Aid or Antagonize: Nuclear Long Noncoding RNAs Regulate Host Responses and Outcomes of Viral Infections
Abstract
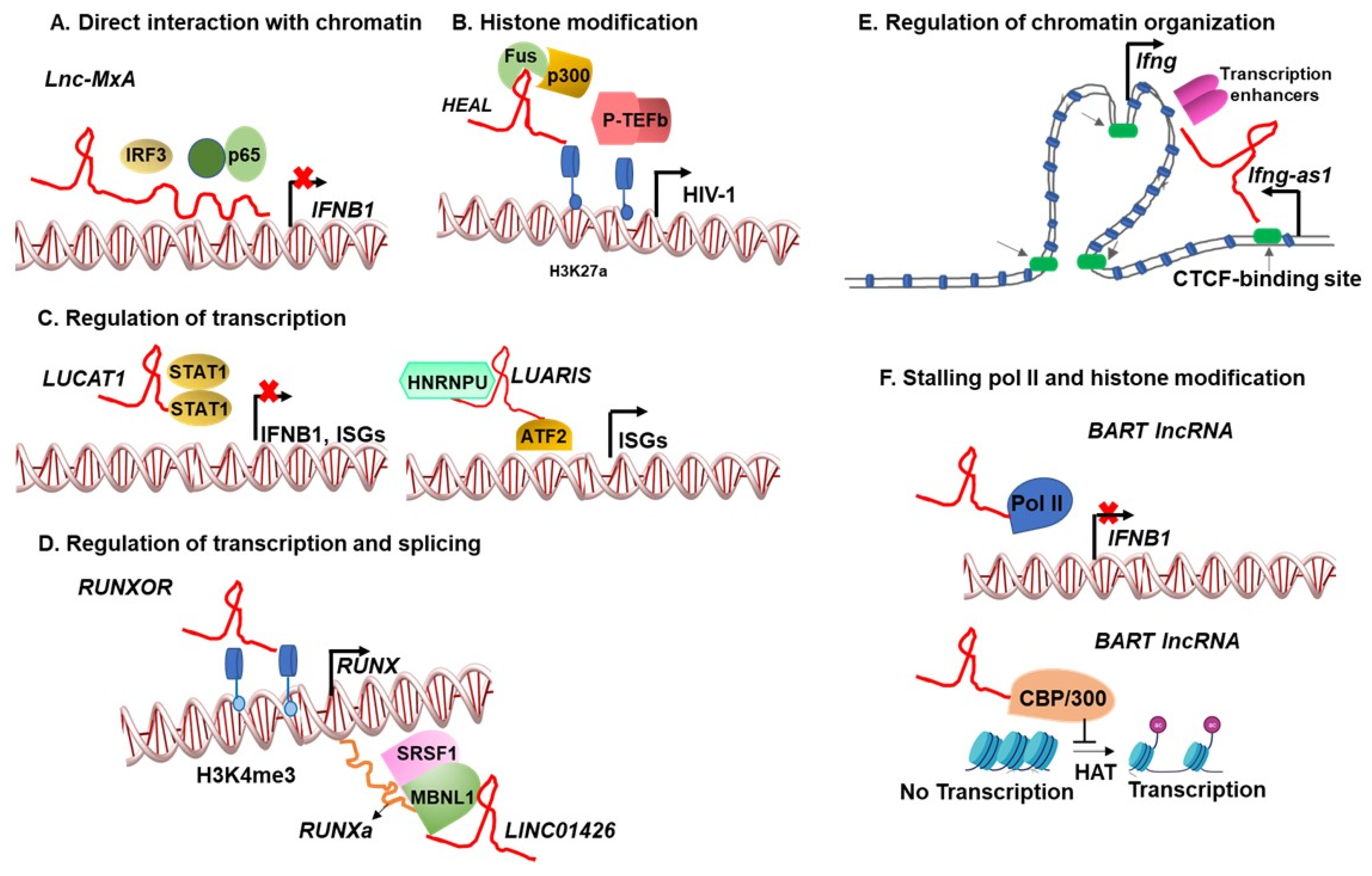
1. Introduction
2. Chromatin–lncRNA Interactions Regulate Viral Infections
3. Virus-Induced lncRNAs Regulate the Transcription and Splicing of Host and Viral Genes
4. Heterogeneity in lncRNA Form, Function, and Phenotype
5. Therapeutic Potential of lncRNAs
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Brass, A.L.; Dykxhoorn, D.M.; Benita, Y.; Yan, N.; Engelman, A.; Xavier, R.J.; Lieberman, J.; Elledge, S.J. Identification of host proteins required for HIV infection through a functional genomic screen. Science 2008, 319, 921–926. [Google Scholar] [CrossRef]
- Brass, A.L.; Huang, I.C.; Benita, Y.; John, S.P.; Krishnan, M.N.; Feeley, E.M.; Ryan, B.J.; Weyer, J.L.; van der Weyden, L.; Fikrig, E.; et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 2009, 139, 1243–1254. [Google Scholar] [CrossRef]
- Cherry, S.; Doukas, T.; Armknecht, S.; Whelan, S.; Wang, H.; Sarnow, P.; Perrimon, N. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes. Dev. 2005, 19, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Deffrasnes, C.; Marsh, G.A.; Foo, C.H.; Rootes, C.L.; Gould, C.M.; Grusovin, J.; Monaghan, P.; Lo, M.K.; Tompkins, S.M.; Adams, T.E.; et al. Genome-wide siRNA Screening at Biosafety Level 4 Reveals a Crucial Role for Fibrillarin in Henipavirus Infection. PLoS Pathog. 2016, 12, e1005478. [Google Scholar] [CrossRef]
- Karlas, A.; Machuy, N.; Shin, Y.; Pleissner, K.P.; Artarini, A.; Heuer, D.; Becker, D.; Khalil, H.; Ogilvie, L.A.; Hess, S.; et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 2010, 463, 818–822. [Google Scholar] [CrossRef]
- Konig, R.; Zhou, Y.; Elleder, D.; Diamond, T.L.; Bonamy, G.M.; Irelan, J.T.; Chiang, C.Y.; Tu, B.P.; De Jesus, P.D.; Lilley, C.E.; et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 2008, 135, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Lipovsky, A.; Popa, A.; Pimienta, G.; Wyler, M.; Bhan, A.; Kuruvilla, L.; Guie, M.A.; Poffenberger, A.C.; Nelson, C.D.; Atwood, W.J.; et al. Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus. Proc. Natl. Acad. Sci. USA 2013, 110, 7452–7457. [Google Scholar] [CrossRef]
- Martin, S.; Chiramel, A.I.; Schmidt, M.L.; Chen, Y.C.; Whitt, N.; Watt, A.; Dunham, E.C.; Shifflett, K.; Traeger, S.; Leske, A.; et al. A genome-wide siRNA screen identifies a druggable host pathway essential for the Ebola virus life cycle. Genome Med. 2018, 10, 58. [Google Scholar] [CrossRef]
- Ooi, Y.S.; Stiles, K.M.; Liu, C.Y.; Taylor, G.M.; Kielian, M. Genome-wide RNAi screen identifies novel host proteins required for alphavirus entry. PLoS Pathog. 2013, 9, e1003835. [Google Scholar] [CrossRef] [PubMed]
- Sivan, G.; Martin, S.E.; Myers, T.G.; Buehler, E.; Szymczyk, K.H.; Ormanoglu, P.; Moss, B. Human genome-wide RNAi screen reveals a role for nuclear pore proteins in poxvirus morphogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 3519–3524. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.X.; Phuektes, P.; Kumar, P.; Goh, G.Y.; Moreau, D.; Chow, V.T.; Bard, F.; Chu, J.J. Human genome-wide RNAi screen reveals host factors required for enterovirus 71 replication. Nat. Commun. 2016, 7, 13150. [Google Scholar] [CrossRef] [PubMed]
- Yasunaga, A.; Hanna, S.L.; Li, J.; Cho, H.; Rose, P.P.; Spiridigliozzi, A.; Gold, B.; Diamond, M.S.; Cherry, S. Genome-wide RNAi screen identifies broadly-acting host factors that inhibit arbovirus infection. PLoS Pathog. 2014, 10, e1003914. [Google Scholar] [CrossRef]
- Yeung, M.L.; Houzet, L.; Yedavalli, V.S.; Jeang, K.T. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J. Biol. Chem. 2009, 284, 19463–19473. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Xu, M.; Huang, Q.; Gates, A.T.; Zhang, X.D.; Castle, J.C.; Stec, E.; Ferrer, M.; Strulovici, B.; Hazuda, D.J.; et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 2008, 4, 495–504. [Google Scholar] [CrossRef]
- Baggen, J.; Persoons, L.; Vanstreels, E.; Jansen, S.; Van Looveren, D.; Boeckx, B.; Geudens, V.; De Man, J.; Jochmans, D.; Wauters, J.; et al. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat. Genet. 2021, 53, 435–444. [Google Scholar] [CrossRef]
- Krasnopolsky, S.; Kuzmina, A.; Taube, R. Genome-wide CRISPR knockout screen identifies ZNF304 as a silencer of HIV transcription that promotes viral latency. PLoS Pathog. 2020, 16, e1008834. [Google Scholar] [CrossRef]
- Kulsuptrakul, J.; Wang, R.; Meyers, N.L.; Ott, M.; Puschnik, A.S. A genome-wide CRISPR screen identifies UFMylation and TRAMP-like complexes as host factors required for hepatitis A virus infection. Cell Rep. 2021, 34, 108859. [Google Scholar] [CrossRef]
- Li, B.; Clohisey, S.M.; Chia, B.S.; Wang, B.; Cui, A.; Eisenhaure, T.; Schweitzer, L.D.; Hoover, P.; Parkinson, N.J.; Nachshon, A.; et al. Genome-wide CRISPR screen identifies host dependency factors for influenza A virus infection. Nat. Commun. 2020, 11, 164. [Google Scholar] [CrossRef]
- Li, Y.; Muffat, J.; Omer Javed, A.; Keys, H.R.; Lungjangwa, T.; Bosch, I.; Khan, M.; Virgilio, M.C.; Gehrke, L.; Sabatini, D.M.; et al. Genome-wide CRISPR screen for Zika virus resistance in human neural cells. Proc. Natl. Acad. Sci. USA 2019, 116, 9527–9532. [Google Scholar] [CrossRef] [PubMed]
- Park, R.J.; Wang, T.; Koundakjian, D.; Hultquist, J.F.; Lamothe-Molina, P.; Monel, B.; Schumann, K.; Yu, H.; Krupzcak, K.M.; Garcia-Beltran, W.; et al. A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors. Nat. Genet. 2017, 49, 193–203. [Google Scholar] [CrossRef]
- Thamamongood, T.; Aebischer, A.; Wagner, V.; Chang, M.W.; Elling, R.; Benner, C.; Garcia-Sastre, A.; Kochs, G.; Beer, M.; Schwemmle, M. A Genome-Wide CRISPR-Cas9 Screen Reveals the Requirement of Host Cell Sulfation for Schmallenberg Virus Infection. J. Virol. 2020, 94, e00752-20. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Feng, F.; Hu, G.; Wang, Y.; Yu, Y.; Zhu, Y.; Xu, W.; Cai, X.; Sun, Z.; Han, W.; et al. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 2021, 12, 961. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P.; Bernstein, B.E.; Birney, E.; Dunham, I.; Green, E.D.; Gunter, C.; Snyder, M. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Fraser, P. No-nonsense functions for long noncoding RNAs. Cell 2011, 145, 178–181. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes. Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef]
- Chen, Y.G.; Satpathy, A.T.; Chang, H.Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 2017, 18, 962–972. [Google Scholar] [CrossRef]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef]
- Winterling, C.; Koch, M.; Koeppel, M.; Garcia-Alcalde, F.; Karlas, A.; Meyer, T.F. Evidence for a crucial role of a host non-coding RNA in influenza A virus replication. RNA Biol. 2014, 11, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Yao, M.; Yuan, L.; Qiao, J. Long-chain non-coding RNA n337374 relieves symptoms of respiratory syncytial virus-induced asthma by inhibiting dendritic cell maturation via the CD86 and the ERK pathway. Allergol. Immunopathol. (Madr) 2021, 49, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Carnero, E.; Barriocanal, M.; Prior, C.; Pablo Unfried, J.; Segura, V.; Guruceaga, E.; Enguita, M.; Smerdou, C.; Gastaminza, P.; Fortes, P. Long noncoding RNA EGOT negatively affects the antiviral response and favors HCV replication. EMBO Rep. 2016, 17, 1013–1028. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, M.; Lind, S.B.; Pettersson, U. Distinct temporal changes in host cell lncRNA expression during the course of an adenovirus infection. Virology 2016, 492, 242–250. [Google Scholar] [CrossRef]
- Liu, H.; Xu, J.; Yang, Y.; Wang, X.; Wu, E.; Majerciak, V.; Zhang, T.; Steenbergen, R.D.M.; Wang, H.K.; Banerjee, N.S.; et al. Oncogenic HPV promotes the expression of the long noncoding RNA lnc-FANCI-2 through E7 and YY1. Proc. Natl. Acad. Sci. USA 2021, 118, e2014195118. [Google Scholar] [CrossRef]
- Kuo, R.L.; Chen, Y.T.; Li, H.A.; Wu, C.C.; Chiang, H.C.; Lin, J.Y.; Huang, H.I.; Shih, S.R.; Chin-Ming Tan, B. Molecular determinants and heterogeneity underlying host response to EV-A71 infection at single-cell resolution. RNA Biol. 2021, 18, 796–808. [Google Scholar] [CrossRef]
- Devadoss, D.; Acharya, A.; Manevski, M.; Pandey, K.; Borchert, G.M.; Nair, M.; Mirsaeidi, M.; Byrareddy, S.N.; Chand, H.S. Distinct Mucoinflammatory Phenotype and the Immunomodulatory Long Noncoding Transcripts Associated with SARS-CoV-2 Airway Infection. medRxiv 2021. [Google Scholar] [CrossRef]
- Laha, S.; Saha, C.; Dutta, S.; Basu, M.; Chatterjee, R.; Ghosh, S.; Bhattacharyya, N.P. In silico analysis of altered expression of long non-coding RNA in SARS-CoV-2 infected cells and their possible regulation by STAT1, STAT3 and interferon regulatory factors. Heliyon 2021, 7, e06395. [Google Scholar] [CrossRef] [PubMed]
- Morenikeji, O.B.; Bernard, K.; Strutton, E.; Wallace, M.; Thomas, B.N. Evolutionarily Conserved Long Non-coding RNA Regulates Gene Expression in Cytokine Storm During COVID-19. Front. Bioeng. Biotechnol. 2020, 8, 582953. [Google Scholar] [CrossRef]
- Mukherjee, S.; Banerjee, B.; Karasik, D.; Frenkel-Morgenstern, M. mRNA-lncRNA Co-Expression Network Analysis Reveals the Role of lncRNAs in Immune Dysfunction during Severe SARS-CoV-2 Infection. Viruses 2021, 13, 402. [Google Scholar] [CrossRef]
- Natarelli, L.; Parca, L.; Mazza, T.; Weber, C.; Virgili, F.; Fratantonio, D. MicroRNAs and Long Non-Coding RNAs as Potential Candidates to Target Specific Motifs of SARS-CoV-2. Noncoding RNA 2021, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Shaath, H.; Alajez, N.M. Identification of PBMC-based molecular signature associational with COVID-19 disease severity. Heliyon 2021, 7, e06866. [Google Scholar] [CrossRef]
- Tang, H.; Gao, Y.; Li, Z.; Miao, Y.; Huang, Z.; Liu, X.; Xie, L.; Li, H.; Wen, W.; Zheng, Y.; et al. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clin. Transl. Med. 2020, 10, e200. [Google Scholar] [CrossRef]
- Turjya, R.R.; Khan, M.A.; Mir Md Khademul Islam, A.B. Perversely expressed long noncoding RNAs can alter host response and viral proliferation in SARS-CoV-2 infection. Future Virol. 2020, 15, 577–593. [Google Scholar] [CrossRef]
- Vishnubalaji, R.; Shaath, H.; Alajez, N.M. Protein Coding and Long Noncoding RNA (lncRNA) Transcriptional Landscape in SARS-CoV-2 Infected Bronchial Epithelial Cells Highlight a Role for Interferon and Inflammatory Response. Genes. 2020, 11, 760. [Google Scholar] [CrossRef]
- Wu, Y.; Zhao, T.; Deng, R.; Xia, X.; Li, B.; Wang, X. A study of differential circRNA and lncRNA expressions in COVID-19-infected peripheral blood. Sci. Rep. 2021, 11, 7991. [Google Scholar] [CrossRef]
- Xiong, Y.; Liu, Y.; Cao, L.; Wang, D.; Guo, M.; Jiang, A.; Guo, D.; Hu, W.; Yang, J.; Tang, Z.; et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020, 9, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.T.; Sova, P.; Peng, X.; Weiss, J.; Law, G.L.; Palermo, R.E.; Katze, M.G. Next-generation sequencing reveals HIV-1-mediated suppression of T cell activation and RNA processing and regulation of noncoding RNA expression in a CD4+ T cell line. MBio 2011, 2, e00134-11. [Google Scholar] [CrossRef]
- Peng, X.; Sova, P.; Green, R.R.; Thomas, M.J.; Korth, M.J.; Proll, S.; Xu, J.; Cheng, Y.; Yi, K.; Chen, L.; et al. Deep sequencing of HIV-infected cells: Insights into nascent transcription and host-directed therapy. J. Virol. 2014, 88, 8768–8782. [Google Scholar] [CrossRef] [PubMed]
- Schynkel, T.; Szaniawski, M.A.; Spivak, A.M.; Bosque, A.; Planelles, V.; Vandekerckhove, L.; Trypsteen, W. Interferon-Mediated Long Non-Coding RNA Response in Macrophages in the Context of HIV. Int. J. Mol. Sci. 2020, 21, 7741. [Google Scholar] [CrossRef] [PubMed]
- Trypsteen, W.; Mohammadi, P.; Van Hecke, C.; Mestdagh, P.; Lefever, S.; Saeys, Y.; De Bleser, P.; Vandesompele, J.; Ciuffi, A.; Vandekerckhove, L.; et al. Differential expression of lncRNAs during the HIV replication cycle: An underestimated layer in the HIV-host interplay. Sci. Rep. 2016, 6, 36111. [Google Scholar] [CrossRef] [PubMed]
- Trypsteen, W.; White, C.H.; Mukim, A.; Spina, C.A.; De Spiegelaere, W.; Lefever, S.; Planelles, V.; Bosque, A.; Woelk, C.H.; Vandekerckhove, L.; et al. Long non-coding RNAs and latent HIV—A search for novel targets for latency reversal. PLoS ONE 2019, 14, e0224879. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, C.Y.; Yedavalli, V.S.; Jeang, K.T. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio 2013, 4, e00596-12. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Hu, J.; Chen, J.L. lncRNAs regulate the innate immune response to viral infection. Wiley Interdiscip. Rev. RNA 2016, 7, 129–143. [Google Scholar] [CrossRef]
- Fan, J.; Cheng, M.; Chi, X.; Liu, X.; Yang, W. A Human Long Non-coding RNA LncATV Promotes Virus Replication Through Restricting RIG-I-Mediated Innate Immunity. Front. Immunol. 2019, 10, 1711. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, S.; Yang, Z.; Lin, H.; Zhu, J.; Liu, L.; Wang, W.; Liu, S.; Liu, W.; Ma, Y.; et al. Self-Recognition of an Inducible Host lncRNA by RIG-I Feedback Restricts Innate Immune Response. Cell 2018, 173, 906–919 e913. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Jiang, M.; Liu, L.; Yang, Z.; Ma, Z.; Liu, S.; Ma, Y.; Zhang, L.; Cao, X. The long noncoding RNA Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response. Nat. Immunol. 2019, 20, 812–823. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Han, P.; Ye, W.; Chen, H.; Zheng, X.; Cheng, L.; Zhang, L.; Yu, L.; Wu, X.; Xu, Z.; et al. The Long Noncoding RNA NEAT1 Exerts Antihantaviral Effects by Acting as Positive Feedback for RIG-I Signaling. J. Virol. 2017, 91, e02250-16. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Zhu, X.; Chen, Y.; Wei, H.; Chen, Q.; Chi, X.; Qi, B.; Zhang, L.; Zhao, Y.; Gao, G.F.; et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 2014, 16, 616–626. [Google Scholar] [CrossRef]
- Xie, Q.; Chen, S.; Tian, R.; Huang, X.; Deng, R.; Xue, B.; Qin, Y.; Xu, Y.; Wang, J.; Guo, M.; et al. Long Noncoding RNA ITPRIP-1 Positively Regulates the Innate Immune Response through Promotion of Oligomerization and Activation of MDA5. J. Virol. 2018, 92, e00507-18. [Google Scholar] [CrossRef]
- Gonzalez-Moro, I.; Olazagoitia-Garmendia, A.; Colli, M.L.; Cobo-Vuilleumier, N.; Postler, T.S.; Marselli, L.; Marchetti, P.; Ghosh, S.; Gauthier, B.R.; Eizirik, D.L.; et al. The T1D-associated lncRNA Lnc13 modulates human pancreatic beta cell inflammation by allele-specific stabilization of STAT1 mRNA. Proc. Natl. Acad. Sci. USA 2020, 117, 9022–9031. [Google Scholar] [CrossRef]
- Hu, P.; Wilhelm, J.; Gerresheim, G.K.; Shalamova, L.A.; Niepmann, M. Lnc-ITM2C-1 and GPR55 Are Proviral Host Factors for Hepatitis C Virus. Viruses 2019, 11, 549. [Google Scholar] [CrossRef]
- Li, X.; Guo, G.; Lu, M.; Chai, W.; Li, Y.; Tong, X.; Li, J.; Jia, X.; Liu, W.; Qi, D.; et al. Long Noncoding RNA Lnc-MxA Inhibits Beta Interferon Transcription by Forming RNA-DNA Triplexes at Its Promoter. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, D.; Feng, W.; Guo, Y.; Sun, X.; Zhang, M.; Guan, Z.; Duan, M. Long noncoding RNA TSPOAP1 antisense RNA 1 negatively modulates type I IFN signaling to facilitate influenza A virus replication. J. Med. Virol. 2019, 94, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Xia, M.; Wang, K.; Lai, C.; Fan, H.; Gu, H.; Yang, P.; Wang, X. A Long Non-coding RNA IVRPIE Promotes Host Antiviral Immune Responses Through Regulating Interferon beta1 and ISG Expression. Front. Microbiol. 2020, 11, 260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chao, T.C.; Patil, V.S.; Qin, Y.; Tiwari, S.K.; Chiou, J.; Dobin, A.; Tsai, C.M.; Li, Z.; Dang, J.; et al. The long noncoding RNA ROCKI regulates inflammatory gene expression. EMBO J. 2019, 38, e100041. [Google Scholar] [CrossRef]
- Kambara, H.; Niazi, F.; Kostadinova, L.; Moonka, D.K.; Siegel, C.T.; Post, A.B.; Carnero, E.; Barriocanal, M.; Fortes, P.; Anthony, D.D.; et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res. 2014, 42, 10668–10680. [Google Scholar] [CrossRef] [PubMed]
- Rapicavoli, N.A.; Qu, K.; Zhang, J.; Mikhail, M.; Laberge, R.M.; Chang, H.Y. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife 2013, 2, e00762. [Google Scholar] [CrossRef]
- Sonkoly, E.; Bata-Csorgo, Z.; Pivarcsi, A.; Polyanka, H.; Kenderessy-Szabo, A.; Molnar, G.; Szentpali, K.; Bari, L.; Megyeri, K.; Mandi, Y.; et al. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J. Biol. Chem. 2005, 280, 24159–24167. [Google Scholar] [CrossRef]
- Yang, W.S.; Yeh, W.W.; Campbell, M.; Chang, L.; Chang, P.C. Long non-coding RNA KIKAT/LINC01061 as a novel epigenetic regulator that relocates KDM4A on chromatin and modulates viral reactivation. PLoS Pathog. 2021, 17, e1009670. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, P.; Li, Z.; Li, Z.; Han, D.; Wen, M.; Zhao, Q.; Zhang, L.; Ma, Y.; Liu, W.; et al. IRF3-binding lncRNA-ISIR strengthens interferon production in viral infection and autoinflammation. Cell Rep. 2021, 37, 109926. [Google Scholar] [CrossRef]
- Qiu, L.; Wang, T.; Tang, Q.; Li, G.; Wu, P.; Chen, K. Long Non-coding RNAs: Regulators of Viral Infection and the Interferon Antiviral Response. Front. Microbiol. 2018, 9, 1621. [Google Scholar] [CrossRef]
- Xiong, Y.; Yuan, J.; Zhang, C.; Zhu, Y.; Kuang, X.; Lan, L.; Wang, X. The STAT3-regulated long non-coding RNA Lethe promote the HCV replication. Biomed. Pharmacother. 2015, 72, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Duan, X.; Holmes, J.A.; Li, W.; Lee, S.H.; Tu, Z.; Zhu, C.; Salloum, S.; Lidofsky, A.; Schaefer, E.A.; et al. A Long Noncoding RNA Regulates Hepatitis C Virus Infection Through Interferon Alpha-Inducible Protein 6. Hepatology 2019, 69, 1004–1019. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Gao, Y.; Liu, X.; Bai, J.; Jiang, P.; Wang, X. Long non-coding RNA LNC_000641 regulates pseudorabies virus replication. Vet. Res. 2021, 52, 52. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fan, P.; Zhao, Y.; Zhang, S.; Lu, J.; Xie, W.; Jiang, Y.; Lei, F.; Xu, N.; Zhang, Y. NEAT1 modulates herpes simplex virus-1 replication by regulating viral gene transcription. Cell Mol. Life Sci. 2017, 74, 1117–1131. [Google Scholar] [CrossRef]
- Wang, Y.; Huo, Z.; Lin, Q.; Lin, Y.; Chen, C.; Huang, Y.; Huang, C.; Zhang, J.; He, J.; Liu, C.; et al. Positive Feedback Loop of Long Noncoding RNA OASL-IT1 and Innate Immune Response Restricts the Replication of Zika Virus in Epithelial A549 Cells. J. Innate Immun. 2021, 13, 179–193. [Google Scholar] [CrossRef]
- Nishitsuji, H.; Ujino, S.; Yoshio, S.; Sugiyama, M.; Mizokami, M.; Kanto, T.; Shimotohno, K. Long noncoding RNA #32 contributes to antiviral responses by controlling interferon-stimulated gene expression. Proc. Natl. Acad. Sci. USA 2016, 113, 10388–10393. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, C.; Ma, X.; Geng, G.; Liu, B.; Zhang, Y.; Zhang, S.; Zhong, F.; Liu, C.; Yin, Y.; et al. Long noncoding RNA NRON contributes to HIV-1 latency by specifically inducing tat protein degradation. Nat. Commun. 2016, 7, 11730. [Google Scholar] [CrossRef]
- Qu, D.; Sun, W.W.; Li, L.; Ma, L.; Sun, L.; Jin, X.; Li, T.; Hou, W.; Wang, J.H. Long noncoding RNA MALAT1 releases epigenetic silencing of HIV-1 replication by displacing the polycomb repressive complex 2 from binding to the LTR promoter. Nucleic Acids Res. 2019, 47, 3013–3027. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Kiss, T.; Michels, A.A.; Bensaude, O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 2001, 414, 322–325. [Google Scholar] [CrossRef] [PubMed]
- Contreras, X.; Barboric, M.; Lenasi, T.; Peterlin, B.M. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 2007, 3, 1459–1469. [Google Scholar] [CrossRef]
- Budhiraja, S.; Famiglietti, M.; Bosque, A.; Planelles, V.; Rice, A.P. Cyclin T1 and CDK9 T-loop phosphorylation are downregulated during establishment of HIV-1 latency in primary resting memory CD4+ T cells. J. Virol. 2013, 87, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Eilebrecht, S.; Benecke, B.J.; Benecke, A.G. Latent HIV-1 TAR Regulates 7SK-responsive P-TEFb Target Genes and Targets Cellular Immune Responses in the Absence of Tat. Genomics Proteomics Bioinformatics 2017, 15, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Huan, C.; Li, Z.; Ning, S.; Wang, H.; Yu, X.F.; Zhang, W. Long Noncoding RNA uc002yug.2 Activates HIV-1 Latency through Regulation of mRNA Levels of Various RUNX1 Isoforms and Increased Tat Expression. J. Virol. 2018, 92, e01844-17. [Google Scholar] [CrossRef] [PubMed]
- Barichievy, S.; Naidoo, J.; Boulle, M.; Scholefield, J.; Parihar, S.P.; Coussens, A.K.; Brombacher, F.; Sigal, A.; Mhlanga, M.M. Viral Apoptosis Evasion via the MAPK Pathway by Use of a Host Long Noncoding RNA. Front. Cell Infect. Microbiol. 2018, 8, 263. [Google Scholar] [CrossRef]
- Chao, T.C.; Zhang, Q.; Li, Z.; Tiwari, S.K.; Qin, Y.; Yau, E.; Sanchez, A.; Singh, G.; Chang, K.; Kaul, M.; et al. The Long Noncoding RNA HEAL Regulates HIV-1 Replication through Epigenetic Regulation of the HIV-1 Promoter. mBio 2019, 10, e02016-19. [Google Scholar] [CrossRef]
- Saayman, S.; Ackley, A.; Turner, A.W.; Famiglietti, M.; Bosque, A.; Clemson, M.; Planelles, V.; Morris, K.V. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol. Ther. 2014, 22, 1164–1175. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, C.C.; Pari, G. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog. 2012, 8, e1002680. [Google Scholar] [CrossRef]
- Rossetto, C.C.; Tarrant-Elorza, M.; Verma, S.; Purushothaman, P.; Pari, G.S. Regulation of viral and cellular gene expression by Kaposi’s sarcoma-associated herpesvirus polyadenylated nuclear RNA. J. Virol. 2013, 87, 5540–5553. [Google Scholar] [CrossRef]
- Rossetto, C.C.; Tarrant-Elorza, M.; Verma, S.; Purushothaman, P.; Pari, G.S. Correction for Rossetto et al., Regulation of Viral and Cellular Gene Expression by Kaposi’s Sarcoma-Associated Herpesvirus Polyadenylated Nuclear RNA. J. Virol. 2016, 90, 4255. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, C.C.; Tarrant-Elorza, M.; Pari, G.S. Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14 (+) monocytes and CD34 (+) cells. PLoS Pathog. 2013, 9, e1003366. [Google Scholar] [CrossRef]
- Marquitz, A.R.; Mathur, A.; Edwards, R.H.; Raab-Traub, N. Host Gene Expression Is Regulated by Two Types of Noncoding RNAs Transcribed from the Epstein-Barr Virus BamHI A Rightward Transcript Region. J. Virol. 2015, 89, 11256–11268. [Google Scholar] [CrossRef]
- Verhoeven, R.J.A.; Tong, S.; Mok, B.W.; Liu, J.; He, S.; Zong, J.; Chen, Y.; Tsao, S.W.; Lung, M.L.; Chen, H. Epstein-Barr Virus BART Long Non-coding RNAs Function as Epigenetic Modulators in Nasopharyngeal Carcinoma. Front. Oncol. 2019, 9, 1120. [Google Scholar] [CrossRef]
- Park, R.; Miller, G. Epstein-Barr Virus-Induced Nodules on Viral Replication Compartments Contain RNA Processing Proteins and a Viral Long Noncoding RNA. J. Virol. 2018, 92, e01254-18. [Google Scholar] [CrossRef] [PubMed]
- Rennekamp, A.J.; Lieberman, P.M. Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA-DNA hybrid molecule at OriLyt. J. Virol. 2011, 85, 2837–2850. [Google Scholar] [CrossRef]
- Peng, X.; Gralinski, L.; Armour, C.D.; Ferris, M.T.; Thomas, M.J.; Proll, S.; Bradel-Tretheway, B.G.; Korth, M.J.; Castle, J.C.; Biery, M.C.; et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio 2010, 1, e00206-10. [Google Scholar] [CrossRef] [PubMed]
- Sui, B.; Chen, D.; Liu, W.; Wu, Q.; Tian, B.; Li, Y.; Hou, J.; Liu, S.; Xie, J.; Jiang, H.; et al. A novel antiviral lncRNA, EDAL, shields a T309 O-GlcNAcylation site to promote EZH2 lysosomal degradation. Genome Biol. 2020, 21, 228. [Google Scholar] [CrossRef]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314. [Google Scholar] [CrossRef] [PubMed]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigo, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. Annotating noncoding RNA genes. Annu. Rev. Genomics Hum. Genet. 2007, 8, 279–298. [Google Scholar] [CrossRef]
- Lagarde, J.; Uszczynska-Ratajczak, B.; Carbonell, S.; Perez-Lluch, S.; Abad, A.; Davis, C.; Gingeras, T.R.; Frankish, A.; Harrow, J.; Guigo, R.; et al. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat. Genet. 2017, 49, 1731–1740. [Google Scholar] [CrossRef]
- Mele, M.; Mattioli, K.; Mallard, W.; Shechner, D.M.; Gerhardinger, C.; Rinn, J.L. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017, 27, 27–37. [Google Scholar] [CrossRef]
- Schlackow, M.; Nojima, T.; Gomes, T.; Dhir, A.; Carmo-Fonseca, M.; Proudfoot, N.J. Distinctive Patterns of Transcription and RNA Processing for Human lincRNAs. Mol. Cell 2017, 65, 25–38. [Google Scholar] [CrossRef]
- West, J.A.; Davis, C.P.; Sunwoo, H.; Simon, M.D.; Sadreyev, R.I.; Wang, P.I.; Tolstorukov, M.Y.; Kingston, R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 2014, 55, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Lubelsky, Y.; Ulitsky, I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature 2018, 555, 107–111. [Google Scholar] [CrossRef]
- Zhang, B.; Gunawardane, L.; Niazi, F.; Jahanbani, F.; Chen, X.; Valadkhan, S. A novel RNA motif mediates the strict nuclear localization of a long noncoding RNA. Mol. Cell Biol. 2014, 34, 2318–2329. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, R.; Tano, K.; Mizuno, R.; Nakamura, Y.; Ijiri, K.; Rakwal, R.; Shibato, J.; Masuo, Y.; Mayeda, A.; Hirose, T.; et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA 2012, 18, 738–751. [Google Scholar] [CrossRef]
- Hacisuleyman, E.; Goff, L.A.; Trapnell, C.; Williams, A.; Henao-Mejia, J.; Sun, L.; McClanahan, P.; Hendrickson, D.G.; Sauvageau, M.; Kelley, D.R.; et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014, 21, 198–206. [Google Scholar] [CrossRef]
- Dumbovic, G.; Braunschweig, U.; Langner, H.K.; Smallegan, M.; Biayna, J.; Hass, E.P.; Jastrzebska, K.; Blencowe, B.; Cech, T.R.; Caruthers, M.H.; et al. Nuclear compartmentalization of TERT mRNA and TUG1 lncRNA is driven by intron retention. Nat. Commun. 2021, 12, 3308. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, I.V.; Dayton, J.; Harger, S.; Rossetto, C.C. The Expression and Nuclear Retention Element of Polyadenylated Nuclear RNA Is Not Required for Productive Lytic Replication of Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2021, 95, e0009621. [Google Scholar] [CrossRef] [PubMed]
- Conrad, N.K.; Mili, S.; Marshall, E.L.; Shu, M.D.; Steitz, J.A. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol. Cell 2006, 24, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Conrad, N.K.; Shu, M.D.; Uyhazi, K.E.; Steitz, J.A. Mutational analysis of a viral RNA element that counteracts rapid RNA decay by interaction with the polyadenylate tail. Proc. Natl. Acad. Sci. USA 2007, 104, 10412–10417. [Google Scholar] [CrossRef]
- Conrad, N.K.; Steitz, J.A. A Kaposi’s sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 2005, 24, 1831–1841. [Google Scholar] [CrossRef]
- Rossetto, C.C.; Pari, G.S. PAN’s Labyrinth: Molecular biology of Kaposi’s sarcoma-associated herpesvirus (KSHV) PAN RNA, a multifunctional long noncoding RNA. Viruses 2014, 6, 4212–4226. [Google Scholar] [CrossRef]
- Sahin, B.B.; Patel, D.; Conrad, N.K. Kaposi’s sarcoma-associated herpesvirus ORF57 protein binds and protects a nuclear noncoding RNA from cellular RNA decay pathways. PLoS Pathog. 2010, 6, e1000799. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Valenstein, M.L.; Yario, T.A.; Tycowski, K.T.; Steitz, J.A. Formation of triple-helical structures by the 3’-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc. Natl. Acad. Sci. USA 2012, 109, 19202–19207. [Google Scholar] [CrossRef]
- Hautbergue, G.M. RNA Nuclear Export: From Neurological Disorders to Cancer. Adv. Exp. Med. Biol. 2017, 1007, 89–109. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Viphakone, N.; Sudbery, I.; Griffith, L.; Heath, C.G.; Sims, D.; Wilson, S.A. Co-transcriptional Loading of RNA Export Factors Shapes the Human Transcriptome. Mol. Cell 2019, 75, 310–323 e318. [Google Scholar] [CrossRef] [PubMed]
- Cohen, H.R.; Panning, B. XIST RNA exhibits nuclear retention and exhibits reduced association with the export factor TAP/NXF1. Chromosoma 2007, 116, 373–383. [Google Scholar] [CrossRef]
- Bergmann, J.H.; Spector, D.L. Long non-coding RNAs: Modulators of nuclear structure and function. Curr. Opin. Cell Biol. 2014, 26, 10–18. [Google Scholar] [CrossRef]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Engreitz, J.M.; Pandya-Jones, A.; McDonel, P.; Shishkin, A.; Sirokman, K.; Surka, C.; Kadri, S.; Xing, J.; Goren, A.; Lander, E.S.; et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 2013, 341, 1237973. [Google Scholar] [CrossRef]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef]
- Mao, Y.S.; Sunwoo, H.; Zhang, B.; Spector, D.L. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 2011, 13, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Landeras-Bueno, S.; Jorba, N.; Perez-Cidoncha, M.; Ortin, J. The splicing factor proline-glutamine rich (SFPQ/PSF) is involved in influenza virus transcription. PLoS Pathog. 2011, 7, e1002397. [Google Scholar] [CrossRef]
- Markaki, Y.; Gan Chong, J.; Wang, Y.; Jacobson, E.C.; Luong, C.; Tan, S.Y.X.; Jachowicz, J.W.; Strehle, M.; Maestrini, D.; Banerjee, A.K.; et al. Xist nucleates local protein gradients to propagate silencing across the X chromosome. Cell 2021, 184, 6174–6192 e6132. [Google Scholar] [CrossRef]
- Quinodoz, S.A.; Jachowicz, J.W.; Bhat, P.; Ollikainen, N.; Banerjee, A.K.; Goronzy, I.N.; Blanco, M.R.; Chovanec, P.; Chow, A.; Markaki, Y.; et al. RNA promotes the formation of spatial compartments in the nucleus. Cell 2021, 184, 5775–5790 e5730. [Google Scholar] [CrossRef]
- Quinodoz, S.A.; Ollikainen, N.; Tabak, B.; Palla, A.; Schmidt, J.M.; Detmar, E.; Lai, M.M.; Shishkin, A.A.; Bhat, P.; Takei, Y.; et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 2018, 174, 744–757 e724. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Wahrisch, S.; Beisaw, A.; Macura, K.; Blass, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Hartana, C.A.; Rassadkina, Y.; Gao, C.; Martin-Gayo, E.; Walker, B.D.; Lichterfeld, M.; Yu, X.G. Long noncoding RNA MIR4435-2HG enhances metabolic function of myeloid dendritic cells from HIV-1 elite controllers. J. Clin. Invest. 2021, 131, e146136. [Google Scholar] [CrossRef] [PubMed]
- Kalwa, M.; Hanzelmann, S.; Otto, S.; Kuo, C.C.; Franzen, J.; Joussen, S.; Fernandez-Rebollo, E.; Rath, B.; Koch, C.; Hofmann, A.; et al. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 2016, 44, 10631–10643. [Google Scholar] [CrossRef] [PubMed]
- Mondal, T.; Subhash, S.; Vaid, R.; Enroth, S.; Uday, S.; Reinius, B.; Mitra, S.; Mohammed, A.; James, A.R.; Hoberg, E.; et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015, 6, 7743. [Google Scholar] [CrossRef]
- O’Leary, V.B.; Ovsepian, S.V.; Carrascosa, L.G.; Buske, F.A.; Radulovic, V.; Niyazi, M.; Moertl, S.; Trau, M.; Atkinson, M.J.; Anastasov, N. PARTICLE, a Triplex-Forming Long ncRNA, Regulates Locus-Specific Methylation in Response to Low-Dose Irradiation. Cell Rep. 2015, 11, 474–485. [Google Scholar] [CrossRef]
- O’Leary, V.B.; Smida, J.; Buske, F.A.; Carrascosa, L.G.; Azimzadeh, O.; Maugg, D.; Hain, S.; Tapio, S.; Heidenreich, W.; Kerr, J.; et al. PARTICLE triplexes cluster in the tumor suppressor WWOX and may extend throughout the human genome. Sci. Rep. 2017, 7, 7163. [Google Scholar] [CrossRef]
- Postepska-Igielska, A.; Giwojna, A.; Gasri-Plotnitsky, L.; Schmitt, N.; Dold, A.; Ginsberg, D.; Grummt, I. LncRNA Khps1 Regulates Expression of the Proto-oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure. Mol. Cell 2015, 60, 626–636. [Google Scholar] [CrossRef]
- Zhao, Z.; Senturk, N.; Song, C.; Grummt, I. lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes. Dev. 2018, 32, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Arab, K.; Karaulanov, E.; Musheev, M.; Trnka, P.; Schafer, A.; Grummt, I.; Niehrs, C. GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat. Genet. 2019, 51, 217–223. [Google Scholar] [CrossRef]
- Schoggins, J.W.; Wilson, S.J.; Panis, M.; Murphy, M.Y.; Jones, C.T.; Bieniasz, P.; Rice, C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 2011, 472, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, S.; Bonasio, R.; Saldana-Meyer, R.; Yoshida, T.; Son, J.; Nishino, K.; Umezawa, A.; Reinberg, D. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell 2014, 53, 290–300. [Google Scholar] [CrossRef]
- Yang, L.; Lin, C.; Jin, C.; Yang, J.C.; Tanasa, B.; Li, W.; Merkurjev, D.; Ohgi, K.A.; Meng, D.; Zhang, J.; et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 2013, 500, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.W.; Flynn, R.A.; Chen, Y.; Qu, K.; Wan, B.; Wang, K.C.; Lei, M.; Chang, H.Y. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. Elife 2014, 3, e02046. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Mitchell, J.A.; Sanz, L.A.; Pauler, F.M.; Ferguson-Smith, A.C.; Feil, R.; Fraser, P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 2008, 322, 1717–1720. [Google Scholar] [CrossRef]
- Yang, L.; Lin, C.; Liu, W.; Zhang, J.; Ohgi, K.A.; Grinstein, J.D.; Dorrestein, P.C.; Rosenfeld, M.G. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 2011, 147, 773–788. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [CrossRef]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Haller, O.; Staeheli, P.; Kochs, G. Protective role of interferon-induced Mx GTPases against influenza viruses. Rev. Sci. Tech. 2009, 28, 219–231. [Google Scholar] [CrossRef]
- Fan, Y.; Shen, B.; Tan, M.; Mu, X.; Qin, Y.; Zhang, F.; Liu, Y. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 2014, 20, 1531–1541. [Google Scholar] [CrossRef] [PubMed]
- Hirata, H.; Hinoda, Y.; Shahryari, V.; Deng, G.; Nakajima, K.; Tabatabai, Z.L.; Ishii, N.; Dahiya, R. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015, 75, 1322–1331. [Google Scholar] [CrossRef]
- Cheng, Y.; Imanirad, P.; Jutooru, I.; Hedrick, E.; Jin, U.H.; Rodrigues Hoffman, A.; Leal de Araujo, J.; Morpurgo, B.; Golovko, A.; Safe, S. Role of metastasis-associated lung adenocarcinoma transcript-1 (MALAT-1) in pancreatic cancer. PLoS ONE 2018, 13, e0192264. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sehgal, L.; Jain, N.; Khashab, T.; Mathur, R.; Samaniego, F. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J. Transl. Med. 2016, 14, 346. [Google Scholar] [CrossRef]
- Chang, P.C.; Fitzgerald, L.D.; Hsia, D.A.; Izumiya, Y.; Wu, C.Y.; Hsieh, W.P.; Lin, S.F.; Campbell, M.; Lam, K.S.; Luciw, P.A.; et al. Histone demethylase JMJD2A regulates Kaposi’s sarcoma-associated herpesvirus replication and is targeted by a viral transcriptional factor. J. Virol. 2011, 85, 3283–3293. [Google Scholar] [CrossRef]
- Yang, W.S.; Campbell, M.; Chang, P.C. SUMO modification of a heterochromatin histone demethylase JMJD2A enables viral gene transactivation and viral replication. PLoS Pathog. 2017, 13, e1006216. [Google Scholar] [CrossRef]
- Kobayashi-Ishihara, M.; Yamagishi, M.; Hara, T.; Matsuda, Y.; Takahashi, R.; Miyake, A.; Nakano, K.; Yamochi, T.; Ishida, T.; Watanabe, T. HIV-1-encoded antisense RNA suppresses viral replication for a prolonged period. Retrovirology 2012, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Zapata, J.C.; Campilongo, F.; Barclay, R.A.; DeMarino, C.; Iglesias-Ussel, M.D.; Kashanchi, F.; Romerio, F. The Human Immunodeficiency Virus 1 ASP RNA promotes viral latency by recruiting the Polycomb Repressor Complex 2 and promoting nucleosome assembly. Virology 2017, 506, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, P.; Dabral, P.; Gupta, N.; Sarkar, R.; Verma, S.C. KSHV Genome Replication and Maintenance. Front. Microbiol. 2016, 7, 54. [Google Scholar] [CrossRef]
- Sun, R.; Lin, S.F.; Gradoville, L.; Miller, G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 1996, 93, 11883–11888. [Google Scholar] [CrossRef]
- Borah, S.; Darricarrere, N.; Darnell, A.; Myoung, J.; Steitz, J.A. A viral nuclear noncoding RNA binds re-localized poly(A) binding protein and is required for late KSHV gene expression. PLoS Pathog. 2011, 7, e1002300. [Google Scholar] [CrossRef]
- Rossetto, C.C.; Pari, G.S. Kaposi’s sarcoma-associated herpesvirus noncoding polyadenylated nuclear RNA interacts with virus- and host cell-encoded proteins and suppresses expression of genes involved in immune modulation. J. Virol. 2011, 85, 13290–13297. [Google Scholar] [CrossRef]
- Song, M.J.; Brown, H.J.; Wu, T.T.; Sun, R. Transcription activation of polyadenylated nuclear rna by rta in human herpesvirus 8/Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2001, 75, 3129–3140. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.; Kim, K.Y.; Chang, P.C.; Huerta, S.; Shevchenko, B.; Wang, D.H.; Izumiya, C.; Kung, H.J.; Izumiya, Y. A lytic viral long noncoding RNA modulates the function of a latent protein. J. Virol. 2014, 88, 1843–1848. [Google Scholar] [CrossRef]
- Kumar, A.; Lyu, Y.; Yanagihashi, Y.; Chantarasrivong, C.; Majerciak, V.; Salemi, M.; Wang, K.H.; Inagaki, T.; Chuang, F.; Davis, R.R.; et al. KSHV episome tethering sites on host chromosomes and regulation of latency-lytic switch by CHD4. Cell Rep. 2022, 39, 110788. [Google Scholar] [CrossRef]
- Hiura, K.; Strahan, R.; Uppal, T.; Prince, B.; Rossetto, C.C.; Verma, S.C. KSHV ORF59 and PAN RNA Recruit Histone Demethylases to the Viral Chromatin during Lytic Reactivation. Viruses 2020, 12, 420. [Google Scholar] [CrossRef]
- Agarwal, S.; Vierbuchen, T.; Ghosh, S.; Chan, J.; Jiang, Z.; Kandasamy, R.K.; Ricci, E.; Fitzgerald, K.A. The long non-coding RNA LUCAT1 is a negative feedback regulator of interferon responses in humans. Nat. Commun. 2020, 11, 6348. [Google Scholar] [CrossRef]
- Aznaourova, M.; Schmerer, N.; Janga, H.; Zhang, Z.; Pauck, K.; Bushe, J.; Volkers, S.M.; Wendisch, D.; Georg, P.; Ntini, E.; et al. Single-cell RNA sequencing uncovers the nuclear decoy lincRNA PIRAT as a regulator of systemic monocyte immunity during COVID-19. Proc. Natl. Acad. Sci. USA 2022, 119, e2120680119. [Google Scholar] [CrossRef]
- Ma, G.; Yasunaga, J.I.; Shimura, K.; Takemoto, K.; Watanabe, M.; Amano, M.; Nakata, H.; Liu, B.; Zuo, X.; Matsuoka, M. Human retroviral antisense mRNAs are retained in the nuclei of infected cells for viral persistence. Proc. Natl. Acad. Sci. USA 2021, 118, e2014783118. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, P.; Ackley, A.; Vidarsdottir, L.; Lui, W.O.; Corcoran, M.; Grander, D.; Morris, K.V. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat. Struct. Mol. Biol. 2013, 20, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Halley, P.; Kadakkuzha, B.M.; Faghihi, M.A.; Magistri, M.; Zeier, Z.; Khorkova, O.; Coito, C.; Hsiao, J.; Lawrence, M.; Wahlestedt, C. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014, 6, 222–230. [Google Scholar] [CrossRef]
- Klase, Z.; Yedavalli, V.S.; Houzet, L.; Perkins, M.; Maldarelli, F.; Brenchley, J.; Strebel, K.; Liu, P.; Jeang, K.T. Activation of HIV-1 from latent infection via synergy of RUNX1 inhibitor Ro5-3335 and SAHA. PLoS Pathog. 2014, 10, e1003997. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Thakuri, B.K.C.; Zhao, J.; Nguyen, L.N.; Nguyen, L.N.T.; Khanal, S.; Cao, D.; Dang, X.; Schank, M.; Lu, Z.; et al. Long Noncoding RNA RUNXOR Promotes Myeloid-Derived Suppressor Cell Expansion and Functions via Enhancing Immunosuppressive Molecule Expressions during Latent HIV Infection. J. Immunol. 2021, 206, 2052–2060. [Google Scholar] [CrossRef] [PubMed]
- Barriocanal, M.; Carnero, E.; Segura, V.; Fortes, P. Long Non-Coding RNA BST2/BISPR is Induced by IFN and Regulates the Expression of the Antiviral Factor Tetherin. Front. Immunol. 2014, 5, 655. [Google Scholar] [CrossRef]
- Kambara, H.; Gunawardane, L.; Zebrowski, E.; Kostadinova, L.; Jobava, R.; Krokowski, D.; Hatzoglou, M.; Anthony, D.D.; Valadkhan, S. Regulation of Interferon-Stimulated Gene BST2 by a lncRNA Transcribed from a Shared Bidirectional Promoter. Front. Immunol. 2014, 5, 676. [Google Scholar] [CrossRef] [PubMed]
- Vigneau, S.; Rohrlich, P.S.; Brahic, M.; Bureau, J.F. Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J. Virol. 2003, 77, 5632–5638. [Google Scholar] [CrossRef]
- Collier, S.P.; Collins, P.L.; Williams, C.L.; Boothby, M.R.; Aune, T.M. Cutting edge: Influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J. Immunol. 2012, 189, 2084–2088. [Google Scholar] [CrossRef]
- Collier, S.P.; Henderson, M.A.; Tossberg, J.T.; Aune, T.M. Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. J. Immunol. 2014, 193, 3959–3965. [Google Scholar] [CrossRef]
- Gomez, J.A.; Wapinski, O.L.; Yang, Y.W.; Bureau, J.F.; Gopinath, S.; Monack, D.M.; Chang, H.Y.; Brahic, M.; Kirkegaard, K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 2013, 152, 743–754. [Google Scholar] [CrossRef]
- Petermann, F.; Pekowska, A.; Johnson, C.A.; Jankovic, D.; Shih, H.Y.; Jiang, K.; Hudson, W.H.; Brooks, S.R.; Sun, H.W.; Villarino, A.V.; et al. The Magnitude of IFN-gamma Responses Is Fine-Tuned by DNA Architecture and the Non-coding Transcript of Ifng-as1. Mol. Cell 2019, 75, 1229–1242 e1225. [Google Scholar] [CrossRef]
- Stein, N.; Berhani, O.; Schmiedel, D.; Duev-Cohen, A.; Seidel, E.; Kol, I.; Tsukerman, P.; Hecht, M.; Reches, A.; Gamliel, M.; et al. IFNG-AS1 Enhances Interferon Gamma Production in Human Natural Killer Cells. iScience 2019, 11, 466–473. [Google Scholar] [CrossRef]
- Goris, A.; Heggarty, S.; Marrosu, M.G.; Graham, C.; Billiau, A.; Vandenbroeck, K. Linkage disequilibrium analysis of chromosome 12q14-15 in multiple sclerosis: Delineation of a 118-kb interval around interferon-gamma (IFNG) that is involved in male versus female differential susceptibility. Genes. Immun. 2002, 3, 470–476. [Google Scholar] [CrossRef]
- Latiano, A.; Palmieri, O.; Latiano, T.; Corritore, G.; Bossa, F.; Martino, G.; Biscaglia, G.; Scimeca, D.; Valvano, M.R.; Pastore, M.; et al. Investigation of multiple susceptibility loci for inflammatory bowel disease in an Italian cohort of patients. PLoS ONE 2011, 6, e22688. [Google Scholar] [CrossRef]
- Silverberg, M.S.; Cho, J.H.; Rioux, J.D.; McGovern, D.P.; Wu, J.; Annese, V.; Achkar, J.P.; Goyette, P.; Scott, R.; Xu, W.; et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat. Genet. 2009, 41, 216–220. [Google Scholar] [CrossRef]
- Xu-Yang, Z.; Pei-Yu, B.; Chuan-Tao, Y.; Wei, Y.; Hong-Wei, M.; Kang, T.; Chun-Mei, Z.; Ying-Feng, L.; Xin, W.; Ping-Zhong, W.; et al. Interferon-Induced Transmembrane Protein 3 Inhibits Hantaan Virus Infection, and Its Single Nucleotide Polymorphism rs12252 Influences the Severity of Hemorrhagic Fever with Renal Syndrome. Front. Immunol. 2016, 7, 535. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, B.; Servaas, N.H.; Rossato, M.; Tamassia, N.; Cassatella, M.A.; Cossu, M.; Beretta, L.; van der Kroef, M.; Radstake, T.; Bazzoni, F. The Long Non-coding RNA NRIR Drives IFN-Response in Monocytes: Implication for Systemic Sclerosis. Front. Immunol. 2019, 10, 100. [Google Scholar] [CrossRef] [PubMed]
- Imamura, K.; Imamachi, N.; Akizuki, G.; Kumakura, M.; Kawaguchi, A.; Nagata, K.; Kato, A.; Kawaguchi, Y.; Sato, H.; Yoneda, M.; et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 2014, 53, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Matthys, V.S.; Cimica, V.; Dalrymple, N.A.; Glennon, N.B.; Bianco, C.; Mackow, E.R. Hantavirus GnT elements mediate TRAF3 binding and inhibit RIG-I/TBK1-directed beta interferon transcription by blocking IRF3 phosphorylation. J. Virol. 2014, 88, 2246–2259. [Google Scholar] [CrossRef] [PubMed]
- Oshiumi, H.; Miyashita, M.; Okamoto, M.; Morioka, Y.; Okabe, M.; Matsumoto, M.; Seya, T. DDX60 Is Involved in RIG-I-Dependent and Independent Antiviral Responses, and Its Function Is Attenuated by Virus-Induced EGFR Activation. Cell Rep. 2015, 11, 1193–1207. [Google Scholar] [CrossRef] [PubMed]
- Boliar, S.; Gludish, D.W.; Jambo, K.C.; Kamng’ona, R.; Mvaya, L.; Mwandumba, H.C.; Russell, D.G. Inhibition of the lncRNA SAF drives activation of apoptotic effector caspases in HIV-1-infected human macrophages. Proc. Natl. Acad. Sci. USA 2019, 116, 7431–7438. [Google Scholar] [CrossRef]
- Young, L.S.; Rickinson, A.B. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 2004, 4, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Yue, W.; Du, S.; Xin, S.; Zhang, J.; Liu, L.; Li, G.; Lu, J. An update: Epstein-Barr virus and immune evasion via microRNA regulation. Virol. Sin. 2017, 32, 175–187. [Google Scholar] [CrossRef]
- Qiu, J.; Smith, P.; Leahy, L.; Thorley-Lawson, D.A. The Epstein-Barr virus encoded BART miRNAs potentiate tumor growth in vivo. PLoS Pathog. 2015, 11, e1004561. [Google Scholar] [CrossRef]
- Chen, H.L.; Lung, M.M.; Sham, J.S.; Choy, D.T.; Griffin, B.E.; Ng, M.H. Transcription of BamHI-A region of the EBV genome in NPC tissues and B cells. Virology 1992, 191, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.R.; de Jesus, O.; Turner, D.; Hollyoake, M.; Karstegl, C.E.; Griffin, B.E.; Karran, L.; Wang, Y.; Hayward, S.D.; Farrell, P.J. Structure and coding content of CST (BART) family RNAs of Epstein-Barr virus. J. Virol. 2000, 74, 3082–3092. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, R.J.; Tong, S.; Zhang, G.; Zong, J.; Chen, Y.; Jin, D.Y.; Chen, M.R.; Pan, J.; Chen, H. NF-kappaB Signaling Regulates Expression of Epstein-Barr Virus BART MicroRNAs and Long Noncoding RNAs in Nasopharyngeal Carcinoma. J. Virol. 2016, 90, 6475–6488. [Google Scholar] [CrossRef]
- Kang, G.H.; Lee, S.; Kim, W.H.; Lee, H.W.; Kim, J.C.; Rhyu, M.G.; Ro, J.Y. Epstein-barr virus-positive gastric carcinoma demonstrates frequent aberrant methylation of multiple genes and constitutes CpG island methylator phenotype-positive gastric carcinoma. Am. J. Pathol. 2002, 160, 787–794. [Google Scholar] [CrossRef]
- Zhang, J.; Li, X.; Hu, J.; Cao, P.; Yan, Q.; Zhang, S.; Dang, W.; Lu, J. Long noncoding RNAs involvement in Epstein-Barr virus infection and tumorigenesis. Virol. J. 2020, 17, 51. [Google Scholar] [CrossRef]
- Tai-Schmiedel, J.; Karniely, S.; Lau, B.; Ezra, A.; Eliyahu, E.; Nachshon, A.; Kerr, K.; Suarez, N.; Schwartz, M.; Davison, A.J.; et al. Human cytomegalovirus long noncoding RNA4.9 regulates viral DNA replication. PLoS Pathog. 2020, 16, e1008390. [Google Scholar] [CrossRef]
- Lee, S.; Kim, H.; Hong, A.; Song, J.; Lee, S.; Kim, M.; Hwang, S.Y.; Jeong, D.; Kim, J.; Son, A.; et al. Functional and molecular dissection of HCMV long non-coding RNAs. Sci. Rep. 2022, 12, 19303. [Google Scholar] [CrossRef]
- Murphy, J.C.; Fischle, W.; Verdin, E.; Sinclair, J.H. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 2002, 21, 1112–1120. [Google Scholar] [CrossRef] [PubMed]
- Reeves, M.B.; Lehner, P.J.; Sissons, J.G.P.; Sinclair, J.H. An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J. Gen. Virol. 2005, 86, 2949–2954. [Google Scholar] [CrossRef] [PubMed]
- Reeves, M.B.; MacAry, P.A.; Lehner, P.J.; Sissons, J.G.; Sinclair, J.H. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. USA 2005, 102, 4140–4145. [Google Scholar] [CrossRef]
- Sinclair, J.; Sissons, P. Latency and reactivation of human cytomegalovirus. J. Gen. Virol. 2006, 87, 1763–1779. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Guo, X.; Zhang, J.; Li, J.; Xu, M.; Wang, Q.; Liu, Z.; Ma, Y.; Qi, Y.; Ruan, Q. Human cytomegalovirus RNA2.7 inhibits RNA polymerase II (Pol II) Serine-2 phosphorylation by reducing the interaction between Pol II and phosphorylated cyclin-dependent kinase 9 (pCDK9). Virol. Sin. 2022, 37, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Reeves, M.B.; Davies, A.A.; McSharry, B.P.; Wilkinson, G.W.; Sinclair, J.H. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science 2007, 316, 1345–1348. [Google Scholar] [CrossRef] [PubMed]
- Perera, M.R.; Sinclair, J.H. The Human Cytomegalovirus beta2.7 Long Non-Coding RNA Prevents Induction of Reactive Oxygen Species to Maintain Viral Gene Silencing during Latency. Int. J. Mol. Sci. 2022, 23, 11017. [Google Scholar] [CrossRef]
- Lau, B.; Kerr, K.; Camiolo, S.; Nightingale, K.; Gu, Q.; Antrobus, R.; Suarez, N.M.; Loney, C.; Stanton, R.J.; Weekes, M.P.; et al. Human Cytomegalovirus RNA2.7 Is Required for Upregulating Multiple Cellular Genes To Promote Cell Motility and Viral Spread Late in Lytic Infection. J. Virol. 2021, 95, e0069821. [Google Scholar] [CrossRef]
- Lau, B.; Kerr, K.; Gu, Q.; Nightingale, K.; Antrobus, R.; Suarez, N.M.; Stanton, R.J.; Wang, E.C.Y.; Weekes, M.P.; Davison, A.J. Human Cytomegalovirus Long Non-coding RNA1.2 Suppresses Extracellular Release of the Pro-inflammatory Cytokine IL-6 by Blocking NF-kappaB Activation. Front. Cell Infect. Microbiol. 2020, 10, 361. [Google Scholar] [CrossRef]
- Amelio, A.L.; Giordani, N.V.; Kubat, N.J.; O’Neil J, E.; Bloom, D.C. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J. Virol. 2006, 80, 2063–2068. [Google Scholar] [CrossRef] [PubMed]
- Cliffe, A.R.; Coen, D.M.; Knipe, D.M. Kinetics of facultative heterochromatin and polycomb group protein association with the herpes simplex viral genome during establishment of latent infection. mBio 2013, 4, e00590-12. [Google Scholar] [CrossRef]
- Cliffe, A.R.; Garber, D.A.; Knipe, D.M. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J. Virol. 2009, 83, 8182–8190. [Google Scholar] [CrossRef]
- Kwiatkowski, D.L.; Thompson, H.W.; Bloom, D.C. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J. Virol. 2009, 83, 8173–8181. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.Y.; Zhou, C.; Johnson, K.E.; Colgrove, R.C.; Coen, D.M.; Knipe, D.M. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. USA 2005, 102, 16055–16059. [Google Scholar] [CrossRef]
- Hancock, M.H.; Skalsky, R.L. Roles of Non-coding RNAs During Herpesvirus Infection. Curr. Top. Microbiol. Immunol. 2018, 419, 243–280. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zeng, L.S.; Wang, J.; Cai, W.Q.; Cui, W.; Song, T.J.; Peng, X.C.; Ma, Z.; Xiang, Y.; Cui, S.Z.; et al. Multifunctional Non-Coding RNAs Mediate Latent Infection and Recurrence of Herpes Simplex Viruses. Infect. Drug. Resist. 2021, 14, 5335–5349. [Google Scholar] [CrossRef] [PubMed]
- Arun, G.; Diermeier, S.D.; Spector, D.L. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol. Med. 2018, 24, 257–277. [Google Scholar] [CrossRef]
- Lennox, K.A.; Behlke, M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016, 44, 863–877. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, K.T.; Corey, D.R. Guidelines for Experiments Using Antisense Oligonucleotides and Double-Stranded RNAs. Nucleic Acid. Ther. 2019, 29, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Konermann, S.; Lotfy, P.; Brideau, N.J.; Oki, J.; Shokhirev, M.N.; Hsu, P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018, 173, 665–676 e614. [Google Scholar] [CrossRef]
- Smargon, A.A.; Cox, D.B.T.; Pyzocha, N.K.; Zheng, K.; Slaymaker, I.M.; Gootenberg, J.S.; Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S.; et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630 e617. [Google Scholar] [CrossRef]
- Xiang, J.F.; Yin, Q.F.; Chen, T.; Zhang, Y.; Zhang, X.O.; Wu, Z.; Zhang, S.; Wang, H.B.; Ge, J.; Lu, X.; et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014, 24, 513–531. [Google Scholar] [CrossRef] [PubMed]
- Bester, A.C.; Lee, J.D.; Chavez, A.; Lee, Y.R.; Nachmani, D.; Vora, S.; Victor, J.; Sauvageau, M.; Monteleone, E.; Rinn, J.L.; et al. An Integrated Genome-wide CRISPRa Approach to Functionalize lncRNAs in Drug Resistance. Cell 2018, 173, 649–664 e620. [Google Scholar] [CrossRef]
- Shechner, D.M.; Hacisuleyman, E.; Younger, S.T.; Rinn, J.L. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat. Methods 2015, 12, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Li, Q.; Zhao, J.; Yi, D.; Ding, J.; Zhao, F.; Hu, S.; Zhou, J.; Deng, T.; et al. Influenza Virus Exploits an Interferon-Independent lncRNA to Preserve Viral RNA Synthesis through Stabilizing Viral RNA Polymerase PB1. Cell Rep. 2019, 27, 3295–3304 e3294. [Google Scholar] [CrossRef] [PubMed]
- Buske, F.A.; Bauer, D.C.; Mattick, J.S.; Bailey, T.L. Triplexator: Detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 2012, 22, 1372–1381. [Google Scholar] [CrossRef]
- Buske, F.A.; Bauer, D.C.; Mattick, J.S.; Bailey, T.L. Triplex-Inspector: An analysis tool for triplex-mediated targeting of genomic loci. Bioinformatics 2013, 29, 1895–1897. [Google Scholar] [CrossRef]
- Kuo, C.C.; Hanzelmann, S.; Senturk Cetin, N.; Frank, S.; Zajzon, B.; Derks, J.P.; Akhade, V.S.; Ahuja, G.; Kanduri, C.; Grummt, I.; et al. Detection of RNA-DNA binding sites in long noncoding RNAs. Nucleic Acids Res. 2019, 47, e32. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Zhang, H.; Liu, H.; Zhu, H. LongTarget: A tool to predict lncRNA DNA-binding motifs and binding sites via Hoogsteen base-pairing analysis. Bioinformatics 2015, 31, 178–186. [Google Scholar] [CrossRef]
- Hon, J.; Martinek, T.; Rajdl, K.; Lexa, M. Triplex: An R/Bioconductor package for identification and visualization of potential intramolecular triplex patterns in DNA sequences. Bioinformatics 2013, 29, 1900–1901. [Google Scholar] [CrossRef] [PubMed]
- Lexa, M.; Martinek, T.; Burgetova, I.; Kopecek, D.; Brazdova, M. A dynamic programming algorithm for identification of triplex-forming sequences. Bioinformatics 2011, 27, 2510–2517. [Google Scholar] [CrossRef]
- Soibam, B. Super-lncRNAs: Identification of lncRNAs that target super-enhancers via RNA:DNA:DNA triplex formation. RNA 2017, 23, 1729–1742. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.L.; Dobi, A.; Srivastava, S. Prostate cancer: Diagnostic performance of the PCA3 urine test. Nature reviews. Urology 2011, 8, 123–124. [Google Scholar] [CrossRef]
- Hanna, N.; Ohana, P.; Konikoff, F.M.; Leichtmann, G.; Hubert, A.; Appelbaum, L.; Kopelman, Y.; Czerniak, A.; Hochberg, A. Phase 1/2a, dose-escalation, safety, pharmacokinetic and preliminary efficacy study of intratumoral administration of BC-819 in patients with unresectable pancreatic cancer. Cancer gene therapy 2012, 19, 374–381. [Google Scholar] [CrossRef]
- Connelly, C.M.; Moon, M.H.; Schneekloth, J.S., Jr. The Emerging Role of RNA as a Therapeutic Target for Small Molecules. Cell Chem. Biol. 2016, 23, 1077–1090. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Lin, H.; Shatabda, S. Locate-R: Subcellular localization of long non-coding RNAs using nucleotide compositions. Genomics 2020, 112, 2583–2589. [Google Scholar] [CrossRef]
- Gudenas, B.L.; Wang, L. Prediction of LncRNA Subcellular Localization with Deep Learning from Sequence Features. Sci. Rep. 2018, 8, 16385. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhao, B.; Yin, R.; Lu, C.; Guo, F.; Zeng, M. GraphLncLoc: Long non-coding RNA subcellular localization prediction using graph convolutional networks based on sequence to graph transformation. Brief. Bioinform. 2022, 24, bbac565. [Google Scholar] [CrossRef]
- Zeng, M.; Wu, Y.; Lu, C.; Zhang, F.; Wu, F.X.; Li, M. DeepLncLoc: A deep learning framework for long non-coding RNA subcellular localization prediction based on subsequence embedding. Brief. Bioinform. 2022, 23, bbab360. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Han, B.; Ding, Y.; Zhang, H.; Chang, S.; Zhang, L.; Zhao, C.; Yang, N.; Song, J. Linc-GALMD1 Regulates Viral Gene Expression in the Chicken. Front. Genet. 2019, 10, 1122. [Google Scholar] [CrossRef] [PubMed]
- Munschauer, M.; Nguyen, C.T.; Sirokman, K.; Hartigan, C.R.; Hogstrom, L.; Engreitz, J.M.; Ulirsch, J.C.; Fulco, C.P.; Subramanian, V.; Chen, J.; et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature 2018, 561, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Liu, S.; Fan, J.; Jin, Y.; Tian, B.; Zheng, X.; Fu, H. Nuclear retention of the lncRNA SNHG1 by doxorubicin attenuates hnRNPC-p53 protein interactions. EMBO Rep. 2017, 18, 536–548. [Google Scholar] [CrossRef]
- Carrieri, C.; Cimatti, L.; Biagioli, M.; Beugnet, A.; Zucchelli, S.; Fedele, S.; Pesce, E.; Ferrer, I.; Collavin, L.; Santoro, C.; et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012, 491, 454–457. [Google Scholar] [CrossRef]
- Cai, R.; Sun, Y.; Qimuge, N.; Wang, G.; Wang, Y.; Chu, G.; Yu, T.; Yang, G.; Pang, W. Adiponectin AS lncRNA inhibits adipogenesis by transferring from nucleus to cytoplasm and attenuating Adiponectin mRNA translation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 420–432. [Google Scholar] [CrossRef]
- Zhang, L.; Zheng, X.; Li, J.; Wang, G.; Hu, Z.; Chen, Y.; Wang, X.; Gu, M.; Gao, R.; Hu, S.; et al. Long noncoding RNA#45 exerts broad inhibitory effect on influenza a virus replication via its stem ring arms. Virulence 2021, 12, 2443–2460. [Google Scholar] [CrossRef]
- Guo, C.J.; Ma, X.K.; Xing, Y.H.; Zheng, C.C.; Xu, Y.F.; Shan, L.; Zhang, J.; Wang, S.; Wang, Y.; Carmichael, G.G.; et al. Distinct Processing of lncRNAs Contributes to Non-conserved Functions in Stem Cells. Cell 2020, 181, 621–636 e622. [Google Scholar] [CrossRef]
- Hezroni, H.; Koppstein, D.; Schwartz, M.G.; Avrutin, A.; Bartel, D.P.; Ulitsky, I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015, 11, 1110–1122. [Google Scholar] [CrossRef]
- Quinn, J.J.; Zhang, Q.C.; Georgiev, P.; Ilik, I.A.; Akhtar, A.; Chang, H.Y. Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes. Dev. 2016, 30, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Burnett, J.C.; Rossi, J.J. RNA-based therapeutics: Current progress and future prospects. Chemistry & biology 2012, 19, 60–71. [Google Scholar] [CrossRef]
| LncRNA | Virus | Mechanism | Reference |
|---|---|---|---|
| Proviral lncRNAs | |||
| NRAV | IAV, SeV, MDRV, HSV | Histone modification and reduction in active transcription marks at ISG. | Ouyang et al., 2014 [57] |
| TSPOAP1-AS1 | IAV | TSPOAP1-AS1 inhibits FNβ1 transcription, ISRE activation, and ISG expression. | Wang et al., 2019 [62] |
| Lnc-MxA | IAV | Lnc-MxA inhibits IFNβ transcription by binding to its promoter and enhances viral replication. | Li et al., 2019 [61] |
| VIN | IAV, VSV | VIN increases virus replication and viral gene expression. Molecular mechanisms are unknown. | Winterling et al., 2014 [29] |
| EGOT | HCV, IAV, SFV | EGOT inhibits the expression of several ISGs and enhances viral replication. Molecular mechanisms are unknown. | Carnero et al., 2016 [31] |
| Lethe | HCV | Lethe inhibits RelA-mediated DNA-binding; inhibits expression of antiviral factors, protein kinase R (PKR), 2′,5′-oligoadenylate synthetase (OAS) proteins, and Interferon Regulatory Factor 1 (IRF1), and enhances HCV replication | Rapicavoli et al., 2013 [66]; Xiong et al., 2015 [71] |
| LncRNA RP11- 288L9.4 | HCV | TSPOAP1-AS1 inhibits expression of IFNα-inducible protein 6 (IFI6) by histone modification and enhances HCV replication. | Liu et al., 2019 [72] |
| NRIR | HCV | NRIR inhibits transcription of several interferon-stimulated genes (ISG) and enhances HCV replication. | Kambara et al., 2014 [65] |
| Lnc_000641 | pseudorabies virus (PRV) | Lnc_000641 inhibits IFNα transcription, phosphorylation of transcription factors (Jak and STAT1), and increases PRV replication. | Fang et al., 2021 [73] |
| NEAT1 | HSV-1 | NEAT1 recruits STAT3 to viral gene promoters to increase viral gene expression. | Wang et al., 2017 [74] |
| Antiviral lncRNA | |||
| IVRPIE | IAV | IVRPIE upregulates IFNβ and several ISGs, including IRF1, IFIT1, IFIT3, Mx1, ISG15, and IFI44L, by affecting histone modification of these genes. | Zhao et al., 2020 [63] |
| OASL-IT1 | ZKIV | OASL-IT1 enhances expression of IFN-β, Mx1, IFITM1 and inhibits ZKIV replication. | Wang et al., 2021 [75] |
| LUARIS | EMCV, HBV, HCV | LUARIS upregulated the level of IFN-stimulated genes through interactions with hnRNPU and ATF2 and suppressed EMCV, HBV, and HCV. | Nishitsuji et al., 2016 [76] |
| NEAT1 | Hantaan virus | NEAT1 relocates SFPQ to paraspeckles, increases RIG-I and DDX60 transcription, increases IFN- γ, and inhibits virus. | Ma et al., 2017 [56] |
| LncRNAs influence the long-term persistence of the virus | |||
| NRON | HIV | NRON mediates degradation of HIV Tat protein. | Li et al., 2016 [77] |
| MALAT1 | HIV | MALAT1 promotes HIV reactivation from latent provirus. | Qu et al., 2019 [78] |
| 7SK | HIV | 7SK promotes HIV latency by inactivating p-TEFb. | Nguyen et al., 2001 [79] Contreras et al., 2007 [80]; Budhiraja et al.,2013 [81]; Eilebrecht et al., 2017 [82] |
| uc002yug.2 | HIV | uc002yug.2 promotes viral reactivation by inhibition of Transcription Repressor RUNX1. | Huan et al., 2018 [83] |
| lincRNA-p21 | HIV | lincRNA-p21 inhibits DSB-induced cell death, promotes viral persistence. | Barichievy et al., 2018 [84] |
| HEAL | HIV | HEAL promotes viral reactivation by recruiting histone acetyltransferase p300 to HIV-1 promoter region. | Chao et al.,2019 [85] |
| NEAT1 | HIV | NEAT1 sequesters unspliced HIV transcripts in nuclear paraspeckle bodies promoting long-term persistence of HIV. | Zhang et al., 2013 [51] |
| HIV antisense lncRNA | HIV | HIV antisense lncRNA recruits chromatin remodeling proteins such as DNMT3a, the enhancer of Zeste 2 (EZH2), and histone deacetylase 1 (HDAC-1) to HIV 5′long terminal repeat. These proteins bring about H3K9 dimethylation, H3K27 trimethylation, and histone deacetylation, resulting in epigenetic silencing of viral transcription. | Saayman et al., 2014 [86] |
| KSHV-encoded PAN RNA | KSHV | PAN RNA binds lysine demethylases UTX and JMJD3, and the lysine methyltransferase MLL2 facilitates the recruitment of histone demethylases to the viral chromatin. | Rossetto et al., 2012 [87] Rossetto, 2013 [88] Rossetto, 2016 [89] |
| HCMV-encoded RNA4.9 | HCMV | RNA4.9 tethers the components of the polycomb repression complex (PRC) to the major immediate early promoter region (MIEP) and represses viral transcription. | Rosseto., 2013 [90] |
| EBV-encoded BART lncRNA | EBV-associated epithelial tumors | BART lncRNAs downregulate the expression of the tumor suppressor gene RASA1 and unfolded protein response (UPR) genes. BART lncRNAs regulate host gene expressions through chromatin modification. | Marquitz.,2015 [91]; Verhoeven, 2019 [92] |
| EBV-encoded lncRNA BHLF1 | EBV-associated epithelial tumors | BHLF1 localizes at the surface of the viral replication compartment and forms an RNA–DNA hybrid at the site of virus transcription. | Park & Miller, 2018 [93]; Rennekamp & Lieberman, 2011 [94] |
| Method | Use | Advantages | Limitation |
|---|---|---|---|
| siRNA/shRNA | Knockdown | Inexpensive, cost-effective for large-scale screening | Nuclear lncRNAs cannot be targeted efficiently by siRNA; structural constraints limit accessibility, large-scale off-target cleavage, and knockdown may be short-lived. |
| Antisense Oligo (ASO) | Knockdown | Efficient degradation of nuclear lncRNA | Structural constraints limit accessibility, large-scale off-target cleavage, and knockdown may be short-lived. |
| CRISPR/Cas9 | Gene knockout or knock-in | Easily programmable to target genes of interest, most definitive | CRISPR/Cas9-mediated frameshift mutations are not helpful for most lncRNAs as their functional sequence motifs are unknown. CRISPR/Cas9 excision of the entire lncRNA gene may disrupt overlapping coding or noncoding RNA region. |
| CRISPRi | Inhibition of transcription | Easily programmable to target genes of interest | CRISPRi may deregulate overlapping coding or noncoding RNA region, the functions of lncRNA transcript from those of promoter or enhancer element encoded within the lncRNA locus or small peptide encoded by the transcript. |
| CRISPR/Cas13d | Knockdown | Easily programmable, independent of PAM, superior RNA knockdown efficiency and dramatically higher specificity than currently available methods, stable long-term expression | Cannot decipher the function of enhancer element encoded within the lncRNA locus or small peptide encoded by the transcript |
| CRISPRa | Activation of transcription | Easily programmable, enhanced lncRNA expression from the endogenous loci | Dependence on protospacer-adjacent motif (PAM); may deregulate overlapping coding or noncoding RNA region; and the functions of lncRNA transcript cannot be distinguished from those of promoter or enhancer element encoded within the lncRNA locus or small peptide encoded by the transcript. |
| CRISPR-display | Easily programmable, allows site-specific delivery of lncRNA transcript to desired genomic loci; this method can be used to test both cis and trans effects of lncRNA transcripts and distinguish them from the act of lncRNA transcription. | Limited by the number of available functional RNA motifs and RNA-binding protein functions |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kulkarni, V.; Jayakumar, S.; Mohan, M.; Kulkarni, S. Aid or Antagonize: Nuclear Long Noncoding RNAs Regulate Host Responses and Outcomes of Viral Infections. Cells 2023, 12, 987. https://doi.org/10.3390/cells12070987
Kulkarni V, Jayakumar S, Mohan M, Kulkarni S. Aid or Antagonize: Nuclear Long Noncoding RNAs Regulate Host Responses and Outcomes of Viral Infections. Cells. 2023; 12(7):987. https://doi.org/10.3390/cells12070987
Chicago/Turabian StyleKulkarni, Viraj, Sahana Jayakumar, Mahesh Mohan, and Smita Kulkarni. 2023. "Aid or Antagonize: Nuclear Long Noncoding RNAs Regulate Host Responses and Outcomes of Viral Infections" Cells 12, no. 7: 987. https://doi.org/10.3390/cells12070987
APA StyleKulkarni, V., Jayakumar, S., Mohan, M., & Kulkarni, S. (2023). Aid or Antagonize: Nuclear Long Noncoding RNAs Regulate Host Responses and Outcomes of Viral Infections. Cells, 12(7), 987. https://doi.org/10.3390/cells12070987






