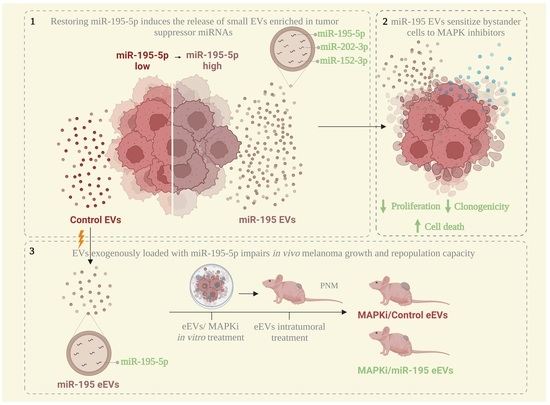
Extracellular Vesicle-Packaged miR-195-5p Sensitizes Melanoma to Targeted Therapy with Kinase Inhibitors
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Mimics Transfection
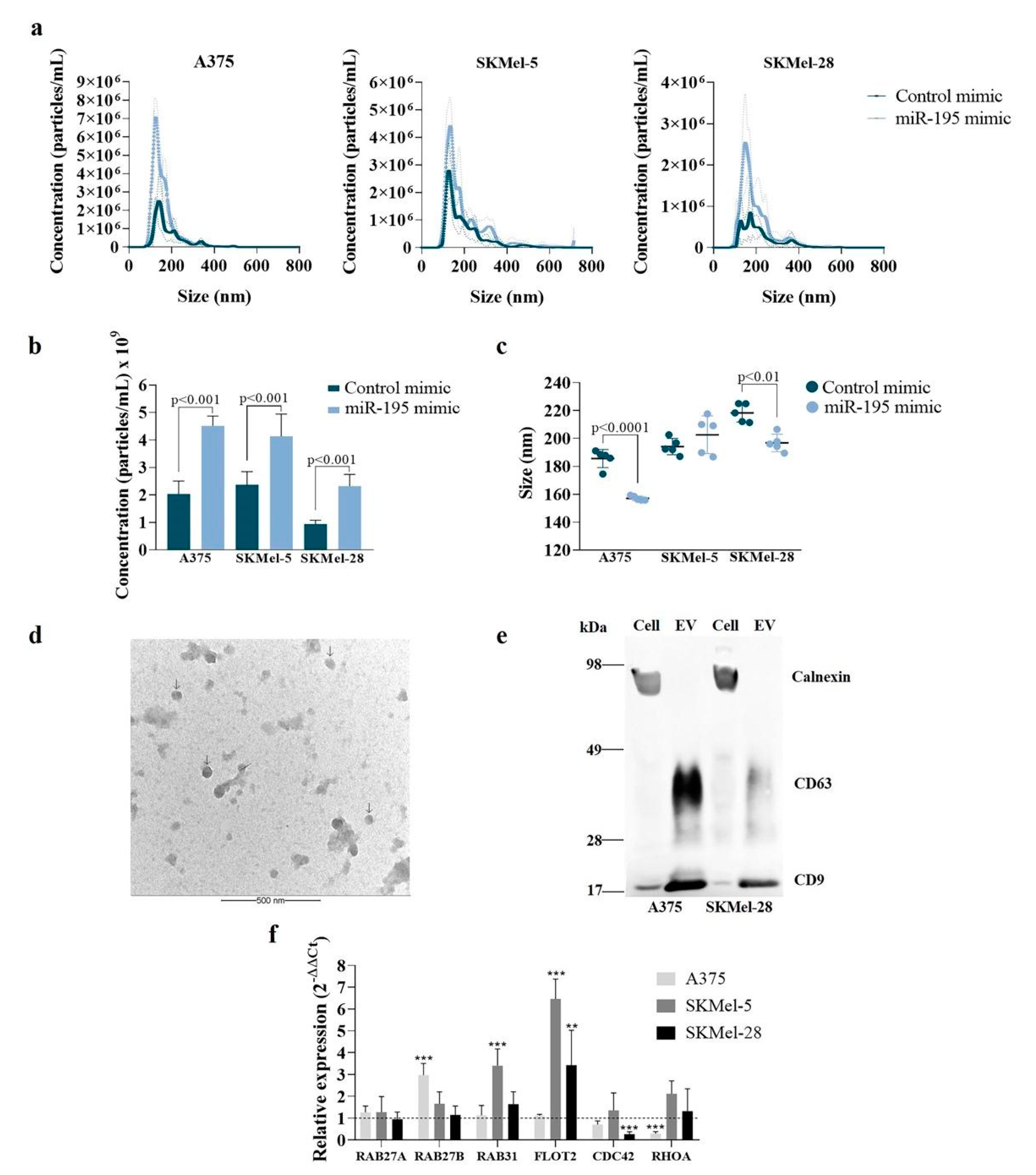
2.3. EVs Isolation and Characterization
Transmission Electron Microscopy (TEM)
2.4. RNA Extraction and RT-qPCR
2.5. Gene Expression Profile
2.6. Western Blotting (WB)
2.7. miRNA Profile
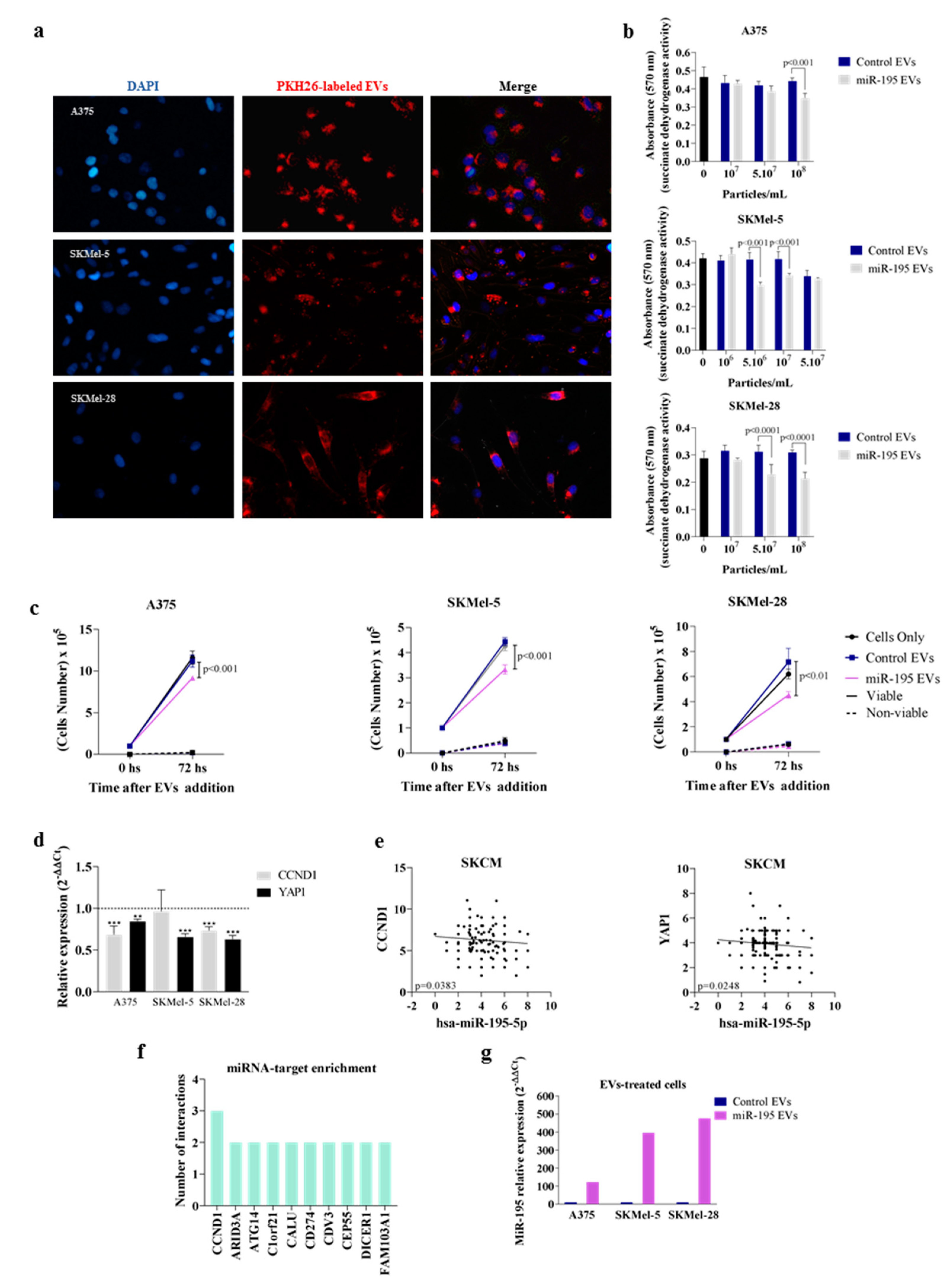
2.8. EV Incorporation Assay
2.9. Functional Assays
2.9.1. Cell Viability Assay
2.9.2. Cell Proliferation Assay
2.9.3. Cell Cycle Profile
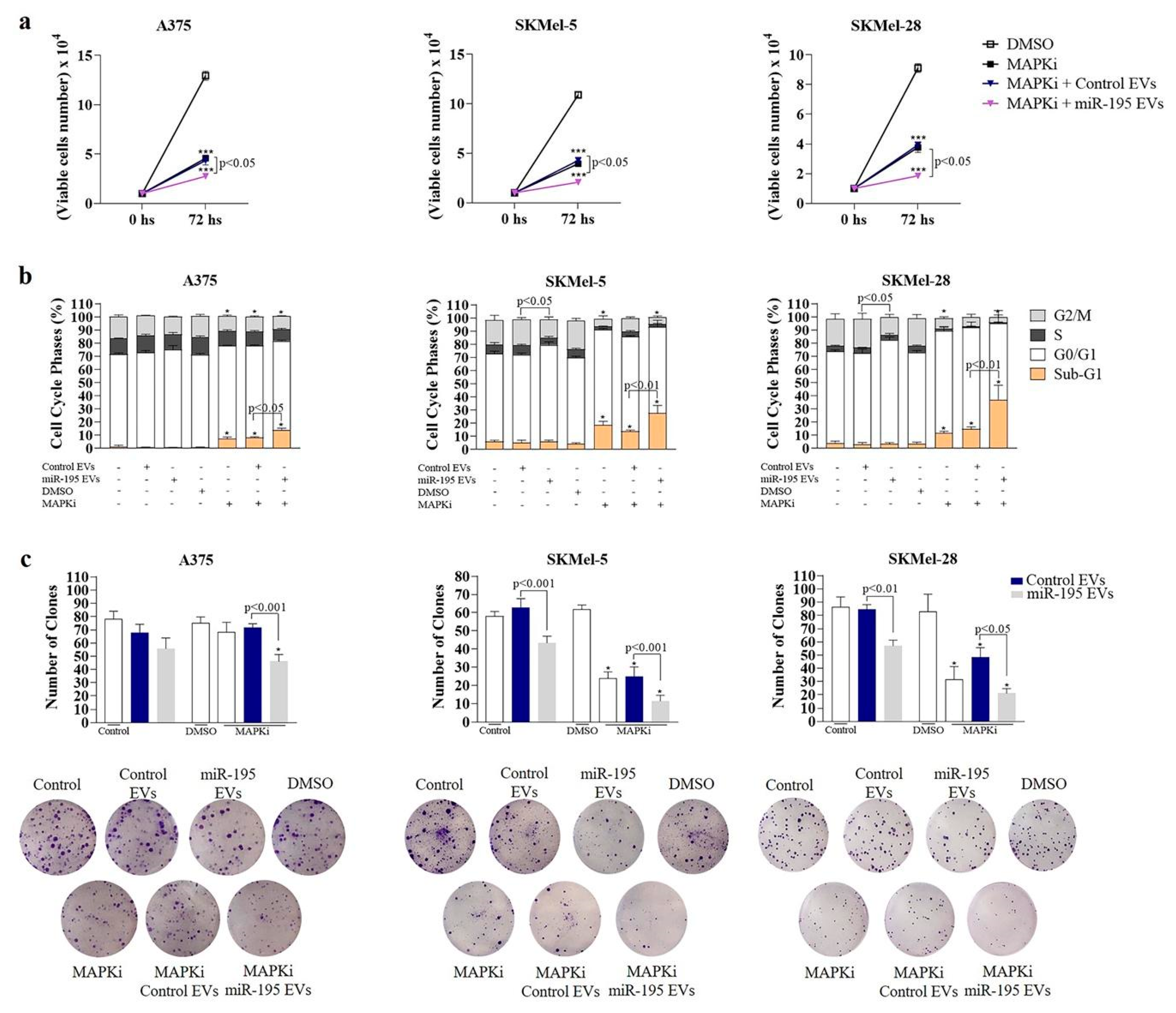
2.9.4. Clonogenic Assay
2.9.5. BRAF and MEK Inhibitors Treatment
2.10. Electroporation of EVs
2.11. In Vivo Tumor Growth
2.12. In Silico Analysis
2.13. Statistical Analysis
3. Results
3.1. MiR-195-5p Overexpression Modulates Melanoma Cells’ Vesiculation Profile
3.2. Overexpressing miR-195-5p Alters EV miRNA Content
3.3. MiR-195 EVs Exerts an Anti-Proliferative Effect in Bystander Tumor Cells
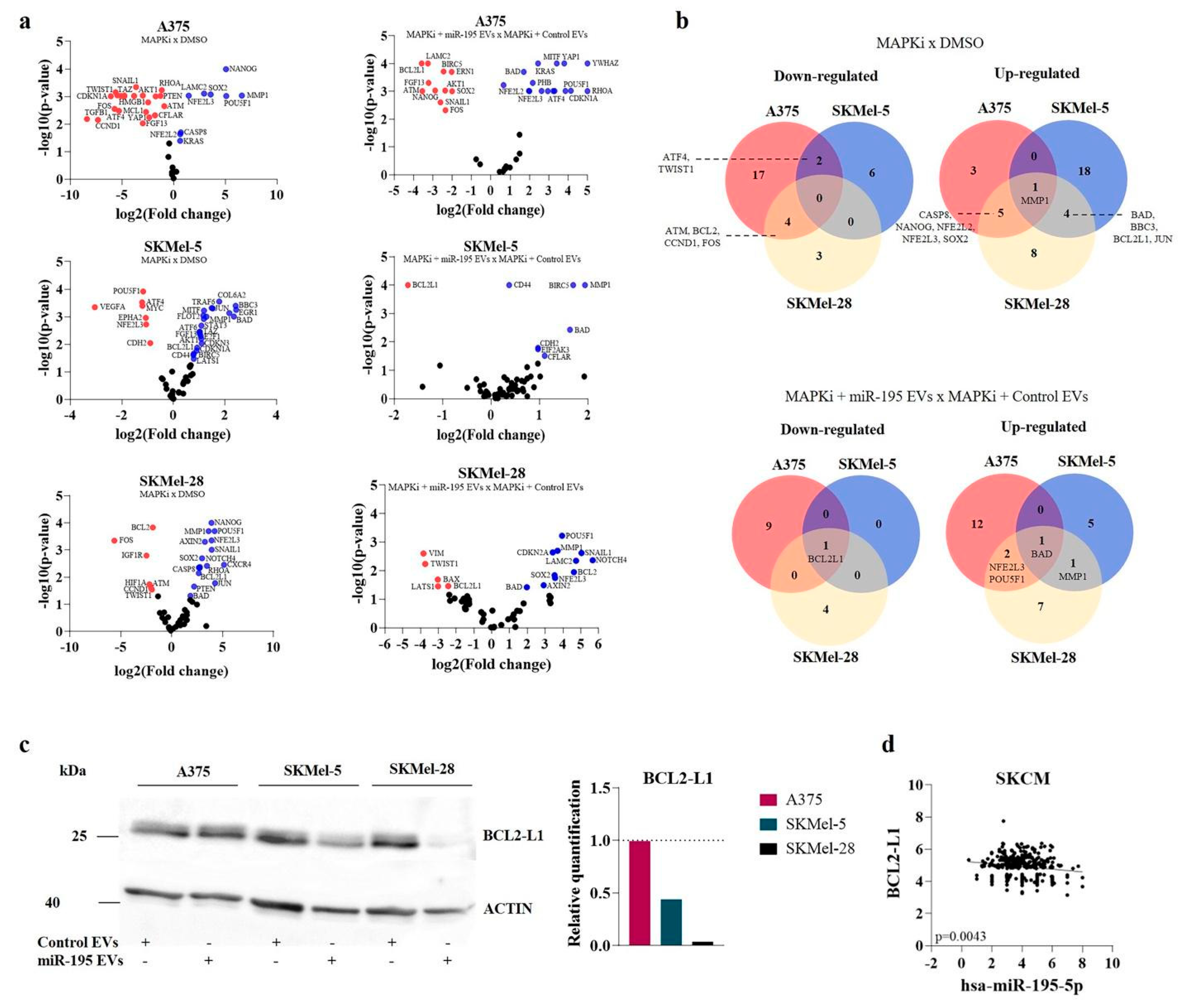
3.4. MiR-195 EVs Sensitize Recipient Cells to Combined Targeted Therapy
3.5. EVs Exogenously Loaded with miR-195-5p Impairs In Vivo Melanoma Growth and Repopulation Capacity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mackiewicz, J.; Mackiewicz, A. BRAF and MEK inhibitors in the era of immunotherapy in melanoma patients. Contemp. Oncol. 2018, 22, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Grob, J.J.; Stroyakovskiy, D.; Karaszewska, B.; Hauschild, A.; Levchenko, E.; Chiarion Sileni, V.; Schachter, J.; Garbe, C.; Bondarenko, I.; et al. Five-Year Outcomes with Dabrafenib plus Trametinib in Metastatic Melanoma. N. Engl. J. Med. 2019, 381, 626–636. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cells 2019, 177, 428–445.e18. [Google Scholar] [CrossRef] [PubMed]
- Santi, A.; Caselli, A.; Ranaldi, F.; Paoli, P.; Mugnaioni, C.; Michelucci, E.; Cirri, P. Cancer associated fibroblasts transfer lipids and proteins to cancer cells through cargo vesicles supporting tumor growth. Biochim. et Biophys. Acta 2015, 1853, 3211–3223. [Google Scholar] [CrossRef]
- Jiang, M.J.; Chen, Y.Y.; Dai, J.J.; Gu, D.N.; Mei, Z.; Liu, F.R.; Huang, Q.; Tian, L. Dying tumor cell-derived exosomal miR-194-5p potentiates survival and repopulation of tumor repopulating cells upon radiotherapy in pancreatic cancer. Mol. Cancer 2020, 19, 68. [Google Scholar] [CrossRef]
- Yoshida, K.; Tsuda, M.; Matsumoto, R.; Semba, S.; Wang, L.; Sugino, H.; Tanino, M.; Kondo, T.; Tanabe, K.; Tanaka, S. Exosomes containing ErbB2/CRK induce vascular growth in premetastatic niches and promote metastasis of bladder cancer. Cancer Sci. 2019, 110, 2119–2132. [Google Scholar] [CrossRef]
- Andrade, L.N.S.; Otake, A.H.; Cardim, S.G.B.; da Silva, F.I.; Ikoma Sakamoto, M.M.; Furuya, T.K.; Uno, M.; Pasini, F.S.; Chammas, R. Extracellular Vesicles Shedding Promotes Melanoma Growth in Response to Chemotherapy. Sci. Rep. 2019, 9, 14482. [Google Scholar] [CrossRef]
- Svedman, F.C.; Lohcharoenkal, W.; Bottai, M.; Brage, S.E.; Sonkoly, E.; Hansson, J.; Pivarcsi, A.; Eriksson, H. Extracellular microvesicle microRNAs as predictive biomarkers for targeted therapy in metastastic cutaneous malignant melanoma. PLoS ONE 2018, 13, e0206942. [Google Scholar] [CrossRef]
- Gad, S.A.; Ali, H.E.A.; Gaballa, R.; Abdelsalam, R.M.; Zerfaoui, M.; Ali, H.I.; Salama, S.H.; Kenawy, S.A.; Kandil, E.; Abd Elmageed, Z.Y. Targeting CDC7 sensitizes resistance melanoma cells to BRAFV600E-specific inhibitor by blocking the CDC7/MCM2-7 pathway. Sci. Rep. 2019, 9, 14197. [Google Scholar] [CrossRef]
- Alegre, E.; Sanmamed, M.F.; Rodriguez, C.; Carranza, O.; Martín-Algarra, S.; González, A. Study of circulating microRNA-125b levels in serum exosomes in advanced melanoma. Arch. Pathol. Lab. Med. 2014, 138, 828–832. [Google Scholar] [CrossRef]
- Lee, J.H.; Dindorf, J.; Eberhardt, M.; Lai, X.; Ostalecki, C.; Koliha, N.; Gross, S.; Blume, K.; Bruns, H.; Wild, S.; et al. Innate extracellular vesicles from melanoma patients suppress β-catenin in tumor cells by miRNA-34a. Life Sci. Alliance 2019, 2, e201800205. [Google Scholar] [CrossRef]
- Cirilo, P.D.R.; de Sousa Andrade, L.N.; Corrêa, B.R.S.; Qiao, M.; Furuya, T.K.; Chammas, R.; Penalva, L.O.F. MicroRNA-195 acts as an anti-proliferative miRNA in human melanoma cells by targeting Prohibitin. BMC Cancer 2017, 17, 750. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatantsand biological fluids. Curr. Protoc. Cell Biol. 2006, 3. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Verweij, F.J.; van Eijndhoven, M.A.; Middeldorp, J.; Pegtel, D.M. Analysis of viral microRNA exchange via exosomes in vitro and in vivo. Methods Mol. Biol. 2013, 1024, 53–68. [Google Scholar]
- Pomatto, M.A.C.; Bussolati, B.; D’Antico, S.; Ghiotto, S.; Tetta, C.; Brizzi, M.F.; Camussi, G. Improved Loading of Plasma-Derived Extracellular Vesicles to Encapsulate Antitumor miRNAs. Mol. Ther. Methods Clin. Dev. 2019, 13, 133–144. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal. Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Dorris, E.R.; Blackshields, G.; Sommerville, G.; Alhashemi, M.; Dias, A.; McEneaney, V.; Smyth, P.; O’Leary, J.J.; Sheils, O. Pluripotency markers are differentially induced by MEK inhibition in thyroid and melanoma BRAFV600E cell lines. Cancer Biol. Ther. 2016, 17, 526–542. [Google Scholar] [CrossRef]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef]
- Rajabi, P.; Neshat, A.; Mokhtari, M.; Rajabi, M.A.; Eftekhari, M.; Tavakoli, P. The role of VEGF in melanoma progression. J. Res. Med. Sci. 2012, 17, 534–539. [Google Scholar]
- Wei, D.; Zhan, W.; Gao, Y.; Huang, L.; Gong, R.; Wang, W.; Zhang, R.; Wu, Y.; Gao, S.; Kang, T. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021, 31, 157–177. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhuang, X.; Greene, K.S.; Si, H.; Antonyak, M.A.; Druso, J.E.; Wilson, K.F.; Cerione, R.A.; Feng, Q.; Wang, H. Cdc42 functions as a regulatory node for tumour-derived microvesicle biogenesis. J. Extracell. Vesicles 2021, 10, e12051. [Google Scholar] [CrossRef] [PubMed]
- Chae, D.K.; Park, J.; Cho, M.; Ban, E.; Jang, M.; Yoo, Y.S.; Kim, E.E.; Baik, J.H.; Song, E.J. MiR-195 and miR-497 suppress tumorigenesis in lung cancer by inhibiting SMURF2-induced TGF-β receptor I ubiquitination. Mol. Oncol. 2019, 13, 2663–2678. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wang, S.; Wang, S. MiR-195 suppresses colon cancer proliferation and metastasis by targeting WNT3A. Mol. Genet. Genom. 2018, 293, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
- Purohit, P.K.; Edwards, R.; Tokatlidis, K.; Saini, N. MiR-195 regulates mitochondrial function by targeting mitofusin-2 in breast cancer cells. RNA Biol. 2019, 16, 918–929. [Google Scholar] [CrossRef]
- Ma, X.; Zou, L.; Chen, Z.; Li, X.; Wei, L.; Wu, X. Demethylation of miR-195 suppresses prostate cancer cell proliferation, migration and invasion. FEBS Open Bio. 2020, 10, 525–534. [Google Scholar] [CrossRef]
- Jiang, X.; Ding, W.; Shen, W.; Jin, J. H19/miR-152-3p/TCF4 axis increases chemosensitivity of gastric cancer cells through suppression of epithelial-mesenchymal transition. Transl. Cancer Res. 2020, 9, 3915–3925. [Google Scholar] [CrossRef]
- Li, B.; Lu, X.; Ma, C.; Sun, S.; Shu, X.; Wang, Z.; Sun, W. Long non-coding RNA NEAT1 promotes human glioma tumor progression via miR-152-3p/CCT6A pathway. Neurosci. Lett. 2020, 732, 135086. [Google Scholar] [CrossRef]
- Zhu, X.; Shen, Z.; Man, D.; Ruan, H.; Huang, S. miR-152-3p Affects the Progression of Colon Cancer via the KLF4/IFITM3 Axis. Comput. Math. Methods Med. 2020, 2020, 8209504. [Google Scholar] [CrossRef]
- Ning, N.; Liu, S.; Liu, X.; Tian, Z.; Jiang, Y.; Yu, N.; Tan, B.; Feng, H.; Feng, X.; Zou, L. Curcumol inhibits the proliferation and metastasis of melanoma via the miR-152-3p/PI3K/AKT and ERK/NF-κB signaling pathways. J. Cancer 2020, 11, 1679–1692. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, J.; Zhang, Q.; Xu, H.; Liu, L. Long Non-Coding RNA HOXA11-AS Modulates Proliferation, Apoptosis, Metastasis and EMT in Cutaneous Melanoma Cells Partly via miR-152-3p/ITGA9 Axis. Cancer Manag. Res. 2021, 13, 925–939. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, Z.; Guo, W.; Ni, S.; Xiao, X.; Wang, L.; Huang, D.; Tan, C.; Xu, Q.; Zha, R.; et al. microRNA-202-3p inhibits cell proliferation by targeting ADP-ribosylation factor-like 5A in human colorectal carcinoma. Clin. Cancer Res. 2014, 20, 1146–1157. [Google Scholar] [CrossRef]
- Chen, J.; Yin, J.; Liu, J.; Zhu, R.X.; Zheng, Y.; Wang, X.L. MiR-202-3p functions as a tumor suppressor and reduces cell migration and invasion in papillary thyroid carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1145–1150. [Google Scholar]
- Harati, R.; Hafezi, S.; Mabondzo, A.; Tlili, A. Silencing miR-202-3p increases MMP-1 and promotes a brain invasive phenotype in metastatic breast cancer cells. PLoS ONE 2020, 15, e0239292. [Google Scholar] [CrossRef]
- Li, H.J.; Sun, X.M.; Li, Z.K.; Yin, Q.W.; Pang, H.; Pan, J.J.; Li, X.; Chen, W. LncRNA UCA1 Promotes Mitochondrial Function of Bladder Cancer via the MiR-195/ARL2 Signaling Pathway. Cell. Physiol. Biochem. 2017, 43, 2548–2561. [Google Scholar] [CrossRef]
- González-Ruiz, L.; González-Moles, M.Á.; González-Ruiz, I.; Ruiz-Ávila, I.; Ramos-García, P. Prognostic and Clinicopathological Significance of CCND1/Cyclin D1 Upregulation in Melanomas: A Systematic Review and Comprehensive Meta-Analysis. Cancers 2021, 13, 1314. [Google Scholar] [CrossRef]
- Yu, C.; Zhang, M.; Song, J.; Zheng, X.; Xu, G.; Bao, Y.; Lan, J.; Luo, D.; Hu, J.; Li, J.J.; et al. Integrin-Src-YAP1 signaling mediates the melanoma acquired resistance to MAPK and PI3K/mTOR dual targeted therapy. Mol. Biomed. 2020, 1, 12. [Google Scholar] [CrossRef]
- Lunavat, T.R.; Cheng, L.; Einarsdottir, B.O.; Olofsson Bagge, R.; Veppil Muralidharan, S.; Sharples, R.A.; Lässer, C.; Gho, Y.S.; Hill, A.F.; Nilsson, J.A.; et al. BRAFV600 inhibition alters the microRNA cargo in the vesicular secretome of malignant melanoma cells. Proc. Natl. Acad. Sci. USA 2017, 114, E5930–E5939. [Google Scholar] [CrossRef]
- Heere-Ress, E.; Thallinger, C.; Lucas, T.; Schlagbauer-Wadl, H.; Wacheck, V.; Monia, B.P.; Wolff, K.; Pehamberger, H.; Jansen, B. Bcl-X(L) is a chemoresistance factor in human melanoma cells that can be inhibited by antisense therapy. Int. J. Cancer 2002, 99, 29–34. [Google Scholar] [CrossRef]
- Wolter, K.G.; Verhaegen, M.; Fernández, Y.; Nikolovska-Coleska, Z.; Riblett, M.; de la Vega, C.M.; Wang, S.; Soengas, M.S. Therapeutic window for melanoma treatment provided by selective effects of the proteasome on Bcl-2 proteins. Cell. Death Differ. 2007, 14, 1605–1616. [Google Scholar] [CrossRef]
- Trisciuoglio, D.; Tupone, M.G.; Desideri, M.; Di Martile, M.; Gabellini, C.; Buglioni, S.; Pallocca, M.; Alessandrini, G.; D’Aguanno, S.; Del Bufalo, D. BCL-XL overexpression promotes tumor progression-associated properties. Cell Death Dis. 2017, 8, 3216. [Google Scholar] [CrossRef] [PubMed]
- Munir, J.; Yoon, J.K.; Ryu, S. Therapeutic miRNA-Enriched Extracellular Vesicles: Current Approaches and Future Prospects. Cells 2020, 9, 2271. [Google Scholar] [CrossRef] [PubMed]


| Control Mimic | miR-195 Mimic | Control EVs | miR-195 EVs | |||||
|---|---|---|---|---|---|---|---|---|
| miRNA | Normilized Count | miRNA | Normilized Count | miRNA | Normilized Count | miRNA | Normilized Count | |
| A375 | hsa-let-7a-5p | 23,086 | hsa-miR-195-5p | 200,788 | hsa-miR-320e | 197 | hsa-miR-195-5p | 14,321 |
| hsa-miR-4454+hsa-miR-7975 | 10,093 | hsa-let-7a-5p | 21,331 | hsa-miR-371a-5p | 179 | hsa-miR-320e | 208 | |
| hsa-miR-125b-5p | 9663 | hsa-miR-125b-5p | 18,233 | hsa-miR-1246 | 169 | hsa-miR-371a-5p | 190 | |
| hsa-miR-29b-3p | 6791 | hsa-miR-4454+hsa-miR-7975 | 14,716 | hsa-miR-1283 | 135 | hsa-miR-630 | 151 | |
| hsa-miR-100-5p | 6705 | hsa-miR-29a-3p | 9444 | hsa-miR-379-5p | 105 | hsa-miR-202-3p | 135 | |
| hsa-miR-23a-3p | 6342 | hsa-miR-100-5p | 9173 | hsa-miR-1253 | 98 | hsa-miR-1283 | 132 | |
| hsa-miR-15b-5p | 4469 | hsa-miR-29b-3p | 7577 | hsa-miR-765 | 96 | hsa-miR-1246 | 117 | |
| hsa-miR-19b-3p | 4347 | hsa-miR-23a-3p | 6398 | hsa-miR-378e | 92 | hsa-miR-152-3p | 102 | |
| hsa-miR-16-5p | 3972 | hsa-miR-19b-3p | 5698 | hsa-miR-1285-5p | 86 | hsa-miR-765 | 93 | |
| hsa-let-7g-5p | 3871 | hsa-miR-15b-5p | 4058 | hsa-miR-630 | 82 | hsa-miR-379-5p | 91 | |
| SKMel-5 | hsa-miR-4454+hsa-miR-7975 | 143,888 | hsa-miR-195-5p | 16,4171 | hsa-miR-320e | 491 | hsa-miR-195-5p | 14,270 |
| hsa-miR-29a-3p | 21,561 | hsa-miR-29a-3p | 42,260 | hsa-miR-1246 | 230 | hsa-miR-320e | 169 | |
| hsa-miR-29b-3p | 19,295 | hsa-miR-4454+hsa-miR-7975 | 30,055 | hsa-miR-1283 | 135 | hsa-miR-1246 | 139 | |
| hsa-let-7a-5p | 9065 | hsa-miR-29b-3p | 26,661 | hsa-miR-1253 | 126 | hsa-miR-202-3p | 123 | |
| hsa-miR-21-5p | 4986 | hsa-let-7a-5p | 13,552 | hsa-miR-378e | 102 | hsa-miR-152-3p | 110 | |
| hsa-let-7g-5p | 4632 | hsa-miR-25-3p | 7336 | hsa-miR-379-5p | 100 | hsa-miR-765 | 100 | |
| hsa-miR-25-3p | 4464 | hsa-miR-23a-3p | 7071 | hsa-miR-371a-5p | 98 | hsa-miR-379-5p | 94 | |
| hsa-miR-146a-5p | 4391 | hsa-miR-21-5p | 6453 | hsa-miR-411-5p | 90 | hsa-miR-1253 | 89 | |
| hsa-miR-16-5p | 4322 | hsa-miR-15b-5p | 6317 | hsa-miR-765 | 89 | hsa-miR-378e | 82 | |
| hsa-miR-15b-5p | 4181 | hsa-let-7g-5p | 5754 | hsa-miR-212-3p | 84 | hsa-miR-411-5p | 81 | |
| SKMel-28 | hsa-miR-29a-3p | 19,593 | hsa-miR-195-5p | 88,671 | hsa-miR-1246 | 174 | hsa-miR-195-5p | 16,913 |
| hsa-miR-29b-3p | 18,515 | hsa-miR-29b-3p | 44,201 | hsa-miR-765 | 136 | hsa-miR-202-3p | 178 | |
| hsa-let-7a-5p | 14,479 | hsa-miR-4454+hsa-miR-7975 | 28,889 | hsa-miR-1283 | 132 | hsa-miR-1283 | 176 | |
| hsa-miR-4454+hsa-miR-7975 | 13,037 | hsa-let-7a-5p | 23,313 | hsa-miR-212-3p | 119 | hsa-miR-1253 | 155 | |
| hsa-miR-16-5p | 8450 | hsa-miR-29a-3p | 12,848 | hsa-miR-28-5p | 105 | hsa-miR-1246 | 137 | |
| hsa-miR-15b-5p | 6679 | hsa-miR-23a-3p | 9675 | hsa-miR-1253 | 100 | hsa-miR-152-3p | 129 | |
| hsa-miR-23a-3p | 5931 | hsa-miR-15b-5p | 4723 | hsa-miR-320e | 92 | hsa-miR-378e | 103 | |
| hsa-let-7i-5p | 3998 | hsa-let-7g-5p | 3906 | hsa-miR-411-5p | 89 | hsa-miR-765 | 101 | |
| hsa-let-7g-5p | 3071 | hsa-miR-25-3p | 3200 | hsa-miR-873-3p | 86 | hsa-miR-371a-5p | 98 | |
| hsa-miR-21-5p | 3036 | hsa-miR-374a-5p | 2586 | hsa-miR-302d-3p | 82 | hsa-miR-379-5p | 84 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, N.L.; Bustos, S.O.; Reis, P.P.; Chammas, R.; Andrade, L.N.S. Extracellular Vesicle-Packaged miR-195-5p Sensitizes Melanoma to Targeted Therapy with Kinase Inhibitors. Cells 2023, 12, 1317. https://doi.org/10.3390/cells12091317
Santos NL, Bustos SO, Reis PP, Chammas R, Andrade LNS. Extracellular Vesicle-Packaged miR-195-5p Sensitizes Melanoma to Targeted Therapy with Kinase Inhibitors. Cells. 2023; 12(9):1317. https://doi.org/10.3390/cells12091317
Chicago/Turabian StyleSantos, Nathalia L., Silvina O. Bustos, Patricia P. Reis, Roger Chammas, and Luciana N. S. Andrade. 2023. "Extracellular Vesicle-Packaged miR-195-5p Sensitizes Melanoma to Targeted Therapy with Kinase Inhibitors" Cells 12, no. 9: 1317. https://doi.org/10.3390/cells12091317
APA StyleSantos, N. L., Bustos, S. O., Reis, P. P., Chammas, R., & Andrade, L. N. S. (2023). Extracellular Vesicle-Packaged miR-195-5p Sensitizes Melanoma to Targeted Therapy with Kinase Inhibitors. Cells, 12(9), 1317. https://doi.org/10.3390/cells12091317