1. Introduction
Striated muscles are elaborately organized machines designated for contraction, and their highly textured structure is a prerequisite for directed force development. Sarcomeres, the smallest functional units of muscle contraction, comprise precisely organized filament systems including thin (actin) and thick (myosin) filaments, titin, and nebulin [
1], which build up the myofibrillar apparatus. The extrasarcomeric cytoskeleton connects the sarcomeres with the sarcolemma and the extracellular matrix through linking the Z-disks to the sub-membrane cytoskeleton, thereby ensuring the transmission of the forces produced by the sarcomeres. Desmin intermediate filaments (IFs), constituting the principal component of the extrasarcomeric cytoskeleton, form a three-dimensional scaffold extending from the nuclear envelope to the sarcolemma and tethering membranous organelles, such as mitochondria and the sarcoplasmic reticulum [
2]. In the heart, desmin IFs are also particularly abundant at the intercalated disks, thereby providing structural and mechanical support to cardiac tissue [
2].
Plectin, a multi-modular cytolinker protein of exceptionally large size (>500 kDa), orchestrates and functionally organizes various cytoskeletal networks, and thereby contributes to fundamental biomechanical properties of stress-bearing tissues, such as skeletal muscle [
3]. Acting as a multi-functional linker protein and signaling scaffold, plectin possesses binding sites for all types of IF subunit proteins, enabling it to anchor IFs to sites of strategic importance for the organization and performance of cells [
4]. In addition, plectin harbors a functional actin-binding domain (ABD), binds to microtubule-associated proteins (MAPs), and interacts with transmembrane receptors, proteins of the sub-membrane cytoskeleton, components of the nuclear envelope, and several kinases with known roles in the migration, proliferation, and energy metabolism of cells [
5,
6]. In the absence of plectin, mechanically stressed tissues, such as skin and muscle, lose their structural integrity, as observed in patients suffering from plectinopathies and in plectin-deficient mice [
7,
8,
9]. Most mutations in the human plectin gene (
PLEC) on chromosome 8q24 lead to epidermolysis bullosa simplex with muscular dystrophy (EBS-MD, MIM #226670), an autosomal recessive skin blistering disorder associated with progressive muscle weakness [
7,
8]. In addition, plectin mutations have been shown to cause EBS-MD with a myasthenic syndrome (EBS-MD-MyS), EBS with pyloric atresia (EBS-PA, MIM #612138), limb-girdle muscular dystrophy (LGMDR17, MIM #613723), EBS with nail dystrophy (EBSND, MIM #616487), or the autosomal dominant variant EBS-Ogna (MIM #131950) [
3,
9]. Emerging cardiac disease manifestations including left ventricular hypertrophy, atrial fibrillation, ventricular non-compaction myopathy, dilated cardiomyopathy, and life-threatening episodes of arrhythmias emphasize the pathological consequences of
PLEC mutations [
3]. Taken together, plectinopathies have emerged as complex multi-systemic disorders, affecting primarily tissues exposed to great mechanical stress, such as skin, skeletal muscle, and the heart, but exhibiting a variety of additional symptoms and disease manifestations.
Plectin’s versatility is in part due to an unusual transcript diversity, where complex splicing events in its N-terminal region give rise to multiple alternatively spliced isoforms containing distinct first exons [
4,
5]. Individual plectin isoforms are differentially expressed in various types of cells and tissues in specific combinations and proportions, where they function as universal IF-docking sites [
5,
6], thereby substantially contributing to fundamental biomechanical properties of stress-bearing tissues. In striated muscle tissue, plectin is predominantly found at the level of Z-disks, where it co-localizes with desmin IFs. The four most prominently expressed isoforms of plectin in muscle are P1d, P1f, P1b, and P1, which are accountable for myofiber integrity by anchoring desmin IFs to Z-disks, costameres, mitochondria, and the nuclear/sarcoplasmic reticulum membrane system, respectively [
10,
11]. With an isoform-specific N-terminal sequence that is just five amino acid residues long, P1d is the only isoform that is exclusively expressed in skeletal muscle and heart, as identified by plectin transcript profiling [
4,
5], and is specifically targeted to Z-disks, as demonstrated in differentiated myotubes and muscle fibers [
12]. Consequently, when skeletal muscle fibers, isolated from isoform-specific P1d-knockout (P1d-KO) mice (lacking just P1d, but expressing the other isoforms [
11]) were immunolabeled using pan-plectin-antibodies (reactive with all plectin isoforms), the Z-disk-specific labeling was lost, and only sarcolemmal staining and dotty remnants in the interior of the fiber became evident. These results indicated that in P1d-KO muscles, P1 and P1f were still preserved in the peripheral, perinuclear, and costameric regions, while Z-disk-associated P1d was absent [
11]. Accordingly, accumulation of desmin-positive protein aggregates was noted in the interior, but not in peripheral areas of P1d-KO muscle fibers. Moreover, P1d-KO muscles disclosed extensive misalignment of myofibrils and disorientation of Z-disks, thereby closely resembling the pathology of muscles completely devoid of plectin, such as those from conditional, skeletal muscle-specific plectin knockout (MCK-Cre/cKO) mice [
11]. However, compared to the latter, the desmin IF network underneath the sarcolemma appeared unaffected in P1d-KO teased fibers, suggesting that P1f still maintains the organization of the costameres [
11]. Finally, when P1d-GFP fusion proteins were expressed in differentiated plectin-deficient myotubes, a cell model closely mimicking the EBS-MD-associated skeletal muscle pathology by displaying impaired Z-disk alignment and desmin IF network collapse, they fully restored sarcomere formation and recruited desmin IFs to the Z-disks [
13]. In contrast, forced expression of any of the other plectin isoforms typical for skeletal muscle (P1, P1b, or P1f) showed no such effect [
13]. Taken together, these experiments highlighted that the striking sarcomere pathology of EBS-MD muscle is primarily due to a loss of function of P1d, the isoform which tethers myofibrils to the desmin IF network at the level of Z-disks.
However, important questions regarding mechanistic aspects of Z-disk-associated plectin and its functionality as well as the pathology arising from plectin deficiency remained unanswered. For instance, neither the targeting mechanisms nor the spatial arrangement of P1d and desmin IFs at Z-disks have been investigated so far, nor is anything known about the biomechanical consequences of P1d-deficiency for individual myofibrils. Moreover, it can be assumed that as a variant of a multifunctional cytolinker protein with a well-established role as versatile cytoplasmic signaling platform [
5,
6], P1d exerts important functions beyond IF anchorage at Z disks in muscle fibers. Addressing these and other contextually relevant issues in the present study, we used high-resolution stimulated emission depletion (STED) microscopy to unravel the spatial arrangement of plectin and desmin networks at the level of individual Z-disks, and yeast two-hybrid (Y2H) screening to identify isoform-specific P1d interaction partners associated with Z-disks. In addition, we assessed the consequences of site-specific (Z-disk-restricted) plectin deficiency on muscle function ex vivo and in vivo by subjecting isoform P1d-specific knockout (P1d-KO) mice, and derivatives thereof, to diverse phenotypic analyses. Among the physiological parameters found to be affected by P1d deficiency were the relaxation kinetics of myofibrillar sarcomeres and the homeostasis of the chaperone-assisted selective autophagy (CASA) proteolytic degradation machinery and its major substrate, the actin-crosslinking protein filamin C. On the organismal level, physical exercise of P1d-KO mice led to myofibrillar lesions, early exhaustion, and lethality. In all, our experiments unequivocally reveal an important role of P1d in the maintenance of muscle structure and provide new insight(s) into the molecular mechanisms underlying plectinopathy-related muscle weakness.
2. Materials and Methods
2.1. Animals
All experiments involving animals were performed according to the Austrian Federal Government laws and regulations. MCK-Cre/cKO, P1d-KO, and P1b-KO mice have been published [
11,
14].
Myofibril bundles for mechanical experiments, as well as bundles and cryosections for STED microscopy, were prepared from MCK-Cre/cKO, P1d KO, or corresponding wild-type mice that were sacrificed by cervical dislocation according to the guidelines and as approved by the ethical committee of the Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen (LANUV).
Behavioral assessments were approved by the Federal Ministry for Science, Research and Economy, Vienna, Austria (BMWFW-66.006/0005-WF/II3b/2014) and performed in cooperation with the Preclinical Phenotyping Facility at Vienna Biocenter Core Facilities (VBCF), a member of Vienna Biocenter (VBC), Austria. During the forced swim test [
15], mice were placed into a round tank filled with pre-warmed water (30–32 °C). Endurance exercises, as described in [
16], were performed according to a ramp protocol starting with 20 min swimming daily, with 10 min increase each day, until 90 min were reached. Mice were exercised daily for two weeks. Animals were continuously observed during the swim test, and any animal that sank below the surface was removed from the water immediately. Mice exhibiting signs of exhaustion during the training session were removed from the water and allowed to rest for 3 min in their home cage before resuming the session. After a workload application, mice were allowed to dry in a warm environment after removal from the water.
2.2. Antibodies
Primary antibodies used for immunofluorescence microscopy (IFM) and/or immunoblotting (IB) are listed in
Table 1. For IFM, primary antibodies were used in combination with goat-anti-mouse IgG Alexa 488, donkey-anti-mouse IgG Rhodamine Red, goat-anti-rat IgG Alexa 488, goat-anti-rabbit IgG Alexa 488, and donkey-anti-rabbit IgG Rhodamine Red at dilutions of 1:500 (all from Jackson ImmunoResearch Laboratories, Cambridge, UK). For IB analyses, HRP-conjugated secondary antibodies were used at dilutions of 1:5000–1:10,000 (Jackson ImmunoResearch Laboratories). Pan-plectin antibodies (rabbit antiserum #9 [
17]) were used for immunoprecipitation. For STED microscopy, primary antibodies were used in combination with the goat-anti-mouse IgG Atto 647N, goat-anti-rabbit IgG Abberior STAR 580, goat-anti-rabbit IgG (H+L) Alexa 488, goat-anti-rabbit IgG (H+L) Alexa 647, and goat-anti-mouse IgG (H+L) Alexa 647 or Zenon anti-mouse IgG
1 488 labelling kits (all from Sigma-Aldrich, St. Louis, MO, USA).
2.3. Preparation of Myofibrils
Muscle strips (0.2–0.3 mm in diameter) were dissected from psoas and soleus muscle and incubated for 2 h in skinning solution (5 mM K-phosphate, 5 mM Na-azide, 3 mM Mg-acetate, 5 mM K
2EGTA, 3 mM Na
2ATP (including 3 mM MgCl
2 and 6 mM KOH), 47 mM Na
2CrP, 2 mM DTT, 0.5 mM 4-(2-aminoethyl)-benzenesulfonylfluoride-HCl, 10 μM leupeptine, 10 μM antipaine, and 5 mg/mL aprotinine, adjusted to pH 6.8 at 20 °C) as described in [
21], supplemented with 30 mM 2,3-butanedione monoxime (BDM) and 0.5% Triton X-100. The solution was replaced by a similar one without detergent and strips, and stored for up to 3 days at 0 °C. For long-term storage, strips were successively incubated (for 30 min each), in skinning solutions containing 0.5 M, 1.0 M, 1.5 M, and 2.0 M sucrose, then rapidly frozen in liquid propane and transferred into liquid nitrogen [
22]. Immediately before mechanical or immunostaining experiments, cryo-conserved strips were thawed and consecutively incubated in skinning solutions containing 2 M, 1.5 M, 1.0 M, 0.5 M, and 0 M sucrose, and then homogenized at 4 °C for 5–10 s with a blender (Ultra-Turrax T25, Janke & Kunkel, Staufen, Germany) at maximum speed.
2.4. Microscopy
For STED microscopy of cryosections, wild-type mouse soleus muscles were dissected in HEPES-buffered, Krebs–Ringer solution, pinned at the tendons, and permeabilized in skinning solution with 1% Triton X-100, 30 mM BDM for 1 h. After washing, the muscle was frozen in liquid nitrogen and stored at −80 °C overnight. Cryosections (10 µm) were then cut, immediately fixed in 3% glyoxal, quenched with 100 mM NH4Cl, and permeabilized in PBS containing 0.1% Triton X-100 and 2.5% bovine serum albumin (BSA). Cryosections were immunolabeled with primary antibodies at dilutions of 1:100 (plectin) or 1:125 (desmin), and with secondary antibodies (Atto 647, Abberior 580) at a dilution of 1:75.
For STED microscopy of myofibril bundles, bundles were sedimented (in skinning solution) onto coverslips for 45 min at 0 °C. Specimens were blocked with 1% BSA and 3% goat serum and incubated with primary antibodies at dilutions 1:100–1:200 (plectin and desmin) and 1:800–1:1000 (α-actinin), and with secondary antibodies at dilutions of 1:500–1:1000. Bundles were immobilized on coverslips either prior to blocking by fixation with 3% paraformaldehyde for 4 min, or after immunolabeling by immersion with PBS containing 10 mM β-mercaptoethylamine.
Confocal and STED images were taken with a TCS SP8 gSTED microscope using a HSX PL Apo ×100/1.40 oil immersion objective (Leica Microsystems, Wetzlar, Germany). The two channels were acquired in a sequential mode. Deconvolution of the images was carried out using the Huygens Professional (Scientific Volume Imaging) software.
Thin sections (5 µm) were obtained from soleus muscle tissue that was frozen in isopentane cooled with liquid nitrogen. Teased muscle fibers were prepared and processed as described previously [
11,
12]. The staining procedure was performed using the M.O.M. Basic Kit (Vector Laboratories, Newark, CA, USA) according to the manufacturer’s instructions, as described in [
13]. Nuclei were stained with Hoechst 33258 (Sigma-Aldrich), and samples were mounted in Mowiol 4-88 (Hoechst, Frankfurt, Germany). Confocal microscopy was performed using a fluorescence laser scanning microscope (LSM710, Zeiss, Oberkochen, Germany) equipped with Plan Apochromat 63 × 1.4 NA and 40 × 1.3 NA objective lenses. Images were recorded using the LSM710 module and the Zeiss ZEN software.
2.5. Transmission Electron Microscopy
Freshly dissected soleus muscles were fixed in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M cacodylate buffer. Specimens were washed three times and post-fixed in 1% OsO4 with 1.5% potassium hexacyanoferrate(II) followed by dehydration in an ascending ethanol series. After embedding in Epon resin, 50 nm ultrathin sections were cut with a Reichert-Jung ULTRACUT microtome and counterstained with lead citrate and uranyl acetate. Images were acquired using an FEI Tecnai20 electron microscope equipped with a 4 K Eagle-CCD camera, and then processed with Adobe Photoshop.
2.6. Force Measurements of Myofibrils
Force measurement were performed at 10 °C using an experimental setup described previously [
21]. Myofibril bundles (diameters of 1.7–3.4 mm) were mounted in relaxing solution (pCa 8) between the tip of an atomic-force cantilever and the tip of a length-driving stiff tungsten needle. After mounting, the slack sarcomere length, overall length, and the diameter of the bundles were determined. The bundles were stretched to a sarcomere length of 2.45 µm and the passive force was determined in relaxing solution (pCa 7). Bundles were rapidly activated and relaxed by a rapid solution change (within 10 ms), and the force kinetics were recorded and thereafter analyzed as described previously [
21,
23]. Each bundle was first activated twice at pCa 4.5 to determine its active force and rate constants of Ca
2+-induced and mechanically-induced force development before eccentric contraction. Then, the bundle was stretched in the third activation for 250 ms with a constant speed of 1 times its length per second, held for 500 ms at the stretched length, released with same speed to the original length, and then force and force kinetic parameters were determined after the eccentric contraction.
2.7. Yeast Two-Hybrid Screening
For the yeast two-hybrid screening, cDNAs encoding plectin fragments P1d-8(2α) and P1f-8 [
10], as well as cDNAs encoding plectin fragments P1d-24(2α) or P1f-24 [
24], were sub-cloned into a modified pEG202 vector (DupLEX-A system, Origene) to obtain the bait plasmids. Bait proteins showing no autonomous activation, and a mouse skeletal muscle cDNA library (DupLEX-A, DLM111, Origene, Rockville, MD, USA) in yeast expression vector pJP4-5, were introduced into yeast strain EGY48 (MATa trp1 his3 ura3 leu2::6 LexAop-LEU2). Transformants were amplified, and positive clones were selected following the protocol recommended in the user manual.
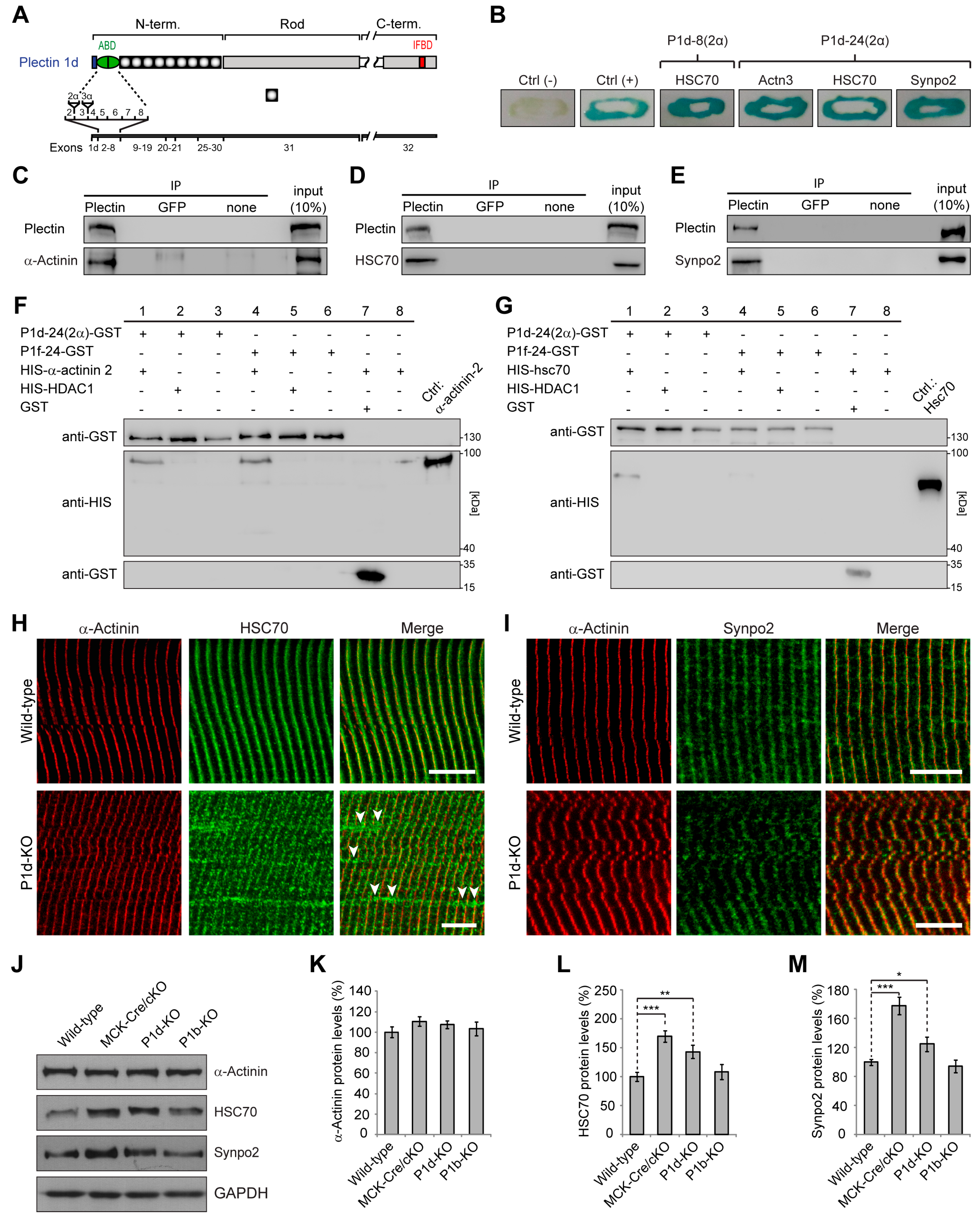
Out of more than 100 positively identified clones, 1 interacting with P1d-8(2α) and P1d-24(2α), but not with P1f-8 or P1f-24, showed 99% identity to a mouse cDNA sequence corresponding to heat shock protein 8/HSC70. Two others, interacting with P1d-24(2α), but not with P1d-8(2α), P1f-8, or P1f-24, showed 99% identity to a mouse cDNA sequence corresponding to α-actinin 3 and synaptopodin 2, respectively.
2.8. Co-Immunoprecipitation
Differentiated wild-type myoblasts were scraped off in lysis buffer [20 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0, 200 mM NaCl, 0.5% Nonidet P-40, 25 µg/mL PMSF, and cOmplete mini protease inhibitor cocktail (from Roche, Basel, Switzerland)], homogenized by pressing the samples through a 27-gauge needle, centrifuged for 10 min at 10,000× g and 4° C, and then the supernatant was used for co-immunoprecipitation. Immunoprecipitation was performed using pan-plectin antibodies and protein A/G sepharose beads (Pierce).
2.9. Protein Expression, Purification, and GST-Pull down Assay
For expression of recombinant plectin fragments in bacteria, cDNAs encoding P1d-24(2α) or P1f-24-specific sequences [
24] were cloned into the expression vector pKG44 (a modified derivate from pBN120 [
25], in which the His-tag was replaced by a GST-tag from pET-42a (Sigma-Aldrich). For bacterial expression of HSC70, the corresponding cDNA was PCR amplified by using primers with EcoRI tails and a mouse skeletal muscle cDNA library (DupLEX-A system, Origene), and cloned into pBN120 (a derivate from pET-15b (Novagene [
25])). cDNA encoding human α-actinin 2, a generous gift from K. N. North (Children’s Hospital at Westmead, University of Sydney, Australia), was subcloned into the bacterial expression vector pBN120. Plasmids expressing His-tagged HDAC1, used as a negative control, were kindly provided by C. Seiser (Department of Cell and Developmental Biology, Center for Anatomy and Cell Biology, Medical University of Vienna, Austria).
Fusion proteins were expressed in the E. coli strain BL21 (DE3)RIL. GST fusion proteins were purified on gluthathione sepharose 4B beads (GE Healthcare, Chicago, IL, USA), as described in the manufacturer’s instructions. His-tagged fusion proteins were purified using nickel affinity chromatography on His•Bind Resin (Merck, Rahway, NJ, USA) according to the manufacturer’s instructions.
For pull-down experiments, GST-fusion proteins were immobilized on glutathione sepharose 4B beads for 30 min at 4 °C, washed with PBS to remove unbound protein, and incubated with 10–20 µg His-tagged fusion proteins in PBS at 4 °C for 1–4 h with gentle agitation. After centrifugation (500× g, 3 min, 4 °C), beads were washed three times with ice-cold wash buffer (20 mM Tris-HCl pH 7.4, 0.1 mM EDTA, 100 mM NaCl, 0.25% Nonidet P-40 for His-tagged α-actinin 2; 20 mM Tris-HCl pH 7.4, 0.1 mM EDTA, 300 mM NaCl, 0.5% Nonidet P-40 for His-tagged HSC70), before being processed for SDS-PAGE and immunoblotting.
2.10. Immunoblotting Analyses
Cell and tissue lysates were prepared as described in [
16]. Dissected muscles were snap frozen in isopentane cooled with liquid nitrogen, ground in a mortar, and homogenized in lysis buffer (5 mM Tris-HCl pH 6.8, 10% SDS, 0.2 M DTT, 1 mM EDTA, 100 mM NaF, 50 mM ß-glycerophosphate, 2 mM Na
3VO
4, 1 mM PMSF, and cOmplete mini protease inhibitor cocktail (Roche)) using a Dounce tissue grinder. The homogenate was centrifuged for 10 min at 10,000×
g, and the supernatant was mixed with 6× SDS sample buffer (500 mM Tris-HCl pH 6.8, 600 mM DDT, 10% SDS, 0.1% bromophenol blue, 30% glycerol). Cells were directly scraped off in 6× SDS sample buffer and DNA was sheared by pressing the samples through a 27-gauge needle. Samples were boiled at 95 °C for 5 min before being subjected to SDS polyacrylamide gel electrophoresis performed under standard conditions. Proteins were transferred to nitrocellulose membranes (Protran 0.2 NC, Amersham Biosciences, Amersham, UK) using a Mini PROTEAN Tetra Cell blot apparatus (Bio-Rad, Hercules, CA, USA). Membranes were scanned, and the amounts of protein contained in individual bands were quantified using ImageJ software (NIH, Bethesda, MD, USA).
2.11. Myoblast Cell Culture
Immortalized skeletal myoblasts were isolated from
Plec−/− and wild-type littermates, both crossed into a p53
−/− background, as described in [
13], and cultivated in Ham’s F10 medium (Life Technologies, Carlsbad, CA, USA), supplemented with 20% fetal calf serum (Sigma-Aldrich), 2.5 ng/mL basic fibroblast growth factor (bFGF, Promega, Madison, WI, USA) and antibiotics on collagen-coated (0.01% collagen in PBS, Nutacon, Leimuiden, Netherlands) culture dishes. To induce differentiation, cultures were switched to Dulbecco’s modified Eagle’s medium (Life Technologies), containing 5% horse serum (Life Technologies) and antibiotics.
2.12. Cell Stretcher
Stretch experiments were carried out on flexible polydimethylsiloxan (PDMS, Sylgard) substrates coated with laminin-1 (Sigma-Aldrich) that were molded into the shape of a cell culture well with a 2.5 cm
2 internal surface, as described in [
13]. Plated myoblasts were differentiated for 10 days by applying differentiation medium. PDMS gels were then attached to a direct current linear motor with an integrated gearbox (RB35, Conrad Electronic SE, Hirschau, Germany). Uniaxial cyclic stretching was performed in an incubator under normal cell culture conditions for 60 min at 30% stretch amplitude (peak-to-peak) and at a frequency of 0.25 Hz with a resting period (dwell time) of 1 s between the lengthening and the shortening phase. After stretching, cells were homogenized in 6× SDS sample buffer and processed for immunoblotting.
2.13. Statistical Analyses
Data analyses and statistical evaluations were performed using Excel 2010 (Microsoft, Redmond, WA, USA) and SPSS Statistics v.19 (IBM, Armonk, NY, USA). Data are given as mean ± SEM. The number of experiments (N) and data points (n) is indicated in the figure legends. Comparisons between the values of two groups were made using an unpaired, two-tailed Student’s t-test (alpha = 0.001–0.05). Comparisons among the values of multiple groups were performed using one-way analysis of variance (ANOVA; alpha = 0.001–0.10). The significance between values of individual groups and controls was subsequently determined using Tukey’s post-hoc test. The p-values are * < 0.05, ** < 0.01, and *** < 0.001; a p-value < 0.05 was considered statistically significant.
4. Discussion
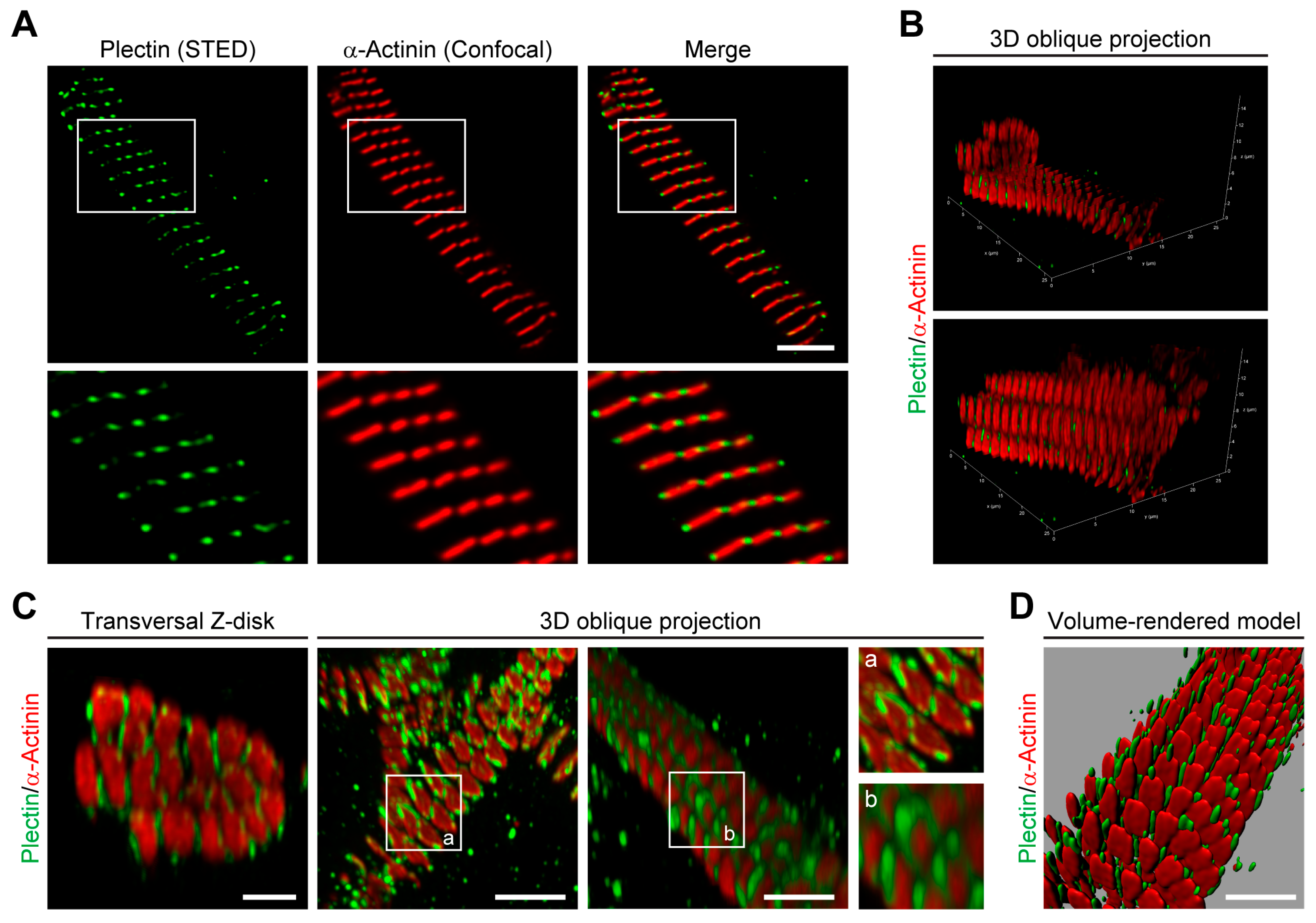
Investigating the spatial arrangement of plectin molecules at the Z-disks of myofibers by super-resolution immunofluorescence microscopy, we show here that plectin is localized in the α-actinin-negative gaps existing between the laterally aligned Z-disks but is missing from the interior of Z-disks. This notion was substantiated by the observation of plectin-positive signals bordering, but not overlapping with, the α-actinin-positive Z-disks. Our data further demonstrate that plectin-positive structures encircle individual (single) myofibrils rather than bundles of multiple myofibrils, whereby the encasing of individual α-actinin-positive Z-disks by plectin molecules became clearly evident by image deconvolution of 3D oblique projections (
Figure 1C). Finally, we could successfully generate volume-rendered models of skeletal muscle myofibrils, illustrating that plectin is located within the gaps between individual α-actinin-positive Z-disks (
Figure 1D). These experiments add to and complement a previous immunogold electron microscopy study, in which fine plectin threads were found to be located between Z-disks and IF networks in skeletal muscle sections [
28]. STED microscopy of transversal muscle cryosections provides direct structural evidence for previous models [
11,
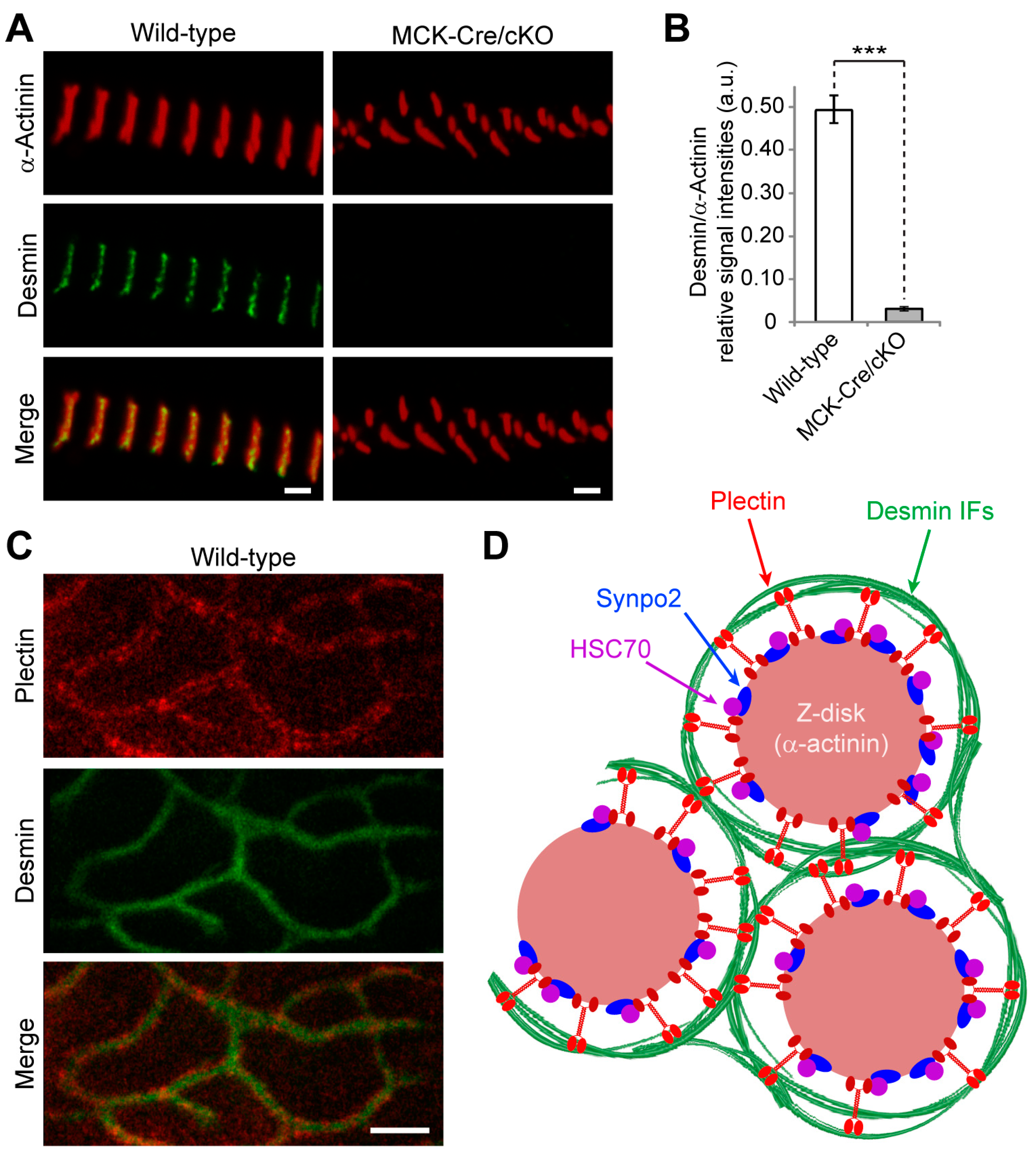
28], suggesting that plectin is located between Z-disks and IFs. Our results are in line with the concept of plectin acting as a universal IF recruiting and anchoring platform. The lack of any desmin-specific staining at the Z-disks of plectin-deficient myofibril bundles confirmed that the association of desmin with Z-disks was plectin-dependent and desmin was dissociated from myofibrillar Z-disks in the absence of plectin linkages. Dispatched desmin filaments consequently collapse and accumulate between plectin-deficient myofibril bundles, thereby generating the desmin-positive protein aggregates observed in EBS-MD patients, MCK-Cre/cKO mice, and plectin-deficient myotubes [
11,
13,
42]. Similar to plectin, missense mutations in desmin have been shown to compromise the structure and maintenance of IF filaments on various levels, frequently causing abnormal cytoplasmic desmin aggregates [
43,
44]. However, while desmin-related myopathies and cardiomyopathies are mostly caused by filament formation defects, in plectinopathies, desmin aggregation is a consequence of missing linkages between the IFs and other structures, such as the Z-disks.
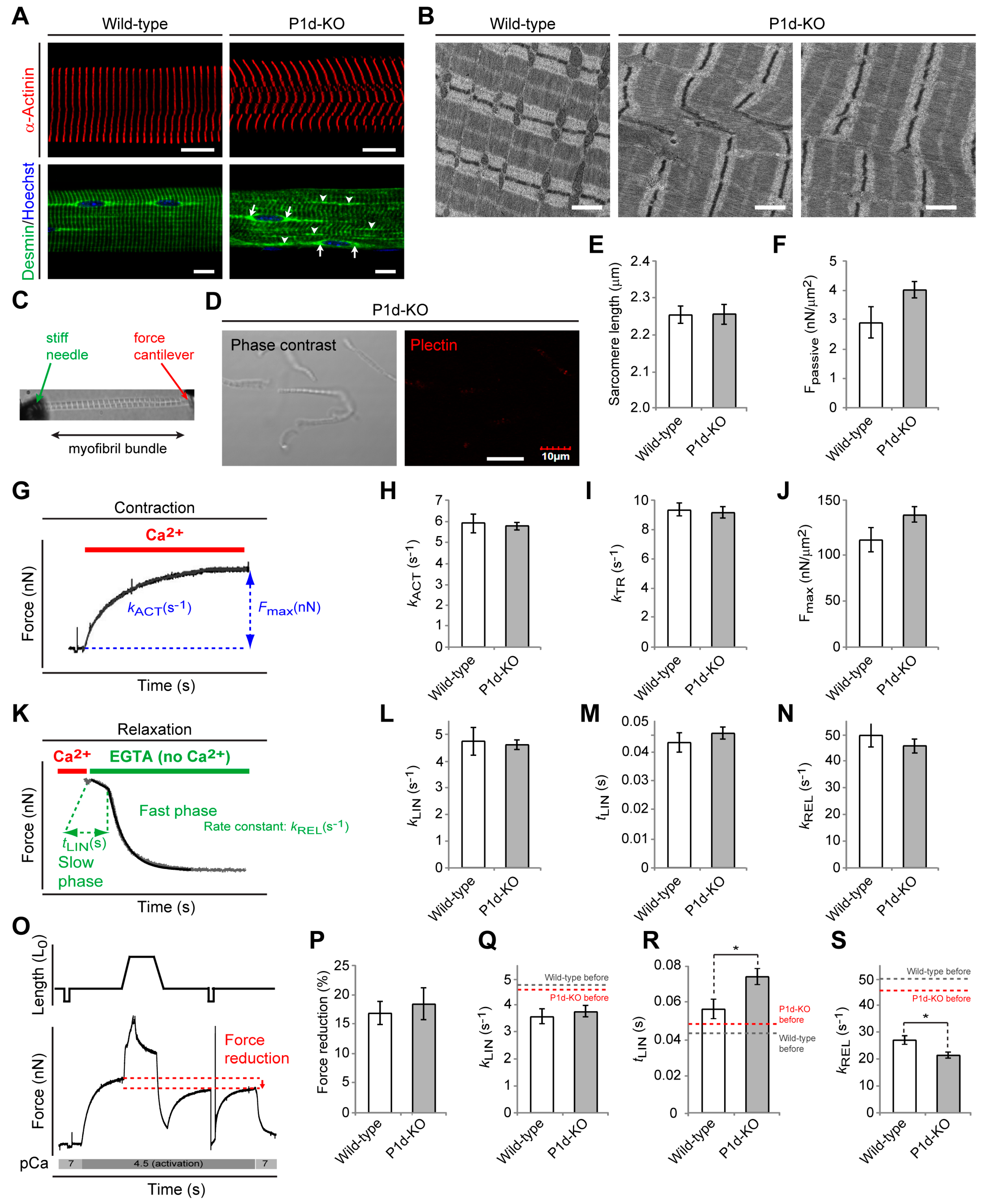
Plectin’s versatility is largely based on the differential cellular targeting of its individual isoforms. As we show here, microscopic and ultrastructural analyses revealed that the loss of Z-disk-associated P1d leads to highly disorganized myofibrillar structures and phase displacement of sarcomeric structures, pointing towards increased inhomogeneity of the muscle structure. Detachment and loss of regularly anchored desmin IFs, as observed in teased muscle fibers from P1d-KO mice, led to the formation of desmin aggregates in interior areas of the fiber. Moreover, peripheral and perinuclear desmin-positive signals indicated that other plectin isoforms expressed in muscle fibers, such as P1f and P1, were still intact. On the other hand, in plectin-stained myofibril bundles from P1d-KO mice, no remaining plectin signals were observed, reassuring us that P1d is the sole Z-disk-associated plectin isoform. In line with previous observations [
11,
13] our analyses emphasize the unique role of isoform P1d in maintaining regular myofibril alignment. The striking sarcomere pathology, as observed in muscle from EBS-MD patients, could, therefore, be attributed primarily to a loss of function of isoform P1d. When we assessed the mechanical performance of myofibrillar bundles, P1d-deficient myofibrils displayed similar passive tension, comparable Ca
2+-dependent contraction abilities, and unaltered biphasic force relaxation compared to wild-type samples. However, when we tested the vulnerability to mechanical stress by eccentric contractions, despite displaying a similar force reduction to wild-type myofibrils, myofibril bundles from P1d-KO revealed a significantly prolonged duration of the slow phase and reduced rate constant of the fast phase after the eccentric contraction. In the context of biomechanical studies that indicate organized inter-sarcomere dynamics as the cause of rapid relaxation [
21,
45,
46], our results suggest that this mechanism of relaxation is impaired upon mechanical stress, particularly when P1d is not available for stabilizing the inter-myofibrillar sarcomere organization.
Previous conventional confocal immunofluorescence microscopy studies on myofibers revealed that of the four major plectin isoforms expressed in mature cells, i.e., P1, P1b, P1d, and P1f, only P1d co-localized with desmin at the level of the Z-disks, while P1f was localized at the periphery of cells along the sarcolemma, P1 in perinuclear areas, and P1b in association with mitochondria. In previous studies, we have identified isoform-specific binding partners that show co-localization with isoforms at their targeting sites, in the cases of P1b, P1f, and P1 [
12,
14,
29], strongly suggesting that isoform-specific interactions are a basic part of the mechanisms underlying isoform-, and, by implication, IF network-targeting. The mechanism of P1d-mediated docking of desmin IFs at a sarcomeric Z-disks-associated platform remained unresolved. The existence of such a platform was implicit, however, based on similar sarcomeric phenotype manifestations of isoform P1d-deficient and desmin-null mice, such as disarrayed disks paired with misaligned myofibrils [
11,
13,
47]. Using Y2H screening combined with co-immunoprecipitation and in vitro pull-down assays, we here identified several Z-disk-associated proteins as direct interaction partners of P1d, among them the bona fide Z-disk constituent protein α-actinin. However, plectin’s binding to α-actinin was found to be isoform-independent, as it also occurred with N-terminal fragments of plectin corresponding to isoforms other than P1d (see
Figure 4F). In fact, there is evidence from previous studies that plectin’s α-actinin-binding domain resides within plectin’s (spectrin repeats-containing) plakin domain that resides downstream of the ABD and precedes the central α-helical rod domain (unpublished data from the authors’ laboratory). Thus, the binding affinity to α-actinin, as important as it might be for P1d’s Z-disk-related functions, is unlikely to be the driving force for its recruitment to Z-disks. On the other hand, our experiments show that the N-terminal fragment of P1d binds in an isoform-specific manner to HSC70, a constitutively expressed chaperone protein involved in diverse cellular processes including protein folding and protein degradation. In mature myofibers, HSC70 is found predominantly at the level of Z-disks (
Figure 4H) and, as we show here, its association with Z-disks is independent of P1d. Thus, HSC70 qualifies as an isoform-specific binding protein that might be instrumental in recruiting P1d, and thereby the desmin network, to Z-disks of the contractile apparatus.
Whether only the five amino acid residue-long isoform-specific protein sequence preceding P1d’s ABD suffices for specific HSC70-binding, or whether the first exon-encoded protein domain acts in combination with the succeeding ABD, remains to be determined. Targeting signals or site-specific interaction domains residing in isoform-specific head domains have been described for a number of isoforms [
10], but only in the case of P1b was the head domain shown to suffice for site-specific targeting [
10,
14]. In most other cases, the experiments reported point towards an involvement and combined action of the specific head domains with the adjacent ABD in determining targeting preferences. To understand the isoform-dependent plasticity of plectin’s ABD and the conformational changes fostering binding to different partners, atomic-level structural analyses of plectin’s diverse N-termini, including that of P1d, will be required. Unfortunately, this approach is hampered by some of the isoform-specific domains having an intrinsically disordered structure [
48]. Once specifically targeted to Z-disks via HSC70, P1d can undergo interactions with a-actinin, sympo2, and likely other Z-disk-associated constituents via interfaces situated in molecular domains other than those forming the P1d-specific N-terminus. Such multiple interactions are expected to strengthen P1d’s affinity to Z-disk structures, thereby consolidating the compactness of IF-docking platforms along individual myofibrils and leading to their physical integration into the mechanotransducing IF network machinery.
Yeast two-hybrid screening revealed that P1d interacts with a wide spectrum of structural and signaling proteins. While its interaction with α-actinin appears to be crucial for the stabilization and lateral arrangement of Z-disks, P1d’s ability to bind to HSC70 and synpo2 suggested that its role goes beyond that of just mechanical support. Since HSC70 was identified in most of the positive transformants showing interaction with the short N-terminal region of P1d, one can assume that it is directly involved in targeting of plectin to the Z-disk. In contrast, α-actinin and synpo2 displayed binding to the longer fragment of P1d, suggesting that the binding site for these proteins is located in the region downstream of plectin’s ABD. This would be consistent with the previous observation that a fragment of the plakin domain binds to α-actinin (see above). Although α-actinin was clearly revealed as a P1d interaction partner using the yeast two-hybrid screening assay, additional in vitro binding studies showed that it can also bind to P1f. These observations imply that both plectin isoforms contain a binding site for α-actinin. While P1d interacts with α-actinin at the Z-disks and in this way stabilizes the contractile apparatus, P1f might be connected with cytoplasmic α-actinin residing underneath the sarcolemma where it supports membrane-associated protein complexes [
11,
12]. However, a specific N-terminus might modulate the remaining part of the molecule in a way that affects the binding affinity of interaction sites common to all isoforms, including the plakin domain-embedded α-actinin-binding site. Furthermore, plectin molecules can contain more than one binding site for a particular protein, as shown for β-dystroglycan [
12]. To specify the location and structurally analyze the binding interfaces with distinct proteins, additional experiments including binding assays using truncated forms of plectin and atomic structure analyses need to be carried out.
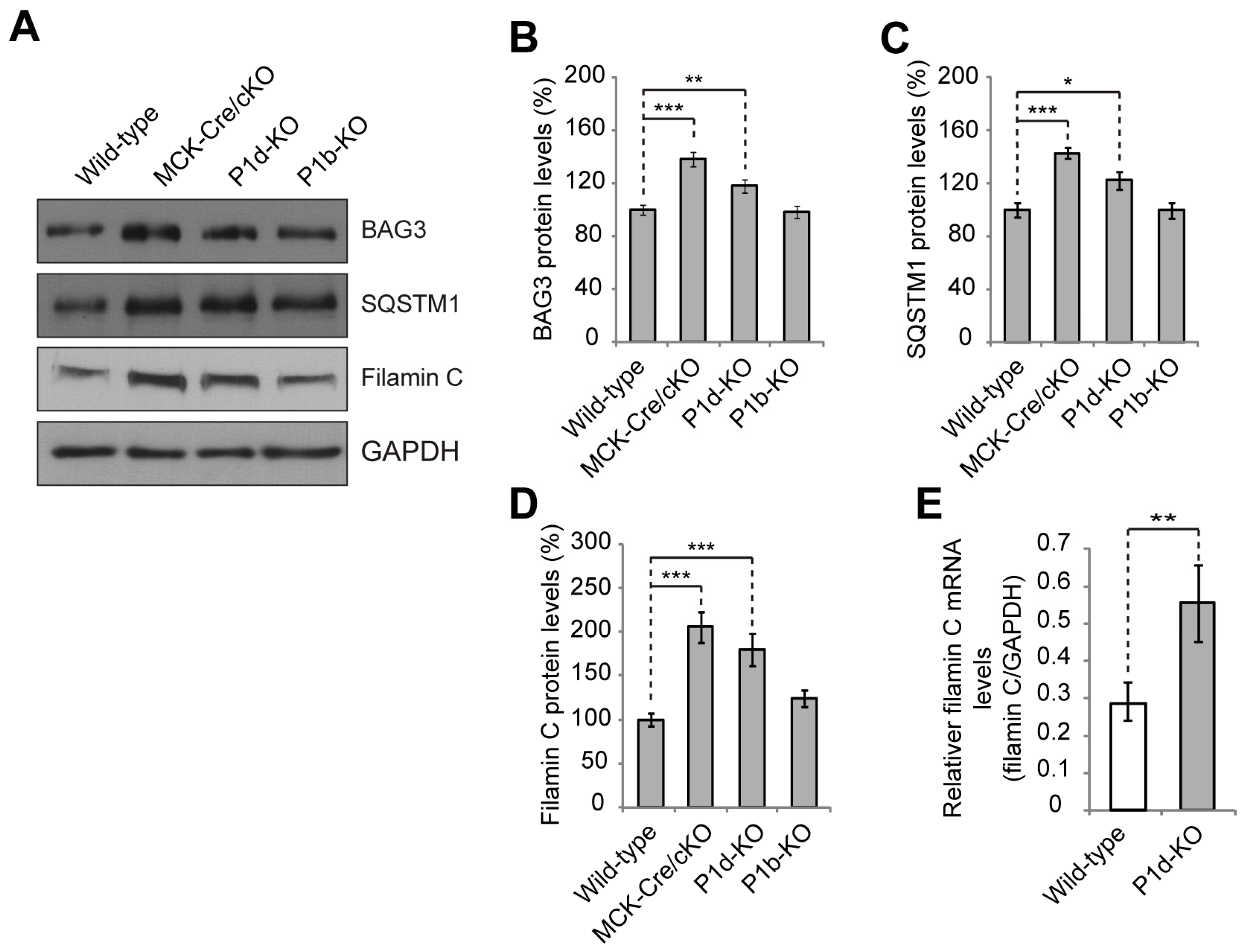
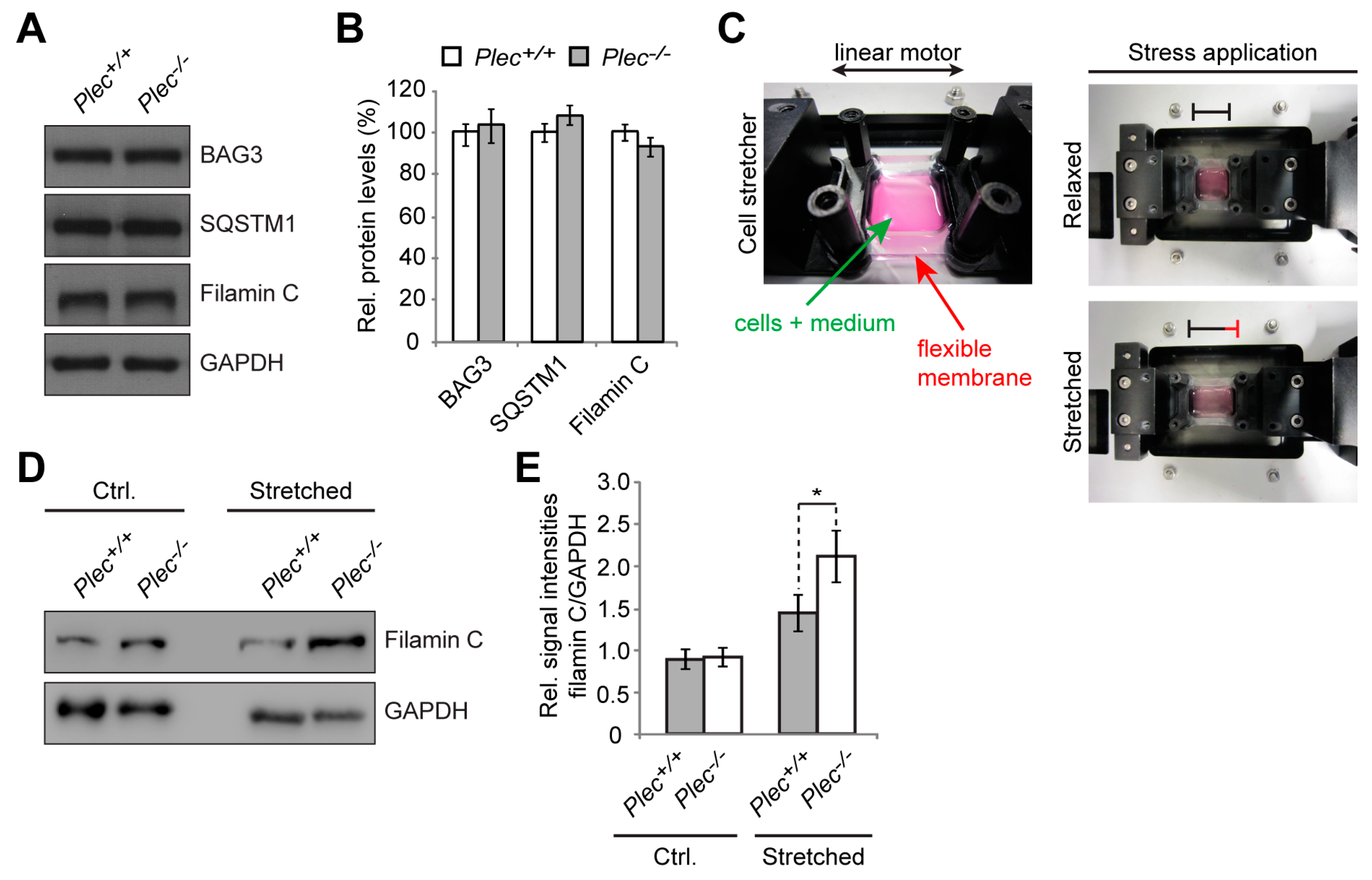
Our study confirms that P1d deficiency causes disorganization and destabilization of Z-disks due to a disrupted IF network–contractile apparatus linkage and the formation of desmin aggregates accumulating in the fiber interior. However, as we show in the present report, P1d plays a role that goes beyond simple structural support by forming a scaffolding platform for the protein degradation machinery CASA. The increased filamin C protein levels observed in P1d-KO and MCK/Cre-cKO myofibers suggested that P1d-mediated homogeneity of Z-disk alignment is crucial for the proper functioning of the CASA machinery. Although many questions regarding the mechanism how P1d regulates CASA remain open, we can conclude from our data that (i) the efficient degradation of filamin C via CASA requires the presence of P1d, and (ii) P1d is crucial for the proper function of the protein degradation machinery that plays an important role in muscle maintenance, especially during intensive contraction. As misfolded filamin C is efficiently degraded and replaced by newly synthesized protein, filamin C is a mechanosensing protein which maintains the structural integrity of the myofibers and preserves the contractile apparatus during muscle contraction. Our analysis shows that the proper alignment of Z-disks seems to be especially important for the efficient removal of unfolded filamin C, as highly upregulated filamin C levels were observed in P1d-KO and MCK-Cre/cKO myofibers. P1d’s interaction with synpo2 suggests that P1d, functioning as a Z-disk-associated scaffolding platform, links the CASA machinery and the autophagic degradation pathway, while P1d-deficiency causes a misalignment of Z-disks and a shift and misplacement of CASA components. Measuring the mechanical strain exerted on cultured myotubes on a stretching device convincingly demonstrated that CASA is a tension-induced protein degradation machinery. As the cell stretcher used for these experiments could indeed mimic the naturally occurring skeletal muscle contraction, it could be used as a convenient tool to analyze multiscale mechanical responses governed by cytoskeletal prestress in ex vivo cell culture systems. Due to the disrupted HSC70-synpo2-Z-disk linkage and the ensuing dysfunction of CASA in plectin (P1d) deficiency, filamin C levels drastically increased upon mechanical strain in
Plec−/− myotubes (see
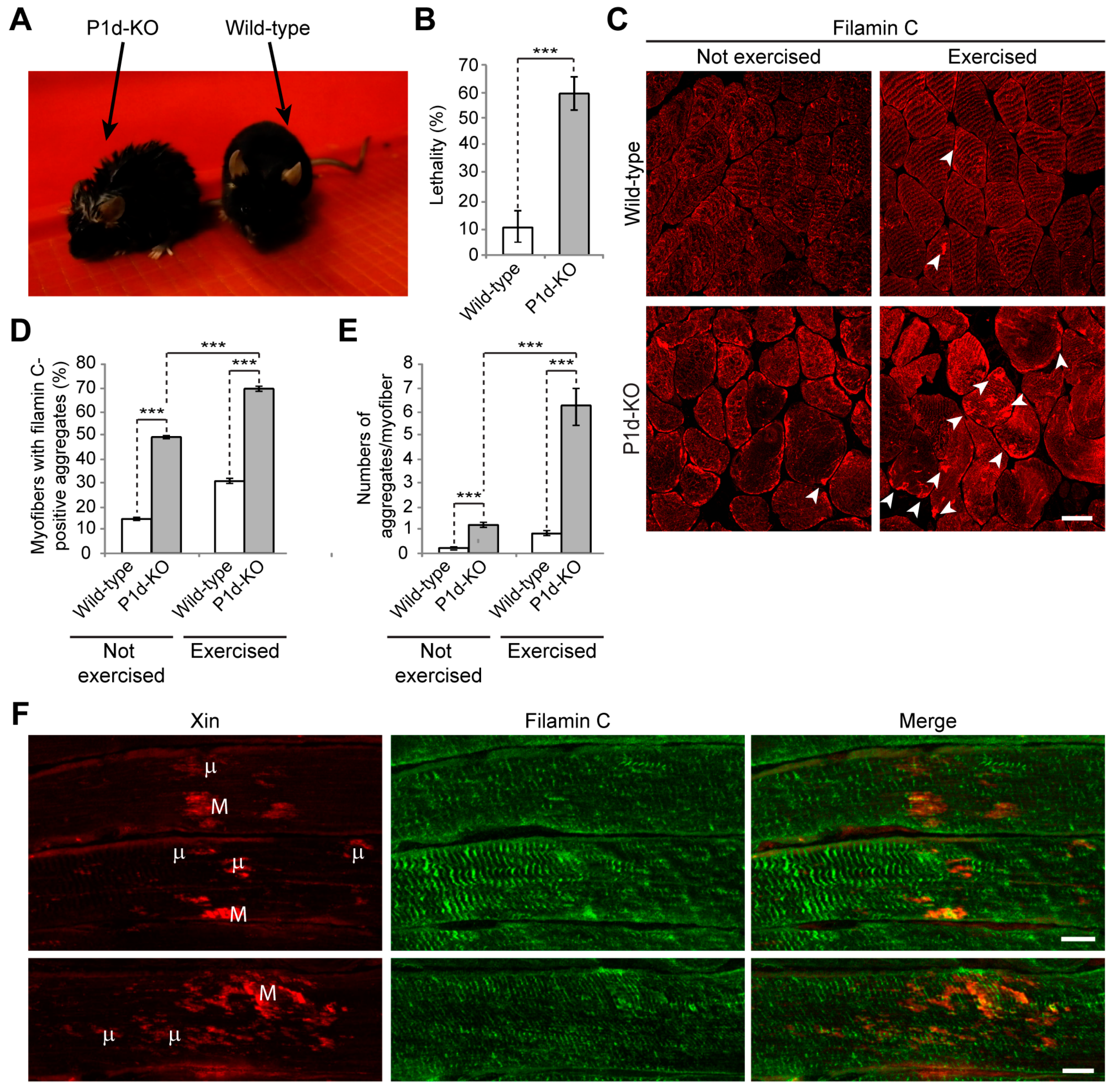
Figure 6E). Moreover, unfolded filamin C was not degraded properly during muscle contraction, but instead accumulated in the form of massive aggregates which further destabilized the internal structure of the myofiber. The forced swim test experiment, showing a striking increase in filamin C aggregate formation and Xin-positive sarcomeric lesions, together with the increased amounts of filamin C upon stretch in
Plec−/− myotubes, indicated that the role of P1d becomes particularly important during intense mechanical strain. Considering that P1d-KO mice became exhausted much faster than wild-type mice, and that many of them died during the experiment, it remains to be tested whether they show any cardiac phenotype, and heart failure was the reason for their frequent death. Future experiments, especially resistance-based evaluations of muscle function in mice or using ex vivo whole muscle preparations in a fatigue protocol, would significantly add to our understanding of the role of P1d in mechanically-induced muscle damage.