hu, D.S.; Hao, Y.W.; Zeng, J.H.; Zhang, Y. How autophagy, a potential therapeutic target, regulates intestinal inflammation. Front. Immunol. 2023, 14, 1087677. [Google Scholar] [CrossRef] [PubMed]
- Larabi, A.; Barnich, N.; Nguyen, H.T.T. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy 2020, 16, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Solanas, G.; Batlle, E. Control of cell adhesion and compartmentalization in the intestinal epithelium. Exp. Cell Res. 2011, 317, 2695–2701. [Google Scholar] [CrossRef] [PubMed]
- Holmberg, J.; Genander, M.; Halford, M.M.; Anneren, C.; Sondell, M.; Chumley, M.J.; Silvany, R.E.; Henkemeyer, M.; Frisen, J. EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell 2006, 125, 1151–1163. [Google Scholar] [CrossRef] [PubMed]
- Anderton, M.; van der Meulen, E.; Blumenthal, M.J.; Schafer, G. The Role of the Eph Receptor Family in Tumorigenesis. Cancers 2021, 13, 206. [Google Scholar] [CrossRef] [PubMed]
- Janes, P.W.; Vail, M.E.; Ernst, M.; Scott, A.M. Eph Receptors in the Immunosuppressive Tumor Microenvironment. Cancer Res. 2021, 81, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Tu, P.; Chen, X.; Guo, S.; Wang, J. Eph Receptors: Actors in Tumor Microenvironment. Crit. Rev. Oncog. 2017, 22, 499–505. [Google Scholar] [CrossRef]
- Cowan, C.A.; Henkemeyer, M. Ephrins in reverse, park and drive. Trends Cell. Biol. 2002, 12, 339–346. [Google Scholar] [CrossRef]
- Merlos-Suarez, A.; Batlle, E. Eph-ephrin signalling in adult tissues and cancer. Curr. Opin. Cell Biol. 2008, 20, 194–200. [Google Scholar] [CrossRef]
- Genander, M.; Frisen, J. Eph receptors tangled up in two: Independent control of cell positioning and proliferation. Cell Cycle 2010, 9, 1865–1866. [Google Scholar] [CrossRef]
- Amessou, M.; Kandouz, M. Role of the Family of Ephs and Ephrins in Cell-Cell Communication in Cancer. In Intercellular Communication in Cancer; Kandouz, M., Ed.; Springer: Dordrecht, The Netherlands, 2015; pp. 255–286. [Google Scholar]
- Kandouz, M. The Eph/Ephrin family in cancer metastasis: Communication at the service of invasion. Cancer Metastasis Rev. 2012, 31, 353–373. [Google Scholar] [CrossRef]
- Pasquale, E.B. Eph receptors and ephrins in cancer progression. Nat. Rev. Cancer 2024, 24, 5–27. [Google Scholar] [CrossRef] [PubMed]
- Batlle, E.; Bacani, J.; Begthel, H.; Jonkheer, S.; Gregorieff, A.; van de Born, M.; Malats, N.; Sancho, E.; Boon, E.; Pawson, T.; et al. EphB receptor activity suppresses colorectal cancer progression. Nature 2005, 435, 1126–1130. [Google Scholar] [CrossRef]
- Batlle, E.; Henderson, J.T.; Beghtel, H.; van den Born, M.M.; Sancho, E.; Huls, G.; Meeldijk, J.; Robertson, J.; van de Wetering, M.; Pawson, T.; et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 2002, 111, 251–263. [Google Scholar] [CrossRef] [PubMed]
- van de Wetering, M.; Sancho, E.; Verweij, C.; de Lau, W.; Oving, I.; Hurlstone, A.; van der Horn, K.; Batlle, E.; Coudreuse, D.; Haramis, A.P.; et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 2002, 111, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Moser, A.R.; Pitot, H.C.; Dove, W.F. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 1990, 247, 322–324. [Google Scholar] [CrossRef] [PubMed]
- Jubb, A.M.; Zhong, F.; Bheddah, S.; Grabsch, H.I.; Frantz, G.D.; Mueller, W.; Kavi, V.; Quirke, P.; Polakis, P.; Koeppen, H. EphB2 is a prognostic factor in colorectal cancer. Clin. Cancer Res. 2005, 11, 5181–5187. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.L.; Zhang, J.; Yuen, S.T.; Tsui, W.Y.; Chan, A.S.; Ho, C.; Ji, J.; Leung, S.Y.; Chen, X. Reduced expression of EphB2 that parallels invasion and metastasis in colorectal tumours. Carcinogenesis 2006, 27, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Lugli, A.; Spichtin, H.; Maurer, R.; Mirlacher, M.; Kiefer, J.; Huusko, P.; Azorsa, D.; Terracciano, L.; Sauter, G.; Kallioniemi, O.P.; et al. EphB2 expression across 138 human tumor types in a tissue microarray: High levels of expression in gastrointestinal cancers. Clin. Cancer Res. 2005, 11, 6450–6458. [Google Scholar] [CrossRef]
- Cortina, C.; Palomo-Ponce, S.; Iglesias, M.; Fernandez-Masip, J.L.; Vivancos, A.; Whissell, G.; Huma, M.; Peiro, N.; Gallego, L.; Jonkheer, S.; et al. EphB-ephrin-B interactions suppress colorectal cancer progression by compartmentalizing tumor cells. Nat. Genet. 2007, 39, 1376–1383. [Google Scholar] [CrossRef]
- Chiu, S.T.; Chang, K.J.; Ting, C.H.; Shen, H.C.; Li, H.; Hsieh, F.J. Over-expression of EphB3 enhances cell-cell contacts and suppresses tumor growth in HT-29 human colon cancer cells. Carcinogenesis 2009, 30, 1475–1486. [Google Scholar] [CrossRef]
- Foerster, E.G.; Mukherjee, T.; Cabral-Fernandes, L.; Rocha, J.D.B.; Girardin, S.E.; Philpott, D.J. How autophagy controls the intestinal epithelial barrier. Autophagy 2022, 18, 86–103. [Google Scholar] [CrossRef]
- Chukkapalli, S.; Amessou, M.; Dilly, A.K.; Dekhil, H.; Zhao, J.; Liu, Q.; Bejna, A.; Thomas, R.D.; Bandyopadhyay, S.; Bismar, T.A.; et al. Role of the EphB2 receptor in autophagy, apoptosis and invasion in human breast cancer cells. Exp. Cell Res. 2014, 320, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Kandouz, M.; Haidara, K.; Zhao, J.; Brisson, M.L.; Batist, G. The EphB2 tumor suppressor induces autophagic cell death via concomitant activation of the ERK1/2 and PI3K pathways. Cell Cycle 2010, 9, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cui, Z.; Cheng, D.; Du, Y.; Guo, X.; Gao, R.; Chen, J.; Sun, W.; He, R.; Ma, X.; et al. RNF186 regulates EFNB1 (ephrin B1)-EPHB2-induced autophagy in the colonic epithelial cells for the maintenance of intestinal homeostasis. Autophagy 2021, 17, 3030–3047. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, H.; Kuribayashi, K.; Tsuji, N.; Tanaka, M.; Kobayashi, D.; Watanabe, N. Sesamin induces autophagy in colon cancer cells by reducing tyrosine phosphorylation of EphA1 and EphB2. Int. J. Oncol. 2011, 39, 33–40. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qiu, P.; Li, D.; Xiao, C.; Xu, F.; Chen, X.; Chang, Y.; Liu, L.; Zhang, L.; Zhao, Q.; Chen, Y. The Eph/ephrin system symphony of gut inflammation. Pharmacol. Res. 2023, 197, 106976. [Google Scholar] [CrossRef]
- Darling, T.K.; Lamb, T.J. Emerging Roles for Eph Receptors and Ephrin Ligands in Immunity. Front. Immunol. 2019, 10, 1473. [Google Scholar] [CrossRef] [PubMed]
- Kuijper, S.; Turner, C.J.; Adams, R.H. Regulation of angiogenesis by Eph-ephrin interactions. Trends Cardiovasc. Med. 2007, 17, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Shiuan, E.; Chen, J. Eph Receptor Tyrosine Kinases in Tumor Immunity. Cancer Res. 2016, 76, 6452–6457. [Google Scholar] [CrossRef]
- El Khoury, F.; Corcos, L.; Durand, S.; Simon, B.; Le Jossic-Corcos, C. Acquisition of anticancer drug resistance is partially associated with cancer stemness in human colon cancer cells. Int. J. Oncol. 2016, 49, 2558–2568. [Google Scholar] [CrossRef]
- Park, S.H.; Jo, M.J.; Kim, B.R.; Jeong, Y.A.; Na, Y.J.; Kim, J.L.; Jeong, S.; Yun, H.K.; Kim, D.Y.; Kim, B.G.; et al. Sonic hedgehog pathway activation is associated with cetuximab resistance and EPHB3 receptor induction in colorectal cancer. Theranostics 2019, 9, 2235–2251. [Google Scholar] [CrossRef]
- Primeaux, M.; Liu, X.; Gowrikumar, S.; Fatima, I.; Fisher, K.W.; Bastola, D.; Vecchio, A.J.; Singh, A.B.; Dhawan, P. Claudin-1 interacts with EPHA2 to promote cancer stemness and chemoresistance in colorectal cancer. Cancer Lett. 2023, 579, 216479. [Google Scholar] [CrossRef] [PubMed]
- DiPrima, M.; Wang, D.; Troster, A.; Maric, D.; Terrades-Garcia, N.; Ha, T.; Kwak, H.; Sanchez-Martin, D.; Kudlinzki, D.; Schwalbe, H.; et al. Identification of Eph receptor signaling as a regulator of autophagy and a therapeutic target in colorectal carcinoma. Mol. Oncol. 2019, 13, 2441–2459. [Google Scholar] [CrossRef] [PubMed]
- Troster, A.; DiPrima, M.; Jores, N.; Kudlinzki, D.; Sreeramulu, S.; Gande, S.L.; Linhard, V.; Ludig, D.; Schug, A.; Saxena, K.; et al. Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor. Chemistry 2023, 29, e202203967. [Google Scholar] [CrossRef] [PubMed]
- Troster, A.; Jores, N.; Mineev, K.S.; Sreeramulu, S.; DiPrima, M.; Tosato, G.; Schwalbe, H. Targeting EPHA2 with Kinase Inhibitors in Colorectal Cancer. ChemMedChem 2023, 18, e202300420. [Google Scholar] [CrossRef]
- Alam, S.K.; Yadav, V.K.; Bajaj, S.; Datta, A.; Dutta, S.K.; Bhattacharyya, M.; Bhattacharya, S.; Debnath, S.; Roy, S.; Boardman, L.A.; et al. DNA damage-induced ephrin-B2 reverse signaling promotes chemoresistance and drives EMT in colorectal carcinoma harboring mutant p53. Cell Death Differ. 2016, 23, 707–722. [Google Scholar] [CrossRef] [PubMed]
- Cioce, M.; Fazio, V.M. EphA2 and EGFR: Friends in Life, Partners in Crime. Can EphA2 Be a Predictive Biomarker of Response to Anti-EGFR Agents? Cancers 2021, 13, 700. [Google Scholar] [CrossRef]
- Strimpakos, A.; Pentheroudakis, G.; Kotoula, V.; De Roock, W.; Kouvatseas, G.; Papakostas, P.; Makatsoris, T.; Papamichael, D.; Andreadou, A.; Sgouros, J.; et al. The prognostic role of ephrin A2 and endothelial growth factor receptor pathway mediators in patients with advanced colorectal cancer treated with cetuximab. Clin. Colorectal. Cancer 2013, 12, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Cuyas, E.; Queralt, B.; Martin-Castillo, B.; Bosch-Barrera, J.; Menendez, J.A. EphA2 receptor activation with ephrin-A1 ligand restores cetuximab efficacy in NRAS-mutant colorectal cancer cells. Oncol. Rep. 2017, 38, 263–270. [Google Scholar] [CrossRef]
- Lau, A.; Le, N.; Nguyen, C.; Kandpal, R.P. Signals transduced by Eph receptors and ephrin ligands converge on MAP kinase and AKT pathways in human cancers. Cell. Signal. 2023, 104, 110579. [Google Scholar] [CrossRef] [PubMed]
- Martini, G.; Cardone, C.; Vitiello, P.P.; Belli, V.; Napolitano, S.; Troiani, T.; Ciardiello, D.; Della Corte, C.M.; Morgillo, F.; Matrone, N.; et al. EPHA2 Is a Predictive Biomarker of Resistance and a Potential Therapeutic Target for Improving Antiepidermal Growth Factor Receptor Therapy in Colorectal Cancer. Mol. Cancer Ther. 2019, 18, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.; Sharma, J.; Bukkapatnam, S.; Oweida, A.; Lennon, S.; Phan, A.; Milner, D.; Uyanga, N.; Jimeno, A.; Raben, D.; et al. Inhibition of EphB4-Ephrin-B2 Signaling Enhances Response to Cetuximab-Radiation Therapy in Head and Neck Cancers. Clin. Cancer Res. 2018, 24, 4539–4550. [Google Scholar] [CrossRef] [PubMed]
- Stahl, S.; Kaminskyy, V.O.; Efazat, G.; Hyrslova Vaculova, A.; Rodriguez-Nieto, S.; Moshfegh, A.; Lewensohn, R.; Viktorsson, K.; Zhivotovsky, B. Inhibition of Ephrin B3-mediated survival signaling contributes to increased cell death response of non-small cell lung carcinoma cells after combined treatment with ionizing radiation and PKC 412. Cell Death Dis. 2013, 4, e454. [Google Scholar] [CrossRef] [PubMed]
- Beekhof, R.; Bertotti, A.; Bottger, F.; Vurchio, V.; Cottino, F.; Zanella, E.R.; Migliardi, G.; Viviani, M.; Grassi, E.; Lupo, B.; et al. Phosphoproteomics of patient-derived xenografts identifies targets and markers associated with sensitivity and resistance to EGFR blockade in colorectal cancer. Sci. Transl. Med. 2023, 15, eabm3687. [Google Scholar] [CrossRef]
- Yang, S.Y.; Sales, K.M.; Fuller, B.; Seifalian, A.M.; Winslet, M.C. Apoptosis and colorectal cancer: Implications for therapy. Trends Mol. Med. 2009, 15, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.J. Apoptosis and colorectal cancer. Gut 2004, 53, 1701–1709. [Google Scholar] [CrossRef] [PubMed]
- Sinicrope, F.A.; Roddey, G.; McDonnell, T.J.; Shen, Y.; Cleary, K.R.; Stephens, L.C. Increased apoptosis accompanies neoplastic development in the human colorectum. Clin. Cancer Res. 1996, 2, 1999–2006. [Google Scholar]
- Manzoor, S.; Muhammad, J.S.; Maghazachi, A.A.; Hamid, Q. Autophagy: A Versatile Player in the Progression of Colorectal Cancer and Drug Resistance. Front. Oncol. 2022, 12, 924290. [Google Scholar] [CrossRef]
- Xie, Q.; Liu, Y.; Li, X. The interaction mechanism between autophagy and apoptosis in colon cancer. Transl. Oncol. 2020, 13, 100871. [Google Scholar] [CrossRef]
- de la Cruz-Morcillo, M.A.; Valero, M.L.; Callejas-Valera, J.L.; Arias-Gonzalez, L.; Melgar-Rojas, P.; Galan-Moya, E.M.; Garcia-Gil, E.; Garcia-Cano, J.; Sanchez-Prieto, R. P38MAPK is a major determinant of the balance between apoptosis and autophagy triggered by 5-fluorouracil: Implication in resistance. Oncogene 2012, 31, 1073–1085. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, S.; Wang, D.; Song, D.; Feng, Y. Colorectal cancer cells are resistant to anti-EGFR monoclonal antibody through adapted autophagy. Am. J. Transl. Res. 2016, 8, 1190–1196. [Google Scholar] [PubMed]
- Koustas, E.; Karamouzis, M.V.; Mihailidou, C.; Schizas, D.; Papavassiliou, A.G. Co-targeting of EGFR and autophagy signaling is an emerging treatment strategy in metastatic colorectal cancer. Cancer Lett. 2017, 396, 94–102. [Google Scholar] [CrossRef]
- Guo, Y.; Shi, W.; Fang, R. miR-18a-5p promotes melanoma cell proliferation and inhibits apoptosis and autophagy by targeting EPHA7 signaling. Mol. Med. Rep. 2021, 23. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Talty, R.; Aladelokun, O.; Bosenberg, M.; Johnson, C.H. Ferroptosis in colorectal cancer: A future target? Br. J. Cancer 2023, 128, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; He, X.; Tong, Y.; Bai, N.; Pu, Y.; Han, K.; Wang, Y. Ferroptosis open a new door for colorectal cancer treatment. Front. Oncol. 2023, 13, 1059520. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Z.; Sun, W.; Zhang, J.; Xu, Q.; Zhou, X.; Mao, L. Ferroptosis in colorectal cancer: Potential mechanisms and effective therapeutic targets. Biomed. Pharmacother. 2022, 153, 113524. [Google Scholar] [CrossRef]
- Chaudhary, N.; Choudhary, B.S.; Shah, S.G.; Khapare, N.; Dwivedi, N.; Gaikwad, A.; Joshi, N.; Raichanna, J.; Basu, S.; Gurjar, M.; et al. Lipocalin 2 expression promotes tumor progression and therapy resistance by inhibiting ferroptosis in colorectal cancer. Int. J. Cancer 2021, 149, 1495–1511. [Google Scholar] [CrossRef]
- Lin, Y.P.; Wu, J.I.; Tseng, C.W.; Chen, H.J.; Wang, L.H. Gjb4 serves as a novel biomarker for lung cancer and promotes metastasis and chemoresistance via Src activation. Oncogene 2019, 38, 822–837. [Google Scholar] [CrossRef]
- Yang, J.; Qin, G.; Luo, M.; Chen, J.; Zhang, Q.; Li, L.; Pan, L.; Qin, S. Reciprocal positive regulation between Cx26 and PI3K/Akt pathway confers acquired gefitinib resistance in NSCLC cells via GJIC-independent induction of EMT. Cell Death Dis. 2015, 6, e1829. [Google Scholar] [CrossRef]
- Gielen, P.R.; Aftab, Q.; Ma, N.; Chen, V.C.; Hong, X.; Lozinsky, S.; Naus, C.C.; Sin, W.C. Connexin43 confers Temozolomide resistance in human glioma cells by modulating the mitochondrial apoptosis pathway. Neuropharmacology 2013, 75, 539–548. [Google Scholar] [CrossRef]
- Ni, C.; Lou, X.; Yao, X.; Wang, L.; Wan, J.; Duan, X.; Liang, J.; Zhang, K.; Yang, Y.; Zhang, L.; et al. ZIP1(+) fibroblasts protect lung cancer against chemotherapy via connexin-43 mediated intercellular Zn(2+) transfer. Nat. Commun. 2022, 13, 5919. [Google Scholar] [CrossRef]
- Arora, S.; Heyza, J.R.; Chalfin, E.C.; Ruch, R.J.; Patrick, S.M. Gap Junction Intercellular Communication Positively Regulates Cisplatin Toxicity by Inducing DNA Damage through Bystander Signaling. Cancers 2018, 10, 368. [Google Scholar] [CrossRef]
- Blondy, S.; David, V.; Verdier, M.; Mathonnet, M.; Perraud, A.; Christou, N. 5-Fluorouracil resistance mechanisms in colorectal cancer: From classical pathways to promising processes. Cancer Sci. 2020, 111, 3142–3154. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, H.; Chen, H.; Tan, T.; Wang, Y.; Yang, H.; Ding, Y.; Wang, S. CX43 down-regulation promotes cell aggressiveness and 5-fluorouracil-resistance by attenuating cell stiffness in colorectal carcinoma. Cancer Biol. Ther. 2023, 24, 2221879. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.W.; Chen, H.J.; Yu, J.L.; Huang, Z.H.; Fang, S.; Lin, X.H. Gap junction composed of connexin43 modulates 5-fluorouracil, oxaliplatin and irinotecan resistance on colorectal cancers. Mol. Med. Rep. 2016, 14, 4893–4900. [Google Scholar] [CrossRef] [PubMed]
- Buhrmann, C.; Shayan, P.; Kraehe, P.; Popper, B.; Goel, A.; Shakibaei, M. Resveratrol induces chemosensitization to 5-fluorouracil through up-regulation of intercellular junctions, Epithelial-to-mesenchymal transition and apoptosis in colorectal cancer. Biochem. Pharmacol. 2015, 98, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Zheng, X.; Wu, X.; Wang, S.; Wang, Y.; Xing, F. All-trans retinoic acid reverses epithelial-mesenchymal transition in paclitaxel-resistant cells by inhibiting nuclear factor kappa B and upregulating gap junctions. Cancer Sci. 2019, 110, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.F.; Varghese, R.T.; Lamouille, S.; Guo, S.; Pridham, K.J.; Kanabur, P.; Osimani, A.M.; Sharma, S.; Jourdan, J.; Rodgers, C.M.; et al. Connexin 43 Inhibition Sensitizes Chemoresistant Glioblastoma Cells to Temozolomide. Cancer Res. 2016, 76, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Munoz, J.L.; Rodriguez-Cruz, V.; Greco, S.J.; Ramkissoon, S.H.; Ligon, K.L.; Rameshwar, P. Temozolomide resistance in glioblastoma cells occurs partly through epidermal growth factor receptor-mediated induction of connexin 43. Cell Death Dis. 2014, 5, e1145. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wang, D.; Du, X.; He, Y.; Chen, S.; Shao, Q.; Ma, C.; Huang, B.; Chen, A.; Zhao, P.; et al. Glioma cells escaped from cytotoxicity of temozolomide and vincristine by communicating with human astrocytes. Med. Oncol. 2015, 32, 43. [Google Scholar] [CrossRef] [PubMed]
- Grek, C.L.; Sheng, Z.; Naus, C.C.; Sin, W.C.; Gourdie, R.G.; Ghatnekar, G.G. Novel approach to temozolomide resistance in malignant glioma: Connexin43-directed therapeutics. Curr. Opin. Pharmacol. 2018, 41, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Iyyathurai, J.; Decuypere, J.P.; Leybaert, L.; D’Hondt, C.; Bultynck, G. Connexins: Substrates and regulators of autophagy. BMC Cell Biol. 2016, 17 (Suppl. S1), 20. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.X.; Tao, L.; Lai, Y.C.; Cai, S.Y.; Zhao, Z.Y.; Yang, F.; Su, R.Y.; Wang, Q. Cx32 promotes autophagy and produces resistance to SN-induced apoptosis via activation of AMPK signalling in cervical cancer. Int. J. Oncol. 2022, 60, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Lai, Y.; Ge, H.; Guo, Y.; Feng, X.; Song, J.; Wang, Q.; Fan, L.; Peng, Y.; Cao, M.; et al. Non-junctional Cx32 mediates anti-apoptotic and pro-tumor effects via epidermal growth factor receptor in human cervical cancer cells. Cell Death Dis. 2017, 8, e2773. [Google Scholar] [CrossRef] [PubMed]
- Lampe, P.D.; Laird, D.W. Recent advances in connexin gap junction biology. Fac. Rev. 2022, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Loewenstein, W.R. Junctional intercellular communication: The cell-to-cell membrane channel. Physiol. Rev. 1981, 61, 829–913. [Google Scholar] [CrossRef]
- Goldberg, G.S.; Valiunas, V.; Brink, P.R. Selective permeability of gap junction channels. Biochim. Biophys. Acta 2004, 1662, 96–101. [Google Scholar] [CrossRef]
- Wei, C.J.; Xu, X.; Lo, C.W. Connexins and cell signaling in development and disease. Annu. Rev. Cell Dev. Biol. 2004, 20, 811–838. [Google Scholar] [CrossRef]
- Evans, W.H.; Martin, P.E. Gap junctions: Structure and function (Review). Mol. Membr. Biol. 2002, 19, 121–136. [Google Scholar] [CrossRef]
- Spray, D.C.; White, R.L.; Mazet, F.; Bennett, M.V. Regulation of gap junctional conductance. Am. J. Physiol. 1985, 248 Pt 2, H753–H764. [Google Scholar] [CrossRef]
- Luo, C.; Yuan, D.; Yao, W.; Cai, J.; Zhou, S.; Zhang, Y.; Hei, Z. Dexmedetomidine protects against apoptosis induced by hypoxia/reoxygenation through the inhibition of gap junctions in NRK-52E cells. Life Sci. 2015, 122, 72–77. [Google Scholar] [CrossRef] [PubMed]
- de Pina-Benabou, M.H.; Szostak, V.; Kyrozis, A.; Rempe, D.; Uziel, D.; Urban-Maldonado, M.; Benabou, S.; Spray, D.C.; Federoff, H.J.; Stanton, P.K.; et al. Blockade of gap junctions in vivo provides neuroprotection after perinatal global ischemia. Stroke 2005, 36, 2232–2237. [Google Scholar] [CrossRef] [PubMed]
- de Rivero Vaccari, J.C.; Corriveau, R.A.; Belousov, A.B. Gap junctions are required for NMDA receptor dependent cell death in developing neurons. J. Neurophysiol. 2007, 98, 2878–2886. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, P.M.; Ryu, S.Y.; Pruzansky, D.P.; Kuriakose, M.; Gilmore, A.; Kinnally, K.W. Mitochondrial apoptosis is amplified through gap junctions. Biochem. Biophys. Res. Commun. 2009, 390, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Saito, C.; Shinzawa, K.; Tsujimoto, Y. Synchronized necrotic death of attached hepatocytes mediated via gap junctions. Sci. Rep. 2014, 4, 5169. [Google Scholar] [CrossRef] [PubMed]
- Satomi, Y.; Nishino, H.; Shibata, S. Glycyrrhetinic acid and related compounds induce G1 arrest and apoptosis in human hepatocellular carcinoma HepG2. Anticancer Res. 2005, 25, 4043–4047. [Google Scholar] [PubMed]
- Yu, J.; Berga, S.L.; Zou, W.; Sun, H.Y.; Johnston-MacAnanny, E.; Yalcinkaya, T.; Sidell, N.; Bagchi, I.C.; Bagchi, M.K.; Taylor, R.N. Gap junction blockade induces apoptosis in human endometrial stromal cells. Mol. Reprod. Dev. 2014, 81, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.J.; Cui, G.Q.; Zhang, J.; Liu, X.M.; Zhang, Z.H.; Jia, Q.; Ng, J.C.; Peng, C.; Bo, C.X.; Shao, H. Inhibition of gap junction intercellular communication is involved in silica nanoparticles-induced H9c2 cardiomyocytes apoptosis via the mitochondrial pathway. Int. J. Nanomed. 2017, 12, 2179–2188. [Google Scholar] [CrossRef]
- Li, C.; Shi, L.; Peng, C.; Yu, G.; Zhang, Y.; Du, Z. Lead-induced cardiomyocytes apoptosis by inhibiting gap junction intercellular communication via autophagy activation. Chem. Biol. Interact. 2021, 337, 109331. [Google Scholar] [CrossRef]
- Wang, Q.; Ma, Y.; Li, Y.; He, Z.; Feng, B. Lead-induced cardiomyocytes apoptosis by inhibiting gap junction intercellular communication via modulating the PKCalpha/Cx43 signaling pathway. Chem. Biol. Interact. 2023, 376, 110451. [Google Scholar] [CrossRef] [PubMed]
- Kandouz, M.; Batist, G. Gap junctions and connexins as therapeutic targets in cancer. Expert Opin. Ther. Targets 2010, 14, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Pitts, J.D. Cancer gene therapy: A bystander effect using the gap junctional pathway. Mol. Carcinog. 1994, 11, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Culver, K.W.; Ram, Z.; Wallbridge, S.; Ishii, H.; Oldfield, E.H.; Blaese, R.M. In vivo gene transfer with retroviral vector-producer cells for treatment of experimental brain tumors. Science 1992, 256, 1550–1552. [Google Scholar] [CrossRef] [PubMed]
- Takamiya, Y.; Short, M.P.; Ezzeddine, Z.D.; Moolten, F.L.; Breakefield, X.O.; Martuza, R.L. Gene therapy of malignant brain tumors: A rat glioma line bearing the herpes simplex virus type 1-thymidine kinase gene and wild type retrovirus kills other tumor cells. J. Neurosci. Res. 1992, 33, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Yahyapour, R.; Motevaseli, E.; Rezaeyan, A.; Abdollahi, H.; Farhood, B.; Cheki, M.; Najafi, M.; Villa, V. Mechanisms of Radiation Bystander and Non-Targeted Effects: Implications to Radiation Carcinogenesis and Radiotherapy. Curr. Radiopharm. 2018, 11, 34–45. [Google Scholar] [CrossRef]
- Mukherjee, S.; Chakraborty, A. Radiation-induced bystander phenomenon: Insight and implications in radiotherapy. Int. J. Radiat. Biol. 2019, 95, 243–263. [Google Scholar] [CrossRef]
- Prise, K.M.; O’Sullivan, J.M. Radiation-induced bystander signalling in cancer therapy. Nat. Rev. Cancer 2009, 9, 351–360. [Google Scholar] [CrossRef]
- Azzam, E.I.; de Toledo, S.M.; Little, J.B. Direct evidence for the participation of gap junction-mediated intercellular communication in the transmission of damage signals from alpha -particle irradiated to nonirradiated cells. Proc. Natl. Acad. Sci. USA 2001, 98, 473–478. [Google Scholar] [CrossRef]
- Reid, R.; Mar, E.C.; Huang, E.S.; Topal, M.D. Insertion and extension of acyclic, dideoxy, and ara nucleotides by herpesviridae, human alpha and human beta polymerases. A unique inhibition mechanism for 9-(1,3-dihydroxy-2-propoxymethyl)guanine triphosphate. J. Biol. Chem. 1988, 263, 3898–3904. [Google Scholar] [CrossRef]
- Beltinger, C.; Fulda, S.; Kammertoens, T.; Meyer, E.; Uckert, W.; Debatin, K.M. Herpes simplex virus thymidine kinase/ganciclovir-induced apoptosis involves ligand-independent death receptor aggregation and activation of caspases. Proc. Natl. Acad. Sci. USA 1999, 96, 8699–8704. [Google Scholar] [CrossRef] [PubMed]
- Morgan, W.F.; Hartmann, A.; Limoli, C.L.; Nagar, S.; Ponnaiya, B. Bystander effects in radiation-induced genomic instability. Mutat. Res. 2002, 504, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Klammer, H.; Mladenov, E.; Li, F.; Iliakis, G. Bystander effects as manifestation of intercellular communication of DNA damage and of the cellular oxidative status. Cancer Lett. 2015, 356, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Jalal, N.; Haq, S.; Anwar, N.; Nazeer, S.; Saeed, U. Radiation induced bystander effect and DNA damage. J. Cancer Res. Ther. 2014, 10, 819–833. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Cai, L.; He, X.; Niu, Z.; Huang, H.; Hu, W.; Bian, H.; Huang, H. Radiation-induced bystander effect and its clinical implications. Front. Oncol. 2023, 13, 1124412. [Google Scholar] [CrossRef] [PubMed]
- Tartier, L.; Gilchrist, S.; Burdak-Rothkamm, S.; Folkard, M.; Prise, K.M. Cytoplasmic irradiation induces mitochondrial-dependent 53BP1 protein relocalization in irradiated and bystander cells. Cancer Res. 2007, 67, 5872–5879. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.B.; Elledge, S.J. The DNA damage response: Putting checkpoints in perspective. Nature 2000, 408, 433–439. [Google Scholar] [CrossRef]
- Batchelor, E.; Mock, C.S.; Bhan, I.; Loewer, A.; Lahav, G. Recurrent initiation: A mechanism for triggering p53 pulses in response to DNA damage. Mol. Cell 2008, 30, 277–289. [Google Scholar] [CrossRef]
- Allocati, N.; Di Ilio, C.; De Laurenzi, V. p63/p73 in the control of cell cycle and cell death. Exp. Cell Res. 2012, 318, 1285–1290. [Google Scholar] [CrossRef]
- Matt, S.; Hofmann, T.G. The DNA damage-induced cell death response: A roadmap to kill cancer cells. Cell Mol. Life Sci. 2016, 73, 2829–2850. [Google Scholar] [CrossRef]
- Shao, C.; Folkard, M.; Michael, B.D.; Prise, K.M. Targeted cytoplasmic irradiation induces bystander responses. Proc. Natl. Acad. Sci. USA 2004, 101, 13495–13500. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.J.; Randers-Pehrson, G.; Xu, A.; Waldren, C.A.; Geard, C.R.; Yu, Z.; Hei, T.K. Targeted cytoplasmic irradiation with alpha particles induces mutations in mammalian cells. Proc. Natl. Acad. Sci. USA 1999, 96, 4959–4964. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, P.K.; Goodwin, E.H.; Lehnert, B.E. Alpha particles initiate biological production of superoxide anions and hydrogen peroxide in human cells. Cancer Res. 1997, 57, 3963–3971. [Google Scholar] [PubMed]
- Nakagami, Y.; Ito, M.; Hara, T.; Inoue, T.; Matsubara, S. Nuclear translocation of DNase II and acid phosphatase during radiation-induced apoptosis in HL60 cells. Acta Oncol. 2003, 42, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Vit, J.P.; Rosselli, F. Role of the ceramide-signaling pathways in ionizing radiation-induced apoptosis. Oncogene 2003, 22, 8645–8652. [Google Scholar] [CrossRef] [PubMed]
- Santana, P.; Pena, L.A.; Haimovitz-Friedman, A.; Martin, S.; Green, D.; McLoughlin, M.; Cordon-Cardo, C.; Schuchman, E.H.; Fuks, Z.; Kolesnick, R. Acid sphingomyelinase-deficient human lymphoblasts and mice are defective in radiation-induced apoptosis. Cell 1996, 86, 189–199. [Google Scholar] [CrossRef]
- Rebillard, A.; Rioux-Leclercq, N.; Muller, C.; Bellaud, P.; Jouan, F.; Meurette, O.; Jouan, E.; Vernhet, L.; Le Quement, C.; Carpinteiro, A.; et al. Acid sphingomyelinase deficiency protects from cisplatin-induced gastrointestinal damage. Oncogene 2008, 27, 6590–6595. [Google Scholar] [CrossRef]
- Smith, E.L.; Schuchman, E.H. The unexpected role of acid sphingomyelinase in cell death and the pathophysiology of common diseases. FASEB J. 2008, 22, 3419–3431. [Google Scholar] [CrossRef]
- Prokhorova, E.A.; Egorshina, A.Y.; Zhivotovsky, B.; Kopeina, G.S. The DNA-damage response and nuclear events as regulators of nonapoptotic forms of cell death. Oncogene 2020, 39, 1–16. [Google Scholar] [CrossRef]
- Vitale, I.; Galluzzi, L.; Castedo, M.; Kroemer, G. Mitotic catastrophe: A mechanism for avoiding genomic instability. Nat. Rev. Mol. Cell Biol. 2011, 12, 385–392. [Google Scholar] [CrossRef]
- Apel, A.; Herr, I.; Schwarz, H.; Rodemann, H.P.; Mayer, A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008, 68, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- Gewirtz, D.A.; Hilliker, M.L.; Wilson, E.N. Promotion of autophagy as a mechanism for radiation sensitization of breast tumor cells. Radiother. Oncol. 2009, 92, 323–328. [Google Scholar] [CrossRef]
- Makowska, A.; Eble, M.; Prescher, K.; Hoss, M.; Kontny, U. Chloroquine Sensitizes Nasopharyngeal Carcinoma Cells but Not Nasoepithelial Cells to Irradiation by Blocking Autophagy. PLoS ONE 2016, 11, e0166766. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, S.; Comincini, S. Autophagy and ionizing radiation in tumors: The “survive or not survive” dilemma. J. Cell Physiol. 2013, 228, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hino, M.; Hamada, N.; Tajika, Y.; Funayama, T.; Morimura, Y.; Sakashita, T.; Yokota, Y.; Fukamoto, K.; Mutou, Y.; Kobayashi, Y.; et al. Heavy ion irradiation induces autophagy in irradiated C2C12 myoblasts and their bystander cells. J. Electron. Microsc. 2010, 59, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, J.; Fu, J.; Wang, J.; Ye, S.; Liu, W.; Shao, C. Role of ROS-mediated autophagy in radiation-induced bystander effect of hepatoma cells. Int. J. Radiat. Biol. 2015, 91, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R.; Kongara, S.; Beaudoin, B.; Karp, C.M.; Bray, K.; Degenhardt, K.; Chen, G.; Jin, S.; White, E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007, 21, 1367–1381. [Google Scholar] [CrossRef]
- Karantza-Wadsworth, V.; Patel, S.; Kravchuk, O.; Chen, G.; Mathew, R.; Jin, S.; White, E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007, 21, 1621–1635. [Google Scholar] [CrossRef]
- Ren, Y.; Yang, P.; Li, C.; Wang, W.A.; Zhang, T.; Li, J.; Li, H.; Dong, C.; Meng, W.; Zhou, H. Ionizing radiation triggers mitophagy to enhance DNA damage in cancer cells. Cell Death Discov. 2023, 9, 267. [Google Scholar] [CrossRef]
- Yang, P.; Li, J.; Zhang, T.; Ren, Y.; Zhang, Q.; Liu, R.; Li, H.; Hua, J.; Wang, W.A.; Wang, J.; et al. Ionizing radiation-induced mitophagy promotes ferroptosis by increasing intracellular free fatty acids. Cell Death Differ. 2023, 30, 2432–2445. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, J.; Kang, R.; Klionsky, D.J.; Tang, D. Ferroptosis: Machinery and regulation. Autophagy 2021, 17, 2054–2081. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Liu, J.; Kang, R.; Klionsky, D.J.; Kroemer, G.; Tang, D. Ferroptosis is a type of autophagy-dependent cell death. Semin. Cancer Biol. 2020, 66, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Conrad, M. The Metabolic Underpinnings of Ferroptosis. Cell Metab. 2020, 32, 920–937. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Minikes, A.M.; Jiang, X. Ferroptosis at the intersection of lipid metabolism and cellular signaling. Mol. Cell 2022, 82, 2215–2227. [Google Scholar] [CrossRef] [PubMed]
- Pope, L.E.; Dixon, S.J. Regulation of ferroptosis by lipid metabolism. Trends Cell Biol. 2023, 33, 1077–1087. [Google Scholar] [CrossRef]
- Yang, W.S.; Stockwell, B.R. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016, 26, 165–176. [Google Scholar] [CrossRef]
- Naowarojna, N.; Wu, T.W.; Pan, Z.; Li, M.; Han, J.R.; Zou, Y. Dynamic Regulation of Ferroptosis by Lipid Metabolism. Antioxid. Redox Signal. 2023, 39, 59–78. [Google Scholar] [CrossRef]
- Zhang, X.; Li, X.; Zheng, C.; Yang, C.; Zhang, R.; Wang, A.; Feng, J.; Hu, X.; Chang, S.; Zhang, H. Ferroptosis, a new form of cell death defined after radiation exposure. Int. J. Radiat. Biol. 2022, 98, 1201–1209. [Google Scholar] [CrossRef]
- Shibata, Y.; Yasui, H.; Higashikawa, K.; Miyamoto, N.; Kuge, Y. Erastin, a ferroptosis-inducing agent, sensitized cancer cells to X-ray irradiation via glutathione starvation in vitro and in vivo. PLoS ONE 2019, 14, e0225931. [Google Scholar] [CrossRef]
- Riegman, M.; Sagie, L.; Galed, C.; Levin, T.; Steinberg, N.; Dixon, S.J.; Wiesner, U.; Bradbury, M.S.; Niethammer, P.; Zaritsky, A.; et al. Ferroptosis occurs through an osmotic mechanism and propagates independently of cell rupture. Nat. Cell Biol. 2020, 22, 1042–1048. [Google Scholar] [CrossRef]
- Jelinek, A.; Heyder, L.; Daude, M.; Plessner, M.; Krippner, S.; Grosse, R.; Diederich, W.E.; Culmsee, C. Mitochondrial rescue prevents glutathione peroxidase-dependent ferroptosis. Free Radic. Biol. Med. 2018, 117, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Yi, J.; Zhu, J.; Minikes, A.M.; Monian, P.; Thompson, C.B.; Jiang, X. Role of Mitochondria in Ferroptosis. Mol. Cell 2019, 73, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Gan, B. Mitochondrial regulation of ferroptosis. J. Cell Biol. 2021, 220, e202105043. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Sha, W.; Gu, Q.; Wang, J.; Zhu, Y.; Xu, T.; Xu, Z.; Yan, F.; Lin, X.; Tian, S. Inhibition of Connexin43 Improves the Recovery of Spinal Cord Injury Against Ferroptosis via the SLC7A11/GPX4 Pathway. Neuroscience 2023, 526, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Losa, D.; Kohler, T.; Bellec, J.; Dudez, T.; Crespin, S.; Bacchetta, M.; Boulanger, P.; Hong, S.S.; Morel, S.; Nguyen, T.H.; et al. Pseudomonas aeruginosa-induced apoptosis in airway epithelial cells is mediated by gap junctional communication in a JNK-dependent manner. J. Immunol. 2014, 192, 4804–4812. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Jiang, X.; Yang, C.; Zhang, J.; Chen, B.; Li, Y.; Yao, S.; Xie, Q.; Gomez, H.; Murugan, R.; et al. Pannexin 1 mediates ferroptosis that contributes to renal ischemia/reperfusion injury. J. Biol. Chem. 2019, 294, 19395–19404. [Google Scholar] [CrossRef]
- Pelegrin, P.; Surprenant, A. Pannexin-1 mediates large pore formation and interleukin-1beta release by the ATP-gated P2X7 receptor. EMBO J. 2006, 25, 5071–5082. [Google Scholar] [CrossRef]
- Iglesias, R.; Dahl, G.; Qiu, F.; Spray, D.C.; Scemes, E. Pannexin 1: The molecular substrate of astrocyte “hemichannels”. J. Neurosci. 2009, 29, 7092–7097. [Google Scholar] [CrossRef]
- Bruzzone, R.; Hormuzdi, S.G.; Barbe, M.T.; Herb, A.; Monyer, H. Pannexins, a family of gap junction proteins expressed in brain. Proc. Natl. Acad. Sci. USA 2003, 100, 13644–13649. [Google Scholar] [CrossRef]
- Linkermann, A.; Skouta, R.; Himmerkus, N.; Mulay, S.R.; Dewitz, C.; De Zen, F.; Prokai, A.; Zuchtriegel, G.; Krombach, F.; Welz, P.S.; et al. Synchronized renal tubular cell death involves ferroptosis. Proc. Natl. Acad. Sci. USA 2014, 111, 16836–16841. [Google Scholar] [CrossRef]
- Li, X.; Peng, X.; Zhou, X.; Li, M.; Chen, G.; Shi, W.; Yu, H.; Zhang, C.; Li, Y.; Feng, Z.; et al. Small extracellular vesicles delivering lncRNA WAC-AS1 aggravate renal allograft ischemia-reperfusion injury by inducing ferroptosis propagation. Cell Death Differ. 2023, 30, 2167–2186. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, N.K.; Suhara, T.; Komai, K.; Shimada, B.K.; Yorichika, N.; Kobayashi, M.; Baba, Y.; Higa, J.K.; Matsui, T. The role of ferroptosis in cell-to-cell propagation of cell death initiated from focal injury in cardiomyocytes. Life Sci. 2023, 332, 122113. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, L.X.; Silva-Almeida, C.; Rondeau, J.D.; Sonveaux, P. Mitochondrial Transfer in Cancer: A Comprehensive Review. Int. J. Mol. Sci. 2021, 22, 3245. [Google Scholar] [CrossRef] [PubMed]
- Torralba, D.; Baixauli, F.; Sanchez-Madrid, F. Mitochondria Know No Boundaries: Mechanisms and Functions of Intercellular Mitochondrial Transfer. Front. Cell Dev. Biol. 2016, 4, 107. [Google Scholar] [CrossRef] [PubMed]
- Rustom, A.; Saffrich, R.; Markovic, I.; Walther, P.; Gerdes, H.H. Nanotubular highways for intercellular organelle transport. Science 2004, 303, 1007–1010. [Google Scholar] [CrossRef] [PubMed]
- Norris, R.P. Transfer of mitochondria and endosomes between cells by gap junction internalization. Traffic 2021, 22, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Rogers, R.S.; Bhattacharya, J. When cells become organelle donors. Physiology 2013, 28, 414–422. [Google Scholar] [CrossRef]
- Murray, L.M.A.; Krasnodembskaya, A.D. Concise Review: Intercellular Communication Via Organelle Transfer in the Biology and Therapeutic Applications of Stem Cells. Stem Cells 2019, 37, 14–25. [Google Scholar] [CrossRef]
- Hayakawa, K.; Esposito, E.; Wang, X.; Terasaki, Y.; Liu, Y.; Xing, C.; Ji, X.; Lo, E.H. Transfer of mitochondria from astrocytes to neurons after stroke. Nature 2016, 535, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.U.; Minhas, P.S.; Liddelow, S.A.; Haileselassie, B.; Andreasson, K.I.; Dorn, G.W., 2nd; Mochly-Rosen, D. Fragmented mitochondria released from microglia trigger A1 astrocytic response and propagate inflammatory neurodegeneration. Nat. Neurosci. 2019, 22, 1635–1648. [Google Scholar] [CrossRef] [PubMed]
- Peruzzotti-Jametti, L.; Bernstock, J.D.; Willis, C.M.; Manferrari, G.; Rogall, R.; Fernandez-Vizarra, E.; Williamson, J.C.; Braga, A.; van den Bosch, A.; Leonardi, T.; et al. Neural stem cells traffic functional mitochondria via extracellular vesicles. PLoS Biol. 2021, 19, e3001166. [Google Scholar] [CrossRef]
- Jin, S.; Cordes, N. ATM controls DNA repair and mitochondria transfer between neighboring cells. Cell Commun. Signal. 2019, 17, 144. [Google Scholar] [CrossRef] [PubMed]
- Watson, D.C.; Bayik, D.; Storevik, S.; Moreino, S.S.; Sprowls, S.A.; Han, J.; Augustsson, M.T.; Lauko, A.; Sravya, P.; Rosland, G.V.; et al. GAP43-dependent mitochondria transfer from astrocytes enhances glioblastoma tumorigenicity. Nat. Cancer 2023, 4, 648–664. [Google Scholar] [CrossRef]
- Walsh, D.W.M.; Siebenwirth, C.; Greubel, C.; Ilicic, K.; Reindl, J.; Girst, S.; Muggiolu, G.; Simon, M.; Barberet, P.; Seznec, H.; et al. Live cell imaging of mitochondria following targeted irradiation in situ reveals rapid and highly localized loss of membrane potential. Sci. Rep. 2017, 7, 46684. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Ryu, S.W.; Lee, S.; Choi, C. Cytosolic irradiation of femtosecond laser induces mitochondria-dependent apoptosis-like cell death via intrinsic reactive oxygen cascades. Sci. Rep. 2015, 5, 8231. [Google Scholar] [CrossRef]
- Santavanond, J.P.; Rutter, S.F.; Atkin-Smith, G.K.; Poon, I.K.H. Apoptotic Bodies: Mechanism of Formation, Isolation and Functional Relevance. Subcell. Biochem. 2021, 97, 61–88. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Gyorgy, B.; Szabo, T.G.; Pasztoi, M.; Pal, Z.; Misjak, P.; Aradi, B.; Laszlo, V.; Pallinger, E.; Pap, E.; Kittel, A.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell Mol. Life Sci. 2011, 68, 2667–2688. [Google Scholar] [CrossRef]
- Tixeira, R.; Caruso, S.; Paone, S.; Baxter, A.A.; Atkin-Smith, G.K.; Hulett, M.D.; Poon, I.K. Defining the morphologic features and products of cell disassembly during apoptosis. Apoptosis 2017, 22, 475–477. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, X.; Gueydan, C.; Han, J. Plasma membrane changes during programmed cell deaths. Cell Res. 2018, 28, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Schiller, M.; Bekeredjian-Ding, I.; Heyder, P.; Blank, N.; Ho, A.D.; Lorenz, H.M. Autoantigens are translocated into small apoptotic bodies during early stages of apoptosis. Cell Death Differ. 2008, 15, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Nieuwland, R.; Falcon-Perez, J.M.; Soekmadji, C.; Boilard, E.; Carter, D.; Buzas, E.I. Essentials of extracellular vesicles: Posters on basic and clinical aspects of extracellular vesicles. J. Extracell. Vesicles 2018, 7, 1548234. [Google Scholar] [CrossRef] [PubMed]
- Tricarico, C.; Clancy, J.; D’Souza-Schorey, C. Biology and biogenesis of shed microvesicles. Small GTPases 2017, 8, 220–232. [Google Scholar] [CrossRef]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Thery, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126 Pt 24, 5553–5565. [Google Scholar] [CrossRef]
- Guescini, M.; Guidolin, D.; Vallorani, L.; Casadei, L.; Gioacchini, A.M.; Tibollo, P.; Battistelli, M.; Falcieri, E.; Battistin, L.; Agnati, L.F.; et al. C2C12 myoblasts release micro-vesicles containing mtDNA and proteins involved in signal transduction. Exp. Cell Res. 2010, 316, 1977–1984. [Google Scholar] [CrossRef]
- Loyer, X.; Vion, A.C.; Tedgui, A.; Boulanger, C.M. Microvesicles as cell-cell messengers in cardiovascular diseases. Circ. Res. 2014, 114, 345–353. [Google Scholar] [CrossRef]
- Simons, M.; Raposo, G. Exosomes—Vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef]
- Ludwig, A.K.; Giebel, B. Exosomes: Small vesicles participating in intercellular communication. Int. J. Biochem. Cell Biol. 2012, 44, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Lopatina, T.; Gai, C.; Deregibus, M.C.; Kholia, S.; Camussi, G. Cross Talk between Cancer and Mesenchymal Stem Cells through Extracellular Vesicles Carrying Nucleic Acids. Front. Oncol. 2016, 6, 125. [Google Scholar] [CrossRef] [PubMed]
- Fruhbeis, C.; Frohlich, D.; Kuo, W.P.; Kramer-Albers, E.M. Extracellular vesicles as mediators of neuron-glia communication. Front. Cell Neurosci. 2013, 7, 182. [Google Scholar] [CrossRef] [PubMed]
- Wendler, F.; Bota-Rabassedas, N.; Franch-Marro, X. Cancer becomes wasteful: Emerging roles of exosomes(dagger) in cell-fate determination. J. Extracell. Vesicles 2013, 2, 22390. [Google Scholar] [CrossRef] [PubMed]
- Wolfers, J.; Lozier, A.; Raposo, G.; Regnault, A.; Thery, C.; Masurier, C.; Flament, C.; Pouzieux, S.; Faure, F.; Tursz, T.; et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med. 2001, 7, 297–303. [Google Scholar] [CrossRef]
- Thery, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Gercel-Taylor, C. Tumour-derived exosomes and their role in cancer-associated T-cell signalling defects. Br. J. Cancer 2005, 92, 305–311. [Google Scholar] [CrossRef]
- Thery, C.; Regnault, A.; Garin, J.; Wolfers, J.; Zitvogel, L.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J. Cell Biol. 1999, 147, 599–610. [Google Scholar] [CrossRef]
- Wilson, C.M.; Naves, T.; Vincent, F.; Melloni, B.; Bonnaud, F.; Lalloue, F.; Jauberteau, M.O. Sortilin mediates the release and transfer of exosomes in concert with two tyrosine kinase receptors. J. Cell Sci. 2014, 127, 3983–3997. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Beer, L.; Zimmermann, M.; Mitterbauer, A.; Ellinger, A.; Gruber, F.; Narzt, M.S.; Zellner, M.; Gyongyosi, M.; Madlener, S.; Simader, E.; et al. Analysis of the Secretome of Apoptotic Peripheral Blood Mononuclear Cells: Impact of Released Proteins and Exosomes for Tissue Regeneration. Sci. Rep. 2015, 5, 16662. [Google Scholar] [CrossRef]
- Du, Y.; Du, S.; Liu, L.; Gan, F.; Jiang, X.; Wangrao, K.; Lyu, P.; Gong, P.; Yao, Y. Radiation-Induced Bystander Effect can be Transmitted Through Exosomes Using miRNAs as Effector Molecules. Radiat. Res. 2020, 194, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Smolarz, M.; Skoczylas, L.; Gawin, M.; Krzyzowska, M.; Pietrowska, M.; Widlak, P. Radiation-Induced Bystander Effect Mediated by Exosomes Involves the Replication Stress in Recipient Cells. Int. J. Mol. Sci. 2022, 23, 4169. [Google Scholar] [CrossRef] [PubMed]
- Al-Mayah, A.; Bright, S.; Chapman, K.; Irons, S.; Luo, P.; Carter, D.; Goodwin, E.; Kadhim, M. The non-targeted effects of radiation are perpetuated by exosomes. Mutat. Res. 2015, 772, 38–45. [Google Scholar] [CrossRef]
- Jella, K.K.; Rani, S.; O’Driscoll, L.; McClean, B.; Byrne, H.J.; Lyng, F.M. Exosomes are involved in mediating radiation induced bystander signaling in human keratinocyte cells. Radiat. Res. 2014, 181, 138–145. [Google Scholar] [CrossRef]
- Al-Mayah, A.H.; Irons, S.L.; Pink, R.C.; Carter, D.R.; Kadhim, M.A. Possible role of exosomes containing RNA in mediating nontargeted effect of ionizing radiation. Radiat. Res. 2012, 177, 539–545. [Google Scholar] [CrossRef]
- Wang, J.; Ma, W.; Si, C.; Zhang, M.; Qian, W.; Park, G.; Zhou, B.; Luo, D. Exosome-mediated miR-4655-3p contributes to UV radiation-induced bystander effects. Exp. Cell Res. 2022, 418, 113247. [Google Scholar] [CrossRef]
- Song, M.; Wang, Y.; Shang, Z.F.; Liu, X.D.; Xie, D.F.; Wang, Q.; Guan, H.; Zhou, P.K. Bystander autophagy mediated by radiation-induced exosomal miR-7-5p in non-targeted human bronchial epithelial cells. Sci. Rep. 2016, 6, 30165. [Google Scholar] [CrossRef]
- Ni, N.; Ma, W.; Tao, Y.; Liu, J.; Hua, H.; Cheng, J.; Wang, J.; Zhou, B.; Luo, D. Exosomal MiR-769-5p Exacerbates Ultraviolet-Induced Bystander Effect by Targeting TGFBR1. Front. Physiol. 2020, 11, 603081. [Google Scholar] [CrossRef]
- Hu, W.; Xu, S.; Yao, B.; Hong, M.; Wu, X.; Pei, H.; Chang, L.; Ding, N.; Gao, X.; Ye, C.; et al. MiR-663 inhibits radiation-induced bystander effects by targeting TGFB1 in a feedback mode. RNA Biol. 2014, 11, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Prise, K.M.; Folkard, M. Signaling factors for irradiated glioma cells induced bystander responses in fibroblasts. Mutat. Res. 2008, 638, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Cocca, B.A.; Cline, A.M.; Radic, M.Z. Blebs and apoptotic bodies are B cell autoantigens. J. Immunol. 2002, 169, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Segura, E.; Guerin, C.; Hogg, N.; Amigorena, S.; Thery, C. CD8+ dendritic cells use LFA-1 to capture MHC-peptide complexes from exosomes in vivo. J. Immunol. 2007, 179, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Duban, L.; Segura, E.; Veron, P.; Lantz, O.; Amigorena, S. Indirect activation of naive CD4+ T cells by dendritic cell-derived exosomes. Nat. Immunol. 2002, 3, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.; Tesse, A.; Hugel, B.; Martinez, M.C.; Morel, O.; Freyssinet, J.M.; Andriantsitohaina, R. Shed membrane particles from T lymphocytes impair endothelial function and regulate endothelial protein expression. Circulation 2004, 109, 1653–1659. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J. Macrophage Polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef]
- Ma, Q.; Liang, M.; Wu, Y.; Luo, F.; Ma, Z.; Dong, S.; Xu, J.; Dou, C. Osteoclast-derived apoptotic bodies couple bone resorption and formation in bone remodeling. Bone Res. 2021, 9, 5. [Google Scholar] [CrossRef]
- Brock, C.K.; Wallin, S.T.; Ruiz, O.E.; Samms, K.M.; Mandal, A.; Sumner, E.A.; Eisenhoffer, G.T. Stem cell proliferation is induced by apoptotic bodies from dying cells during epithelial tissue maintenance. Nat. Commun. 2019, 10, 1044. [Google Scholar] [CrossRef]
- Zhou, M.; Li, Y.J.; Tang, Y.C.; Hao, X.Y.; Xu, W.J.; Xiang, D.X.; Wu, J.Y. Apoptotic bodies for advanced drug delivery and therapy. J. Control. Release 2022, 351, 394–406. [Google Scholar] [CrossRef]
- Kalra, H.; Drummen, G.P.; Mathivanan, S. Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef] [PubMed]
- Freeman, S.M.; Abboud, C.N.; Whartenby, K.A.; Packman, C.H.; Koeplin, D.S.; Moolten, F.L.; Abraham, G.N. The “bystander effect”: Tumor regression when a fraction of the tumor mass is genetically modified. Cancer Res. 1993, 53, 5274–5283. [Google Scholar] [PubMed]
- Matejka, N.; Reindl, J. Influence of alpha-Particle Radiation on Intercellular Communication Networks of Tunneling Nanotubes in U87 Glioblastoma Cells. Front. Oncol. 2020, 10, 1691. [Google Scholar] [CrossRef] [PubMed]
- Heeran, A.B.; Berrigan, H.P.; O’Sullivan, J. The Radiation-Induced Bystander Effect (RIBE) and its Connections with the Hallmarks of Cancer. Radiat. Res. 2019, 192, 668–679. [Google Scholar] [CrossRef]
- Mancuso, M.; Pasquali, E.; Giardullo, P.; Leonardi, S.; Tanori, M.; Di Majo, V.; Pazzaglia, S.; Saran, A. The radiation bystander effect and its potential implications for human health. Curr. Mol. Med. 2012, 12, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Kandouz, M. (Ed.) Intercellular Communication in Cancer; Springer: Dordrecht, The Netherlands, 2015. [Google Scholar]
- Nagata, S.; Hanayama, R.; Kawane, K. Autoimmunity and the clearance of dead cells. Cell 2010, 140, 619–630. [Google Scholar] [CrossRef]
- Kawamoto, Y.; Nakajima, Y.I.; Kuranaga, E. Apoptosis in Cellular Society: Communication between Apoptotic Cells and Their Neighbors. Int. J. Mol. Sci. 2016, 17, 2144. [Google Scholar] [CrossRef]
- Reddien, P.W.; Cameron, S.; Horvitz, H.R. Phagocytosis promotes programmed cell death in C. elegans. Nature 2001, 412, 198–202. [Google Scholar] [CrossRef]
- Hoeppner, D.J.; Hengartner, M.O.; Schnabel, R. Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature 2001, 412, 202–206. [Google Scholar] [CrossRef]
- Chakraborty, S.; Lambie, E.J.; Bindu, S.; Mikeladze-Dvali, T.; Conradt, B. Engulfment pathways promote programmed cell death by enhancing the unequal segregation of apoptotic potential. Nat. Commun. 2015, 6, 10126. [Google Scholar] [CrossRef]

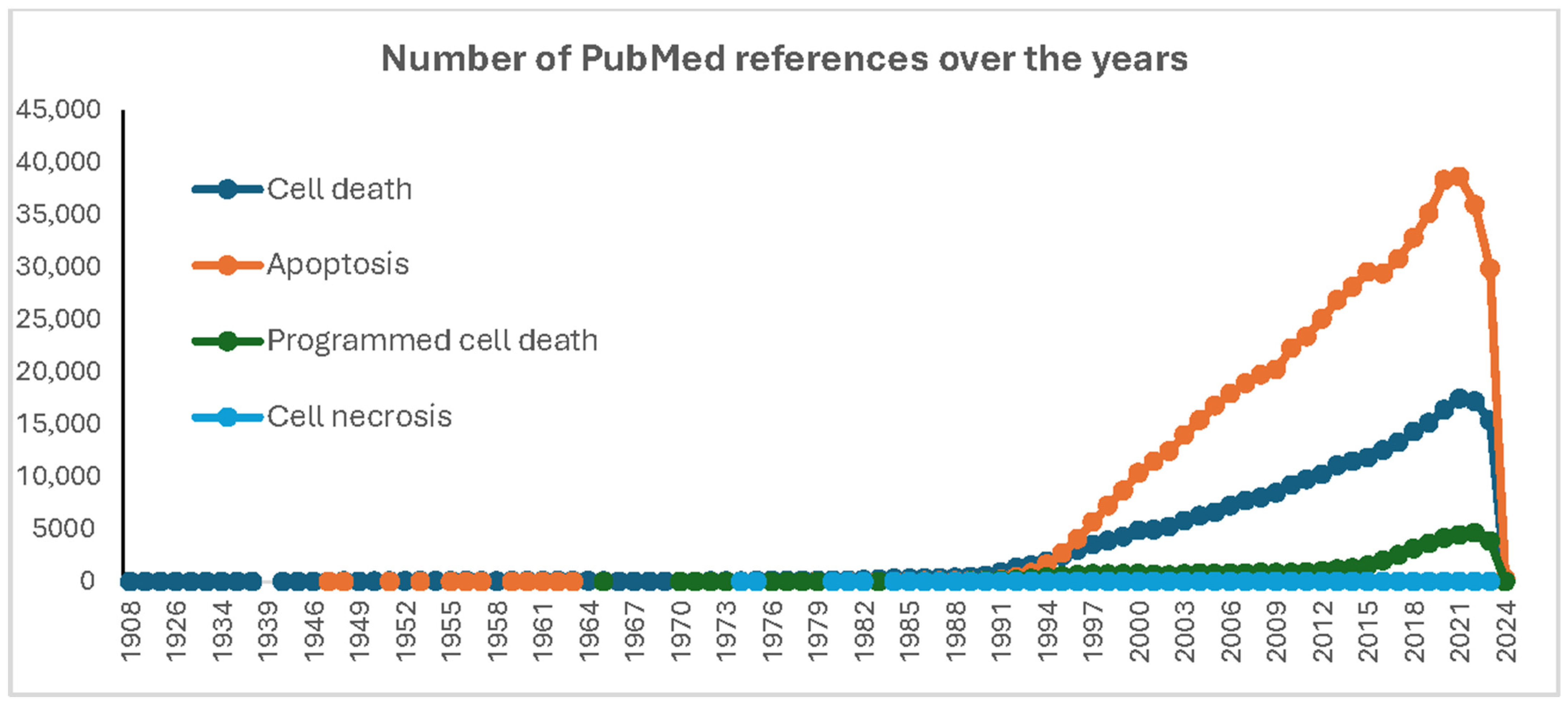
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kandouz, M. Cell Death, by Any Other Name…. Cells 2024, 13, 325. https://doi.org/10.3390/cells13040325
Kandouz M. Cell Death, by Any Other Name…. Cells. 2024; 13(4):325. https://doi.org/10.3390/cells13040325
Chicago/Turabian StyleKandouz, Mustapha. 2024. "Cell Death, by Any Other Name…" Cells 13, no. 4: 325. https://doi.org/10.3390/cells13040325
APA StyleKandouz, M. (2024). Cell Death, by Any Other Name…. Cells, 13(4), 325. https://doi.org/10.3390/cells13040325




