Potential Transcript-Based Biomarkers Predicting Clinical Outcomes of HPV-Positive Head and Neck Squamous Cell Carcinoma Patients
Abstract
1. Introduction
2. Human Papillomavirus as a Determining Factor in the Establishment of HNSCC
3. Proposed Transcript-Based Prognostic Biomarkers for HPV-Positive HNSCC
3.1. lncRNAs
3.2. mRNAs
3.3. miRNAs
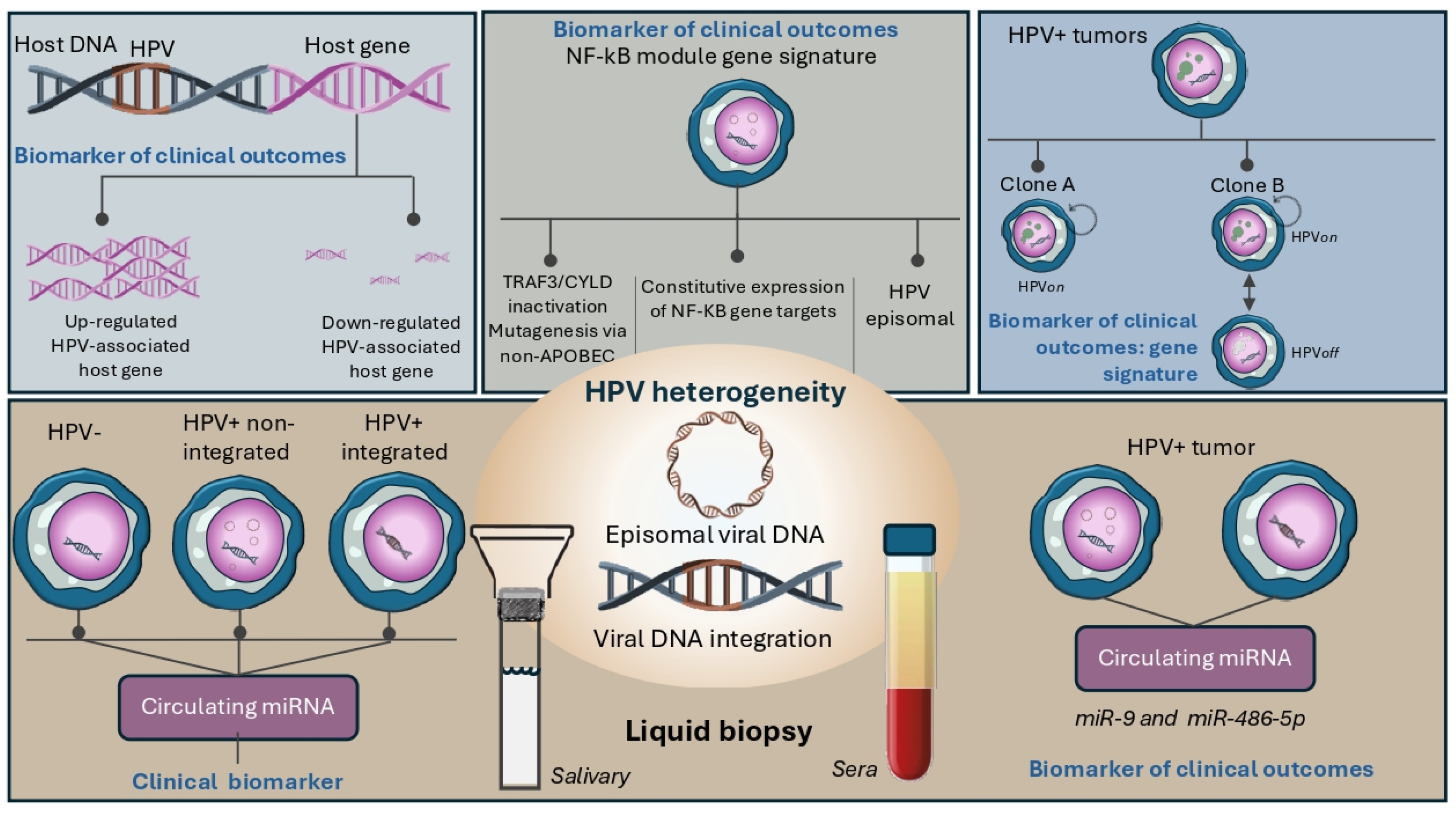
4. Future Directions and Perspectives
4.1. Heterogeneity of HPV-Associated Biology as a Source of New Clinical Biomarkers
4.2. Unraveling Tumor Biology by HPV-Associated Circulating miRNAs
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and Neck Squamous Cell Carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Research on Cancer Cancer Today. Available online: https://gco.iarc.fr/today/en (accessed on 20 April 2024).
- Pai, S.I.; Westra, W.H. Molecular Pathology of Head and Neck Cancer: Implications for Diagnosis, Prognosis, and Treatment. Annu. Rev. Pathol. 2009, 4, 49–70. [Google Scholar] [CrossRef] [PubMed]
- Gormley, M.; Creaney, G.; Schache, A.; Ingarfield, K.; Conway, D.I. Reviewing the Epidemiology of Head and Neck Cancer: Definitions, Trends and Risk Factors. Br. Dent. J. 2022, 233, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Bagnardi, V.; Rota, M.; Botteri, E.; Tramacere, I.; Islami, F.; Fedirko, V.; Scotti, L.; Jenab, M.; Turati, F.; Pasquali, E.; et al. Alcohol Consumption and Site-Specific Cancer Risk: A Comprehensive Dose-Response Meta-Analysis. Br. J. Cancer 2015, 112, 580–593. [Google Scholar] [CrossRef] [PubMed]
- Di Credico, G.; Polesel, J.; Dal Maso, L.; Pauli, F.; Torelli, N.; Luce, D.; Radoï, L.; Matsuo, K.; Serraino, D.; Brennan, P.; et al. Alcohol Drinking and Head and Neck Cancer Risk: The Joint Effect of Intensity and Duration. Br. J. Cancer 2020, 123, 1456–1463. [Google Scholar] [CrossRef] [PubMed]
- Berthiller, J.; Straif, K.; Agudo, A.; Ahrens, W.; Dos Santos, A.B.; Boccia, S.; Cadoni, G.; Canova, C.; Castellsague, X.; Chen, C.; et al. Low Frequency of Cigarette Smoking and the Risk of Head and Neck Cancer in the INHANCE Consortium Pooled Analysis. Int. J. Epidemiol. 2016, 45, 835–845. [Google Scholar] [CrossRef]
- Dal Maso, L.; Torelli, N.; Biancotto, E.; Di Maso, M.; Gini, A.; Franchin, G.; Levi, F.; La Vecchia, C.; Serraino, D.; Polesel, J. Combined Effect of Tobacco Smoking and Alcohol Drinking in the Risk of Head and Neck Cancers: A Re-Analysis of Case-Control Studies Using Bi-Dimensional Spline Models. Eur. J. Epidemiol. 2016, 31, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Guha, N.; Warnakulasuriya, S.; Vlaanderen, J.; Straif, K. Betel Quid Chewing and the Risk of Oral and Oropharyngeal Cancers: A Meta-Analysis with Implications for Cancer Control. Int. J. Cancer 2014, 135, 1433–1443. [Google Scholar] [CrossRef]
- Mork, J.; Lie, A.K.; Glattre, E.; Clark, S.; Hallmans, G.; Jellum, E.; Koskela, P.; Møller, B.; Pukkala, E.; Schiller, J.T.; et al. Human Papillomavirus Infection as a Risk Factor for Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2001, 344, 1125–1131. [Google Scholar] [CrossRef]
- Mody, M.D.; Rocco, J.W.; Yom, S.S.; Haddad, R.I.; Saba, N.F. Head and Neck Cancer. Lancet 2021, 398, 2289–2299. [Google Scholar] [CrossRef]
- Yang, Z.; Sun, P.; Dahlstrom, K.R.; Gross, N.; Li, G. Joint Effect of Human Papillomavirus Exposure, Smoking and Alcohol on Risk of Oral Squamous Cell Carcinoma. BMC Cancer 2023, 23, 457. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.M.; Daling, J.R.; Doody, D.R.; Wipf, G.C.; Carter, J.J.; Madeleine, M.M.; Mao, E.J.; Fitzgibbons, E.D.; Huang, S.; Beckmann, A.M.; et al. Oral Cancer Risk in Relation to Sexual History and Evidence of Human Papillomavirus Infection. J. Natl. Cancer Inst. 1998, 90, 1626–1636. [Google Scholar] [CrossRef] [PubMed]
- Kreimer, A.R.; Clifford, G.M.; Boyle, P.; Franceschi, S. Human Papillomavirus Types in Head and Neck Squamous Cell Carcinomas Worldwide: A Systematic Review. Cancer Epidemiol. Biomark. Prev. 2005, 14, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.C.; de Lima, I.C.C.; Marques, V.M.F.; de Araújo, W.H.A.; de Campos Ferreira, C. Human Papillomavirus Prevalence in Oral and Oropharyngeal Squamous Cell Carcinoma in South America: A Systematic Review and Meta-Analysis. Oncol. Rev. 2022, 16, 552. [Google Scholar] [CrossRef] [PubMed]
- Gillison, M.L.; Koch, W.M.; Capone, R.B.; Spafford, M.; Westra, W.H.; Wu, L.; Zahurak, M.L.; Daniel, R.W.; Viglione, M.; Symer, D.E.; et al. Evidence for a Causal Association between Human Papillomavirus and a Subset of Head and Neck Cancers. J. Natl. Cancer Inst. 2000, 92, 709–720. [Google Scholar] [CrossRef] [PubMed]
- de Sanjose, S.; Serrano, B.; Tous, S.; Alejo, M.; Loveras, B.L.; Quiros, B.; Clavero, O.; Vidal, A.; Ferrandiz-Pulido, C.; Pavon, M.A.; et al. Burden of Human Papillomavirus (HPV)-Related Cancers Attributable to HPVs 6/11/16/18/31/33/45/52 and 58. JNCI Cancer Spectr. 2019, 2, pky045. [Google Scholar] [CrossRef] [PubMed]
- Hussein, A.A.; Helder, M.N.; de Visscher, J.G.; Leemans, C.R.; Braakhuis, B.J.; de Vet, H.C.W.; Forouzanfar, T. Global Incidence of Oral and Oropharynx Cancer in Patients Younger than 45 Years versus Older Patients: A Systematic Review. Eur. J. Cancer 2017, 82, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Deschler, D.G.; Richmon, J.D.; Khariwala, S.S.; Ferris, R.L.; Wang, M.B. The “New” Head and Neck Cancer Patient-Young, Nonsmoker, Nondrinker, and HPV Positive: Evaluation. Otolaryngol. Head Neck Surg. 2014, 151, 375–380. [Google Scholar] [CrossRef]
- Chaturvedi, A.K.; Engels, E.A.; Pfeiffer, R.M.; Hernandez, B.Y.; Xiao, W.; Kim, E.; Jiang, B.; Goodman, M.T.; Sibug-Saber, M.; Cozen, W.; et al. Human Papillomavirus and Rising Oropharyngeal Cancer Incidence in the United States. J. Clin. Oncol. 2023, 41, 3081–3088. [Google Scholar] [CrossRef]
- Hammarstedt, L.; Lindquist, D.; Dahlstrand, H.; Romanitan, M.; Onelöv, L.; Joneberg, J.; Creson, N.; Lindholm, J.; Ye, W.; Dalianis, T.; et al. Human Papillomavirus as a Risk Factor for the Increase in Incidence of Tonsillar Cancer. Int. J. Cancer 2006, 119, 2620–2623. [Google Scholar] [CrossRef]
- Zhang, Y.; Fakhry, C.; D’Souza, G. Projected Association of Human Papillomavirus Vaccination With Oropharynx Cancer Incidence in the US, 2020–2045. JAMA Oncol. 2021, 7, E212907. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, Z.; Qiu, S.; Wang, R. Therapeutic Strategies of Different HPV Status in Head and Neck Squamous Cell Carcinoma. Int. J. Biol. Sci. 2021, 17, 1104–1118. [Google Scholar] [CrossRef] [PubMed]
- Wuerdemann, N.; Wittekindt, C.; Sharma, S.J.; Prigge, E.S.; Reuschenbach, M.; Gattenlöhner, S.; Klussmann, J.P.; Wagner, S. Risk Factors for Overall Survival Outcome in Surgically Treated Human Papillomavirus-Negative and Positive Patients with Oropharyngeal Cancer. Oncol. Res. Treat. 2017, 40, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to Build a Bridge from a Population-Based to a More “Personalized” Approach to Cancer Staging. CA A Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Sabatini, M.E.; Chiocca, S. Human Papillomavirus as a Driver of Head and Neck Cancers. Br. J. Cancer 2020, 122, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Ang, K.K.; Zhang, Q.; Rosenthal, D.I.; Nguyen-Tan, P.F.; Sherman, E.J.; Weber, R.S.; Galvin, J.M.; Bonner, J.A.; Harris, J.; El-Naggar, A.K.; et al. Randomized Phase III Trial of Concurrent Accelerated Radiation plus Cisplatin with or without Cetuximab for Stage III to IV Head and Neck Carcinoma: RTOG 0522. J. Clin. Oncol. 2014, 32, 2940–2950. [Google Scholar] [CrossRef] [PubMed]
- Christopherson, K.M.; Moreno, A.C.; Elgohari, B.; Gross, N.; Ferrarotto, R.; Mohamed, A.S.R.; Brandon Gunn, G.; Goepfert, R.P.; Mott, F.E.; Shah, S.J.; et al. Outcomes after Salvage for HPV-Positive Recurrent Oropharyngeal Cancer Treated with Primary Radiation. Oral Oncol. 2021, 113, 105125. [Google Scholar] [CrossRef]
- Lechner, M.; Liu, J.; Masterson, L.; Fenton, T.R. HPV-Associated Oropharyngeal Cancer: Epidemiology, Molecular Biology and Clinical Management. Nat. Rev. Clin. Oncol. 2022, 19, 306–327. [Google Scholar] [CrossRef]
- Egawa, N.; Egawa, K.; Griffin, H.; Doorbar, J. Human Papillomaviruses; Epithelial Tropisms, and the Development of Neoplasia. Viruses 2015, 7, 3863–3890. [Google Scholar] [CrossRef]
- National Institute of Allergy and Infectious Diseases PaVE: The Papillomavirus Episteme. Available online: https://pave.niaid.nih.gov/ (accessed on 20 April 2024).
- de Martel, C.; Plummer, M.; Vignat, J.; Franceschi, S. Worldwide Burden of Cancer Attributable to HPV by Site, Country and HPV Type. Int. J. Cancer 2017, 141, 664–670. [Google Scholar] [CrossRef]
- Bouvard, V.; Baan, R.; Straif, K.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Freeman, C.; Galichet, L.; et al. A Review of Human Carcinogens—Part B: Biological Agents. Lancet Oncol. 2009, 10, 321–322. [Google Scholar] [CrossRef] [PubMed]
- Vani, N.V.; Rama, R.; Madhanagopal, R.; Vijayalakshmi, R.; Swaminathan, R. Human Papillomavirus-Attributable Head and Neck Cancers in India-A Systematic Review and Meta-Analysis. JCO Glob. Oncol. 2024, 10, e2300464. [Google Scholar] [CrossRef] [PubMed]
- Parfenov, M.; Pedamallu, C.S.; Gehlenborg, N.; Freeman, S.S.; Danilova, L.; Bristow, C.A.; Lee, S.; Hadjipanayis, A.G.; Ivanova, E.V.; Wilkerson, M.D.; et al. Characterization of HPV and Host Genome Interactions in Primary Head and Neck Cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 15544–15549. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Yang, M.; Li, S.; Zhu, W.; Chen, M.; Pan, J.; Long, D.; Liu, Z.; Zhang, C. Expression and Molecular Regulation of Non-Coding RNAs in HPV-Positive Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2023, 13, 1122982. [Google Scholar] [CrossRef] [PubMed]
- Slebos, R.J.C.; Yi, Y.; Ely, K.; Carter, J.; Evjen, A.; Zhang, X.; Shyr, Y.; Murphy, B.M.; Cmelak, A.J.; Burkey, B.B.; et al. Gene Expression Differences Associated with Human Papillomavirus Status in Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 2006, 12, 701–709. [Google Scholar] [CrossRef]
- Cheng, H.; Yang, X.; Si, H.; Saleh, A.D.; Xiao, W.; Coupar, J.; Gollin, S.M.; Ferris, R.L.; Issaeva, N.; Yarbrough, W.G.; et al. Genomic and Transcriptomic Characterization Links Cell Lines with Aggressive Head and Neck Cancers. Cell Rep. 2018, 25, 1332–1345.e5. [Google Scholar] [CrossRef]
- Das, S.; Dey, M.K.; Devireddy, R.; Gartia, M.R. Biomarkers in Cancer Detection, Diagnosis, and Prognosis. Sensors 2023, 24, 37. [Google Scholar] [CrossRef] [PubMed]
- Begliarzade, S.; Beilerli, A.; Sufianov, A.; Tamrazov, R.; Kudriashov, V.; Ilyasova, T.; Liang, Y.; Beylerli, O. Long Non-Coding RNAs as Promising Biomarkers and Therapeutic Targets in Cervical Cancer. Noncoding RNA Res. 2023, 8, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Beane, J.; Campbell, J.D.; Lel, J.; Vick, J.; Spira, A. Genomic Approaches to Accelerate Cancer Interception. Lancet Oncol. 2017, 18, e494–e502. [Google Scholar] [CrossRef]
- Bhattacharjee, R.; Prabhakar, N.; Kumar, L.; Bhattacharjee, A.; Kar, S.; Malik, S.; Kumar, D.; Ruokolainen, J.; Negi, A.; Jha, N.K.; et al. Crosstalk between Long Noncoding RNA and MicroRNA in Cancer. Cell. Oncol. 2023, 46, 885–908. [Google Scholar] [CrossRef]
- Kopczyńska, M.; Kolenda, T.; Guglas, K.; Sobocińska, J.; Teresiak, A.; Bliźniak, R.; Mackiewicz, A.; Mackiewicz, J.; Lamperska, K. PRINS LncRNA Is a New Biomarker Candidate for HPV Infection and Prognosis of Head and Neck Squamous Cell Carcinomas. Diagnostics 2020, 10, 762. [Google Scholar] [CrossRef] [PubMed]
- Zeng, P.Y.F.; Cecchini, M.J.; Barrett, J.W.; Shammas-Toma, M.; De Cecco, L.; Serafini, M.S.; Cavalieri, S.; Licitra, L.; Hoebers, F.; Brakenhoff, R.H.; et al. Immune-Based Classification of HPV-Associated Oropharyngeal Cancer with Implications for Biomarker-Driven Treatment de-Intensification. eBioMedicine 2022, 86, 104373. [Google Scholar] [CrossRef] [PubMed]
- Mei, Z.; Zhengdong, L.; Shupeng, L.; Xin, Z.; Lei, W.; Wang, C. Identification of an 8 HPV-Related RNA Signature as a Novel Prognostic Biomarker for Squamous Cell Carcinoma of the Head and Neck. Medicine 2024, 103, e36448. [Google Scholar] [CrossRef] [PubMed]
- Mourtada, J.; Lony, C.; Nicol, A.; De Azevedo, J.; Bour, C.; Macabre, C.; Roncarati, P.; Ledrappier, S.; Schultz, P.; Borel, C.; et al. A Novel ΔNp63-Dependent Immune Mechanism Improves Prognosis of HPV-Related Head and Neck Cancer. Front. Immunol. 2023, 14, 1264093. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, P.S.; Bovilla, V.R.; Swamy, V.H.; Manoli, N.N.; Dasegowda, K.B.; Siddegowda, S.M.; Chandrashekarappa, S.; Somasundara, V.M.; Kabekkodu, S.P.; Rajesh, R.; et al. Human Papillomavirus-Driven Repression of NRF2 Signalling Confers Chemo-Radio Sensitivity and Predicts Prognosis in Head and Neck Squamous Cell Carcinoma. Free. Radic. Biol. Med. 2023, 205, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Kori, M.; Arga, K.Y. HPV16 Status Predicts Potential Protein Biomarkers and Therapeutics in Head and Neck Squamous Cell Carcinoma. Virology 2023, 582, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Sobocińska, J.; Nowakowska, J.; Molenda, S.; Olechnowicz, A.; Guglas, K.; Kozłowska-Masłoń, J.; Kazimierczak, U.; Machnik, M.; Oleksiewicz, U.; Teresiak, A.; et al. Zinc Finger Proteins in Head and Neck Squamous Cell Carcinomas: ZNF540 May Serve as a Biomarker. Curr. Oncol. 2022, 29, 9896–9915. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Shao, T.; Huang, G.; Zheng, Z.; Jiang, Y.; Zeng, W.; Lv, X. FDCSP Is an Immune-Associated Prognostic Biomarker in HPV-Positive Head and Neck Squamous Carcinoma. Biomolecules 2022, 12, 1458. [Google Scholar] [CrossRef]
- Yang, Y.; Feng, L.; Wang, R.; Ma, H.; He, S.; Fang, J. Integrated Analysis of LncRNA-Associated CeRNA Network in P16-Positive and P16-Negative Head and Neck Squamous Cell Carcinoma. Medicine 2022, 101, e26120. [Google Scholar] [CrossRef]
- Méndez-Matías, G.; Velázquez-Velázquez, C.; Castro-Oropeza, R.; Mantilla-Morales, A.; Ocampo-Sandoval, D.; Burgos-González, A.; Heredia-Gutiérrez, C.; Alvarado-Cabrero, I.; Sánchez-Sandoval, R.; Barco-Bazán, A.; et al. Prevalence of HPV in Mexican Patients with Head and Neck Squamous Carcinoma and Identification of Potential Prognostic Biomarkers. Cancers 2021, 13, 5602. [Google Scholar] [CrossRef]
- Zhang, Z.; Bao, Y.; Zhou, L.; Ye, Y.; Fu, W.; Sun, C. DOCK8 Serves as a Prognostic Biomarker and Is Related to Immune Infiltration in Patients With HPV Positive Head and Neck Squamous Cell Carcinoma. Cancer Control 2021, 28, 10732748211011951. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, P.; Chernock, R.D.; Kuhs, K.A.L.; Lewis, J.S.; Luo, J.; Gay, H.A.; Thorstad, W.L.; Wang, X. A Prognostic Gene Expression Signature for Oropharyngeal Squamous Cell Carcinoma. eBioMedicine 2020, 61, 102805. [Google Scholar] [CrossRef] [PubMed]
- Prusinkiewicz, M.A.; Gameiro, S.F.; Ghasemi, F.; Dodge, M.J.; Zeng, P.Y.F.; Maekebay, H.; Barrett, J.W.; Nichols, A.C.; Mymryk, J.S. Survival-Associated Metabolic Genes in Human Papillomavirus-Positive Head and Neck Cancers. Cancers 2020, 12, 253. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Bai, J.; Wei, Y.; Wang, G.; Li, Q.; Zhang, R.; Duan, W.; Yang, S.; Du, M.; Zhao, Y.; et al. A Seven-Gene Prognostic Signature for Rapid Determination of Head and Neck Squamous Cell Carcinoma Survival. Oncol. Rep. 2017, 38, 3403–3411. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.J.; Zheng, M.; Cao, M.X.; Zhang, W.L.; Huang, M.C.; Dai, L.; Tang, Y.L.; Liang, X.H. Distinguishable Prognostic MiRNA Signatures of Head and Neck Squamous Cell Cancer With or Without HPV Infection. Front. Oncol. 2021, 10, 614487. [Google Scholar] [CrossRef] [PubMed]
- Bersani, C.; Mints, M.; Tertipis, N.; Haeggblom, L.; Näsman, A.; Romanitan, M.; Dalianis, T.; Ramqvist, T. MicroRNA-155, -185 and -193b as Biomarkers in Human Papillomavirus Positive and Negative Tonsillar and Base of Tongue Squamous Cell Carcinoma. Oral Oncol. 2018, 82, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, P.; Chernock, R.D.; Yang, Z.; Kuhs, K.A.L.; Lewis, J.S.; Luo, J.; Li, H.; Gay, H.A.; Thorstad, W.L.; et al. A MicroRNA Expression Signature as Prognostic Marker for Oropharyngeal Squamous Cell Carcinoma. J. Natl. Cancer Inst. 2021, 113, 752–759. [Google Scholar] [CrossRef] [PubMed]
- Nulton, T.J.; Kim, N.K.; DiNardo, L.J.; Morgan, I.M.; Windle, B. Patients with Integrated HPV16 in Head and Neck Cancer Show Poor Survival. Oral Oncol. 2018, 80, 52–55. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Torabi, S.J.; Yarbrough, W.G.; Mehra, S.; Osborn, H.A.; Judson, B. Association of Human Papillomavirus Status at Head and Neck Carcinoma Subsites With Overall Survival. JAMA Otolaryngol. Head Neck Surg. 2018, 144, 519–525. [Google Scholar] [CrossRef]
- Kala, P.S.; Thapliyal, N.; Pant, B.; Sharma, N.; Pandey, H.S. Prognostic Role of PD-L1 Expression in Head and Neck Squamous Cell Carcinoma: An Institutional Experience from India. Pathol. Res. Pract. 2024, 254, 155133. [Google Scholar] [CrossRef]
- Yang, S.-M.; Wu, M.; Han, F.-Y.; Sun, Y.-M.; Yang, J.-Q.; Liu, H.-X. Role of HPV Status and PD-L1 Expression in Prognosis of Laryngeal Squamous Cell Carcinoma. Int. J. Clin. Exp. Pathol. 2021, 14, 107. [Google Scholar]
- Glathar, A.R.; Oyelakin, A.; Gluck, C.; Bard, J.; Sinha, S. P63 Directs Subtype-Specific Gene Expression in HPV+ Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2022, 12, 879054. [Google Scholar] [CrossRef]
- Rocco, J.W.; Leong, C.O.; Kuperwasser, N.; DeYoung, M.P.; Ellisen, L.W. P63 Mediates Survival in Squamous Cell Carcinoma by Suppression of P73-Dependent Apoptosis. Cancer Cell 2006, 9, 45–56. [Google Scholar] [CrossRef]
- Koneva, L.A.; Zhang, Y.; Virani, S.; Hall, P.B.; McHugh, J.B.; Chepeha, D.B.; Wolf, G.T.; Carey, T.E.; Rozek, L.S.; Sartor, M.A. HPV Integration in HNSCC Correlates with Survival Outcomes, Immune Response Signatures, and Candidate Drivers. Mol. Cancer Res. 2018, 16, 90–102. [Google Scholar] [CrossRef]
- Schrank, T.P.; Kothari, A.; Weir, W.H.; Stepp, W.H.; Rehmani, H.; Liu, X.; Wang, X.; Sewell, A.; Li, X.; Tasoulas, J.; et al. Noncanonical HPV Carcinogenesis Drives Radiosensitization of Head and Neck Tumors. Proc. Natl. Acad. Sci. USA 2023, 120, e2216532120. [Google Scholar] [CrossRef] [PubMed]
- Tabatabaeian, H.; Bai, Y.; Huang, R.; Chaurasia, A.; Darido, C. Navigating Therapeutic Strategies: HPV Classification in Head and Neck Cancer. Br. J. Cancer 2024. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Ruales, C.; Arguello, J.V.; López-Cortés, A.; Cabrera-Andrade, A.; García-Cárdenas, J.M.; Guevara-Ramírez, P.; Peralta, P.; Leone, P.E.; Paz-Y-Miño, C. Salivary MicroRNAs for Early Detection of Head and Neck Squamous Cell Carcinoma: A Case-Control Study in the High Altitude Mestizo Ecuadorian Population. BioMed Res. Int. 2018, 2018, 9792730. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Vagenas, D.; Salazar, C.; Kenny, L.; Perry, C.; Calvopiña, D.; Punyadeera, C. Salivary MiRNA Panel to Detect HPV-Positive and HPV-Negative Head and Neck Cancer Patients. Oncotarget 2017, 8, 99990–100001. [Google Scholar] [CrossRef] [PubMed]
- Langevin, S.; Kuhnell, D.; Parry, T.; Biesiada, J.; Huang, S.; Wise-Draper, T.; Casper, K.; Zhang, X.; Medvedovic, M.; Kasper, S. Comprehensive MicroRNA-Sequencing of Exosomes Derived from Head and Neck Carcinoma Cells in Vitro Reveals Common Secretion Profiles and Potential Utility as Salivary Biomarkers. Oncotarget 2017, 8, 82459–82474. [Google Scholar] [CrossRef] [PubMed]
- Peacock, B.; Rigby, A.; Bradford, J.; Pink, R.; Hunter, K.; Lambert, D.; Hunt, S. Extracellular Vesicle MicroRNA Cargo Is Correlated with HPV Status in Oropharyngeal Carcinoma. J. Oral Pathol. Med. 2018, 47, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Liu, L.; Fu, H.; Wang, Q.; Shi, Y. Association of Decreased Expression of Serum MiR-9 with Poor Prognosis of Oral Squamous Cell Carcinoma Patients. Med. Sci. Monit. 2016, 22, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zheng, X.; Li, W.; Bai, F.; Lyu, J.; Meng, Q.H. Serum MiR-486-5p as a Diagnostic Marker in Cervical Cancer: With Investigation of Potential Mechanisms. BMC Cancer 2018, 18, 61. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liang, H.; Zhang, Q.; Ma, S. Replicability in Cancer Omics Data Analysis: Measures and Empirical Explorations. Brief. Bioinform. 2022, 23, bbac304. [Google Scholar] [CrossRef] [PubMed]
| Name | Level | Biomolecule | Cancer Type | Clinical Outcome Impact | References |
|---|---|---|---|---|---|
PRINS (Expression) | High | lncRNA | HNSCC | Improved OS and DFS | [43] |
CD3E, IRF4, ZAP70 (UWO3 Risk score) | Low | mRNA | OPSCC | Improved OS and DFS | [44] |
Clorf105, CGA, CHRNA2, CRIP3, CTAG2, ENPP6, NEFH, RNF212 (Risk score) | High | mRNA | HNSCC | Worse OS and DFS | [45] |
S100A9 THBS4 (Expression) | Low High | mRNA | OPSCC | Worse MFS | [46] |
NRF2 NQO1 TP53 KEAP1 (Expression) | Low Low Low High | mRNA | HNSCC | Improved OS | [47] |
mCBX5, mCDKN2A, mMCM5, mMCM6, mRBBP7, mTMPO, mVCAM1 (Risk score) | High | mRNA | HNSCC | Worse OS | [48] |
ZNF540 (Expression) | High | mRNA | HNSCC | Improved OS | [49] |
FDCSP NEFH FAM3B (Expression) | High High High | mRNA | HNSCC | Improved OS | [50] |
FDCSP (Expression) CD8+ T cells (Infiltration) | High High | mRNA | HNSCC | Improved OS | [50] |
U62317.3 hsa-miR-375 KLHD (Expression) | High Low High | lncRNA miRNA mRNA | HNSCC | Improved OS | [51] |
PDLIM5 USP25 SLMAP (Expression) | Low Low Low | mRNA | HNSCC | Worse OS | [51] |
SLC25A39 GJB2 (Expression) | High High | mRNA | HNSCC | Worse OS | [52] |
DOCK8 (Expression) | High | mRNA | HNSCC | Improved OS and DFS | [53] |
RASSF8, LGALSL, TRIB3, FAM106A, MICAL3, STIP1, ZNF146, ZNF284, TPST1, PSMG3, CPEB2, SH3D21, CNOT2, ENTPD6, PPIAL4C, SARS, SULT1E1, EAF1, LOC100128108, PLPPR2, OTUD4, MUL1, BCL2L13, GUCY1B1, MRPL45, GALK1, RSU1, AVPI1, PDPR, ALKBH6, HCK, ATP6V1A, TAF5L, BUD13, TNFRSF6B, ACOT11, RNF167, ORAI1, CRYBG3, HDDC3, CREB3L4, TMEM246, PEX16, HAUS1, NUP214, AURKB, OGN, FBXO41, SLFN13, CXCL13, COMMD3, FOXRED2, FPGS, UCP2, GLUL, LYN, MEI1, CYBA, NUP210, ARHGAP4 (Risk score) | Low | mRNA | OPSCC | Improved OS, RFS and MFS | [54] |
SDHC COX7A1 COX16 COX17 ELOVL6 GOT2 SLC16A2 (Expression) | Low Low Low Low Low Low Low | mRNA | HNSCC | Improved OS | [55] |
AATF, APP, GNPDA1, HPRT1, LASP1, P4HA1, ILF3 (Risk score) | High | mRNA | HNSCC | Worse OS | [56] |
hsa-miR-380-5p, hsa-miR-493-3p, hsa-miR-454-5p, hsa-miR-376c-3p, hsa-miR-338-5p, hsa-miR-16-1-3p, hsa-miR-378a-3p (Risk score) | Low | miRNA | HNSCC | Improved OS | [57] |
miR-155 (Expression) | High | miRNA | TSCC/BOTSCC | Improved OS and PFS | [58] |
hsa-miR-27a-3p, hsa-miR-455-5p, hsa-miR-203a-3p, hsa-miR-584-5p, hsa-miR-24-3p, hsa-miR-548k, hsa-miR-126-3p, hsa-miR-126-5p, hsa-miR-365a-5p, hsa-miR-98-5p, hsa-miR-151b, hsa-miR-361-3p, hsa-miR-374c-5p, hsa-miR-150-5p, hsa-miR-374b-5p, hsa-miR-107, hsa-miR-125b-5p, hsa-miR-1287-5p, hsa-miR-146a-5p, hsa-miR-106a-5p, hsa-miR-15b-5p, hsa-miR-20b-5p, hsa-miR-532-3p, hsa-miR-361-5p, hsa-miR-363-3p, hsa-miR-625-3p (Risk score) | High | miRNA | OPSCC | Worse OS, RFS and MFS | [59] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muñoz-Bello, J.O.; Romero-Córdoba, S.L.; García-Chávez, J.N.; González-Espinosa, C.; Langley, E.; Lizano, M. Potential Transcript-Based Biomarkers Predicting Clinical Outcomes of HPV-Positive Head and Neck Squamous Cell Carcinoma Patients. Cells 2024, 13, 1107. https://doi.org/10.3390/cells13131107
Muñoz-Bello JO, Romero-Córdoba SL, García-Chávez JN, González-Espinosa C, Langley E, Lizano M. Potential Transcript-Based Biomarkers Predicting Clinical Outcomes of HPV-Positive Head and Neck Squamous Cell Carcinoma Patients. Cells. 2024; 13(13):1107. https://doi.org/10.3390/cells13131107
Chicago/Turabian StyleMuñoz-Bello, J. Omar, Sandra L. Romero-Córdoba, J. Noé García-Chávez, Claudia González-Espinosa, Elizabeth Langley, and Marcela Lizano. 2024. "Potential Transcript-Based Biomarkers Predicting Clinical Outcomes of HPV-Positive Head and Neck Squamous Cell Carcinoma Patients" Cells 13, no. 13: 1107. https://doi.org/10.3390/cells13131107
APA StyleMuñoz-Bello, J. O., Romero-Córdoba, S. L., García-Chávez, J. N., González-Espinosa, C., Langley, E., & Lizano, M. (2024). Potential Transcript-Based Biomarkers Predicting Clinical Outcomes of HPV-Positive Head and Neck Squamous Cell Carcinoma Patients. Cells, 13(13), 1107. https://doi.org/10.3390/cells13131107







