Unraveling the Oncogenic Potential of VAV1 in Human Cancer: Lessons from Mouse Models
Abstract
1. Introduction
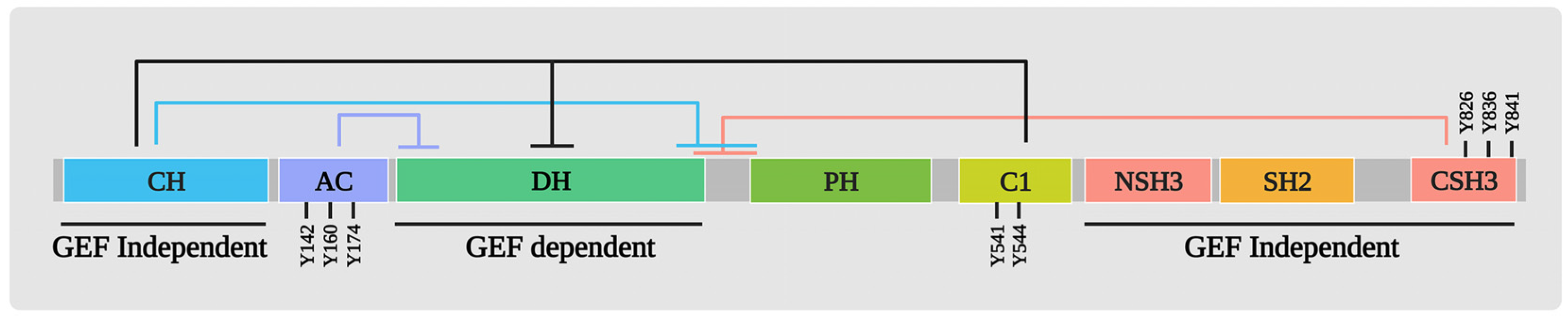
2. Structure/Function of VAV1
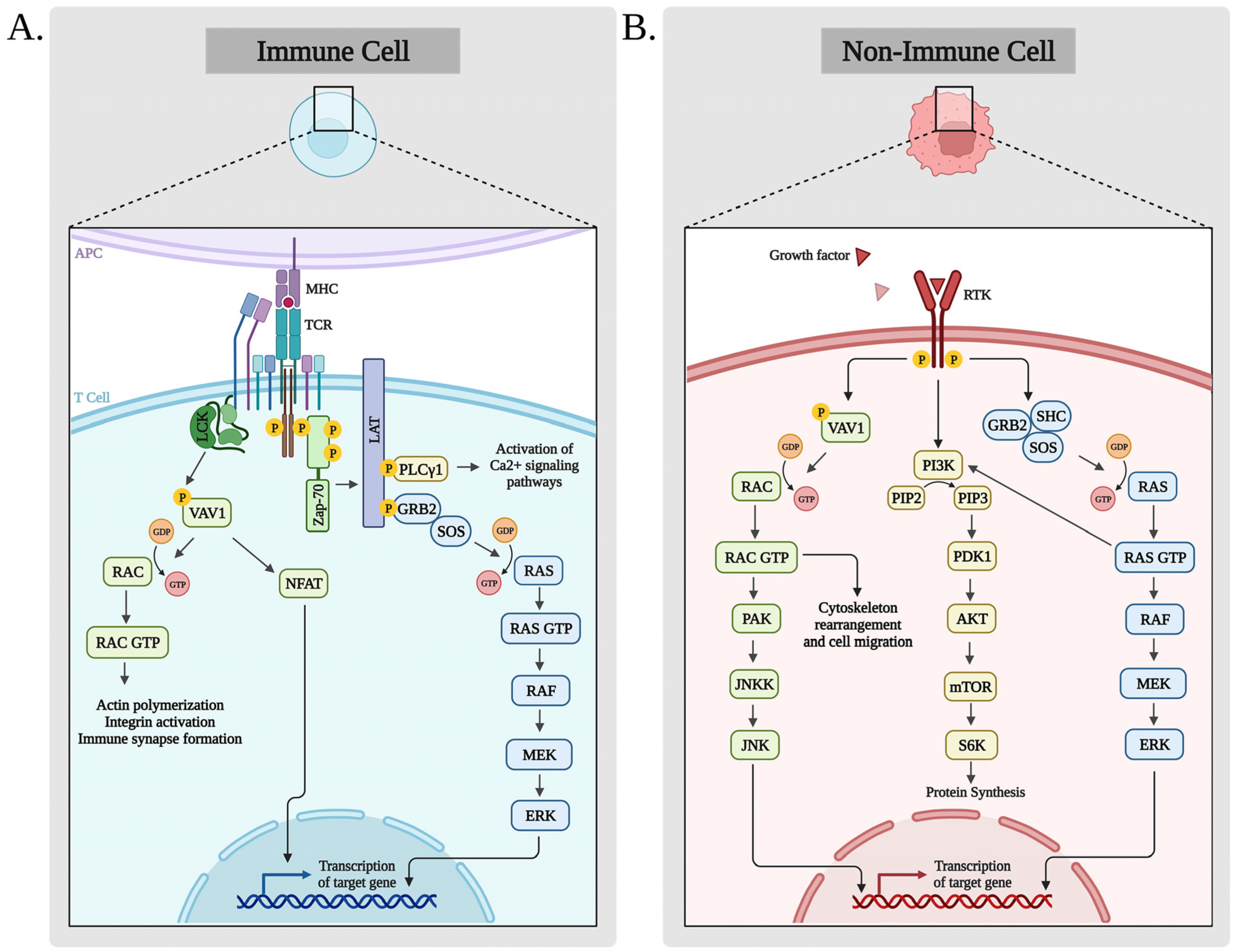
3. Biological Functions of VAV1
4. VAV1 in Human Cancer
5. VAV1 Ectopic Expression in Human Tumors
6. Mutations of VAV1 in Human Cancer
6.1. VAV1 Mutants in Human Solid Tumors

6.2. VAV1 Mutants in Human Hematological Malignancies
6.3. Differences in Mutations in VAV1 between Solid and Hematopoietic Human Malignancies
6.4. Function of VAV1 Mutants in Human Cancer
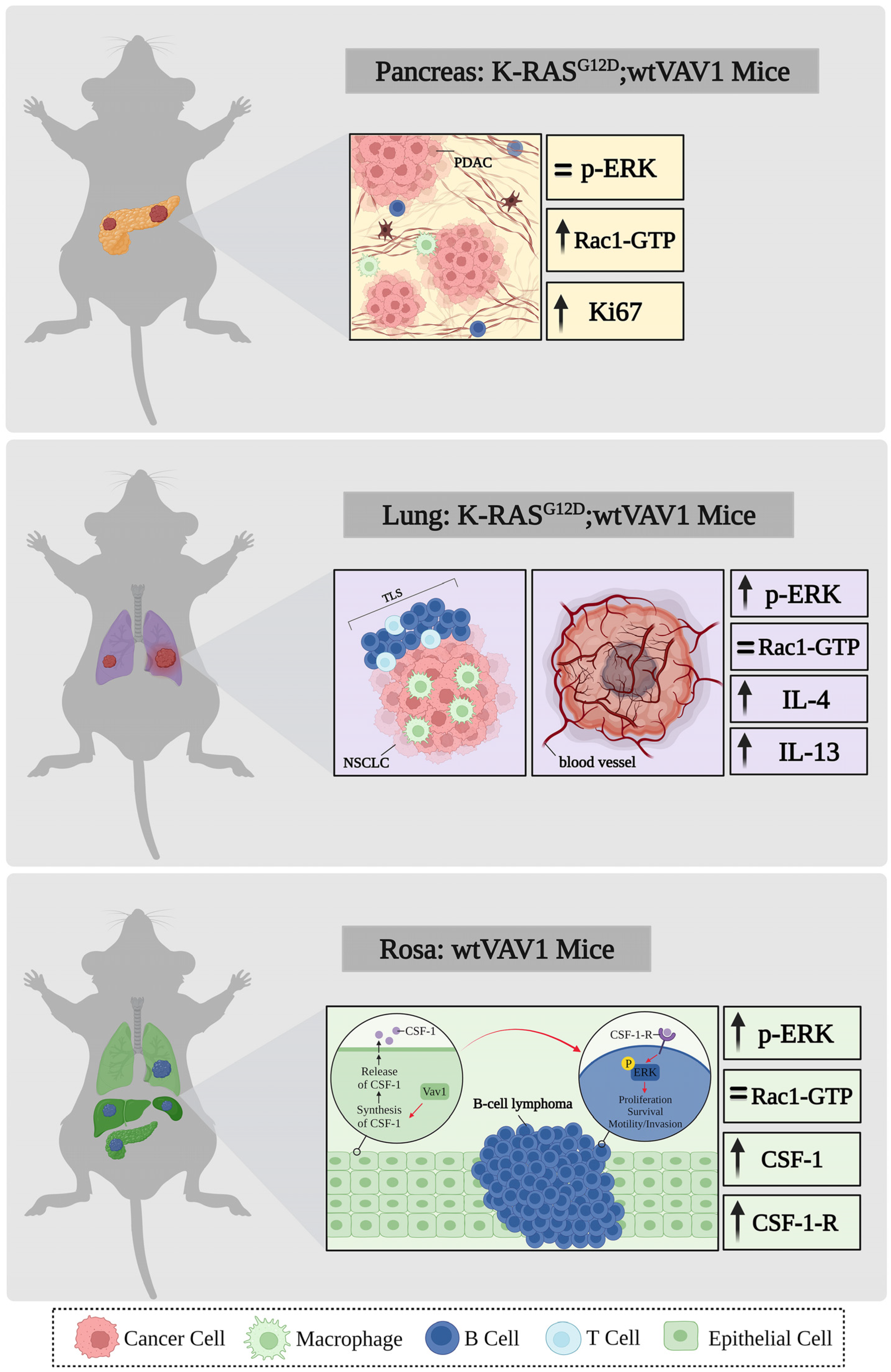
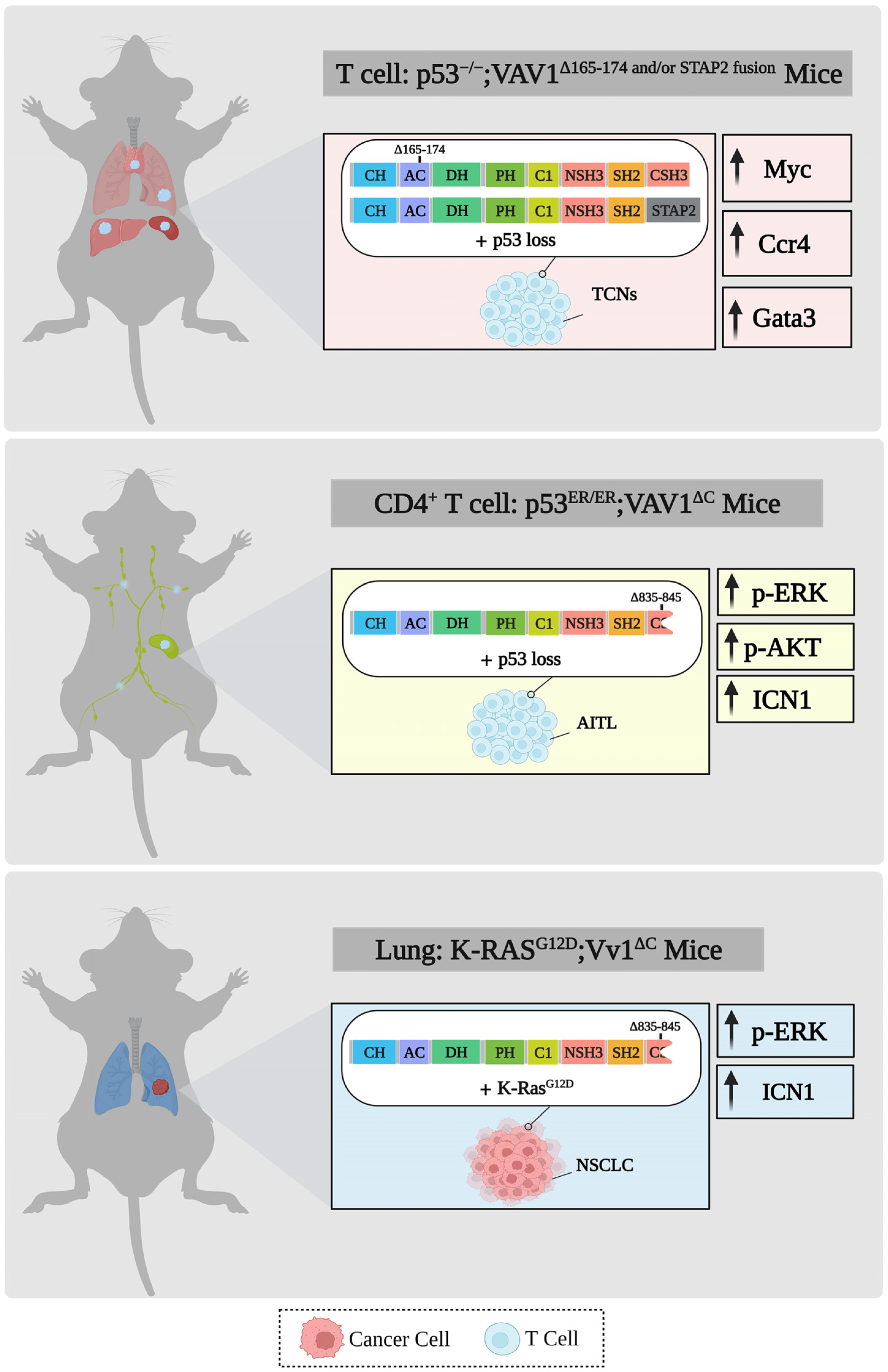
7. VAV1 and Genetically-Engineered Mouse Models (GEMMs)
8. Open Questions and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Katzav, S.; Martin-Zanca, D.; Barbacid, M. vav, a novel human oncogene derived from a locus ubiquitously expressed in hematopoietic cells. EMBO J. 1989, 8, 2283–2290. [Google Scholar] [CrossRef] [PubMed]
- Katzav, S. Vav1: An oncogene that regulates specific transcriptional activation of T cells. Blood 2004, 103, 2443–2451. [Google Scholar] [CrossRef]
- Crespo, P.; Schuebel, K.E.; Ostrom, A.A.; Gutkind, J.S.; Bustelo, X.R. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature 1997, 385, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Sini, P.; Cannas, A.; Koleske, A.J.; Di Fiore, P.P.; Scita, G. Abl-dependent tyrosine phosphorylation of Sos-1 mediates growth-factor-induced Rac activation. Nat. Cell Biol. 2004, 6, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Herrero, S.; Fernández-Infante, C.; Hernández-Cano, L.; Ortiz-Rivero, S.; Guijas, C.; Martín-Granado, V.; González-Porras, J.R.; Balsinde, J.; Porras, A.; Guerrero, C. C3G contributes to platelet activation and aggregation by regulating major signaling pathways. Signal Transduct. Target. Ther. 2020, 5, 29. [Google Scholar] [CrossRef]
- Schiller, M.R. Coupling receptor tyrosine kinases to Rho GTPases—GEFs what’s the link. Cell. Signal. 2006, 18, 1834–1843. [Google Scholar] [CrossRef]
- Katzav, S. Flesh and blood: The story of Vav1, a gene that signals in hematopoietic cells but can be transforming in human malignancies. Cancer Lett. 2007, 255, 241–254. [Google Scholar] [CrossRef]
- Tybulewicz, V.L.J. Vav-family proteins in T-cell signalling. Curr. Opin. Immunol. 2005, 17, 267–274. [Google Scholar] [CrossRef]
- Zugaza, J.L.; López-Lago, M.A.; Caloca, M.J.; Dosil, M.; Movilla, N.; Bustelo, X.R. Structural determinants for the biological activity of Vav proteins. J. Biol. Chem. 2002, 277, 45377–45392. [Google Scholar] [CrossRef]
- Aghazadeh, B.; Lowry, W.E.; Huang, X.Y.; Rosen, M.K. Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell 2000, 102, 625–633. [Google Scholar] [CrossRef]
- Galland, F.; Katzav, S.; Birnbaum, D. The products of the mcf-2 and vav proto-oncogenes and of the yeast gene cdc-24 share sequence similarities. Oncogene 1992, 7, 585–587. [Google Scholar] [PubMed]
- Prisco, A.; Vanes, L.; Ruf, S.; Trigueros, C.; Tybulewicz, V.L.J. Lineage-specific requirement for the PH domain of Vav1 in the activation of CD4+ but not CD8+ T cells. Immunity 2005, 23, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.M.; Houston, H.; Allen, J.; Lints, T.; Harvey, R. The hematopoietically expressed vav proto-oncogene shares homology with the dbl GDP-GTP exchange factor, the bcr gene and a yeast gene (CDC24) involved in cytoskeletal organization. Oncogene 1992, 7, 611–618. [Google Scholar] [PubMed]
- Rodríguez-Fdez, S.; Citterio, C.; Lorenzo-Martín, L.F.; Baltanás-Copado, J.; Llorente-González, C.; Corbalán-García, S.; Vicente-Manzanares, M.; Bustelo, X.R. Phosphatidylinositol monophosphates regulate optimal vav1 signaling output. Cells 2019, 8, 1649. [Google Scholar] [CrossRef]
- Colón-González, F.; Kazanietz, M.G. C1 domains exposed: From diacylglycerol binding to protein-protein interactions. Biochim. Biophys. Acta 2006, 1761, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Rapley, J.; Tybulewicz, V.L.J.; Rittinger, K. Crucial structural role for the PH and C1 domains of the Vav1 exchange factor. EMBO Rep. 2008, 9, 655–661. [Google Scholar] [CrossRef]
- Ramos-Morales, F.; Romero, F.; Schweighoffer, F.; Bismuth, G.; Camonis, J.; Tortolero, M.; Fischer, S. The proline-rich region of Vav binds to Grb2 and Grb3-3. Oncogene 1995, 11, 1665–1669. [Google Scholar]
- Margolis, B.; Hu, P.; Katzav, S.; Li, W.; Oliver, J.M.; Ullrich, A.; Weiss, A.; Schlessinger, J. Tyrosine phosphorylation of vav proto-oncogene product containing SH2 domain and transcription factor motifs. Nature 1992, 356, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Bustelo, X.R.; Ledbetter, J.A.; Barbacid, M. Product of vav proto-oncogene defines a new class of tyrosine protein kinase substrates. Nature 1992, 356, 68–71. [Google Scholar] [CrossRef]
- Lazer, G.; Pe’er, L.; Farago, M.; Machida, K.; Mayer, B.J.; Katzav, S. Tyrosine residues at the carboxyl terminus of Vav1 play an important role in regulation of its biological activity. J. Biol. Chem. 2010, 285, 23075–23085. [Google Scholar] [CrossRef] [PubMed]
- Robles-Valero, J.; Lorenzo-Martín, L.F.; Menacho-Márquez, M.; Fernández-Pisonero, I.; Abad, A.; Camós, M.; Toribio, M.L.; Espinosa, L.; Bigas, A.; Bustelo, X.R. A Paradoxical Tumor-Suppressor Role for the Rac1 Exchange Factor Vav1 in T Cell Acute Lymphoblastic Leukemia. Cancer Cell 2017, 32, 608–623.e9. [Google Scholar] [CrossRef] [PubMed]
- Katzav, S. Vav1: A hematopoietic signal transduction molecule involved in human malignancies. Int. J. Biochem. Cell Biol. 2009, 41, 1245–1248. [Google Scholar] [CrossRef] [PubMed]
- Katzav, S. Vav1: A Dr. Jekyll and Mr. Hyde protein—Good for the hematopoietic system, bad for cancer. Oncotarget 2015, 6, 28731–28742. [Google Scholar] [CrossRef]
- Bustelo, X.R. Regulatory and signaling properties of the Vav family. Mol. Cell. Biol. 2000, 20, 1461–1477. [Google Scholar] [CrossRef]
- Movilla, N.; Dosil, M.; Zheng, Y.; Bustelo, X.R. How Vav proteins discriminate the GTPases Rac1 and RhoA from Cdc42. Oncogene 2001, 20, 8057–8065. [Google Scholar] [CrossRef]
- Schuebel, K.E.; Movilla, N.; Rosa, J.L.; Bustelo, X.R. Phosphorylation-dependent and constitutive activation of Rho proteins by wild-type and oncogenic Vav-2. EMBO J. 1998, 17, 6608–6621. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Rossman, K.L.; Liu, B.; Ritola, K.D.; Chiang, D.; Campbell, S.L.; Burridge, K.; Der, C.J. Vav2 is an activator of Cdc42, Rac1, and RhoA. J. Biol. Chem. 2000, 275, 10141–10149. [Google Scholar] [CrossRef] [PubMed]
- Bustelo, X.R.; Barbacid, M. Tyrosine phosphorylation of the vav proto-oncogene product in activated B cells. Science 1992, 256, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Manetz, T.S.; Gonzalez-Espinosa, C.; Arudchandran, R.; Xirasagar, S.; Tybulewicz, V.; Rivera, J. Vav1 regulates phospholipase cgamma activation and calcium responses in mast cells. Mol. Cell. Biol. 2001, 21, 3763–3774. [Google Scholar] [CrossRef]
- Evans, G.A.; Howard, O.M.; Erwin, R.; Farrar, W.L. Interleukin-2 induces tyrosine phosphorylation of the vav proto-oncogene product in human T cells: Lack of requirement for the tyrosine kinase lck. Biochem. J. 1993, 294 Pt 2, 339–342. [Google Scholar] [CrossRef]
- Galandrini, R.; Palmieri, G.; Piccoli, M.; Frati, L.; Santoni, A. Role for the Rac1 exchange factor Vav in the signaling pathways leading to NK cell cytotoxicity. J. Immunol. 1999, 162, 3148–3152. [Google Scholar] [CrossRef]
- García-Bernal, D.; Wright, N.; Sotillo-Mallo, E.; Nombela-Arrieta, C.; Stein, J.V.; Bustelo, X.R.; Teixidó, J. Vav1 and Rac control chemokine-promoted T lymphocyte adhesion mediated by the integrin alpha4beta1. Mol. Biol. Cell 2005, 16, 3223–3235. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Sjölander, A.; Eckerdal, J.; Andersson, T. Antibody-induced engagement of beta 2 integrins on adherent human neutrophils triggers activation of p21ras through tyrosine phosphorylation of the protooncogene product Vav. Proc. Natl. Acad. Sci. USA 1996, 93, 8431–8436. [Google Scholar] [CrossRef]
- Krawczyk, C.; Oliveira-dos-Santos, A.; Sasaki, T.; Griffiths, E.; Ohashi, P.S.; Snapper, S.; Alt, F.; Penninger, J.M. Vav1 controls integrin clustering and MHC/peptide-specific cell adhesion to antigen-presenting cells. Immunity 2002, 16, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Arana, E.; Vehlow, A.; Harwood, N.E.; Vigorito, E.; Henderson, R.; Turner, M.; Tybulewicz, V.L.J.; Batista, F.D. Activation of the small GTPase Rac2 via the B cell receptor regulates B cell adhesion and immunological-synapse formation. Immunity 2008, 28, 88–99. [Google Scholar] [CrossRef]
- Fischer, K.D.; Kong, Y.Y.; Nishina, H.; Tedford, K.; Marengère, L.E.; Kozieradzki, I.; Sasaki, T.; Starr, M.; Chan, G.; Gardener, S.; et al. Vav is a regulator of cytoskeletal reorganization mediated by the T-cell receptor. Curr. Biol. 1998, 8, 554–562. [Google Scholar] [CrossRef]
- Holsinger, L.J.; Graef, I.A.; Swat, W.; Chi, T.; Bautista, D.M.; Davidson, L.; Lewis, R.S.; Alt, F.W.; Crabtree, G.R. Defects in actin-cap formation in Vav-deficient mice implicate an actin requirement for lymphocyte signal transduction. Curr. Biol. 1998, 8, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Billadeau, D.D.; Brumbaugh, K.M.; Dick, C.J.; Schoon, R.A.; Bustelo, X.R.; Leibson, P.J. The Vav-Rac1 pathway in cytotoxic lymphocytes regulates the generation of cell-mediated killing. J. Exp. Med. 1998, 188, 549–559. [Google Scholar] [CrossRef]
- Hall, A.B.; Gakidis, M.A.M.; Glogauer, M.; Wilsbacher, J.L.; Gao, S.; Swat, W.; Brugge, J.S. Requirements for Vav guanine nucleotide exchange factors and Rho GTPases in FcgammaR- and complement-mediated phagocytosis. Immunity 2006, 24, 305–316. [Google Scholar] [CrossRef]
- Wells, C.M.; Walmsley, M.; Ooi, S.; Tybulewicz, V.; Ridley, A.J. Rac1-deficient macrophages exhibit defects in cell spreading and membrane ruffling but not migration. J. Cell Sci. 2004, 117, 1259–1268. [Google Scholar] [CrossRef]
- Vedham, V.; Phee, H.; Coggeshall, K.M. Vav activation and function as a rac guanine nucleotide exchange factor in macrophage colony-stimulating factor-induced macrophage chemotaxis. Mol. Cell. Biol. 2005, 25, 4211–4220. [Google Scholar] [CrossRef] [PubMed]
- Razidlo, G.L.; Wang, Y.; Chen, J.; Krueger, E.W.; Billadeau, D.D.; McNiven, M.A. Dynamin 2 potentiates invasive migration of pancreatic tumor cells through stabilization of the Rac1 GEF Vav1. Dev. Cell 2013, 24, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Razidlo, G.L.; Schroeder, B.; Chen, J.; Billadeau, D.D.; McNiven, M.A. Vav1 as a central regulator of invadopodia assembly. Curr. Biol. 2014, 24, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Katzav, S.; Weiss, A. A functional T-cell receptor signaling pathway is required for p95vav activity. Mol. Cell. Biol. 1995, 15, 4337–4346. [Google Scholar] [CrossRef] [PubMed]
- Costello, P.S.; Walters, A.E.; Mee, P.J.; Turner, M.; Reynolds, L.F.; Prisco, A.; Sarner, N.; Zamoyska, R.; Tybulewicz, V.L. The Rho-family GTP exchange factor Vav is a critical transducer of T cell receptor signals to the calcium, ERK, and NF-kappaB pathways. Proc. Natl. Acad. Sci. USA 1999, 96, 3035–3040. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.Y.; Fischer, K.D.; Bachmann, M.F.; Mariathasan, S.; Kozieradzki, I.; Nghiem, M.P.; Bouchard, D.; Bernstein, A.; Ohashi, P.S.; Penninger, J.M. Vav regulates peptide-specific apoptosis in thymocytes. J. Exp. Med. 1998, 188, 2099–2111. [Google Scholar] [CrossRef]
- Huang, M.-Y.; Wang, J.-Y.; Chang, H.-J.; Kuo, C.-W.; Tok, T.-S.; Lin, S.-R. CDC25A, VAV1, TP73, BRCA1 and ZAP70 gene overexpression correlates with radiation response in colorectal cancer. Oncol. Rep. 2011, 25, 1297–1306. [Google Scholar] [CrossRef]
- Zhao, T.; Benard, V.; Bohl, B.P.; Bokoch, G.M. The molecular basis for adhesion-mediated suppression of reactive oxygen species generation by human neutrophils. J. Clin. Investig. 2003, 112, 1732–1740. [Google Scholar] [CrossRef] [PubMed]
- Kaminuma, O.; Deckert, M.; Elly, C.; Liu, Y.C.; Altman, A. Vav-Rac1-mediated activation of the c-Jun N-terminal kinase/c-Jun/AP-1 pathway plays a major role in stimulation of the distal NFAT site in the interleukin-2 gene promoter. Mol. Cell. Biol. 2001, 21, 3126–3136. [Google Scholar] [CrossRef] [PubMed]
- Villalba, M.; Coudronniere, N.; Deckert, M.; Teixeiro, E.; Mas, P.; Altman, A. A novel functional interaction between Vav and PKCtheta is required for TCR-induced T cell activation. Immunity 2000, 12, 151–160. [Google Scholar] [CrossRef]
- Reynolds, L.F.; de Bettignies, C.; Norton, T.; Beeser, A.; Chernoff, J.; Tybulewicz, V.L.J. Vav1 transduces T cell receptor signals to the activation of the Ras/ERK pathway via LAT, Sos, and RasGRP1. J. Biol. Chem. 2004, 279, 18239–18246. [Google Scholar] [CrossRef]
- Katzav, S.; Cleveland, J.L.; Heslop, H.E.; Pulido, D. Loss of the amino-terminal helix-loop-helix domain of the vav proto-oncogene activates its transforming potential. Mol. Cell. Biol. 1991, 11, 1912–1920. [Google Scholar] [CrossRef]
- Coppola, J.; Bryant, S.; Koda, T.; Conway, D.; Barbacid, M. Mechanism of activation of the vav protooncogene. Cell Growth Differ. 1991, 2, 95–105. [Google Scholar]
- Yu, B.; Martins, I.R.S.; Li, P.; Amarasinghe, G.K.; Umetani, J.; Fernandez-Zapico, M.E.; Billadeau, D.D.; Machius, M.; Tomchick, D.R.; Rosen, M.K. Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell 2010, 140, 246–256. [Google Scholar] [CrossRef]
- Hornstein, I.; Pikarsky, E.; Groysman, M.; Amir, G.; Peylan-Ramu, N.; Katzav, S. The haematopoietic specific signal transducer Vav1 is expressed in a subset of human neuroblastomas. J. Pathol. 2003, 199, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Zapico, M.E.; Gonzalez-Paz, N.C.; Weiss, E.; Savoy, D.N.; Molina, J.R.; Fonseca, R.; Smyrk, T.C.; Chari, S.T.; Urrutia, R.; Billadeau, D.D. Ectopic expression of VAV1 reveals an unexpected role in pancreatic cancer tumorigenesis. Cancer Cell 2005, 7, 39–49. [Google Scholar] [CrossRef]
- Lazer, G.; Idelchuk, Y.; Schapira, V.; Pikarsky, E.; Katzav, S. The haematopoietic specific signal transducer Vav1 is aberrantly expressed in lung cancer and plays a role in tumourigenesis. J. Pathol. 2009, 219, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Sebban, S.; Farago, M.; Rabinovich, S.; Lazer, G.; Idelchuck, Y.; Ilan, L.; Pikarsky, E.; Katzav, S. Vav1 promotes lung cancer growth by instigating tumor-microenvironment cross-talk via growth factor secretion. Oncotarget 2014, 5, 9214–9226. [Google Scholar] [CrossRef]
- Grassilli, S.; Brugnoli, F.; Lattanzio, R.; Rossi, C.; Perracchio, L.; Mottolese, M.; Marchisio, M.; Palomba, M.; Nika, E.; Natali, P.G.; et al. High nuclear level of Vav1 is a positive prognostic factor in early invasive breast tumors: A role in modulating genes related to the efficiency of metastatic process. Oncotarget 2014, 5, 4320–4336. [Google Scholar] [CrossRef]
- Wakahashi, S.; Sudo, T.; Oka, N.; Ueno, S.; Yamaguchi, S.; Fujiwara, K.; Ohbayashi, C.; Nishimura, R. VAV1 represses E-cadherin expression through the transactivation of Snail and Slug: A potential mechanism for aberrant epithelial to mesenchymal transition in human epithelial ovarian cancer. Transl. Res. 2013, 162, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Jin, H.; Xia, Z.; Wu, X.; Yang, M.; Zhang, H.; Shang, X.; Cheng, R.; Zhan, Z.; Yu, Z. Vav1 expression is increased in esophageal squamous cell carcinoma and indicates poor prognosis. Biochem. Biophys. Res. Commun. 2017, 486, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, J.C.; Kawauchi, D.; Schwalbe, E.C.; Solecki, D.J.; Selby, M.P.; McKinnon, P.J.; Olson, J.M.; Hayden, J.T.; Grundy, R.G.; Ellison, D.W.; et al. Cross-species epigenetics identifies a critical role for VAV1 in SHH subgroup medulloblastoma maintenance. Oncogene 2015, 34, 4746–4757. [Google Scholar] [CrossRef]
- Kang, L.; Hao, X.; Tang, Y.; Zhao, Z.; Zhang, H.; Gong, Y. Elevated level of Vav1 was correlated with advanced biological behavior and poor prognosis in patients with gastric cancer. Int. J. Clin. Exp. Pathol. 2018, 11, 391–398. [Google Scholar]
- Prieto-Sánchez, R.M.; Hernández, J.A.; García, J.L.; Gutiérrez, N.C.; San Miguel, J.; Bustelo, X.R.; Hernández, J.M. Overexpression of the VAV proto-oncogene product is associated with B-cell chronic lymphocytic leukaemia displaying loss on 13q. Br. J. Haematol. 2006, 133, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Hollmann, A.; Aloyz, R.; Baker, K.; Dirnhofer, S.; Owens, T.; Sladek, R.; Tzankov, A. Vav-1 expression correlates with NFκB activation and CD40-mediated cell death in diffuse large B-cell lymphoma cell lines. Hematol. Oncol. 2010, 28, 142–150. [Google Scholar] [CrossRef]
- Mu, D.; Long, S.; Guo, L.; Liu, W. High expression of VAV gene family predicts poor prognosis of acute myeloid leukemia. Technol. Cancer Res. Treat. 2021, 20, 15330338211065876. [Google Scholar] [CrossRef] [PubMed]
- Abate, F.; da Silva-Almeida, A.C.; Zairis, S.; Robles-Valero, J.; Couronne, L.; Khiabanian, H.; Quinn, S.A.; Kim, M.-Y.; Laginestra, M.A.; Kim, C.; et al. Activating mutations and translocations in the guanine exchange factor VAV1 in peripheral T-cell lymphomas. Proc. Natl. Acad. Sci. USA 2017, 114, 764–769. [Google Scholar] [CrossRef]
- Huang, P.H.; Lu, P.J.; Ding, L.Y.; Chu, P.C.; Hsu, W.Y.; Chen, C.S.; Tsao, C.C.; Chen, B.H.; Lee, C.T.; Shan, Y.S.; et al. TGFβ promotes mesenchymal phenotype of pancreatic cancer cells, in part, through epigenetic activation of VAV1. Oncogene 2017, 36, 2202–2214. [Google Scholar] [CrossRef]
- Ilan, L.; Katzav, S. Human Vav1 expression in hematopoietic and cancer cell lines is regulated by c-Myb and by CpG methylation. PLoS ONE 2012, 7, e29939. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Das, B.; Wei, W.; Van Aelst, L.; Mosteller, R.D.; Khosravi-Far, R.; Westwick, J.K.; Der, C.J.; Broek, D. Lck regulates Vav activation of members of the Rho family of GTPases. Mol. Cell. Biol. 1997, 17, 1346–1353. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Fontana, L.I.; Barr, V.; Samelson, L.E.; Bierer, B.E. CD28 engagement promotes actin polymerization through the activation of the small Rho GTPase Cdc42 in human T cells. J. Immunol. 2003, 171, 2225–2232. [Google Scholar] [CrossRef]
- Godambe, S.A.; Knapp, K.M.; Meals, E.A.; English, B.K. Role of vav1 in the lipopolysaccharide-mediated upregulation of inducible nitric oxide synthase production and nuclear factor for interleukin-6 expression activity in murine macrophages. Clin. Diagn. Lab. Immunol. 2004, 11, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Darby, C.; Geahlen, R.L.; Schreiber, A.D. Stimulation of macrophage Fc gamma RIIIA activates the receptor-associated protein tyrosine kinase Syk and induces phosphorylation of multiple proteins including p95Vav and p62/GAP-associated protein. J. Immunol. 1994, 152, 5429–5437. [Google Scholar] [CrossRef]
- Sakai, H.; Chen, Y.; Itokawa, T.; Yu, K.-P.; Zhu, M.-L.; Insogna, K. Activated c-Fms recruits Vav and Rac during CSF-1-induced cytoskeletal remodeling and spreading in osteoclasts. Bone 2006, 39, 1290–1301. [Google Scholar] [CrossRef] [PubMed]
- Alai, M.; Mui, A.L.; Cutler, R.L.; Bustelo, X.R.; Barbacid, M.; Krystal, G. Steel factor stimulates the tyrosine phosphorylation of the proto-oncogene product, p95vav, in human hemopoietic cells. J. Biol. Chem. 1992, 267, 18021–18025. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, C.P.; Benzing, J.; Schmitt, T.; Erz, D.H.R.; Tewes, M.; Bartram, C.R.; Janssen, J.W.G. Novel interaction partners of the TPR/MET tyrosine kinase. FASEB J. 2005, 19, 267–269. [Google Scholar] [CrossRef]
- Hornstein, I.; Alcover, A.; Katzav, S. Vav proteins, masters of the world of cytoskeleton organization. Cell. Signal. 2004, 16, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sebban, S.; Farago, M.; Gashai, D.; Ilan, L.; Pikarsky, E.; Ben-Porath, I.; Katzav, S. Vav1 fine tunes p53 control of apoptosis versus proliferation in breast cancer. PLoS ONE 2013, 8, e54321. [Google Scholar] [CrossRef] [PubMed]
- Schapira, V.; Lazer, G.; Katzav, S. Osteopontin is an oncogenic Vav1- but not wild-type Vav1-responsive gene: Implications for fibroblast transformation. Cancer Res. 2006, 66, 6183–6191. [Google Scholar] [CrossRef]
- Wilsbacher, J.L.; Moores, S.L.; Brugge, J.S. An active form of Vav1 induces migration of mammary epithelial cells by stimulating secretion of an epidermal growth factor receptor ligand. Cell Commun. Signal. 2006, 4, 5. [Google Scholar] [CrossRef]
- Salaymeh, Y.; Farago, M.; Sebban, S.; Shalom, B.; Pikarsky, E.; Katzav, S. Vav1 and mutant K-Ras synergize in the early development of pancreatic ductal adenocarcinoma in mice. Life Sci. Alliance 2020, 3. [Google Scholar] [CrossRef] [PubMed]
- Shalom, B.; Farago, M.; Salaymeh, Y.; Sebban, S.; Pikarsky, E.; Katzav, S. Vav1 Promotes B-Cell Lymphoma Development. Cells 2022, 11, 949. [Google Scholar] [CrossRef] [PubMed]
- Shalom, B.; Farago, M.; Salaymeh, Y.; Sebban, S.; Risling, M.; Pikarsky, E.; Katzav, S. Vav1 accelerates Ras-driven lung cancer and modulates its tumor microenvironment. Cell. Signal. 2022, 97, 110395. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Kitazawa, M.; Miyagawa, Y.; Koyama, M.; Miyazaki, S.; Hondo, N.; Muranaka, F.; Tokumaru, S.; Yamamoto, Y.; Ehara, T.; et al. RhoA G17E/Vav1 Signaling Induces Cancer Invasion via Matrix Metalloproteinase-9 in Gastric Cancer. Technol. Cancer Res. Treat. 2023, 22, 15330338221146024. [Google Scholar] [CrossRef] [PubMed]
- Katzav, S. Single point mutations in the SH2 domain impair the transforming potential of vav and fail to activate proto-vav. Oncogene 1993, 8, 1757–1763. [Google Scholar]
- Groysman, M.; Nagano, M.; Shaanan, B.; Katzav, S. Mutagenic analysis of Vav reveals that an intact SH3 domain is required for transformation. Oncogene 1998, 17, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Razanadrakoto, L.; Cormier, F.; Laurienté, V.; Dondi, E.; Gardano, L.; Katzav, S.; Guittat, L.; Varin-Blank, N. Mutation of Vav1 adaptor region reveals a new oncogenic activation. Oncotarget 2015, 6, 2524–2537. [Google Scholar] [CrossRef]
- Campbell, J.D.; Alexandrov, A.; Kim, J.; Wala, J.; Berger, A.H.; Pedamallu, C.S.; Shukla, S.A.; Guo, G.; Brooks, A.N.; Murray, B.A.; et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 2016, 48, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, M.; Sakata-Yanagimoto, M.; Nishizawa, S.; Komori, D.; Gershon, P.; Kiryu, M.; Tanzima, S.; Fukumoto, K.; Enami, T.; Muratani, M.; et al. Activation of RHOA-VAV1 signaling in angioimmunoblastic T-cell lymphoma. Leukemia 2018, 32, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Shalom, B.; Farago, M.; Pikarsky, E.; Katzav, S. Vav1 mutations identified in human cancers give rise to different oncogenic phenotypes. Oncogenesis 2018, 7, 80. [Google Scholar] [CrossRef] [PubMed]
- Farago, M.; Yarnitzky, T.; Shalom, B.; Katzav, S. Vav1 mutations: What makes them oncogenic? Cell. Signal. 2020, 65, 109438. [Google Scholar] [CrossRef] [PubMed]
- Boddicker, R.L.; Razidlo, G.L.; Dasari, S.; Zeng, Y.; Hu, G.; Knudson, R.A.; Greipp, P.T.; Davila, J.I.; Johnson, S.H.; Porcher, J.C.; et al. Integrated mate-pair and RNA sequencing identifies novel, targetable gene fusions in peripheral T-cell lymphoma. Blood 2016, 128, 1234–1245. [Google Scholar] [CrossRef]
- Watatani, Y.; Sato, Y.; Miyoshi, H.; Sakamoto, K.; Nishida, K.; Gion, Y.; Nagata, Y.; Shiraishi, Y.; Chiba, K.; Tanaka, H.; et al. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia 2019, 33, 2867–2883. [Google Scholar] [CrossRef]
- Kataoka, K.; Nagata, Y.; Kitanaka, A.; Shiraishi, Y.; Shimamura, T.; Yasunaga, J.-I.; Totoki, Y.; Chiba, K.; Sato-Otsubo, A.; Nagae, G.; et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat. Genet. 2015, 47, 1304–1315. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ning, J.; Liu, X.; Chan, W.C.J. Mutations Affecting Genes in the Proximal T-Cell Receptor Signaling Pathway in Peripheral T-Cell Lymphoma. Cancers 2022, 14, 3716. [Google Scholar] [CrossRef] [PubMed]
- Drieux, F.; Ruminy, P.; Sater, V.; Marchand, V.; Fataccioli, V.; Lanic, M.-D.; Viennot, M.; Viailly, P.-J.; Sako, N.; Robe, C.; et al. Detection of Gene Fusion Transcripts in Peripheral T-Cell Lymphoma Using a Multiplexed Targeted Sequencing Assay. J. Mol. Diagn. 2021, 23, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Eckert, R.L.; Lee, K.C. S100A7 (Psoriasin): A story of mice and men. J. Investig. Dermatol. 2006, 126, 1442–1444. [Google Scholar] [CrossRef]
- Bianchetti, C.M.; Blouin, G.C.; Bitto, E.; Olson, J.S.; Phillips, G.N. The structure and NO binding properties of the nitrophorin-like heme-binding protein from Arabidopsis thaliana gene locus At1g79260.1. Proteins 2010, 78, 917–931. [Google Scholar] [CrossRef]
- Sakata-Yanagimoto, M.; Enami, T.; Yoshida, K.; Shiraishi, Y.; Ishii, R.; Miyake, Y.; Muto, H.; Tsuyama, N.; Sato-Otsubo, A.; Okuno, Y.; et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat. Genet. 2014, 46, 171–175. [Google Scholar] [CrossRef]
- Palomero, T.; Couronné, L.; Khiabanian, H.; Kim, M.-Y.; Ambesi-Impiombato, A.; Perez-Garcia, A.; Carpenter, Z.; Abate, F.; Allegretta, M.; Haydu, J.E.; et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat. Genet. 2014, 46, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Ikezoe, T.; Yang, J.; Nishioka, C.; Yokoyama, A. p53 is critical for the Aurora B kinase inhibitor-mediated apoptosis in acute myelogenous leukemia cells. Int. J. Hematol. 2010, 91, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Zenz, T.; Vollmer, D.; Trbusek, M.; Smardova, J.; Benner, A.; Soussi, T.; Helfrich, H.; Heuberger, M.; Hoth, P.; Fuge, M.; et al. TP53 mutation profile in chronic lymphocytic leukemia: Evidence for a disease specific profile from a comprehensive analysis of 268 mutations. Leukemia 2010, 24, 2072–2079. [Google Scholar] [CrossRef]
- Sande, C.M.; Chang, B.; Monga, V.; Bossler, A.D.; Ma, D. Biallelic TP53 gain of function mutations in rapidly progressing solid tumors. Cancer Genet. 2018, 222–223, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Foulkes, W.D.; Stefansson, I.M.; Chappuis, P.O.; Bégin, L.R.; Goffin, J.R.; Wong, N.; Trudel, M.; Akslen, L.A. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J. Natl. Cancer Inst. 2003, 95, 1482–1485. [Google Scholar] [CrossRef] [PubMed]
- Daver, N.G.; Maiti, A.; Kadia, T.M.; Vyas, P.; Majeti, R.; Wei, A.H.; Garcia-Manero, G.; Craddock, C.; Sallman, D.A.; Kantarjian, H.M. TP53-Mutated Myelodysplastic Syndrome and Acute Myeloid Leukemia: Biology, Current Therapy, and Future Directions. Cancer Discov. 2022, 12, 2516–2529. [Google Scholar] [CrossRef]
- Baugh, E.H.; Ke, H.; Levine, A.J.; Bonneau, R.A.; Chan, C.S. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018, 25, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Cook, J.H.; Melloni, G.E.M.; Gulhan, D.C.; Park, P.J.; Haigis, K.M. The origins and genetic interactions of KRAS mutations are allele- and tissue-specific. Nat. Commun. 2021, 12, 1808. [Google Scholar] [CrossRef] [PubMed]
- Robles-Valero, J.; Fernández-Nevado, L.; Lorenzo-Martín, L.F.; Cuadrado, M.; Fernández-Pisonero, I.; Rodríguez-Fdez, S.; Astorga-Simón, E.N.; Abad, A.; Caloto, R.; Bustelo, X.R. Cancer-associated mutations in VAV1 trigger variegated signaling outputs and T-cell lymphomagenesis. EMBO J. 2021, 40, e108125. [Google Scholar] [CrossRef]
- Barreira, M.; Fabbiano, S.; Couceiro, J.R.; Torreira, E.; Martínez-Torrecuadrada, J.L.; Montoya, G.; Llorca, O.; Bustelo, X.R. The C-terminal SH3 domain contributes to the intramolecular inhibition of Vav family proteins. Sci. Signal. 2014, 7, ra35. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, K.; Sakata-Yanagimoto, M.; Fujisawa, M.; Sakamoto, T.; Miyoshi, H.; Suehara, Y.; Nguyen, T.B.; Suma, S.; Yanagimoto, S.; Shiraishi, Y.; et al. VAV1 mutations contribute to development of T-cell neoplasms in mice. Blood 2020, 136, 3018–3032. [Google Scholar] [CrossRef] [PubMed]
- Robles-Valero, J.; Fernández-Nevado, L.; Cuadrado, M.; Lorenzo-Martín, L.F.; Fernández-Pisonero, I.; Abad, A.; Redín, E.; Montuenga, L.; Martín-Zanca, D.; Bigas, A.; et al. Characterization of the spectrum of trivalent VAV1-mutation-driven tumours using a gene-edited mouse model. Mol. Oncol. 2022, 16, 3533–3553. [Google Scholar] [CrossRef] [PubMed]
- Razidlo, G.L.; Magnine, C.; Sletten, A.C.; Hurley, R.M.; Almada, L.L.; Fernandez-Zapico, M.E.; Ji, B.; McNiven, M.A. Targeting pancreatic cancer metastasis by inhibition of vav1, a driver of tumor cell invasion. Cancer Res. 2015, 75, 2907–2915. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shalom, B.; Salaymeh, Y.; Risling, M.; Katzav, S. Unraveling the Oncogenic Potential of VAV1 in Human Cancer: Lessons from Mouse Models. Cells 2023, 12, 1276. https://doi.org/10.3390/cells12091276
Shalom B, Salaymeh Y, Risling M, Katzav S. Unraveling the Oncogenic Potential of VAV1 in Human Cancer: Lessons from Mouse Models. Cells. 2023; 12(9):1276. https://doi.org/10.3390/cells12091276
Chicago/Turabian StyleShalom, Batel, Yaser Salaymeh, Matan Risling, and Shulamit Katzav. 2023. "Unraveling the Oncogenic Potential of VAV1 in Human Cancer: Lessons from Mouse Models" Cells 12, no. 9: 1276. https://doi.org/10.3390/cells12091276
APA StyleShalom, B., Salaymeh, Y., Risling, M., & Katzav, S. (2023). Unraveling the Oncogenic Potential of VAV1 in Human Cancer: Lessons from Mouse Models. Cells, 12(9), 1276. https://doi.org/10.3390/cells12091276






