Abstract
Genetic sensorineural hearing loss and Meniere disease have been associated with rare variations in the coding and non-coding region of the human genome. Most of these variants were classified as likely pathogenic or variants of unknown significance and require functional validation in cellular or animal models. Given the difficulties to obtain human samples and the raising concerns about animal experimentation, human-induced pluripotent stem cells emerged as cellular models to investigate the interaction of genetic and environmental factors in the pathogenesis of inner ear disorders. The generation of human sensory epithelia and neuron-like cells carrying the variants of interest may facilitate a better understanding of their role during differentiation. These cellular models will allow us to explore new strategies for restoring hearing and vestibular sensory epithelia as well as neurons. This review summarized the use of human-induced pluripotent stem cells in sensorineural hearing loss and Meniere disease and proposed some strategies for its application in clinical practice.
1. Introduction
Stem cell culture is essential in biomedical research to study cellular and developmental biology [1,2]. Several biological phenomena specific to humans, such as brain development and inner ear formation amongst others, are not reproduced in animal models. This limitation promoted the generation of cellular models to reproduce more exactly human biology and development [1], leading to a better understanding of genetic disorders.
Stem cell science presents a highly promising direction in translational medicine, allowing the proliferation and differentiation of any cell type. Stem cell models are more clinically appropriate to study the molecular pathophysiology of certain diseases, and further develop better therapeutic strategies [2]. Therefore, human pluripotent stem cells (hPSCs) are progressively used to model rare diseases, as an alternative to animal models which may raise research costs and ethical issues [1]. Disease modeling of human disorders may allow the development of new therapies for rare diseases [1]. Henceforth, human organoids provide a unique opportunity to study them in a multicellular environment and complement animal models [3].
The most common sensory disorder is hearing loss, affecting around 15% of the population [4]. Sensorineural hearing loss (SNHL) and vestibular disorders are caused by damage to the sensory epithelia and neurons, which do not regenerate in humans. These could originate from a variety of causes, such as genetic and environmental factors, aging, and ototoxic drugs [5].
Extensive studies using animal models expanded our knowledge of the human inner ear function and disease, although access to the inner ear in mammalian animal models is limited, slowing research progress in the field [6]. Access to the human inner ear is also strictly restricted since tissue sampling may lead to irreversible damage and profound hearing loss or vestibular impairment. In addition, non-invasive clinical imaging techniques such as computed tomography or MRI, do not provide sufficient resolution to investigate most pathologies of the inner ear at the cellular and molecular levels [7].
Researchers successfully derived inner ear progenitors and sensory cells from hPSCs [8,9,10,11] using different induction protocols to form hair cells-like cells. Many of these protocols were carried out entirely in 2D culture providing a homogeneous cell population [12,13,14]. In contrast, 3D organoid systems containing multiple cell types can better recapitulate in vivo composition of the organ [15,16,17,18]. Since hair cells (HCs), supporting cells (SCs), and their associated neurons are damaged in SNHL, gene and cell-based therapies may restore hearing and vestibular impairment.
This scoping review describes the major achievements, gaps in research, and challenges to translating the application of hPSCs into clinical practice. Furthermore, we present their application in monogenic deafness and the potential use to model Meniere disease (MD).
2. Generation of Human Pluripotent Stem Cells
Human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) and hiPSCs, emerged as a new model system that offers unique advantages for disease modeling [2].
hESCs are derived from the inner cell mass of the blastocyst of an embryo, while hiPSCs are derived from adult somatic tissues. Since their discovery in 2007 [19], hiPSCs emerged as an alternative to hESCs, because they can be obtained from individual patients or healthy donors and immune rejection can be avoided when they are transplanted autologously [20,21]. Since then, researchers showed the potential of reprogramming to convert a given somatic cell type to an iPSC state.
Reprogramming methods may be divided into two categories: integrative and non-integrative (Table 1). Integrative reprogramming approaches generate heterogeneous hiPSCs lines and present genomic mutation risks, which could obscure comparative analysis between lines. So, to avoid the risk of tumorigenesis, non-integrative reprogramming methods were designed, including non-integrating viral vectors such as Sendai virus (SeV), adenovirus, and non-viral vectors such as episomal DNA, Piggy-Bac transposons, modified synthetic mRNA (modRNA), microRNA, and recombinant proteins.

Table 1.
Reprogramming methods used in human somatic cells.
The most commonly used reprogramming methods to generate hiPSCs are SeV, synthetic mRNAs, and episomal vectors, due to their high reprogramming efficiency and their wide application to different cell types [20,22,23,24,25,50,51,52,53]. In addition, SeV reprogramming is highly effective, with a lower workload and no non-appearance of viral sequences in most cell lines at higher passages in culture. Moreover, the combination of bFGF, ascorbic acid (AA), and Y-27632 dihydrochloride-ROCK inhibitor in the culture medium increase the reprogramming efficiency [54,55,56,57] (Figure 1). However, in recent years, modRNA-based somatic reprogramming became a more effective alternative to the SeV system, since the modRNA molecules encode multiple pluripotent factors and can be applied successfully in reprogramming somatic cells [24,25,29].
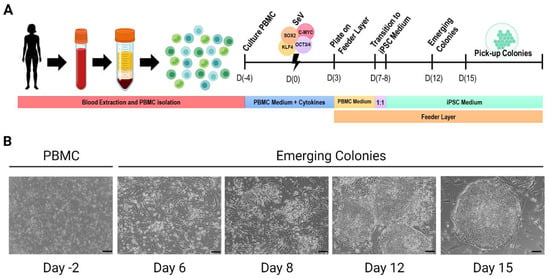
Figure 1.
Cell reprogramming of human PBMC. (A). Generation of hiPSC lines by SeV cell reprogramming technology. (B). Images of cell reprogramming procedure. Scale bar = 100 μm. (Created with BioRender.com, accessed on 1 December 2022).
Selecting the most suitable reprogramming method and implementing good manufacturing procedures for hiPSCs generation is key to obtaining high-quality hiPSCs cell lines.
3. Disease Modeling by Human-Induced Pluripotent Stem Cells
The advantages of hiPSCs are well-known at many distinct levels such as their capacity to self-renew indefinitely in culture and differentiate to any cell type in the human body. So, hiPSCs are the ideal source for generating a patient-derived personalized disease model [1,2].
Experimental human modeling of inherited rare disorders provides insight into the cellular and molecular mechanisms involved in the disease. It is worth mentioning the early success history of disease modeling using hiPSCs, which was based on neurodegenerative diseases [58]. Neuronal differentiation was one of the initially differentiated target tissues from several sources of stem cells due to their potential future therapeutic usage [59,60,61]. Neurodegenerative disorders such as amyotrophic lateral sclerosis, Huntington’s disease, Parkinson’s disease, and Alzheimer’s disease were among the most studied [62,63,64].
Recent advances in organoid technology revolutionized the in vitro culture tools for biomedical research by creating powerful 3D models to recapitulate the cellular heterogeneity, structure, and functions of the primary tissues, compared with 2D cell models. Hence, it is a methodology with many translational applications such as regenerative medicine, drug discovery, and precision medicine. In 2011, the first organoids from hPSCs were generated to recreate intestinal tissue in vitro [65]. In 2012, the first retinal organoids were generated from hPSCs [66]. These human retinal organoids are larger than mouse organoids and can grow into multilayered tissue containing both rods and cones [66,67]. In 2013, Lancaster et al. further established the 3D cerebral organoids containing different brain regions within single organoids [68].
4. Clinical Applications in Genetic Deafness and Vestibular Disorders
According to the World Health Organization (WHO), hearing impairment, the partial or total inability to hear sounds, is among the top 10 disabilities of today’s society. (https://www.who.int/newsroom/factsheets/detail/deafness-and-hearing-loss, accessed on 17 November 2022).
Congenital SNHL is estimated to have a genetic origin in 70% of cases and, mutations can affect the organ of Corti, the spiral ganglion, and almost any part of the auditory pathway [41,42]. Monogenic SNHL are considered rare disorders [43,44]. There are around 126 genes associated with non-syndromic SNHL (https://hereditaryhearingloss.org/dominant, accessed on 10 November 2022) and many of them are related to inner ear homeostasis and mechano-electrical transduction [69,70,71,72].
Several otologic conditions may show fluctuating SNHL, including Meniere Disease (MD), a debilitating condition, characterized by episodes of spontaneous vertigo, tinnitus, and aural fullness [73,74]. It is associated with the progressive accumulation of endolymph in the cochlear duct [75]. The diagnosis is based on the characteristic presentation of the different clinical symptoms mentioned during the vertigo attacks [76]. MD prevalence is higher in European than Asian and African populations with a range of 10–225 cases/100,000 individuals. Most patients suffer from SNHL in one ear, but 25–40% of these patients with unilateral SNHL may develop hearing loss in the contralateral ear after several years [77]. MD is known to have a genetic predisposition [74] and familial MD is found in around 8–10% of cases with several genes involved [78,79,80,81,82]. Some forms of monogenic SNHL and familial MD are rare diseases which may benefit from gene and cellular therapy.
Despite the extensive occurrence of genetic SNHL in the world, there are no Food and Drug Administration (FDA)-approved cellular or molecular therapies [4,50]. Current treatments for human SNHL and MD are medical therapy using steroids, hearing aids, surgery to correct the cause of the hearing loss, or cochlear implants [83,84,85,86,87,88,89]. Though these devices offer significant relief of the moderate and severe SNHL by amplifying sound or directly electrically stimulating the auditory nerve, they have significant limitations in terms of speech discrimination in complex acoustic environments [90]. These medical devices require the presence of functional auditory neurons in the inner ear. Therefore, in recent years, new studies focused on possibilities for neuronal replacement, including exogenous stem cell transplantation and endogenous cell source replacement. Several studies proved that neural stem cell transplantation in the inner ear has an important therapeutic effect on the activation and regeneration of cells, restoring damaged neurons [91,92,93]. However, more research is still needed to improve and standardize the protocol for differentiating stem cells into inner ear HCs and neurons.
Transcriptional networks are key in governing the regeneration or replacement of auditory neurons from stem cells. Development of the inner ear is an organized molecular transformation of a set of epidermal cells (the otic placode) into the fully developed ear with its neurosensory component, necessary for signal extraction and transmission, and the non-sensory component, forming the labyrinth necessary for directing sensory stimuli to specific sensory epithelia [6,94]. The main genes involved in neurosensory development in the inner ear are MYO7A, HES5, SOX2, NEUROG1, NEUROD1, and POU4F1 [80,95,96,97,98].
Since animal models are only able to represent the chronic end stages of the disease when permanent damage of sensory epithelia occurred, understanding, and identifying the transcription factors involved in the development and survival of auditory neurons is vital for the generation of disease models and the identification of more effective treatments for hearing loss in the future.
4.1. Modeling Inner Ear Disorders: 2D and 3D Cell Culture
The inner ear is highly complex; both the anterior and posterior labyrinth are connected, and the extent of involvement in each organ may vary, resulting in hearing or vestibular disorders. Consequently, inner ear disorders may be caused by damage to sensory epithelia and neurons, which do not regenerate to any clinically relevant extent in humans. Since the pathophysiology of certain types of SNHL and MD have not yet been explained at cellular and molecular levels, it is difficult to generate an animal model that accurately reflects these diseases.
Nevertheless, extensive studies used animal models, including frog, zebrafish, chick, and several species of mammals that expanded our knowledge of the human inner ear function and disease, although access to the inner ear in mammalian animal models is limited, slowing progress in the field. Additionally, these animal models are only able to represent the chronic end stages of disease with permanent loss of hearing and vestibular function [5].
Access to the human inner ear is also strictly restricted since tissue sampling is challenging and leads to irreversible damage. In addition, non-invasive clinical imaging techniques, such as computed tomography or MRI, do not provide enough resolution to investigate most pathologies of the inner ear at the cellular and molecular levels [5,7].
For this reason, the best option to study the inner ear development and disease is to generate human cell models. Several protocols were devised to direct hPSCs into inner ear HCs and neuron-like cells [99,100,101,102]. The efficiency, reproducibility, and scalability of these protocols were enhanced by incorporating knowledge of inner ear development [13,17,18,67,68,69,70]. Early studies on the transplantation of hPSCs-derived otic progenitors were successful in certain animal models [103], but the hearing was transiently restored, and long-term cell survival continues to be a major challenge. Understanding the complex sequence of transcriptional changes and signaling pathways in vivo in inner ear development is critical to the successful differentiation of hPSCs into inner ear tissues, such as HCs, SCs, and neurons in vitro.
Recent years saw a surge in the number of studies that were conducted in vitro 2D and 3D hiPSC models to study auditory and vestibular disorders.
4.1.1. Sensory Epithelia
Several induction protocols were developed to differentiate hPSCs into HC-like cells. Many of these protocols start with the generation of floating embryoid bodies (EBs) followed by the combination of small molecules and/or recombinant proteins that become adhesive 2D cell cultures after approximately 5 days and other protocols were carried out entirely in 2D, which can provide a homogeneous cell population [12,13,14,67,72]. In contrast, 3D organoid systems contain multiple cell types and more exactly recapitulate in vivo composition of an organ.
The initial steps of inner ear development require the formation of the ectodermal germ layer followed by the generation of the pre-placodal ectoderm (PPE). Protocols first inhibit transforming growth factor β and WNT signaling and activate BMP to promote non-neural ectoderm (NNE) development, while reducing mesoderm development [13,17,72]. Insulin-like growth factor 1 promotes the fate of the anterior ectoderm, where the PPE emerges [104]. The PPE gives rise to most of the cranial placodes, including the otic placode. Physical environmental signals provided by extracellular matrices, such as Matrigel, improve the efficiency of differentiation from hPSCs, as well as the resulting cellular assemblage [15,17,18]. The addition of Matrigel in the 3D inner ear organoid systems facilitates the formation of fluid-filled vesicles containing HCs and SC-like cells [17]. However, vestibular tissue-like organoids derived from hPSCs using the rotary cell culture system form HC-like cells on the surface of the organoids [18]. To date, hPSCs-derived HC-like cells display molecular markers and electrophysiological properties of vestibular HCs, not cochlear HCs. The discovery of alternative small molecules or culture conditions to improve the generation of cochlear HCs from hPSCs is still needed.
SCs play an active role in ion metabolism necessary for HCs’ function. The connexin proteins are the most abundant gap junction proteins expressed in the SCs. Mutations in the GJB2 gene, encoding connexin 26 (CX26), are the most common cause of autosomal recessive non-syndromic SNHL [105]. An in vitro model for the homozygous 235delC mutation in GJB2 was developed from hPSC to develop a therapy for deafness [106] (Figure 2).

Figure 2.
2D and 3D hPSCs-derived inner ear hair cells protocols [12,13,14,15,17,18,101,104]. (Created with BioRender.com, accessed on 22 March 2023).
Frejo et al. generated a hiPSC line derived from an MD patient. This model was differentiated into HCs by a 2D protocol based on Boddy et al. [101]. This method consisted of the generation of otic epithelial progenitors (OEPs) and, consequently, differentiation to HCs-like cells (Figure 3).
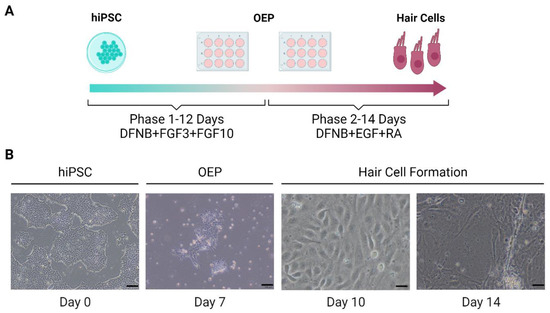
Figure 3.
Schematic representation of 2D differentiation protocol of hPSCs-derived hair cells. (A). Protocol of the generation of inner ear hair cells from hPSCs. (B). Images obtained by microscopy of hair cell formation. Scale bar at day 0 and 7 =100 μm and at day 10 and 14 =20 μm. OEP: Otic epithelial progenitor. (Created with BioRender.com, accessed on 4 December 2022).
Moreover, we started to generate inner ear organoids from the hiPSC-MD model derived from a patient with mutations in DTNA and FAM136A genes (https://hpscreg.eu/cell-line/GENYOi007-A, accessed on 15 March 2023) for studying the development of the inner ear in this patient. This model could explain how these mutations found in DTNA and FAM136A genes in a Spanish family with three affected women in three consecutive generations (autosomal dominant inheritance pattern) affect the development and functionality of the system itself when sensory organs mature [75,76,82,107,108]. Koehler et al. described the protocol [17] to generate inner ear sensory epithelium harboring HCs using an in vitro 3D differentiation system from hiPSCs. Cells were treated with recombinant proteins that modulate BMP, FGF, and WNT signaling pathways to induce the sequential formation of NNE, otic-epibranchial progenitor domain (OEPD), and otic placodes. The otic placodes subsequently underwent self-guided morphogenesis to form inner ear HCs and SCs (Figure 4).

Figure 4.
Generation of inner ear organoids from hPSCs (modified from Nie and Hashino 2020 [109]). (A). 3D differentiation protocol of hPSCs into inner ear organoids from Koehler et al. [17]. (B). Images of inner ear organoids generation. NNE: Non-neural ectoderm. OEDP: Otic-epibranchial progenitor domain. OTIC: Otic Placodes. (Created with BioRender.com, accessed on 22 March 2023).
4.1.2. Sensory Neuron
Early neural induction protocols from PSCs using stromal cells derived from skull bone marrow, resulted in efficient dopaminergic neuron production [110]. These protocols were modified to derive inner ear sensory neurons and glial cells from PSCs. However, many of the available protocols were tailored to generate hPSCs-dopaminergic neurons due to their role in neurodegenerative disorders, such as Parkinson’s disease [111]. In contrast, glutamate was the main neurotransmitter for the synaptic transmission between HCs and afferent sensory neurons within the inner ear. So, the derivation of glutamatergic neurons was a key stage for recapitulating afferent neural transmission in the inner ear [112].
As with the sensory epithelia, inner ear sensory neurons are derived from the otic placode. Therefore, some of the early induction steps for hPSCs-derived sensory neurons are based on known developmental pathways of the otic placode. Small molecules, such as FGFs, BMP, SHH, and noggin, are used to support neuronal outgrowth from the otic placode [113,114,115]. POU4F1 and β-III tubulin are commonly used to verify neuron formation, with glutamate receptors and transporters subsequently used to confirm subtype-specific derivatives. Matsuoka et al. showed that ~90% of neuron-like cells were peripherin+ and β-III tubulin+, but only ~46% were POU4F1+ [114]. To recapitulate the peripheral neural circuit in vitro, the model must contain both sensory epithelia and innervating neurons.
Two systems potentially can generate both tissues simultaneously, providing complementary research tools for disease modeling: 2D and 3D cell cultures (Figure 5).
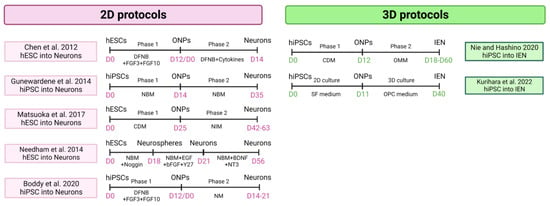
Figure 5.
Cellular models of hPSC-derived 2D and 3D inner ear neurons protocols [100,101,103,109,113,114,115]. NBM: neurobasal medium. CDM: chemically defined medium. NIM: neural inducing medium. NM: neuralizing medium. (Created with BioRender.com, accessed on 22 March 2023).
The treatment of hESCs with FGF3 and FGF10 for 10–12 days in 2D culture gives rise to both otic epithelial progenitors (OEPs) and otic neural progenitors (ONPs), which can be distinguished based on their morphology [103]. HC-like cells can be derived from OEP after inhibiting Notch and supplementing with RA and epidermal growth factor (EGF) [67,71]. Sensory neuron-like cells can be derived from ONPs using bFGF and SHH followed by BDNF and NT3 supplementation [103]. Co-culture of these derived HCs and sensory neurons form neural connections in vitro. However, this method required separate induction protocols before co-culturing them [116].
The 3D inner ear organoids contain sensory epithelia and sensory neuron-like cells [15,16,17]. Furthermore, several studies replicated these protocols and used the systems for different applications, such as electrophysiological studies and disease modeling [117]. The inner ear organoid system is a powerful tool to study peripheral sensory neural networks in the inner ear in vitro. Human inner ear tissue derived from hPSCs also offers the chance to explore developmental biology and understand the differences between mice and human inner ear development. Moreover, it would enable both in vitro screening of drug candidates for the treatment of hearing loss and balance dysfunction and a source of cells for cell-based therapies of the inner ear. Nie and Hashino described a 3D protocol to form inner ear neurons from hPSC [109]. Other studies generated otic organoids with neuron-like cells from hiPSC models combining the 2D and 3D systems in the same differentiation protocol [100].
Stem cell science was also applied to generate a hiPSCs model obtained from patients with severe tinnitus and rare variants in the ANK2 gene, and differentiating them to inner ear neurons using a 2D system [101]. This protocol consists of two phases, a first phase in which hiPSC-derived otic neural progenitors (ONPs) are generated by inhibiting Wnt signaling, which is accompanied by subsequent activation of this pathway, and a second phase consisting of ONPs expansion and enrichment, which will later differentiate into inner ear neurons (Figure 6).
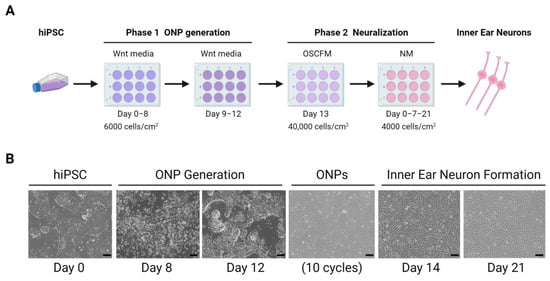
Figure 6.
Inner ear neurons generation from hiPSCs. (A). Differentiation protocol of hPSC-derived inner ear neurons in 2D culture from Boddy et al. [101]. (B). Images of inner ear neurons culture derived from hiPSC. Scale bar = 100 μm. ONP: otic neural progenitor. OSCFM: otic stem cell full media. NM: neuralizing media. (Created with BioRender.com, accessed on 22 March 2023).
5. Clinical Trials in Inner Ear Disorders
Several clinical trials were performed using human stem cells to treat SNHL. Autologous human umbilical cord blood stem cells were used for over twenty years, with excellent safety records. A study in 2015 used umbilical cord blood stem cells to improve inner ear function, audition, and language in children [118]. The potential of human mesenchymal stem cells was evaluated for years for regenerating inner ear HCs [119].
The major achievements in disease modeling using a hiPSC-derived inner ear for genetic SNHL include the genes MYO7A, MYO15, and MERRF in HCs [70,89,90], and GJB2 and SLC26A4 (Pendred syndrome) in SCs [106,120,121,122,123,124] (Table 2). These studies defined the molecular mechanisms involving each gene and showed the cellular effects of each mutation. Recently, a clinical trial using iPSC derived from patients with Pendred syndrome to generate cochlear cells allowed the study of the effects of low-dose oral administration of sirolimus for fluctuating and progressive hearing loss [125].

Table 2.
Genetic mutations causing inner ear disorders modeled using hPSCs.
However, many research gaps were not addressed and more than 150 genes in SNHL and familial MD remain to be modeled (https://deafnessvariationdatabase.org/, accessed on 4 December 2022). Some of the current limitations of the use of hPSCs to investigate inner ear disorders include laborious culture protocols that are sometimes time-consuming and not reproducible, variable efficiency of tissue derivation, and limited differentiation and integration in host tissues. Deciphering the disease molecular mechanisms is the first step to finding a drug target that may offer new opportunities for therapy.
6. Conclusions
Human iPSCs are a reliable model to study the functional consequences of rare variants in early stages of inner ear disease development and design novel gene therapies to restore the phenotype. The use of 3D models to generate patient-specific organoids allows us to elucidate the effect of rare variants in genetic deafness and vestibular disorders, investigate the molecular mechanisms underlying the disease, and analyze the effects of genetic mutations on the phenotype at the cellular and organoid levels. Modeling inner ear diseases by differentiating hiPSCs is a promising tool for designing novel therapies for the treatment of SNHL and MD.
Author Contributions
M.L., paper search, literature review, writing and review; L.F., design and search methods, literature review, writing and review; A.G.-M., literature review, writing, and review; J.A.L.-E.: design and search methods, literature review, writing and review. All authors have read and agreed to the published version of the manuscript.
Funding
M.L. is funded by a research grant RH-0073-2021 from Andalusian Health Department. L.F. has received funds from Sara Borrell’s postdoctoral Fellowship (ISCIII; grant code: CD20/00153). A.G.-M. is funded by a postdoctoral grant from Economic Change, Industry, Knowledge and Universities Department (DOC_01677). J.A.L.-E. and L.F. also received funds for stem cell research from ibs.GRANADA (INTRAIBS-2021-07).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Avior, Y.; Sagi, I.; Benvenisty, N. Pluripotent Stem Cells in Disease Modelling and Drug Discovery. Nat. Rev. Mol. Cell Biol. 2016, 17, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Bai, X. Stem Cell-Based Disease Modeling and Cell Therapy. Cells 2020, 9, 2193. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human Organoids: Model Systems for Human Biology and Medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, K.; Zhang, X.; Peng, K.A. Fluctuating Sensorineural Hearing Loss. Audiol. Neurootol. 2019, 24, 109–116. [Google Scholar] [CrossRef]
- Tang, P.-C.; Hashino, E.; Nelson, R.F. Progress in Modeling and Targeting Inner Ear Disorders with Pluripotent Stem Cells. Stem Cell Rep. 2020, 14, 996–1008. [Google Scholar] [CrossRef]
- Toyoda, S.; Shiraki, N.; Yamada, S.; Uwabe, C.; Imai, H.; Matsuda, T.; Yoneyama, A.; Takeda, T.; Takakuwa, T. Morphogenesis of the Inner Ear at Different Stages of Normal Human Development. Anat. Rec. 2015, 298, 2081–2090. [Google Scholar] [CrossRef]
- Kayyali, M.N.; Wright, A.C.; Ramsey, A.J.; Brant, J.A.; Stein, J.M.; O’Malley, B.W.; Li, D. Challenges and Opportunities in Developing Targeted Molecular Imaging to Determine Inner Ear Defects of Sensorineural Hearing Loss. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 397–404. [Google Scholar] [CrossRef]
- Boddy, S.L.; Chen, W.; Romero-Guevara, R.; Kottam, L.; Bellantuono, I.; Rivolta, M.N. Inner Ear Progenitor Cells Can Be Generated in Vitro from Human Bone Marrow Mesenchymal Stem Cells. Regen. Med. 2012, 7, 757–767. [Google Scholar] [CrossRef]
- Durán Alonso, M.B.; Feijoo-Redondo, A.; Conde de Felipe, M.; Carnicero, E.; García, A.S.; García-Sancho, J.; Rivolta, M.N.; Giráldez, F.; Schimmang, T. Generation of Inner Ear Sensory Cells from Bone Marrow-Derived Human Mesenchymal Stem Cells. Regen. Med. 2012, 7, 769–783. [Google Scholar] [CrossRef]
- Mittal, R.; Ocak, E.; Zhu, A.; Perdomo, M.M.; Pena, S.A.; Mittal, J.; Bohorquez, J.; Eshraghi, A.A. Effect of Bone Marrow-Derived Mesenchymal Stem Cells on Cochlear Function in an Experimental Rat Model. Anat. Rec. Hoboken NJ 2007 2020, 303, 487–493. [Google Scholar] [CrossRef]
- Xu, Y.-P.; Shan, X.-D.; Liu, Y.-Y.; Pu, Y.; Wang, C.-Y.; Tao, Q.-L.; Deng, Y.; Cheng, Y.; Fan, J.-P. Olfactory Epithelium Neural Stem Cell Implantation Restores Noise-Induced Hearing Loss in Rats. Neurosci. Lett. 2016, 616, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Tang, Z.; Chen, J.; Shi, H.; Chen, J.; Wang, C.; Zhang, C.; Li, L.; Chen, P.; Wang, J. Induction of Differentiation of Human Embryonic Stem Cells into Functional Hair-Cell-like Cells in the Absence of Stromal Cells. Int. J. Biochem. Cell Biol. 2016, 81, 208–222. [Google Scholar] [CrossRef] [PubMed]
- Lahlou, H.; Lopez-Juarez, A.; Fontbonne, A.; Nivet, E.; Zine, A. Modeling Human Early Otic Sensory Cell Development with Induced Pluripotent Stem Cells. PLoS ONE 2018, 13, e0198954. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, H.; Skerleva, D.; Kitajiri, S.; Sakamoto, T.; Yamamoto, N.; Ito, J.; Nakagawa, T. Limited Hair Cell Induction from Human Induced Pluripotent Stem Cells Using a Simple Stepwise Method. Neurosci. Lett. 2015, 599, 49–54. [Google Scholar] [CrossRef]
- Jeong, M.; O’Reilly, M.; Kirkwood, N.K.; Al-Aama, J.; Lako, M.; Kros, C.J.; Armstrong, L. Generating Inner Ear Organoids Containing Putative Cochlear Hair Cells from Human Pluripotent Stem Cells. Cell Death Dis. 2018, 9, 922. [Google Scholar] [CrossRef]
- Koehler, K.R.; Mikosz, A.M.; Molosh, A.I.; Patel, D.; Hashino, E. Generation of Inner Ear Sensory Epithelia from Pluripotent Stem Cells in 3D Culture. Nature 2013, 500, 217–221. [Google Scholar] [CrossRef]
- Koehler, K.R.; Nie, J.; Longworth-Mills, E.; Liu, X.-P.; Lee, J.; Holt, J.R.; Hashino, E. Generation of Inner Ear Organoids Containing Functional Hair Cells from Human Pluripotent Stem Cells. Nat. Biotechnol. 2017, 35, 583–589. [Google Scholar] [CrossRef]
- Mattei, C.; Lim, R.; Drury, H.; Nasr, B.; Li, Z.; Tadros, M.A.; D’Abaco, G.M.; Stok, K.S.; Nayagam, B.A.; Dottori, M. Generation of Vestibular Tissue-Like Organoids From Human Pluripotent Stem Cells Using the Rotary Cell Culture System. Front. Cell Dev. Biol. 2019, 7, 25. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Choi, J.; Lee, S.; Mallard, W.; Clement, K.; Tagliazucchi, G.M.; Lim, H.; Choi, I.Y.; Ferrari, F.; Tsankov, A.M.; Pop, R.; et al. A Comparison of Genetically Matched Cell Lines Reveals the Equivalence of Human IPSCs and ESCs. Nat. Biotechnol. 2015, 33, 1173–1181. [Google Scholar] [CrossRef]
- Ortuño-Costela, M.D.C.; Cerrada, V.; García-López, M.; Gallardo, M.E. The Challenge of Bringing IPSCs to the Patient. Int. J. Mol. Sci. 2019, 20, 6305. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T.; Raya, A.; Barrero, M.J.; Garreta, E.; Consiglio, A.; Gonzalez, F.; Vassena, R.; Bilić, J.; Pekarik, V.; Tiscornia, G.; et al. Efficient and Rapid Generation of Induced Pluripotent Stem Cells from Human Keratinocytes. Nat. Biotechnol. 2008, 26, 1276–1284. [Google Scholar] [CrossRef] [PubMed]
- Esteban, M.A.; Wang, T.; Qin, B.; Yang, J.; Qin, D.; Cai, J.; Li, W.; Weng, Z.; Chen, J.; Ni, S.; et al. Vitamin C Enhances the Generation of Mouse and Human Induced Pluripotent Stem Cells. Cell Stem Cell 2010, 6, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Loh, Y.-H.; Agarwal, S.; Park, I.-H.; Urbach, A.; Huo, H.; Heffner, G.C.; Kim, K.; Miller, J.D.; Ng, K.; Daley, G.Q. Generation of Induced Pluripotent Stem Cells from Human Blood. Blood 2009, 113, 5476–5479. [Google Scholar] [CrossRef]
- Nakagawa, M.; Koyanagi, M.; Tanabe, K.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Okita, K.; Mochiduki, Y.; Takizawa, N.; Yamanaka, S. Generation of Induced Pluripotent Stem Cells without Myc from Mouse and Human Fibroblasts. Nat. Biotechnol. 2008, 26, 101–106. [Google Scholar] [CrossRef]
- Subramanyam, D.; Lamouille, S.; Judson, R.L.; Liu, J.Y.; Bucay, N.; Derynck, R.; Blelloch, R. Multiple Targets of MiR-302 and MiR-372 Promote Reprogramming of Human Fibroblasts to Induced Pluripotent Stem Cells. Nat. Biotechnol. 2011, 29, 443–448. [Google Scholar] [CrossRef]
- Li, W.; Zhou, H.; Abujarour, R.; Zhu, S.; Young Joo, J.; Lin, T.; Hao, E.; Schöler, H.R.; Hayek, A.; Ding, S. Generation of Human-Induced Pluripotent Stem Cells in the Absence of Exogenous Sox2. Stem Cells Dayt. Ohio 2009, 27, 2992–3000. [Google Scholar] [CrossRef]
- Lin, S.-L.; Chang, D.C.; Chang-Lin, S.; Lin, C.-H.; Wu, D.T.S.; Chen, D.T.; Ying, S.-Y. Mir-302 Reprograms Human Skin Cancer Cells into a Pluripotent ES-Cell-like State. RNA 2008, 14, 2115–2124. [Google Scholar] [CrossRef]
- Zhao, Y.; Yin, X.; Qin, H.; Zhu, F.; Liu, H.; Yang, W.; Zhang, Q.; Xiang, C.; Hou, P.; Song, Z.; et al. Two Supporting Factors Greatly Improve the Efficiency of Human IPSC Generation. Cell Stem Cell 2008, 3, 475–479. [Google Scholar] [CrossRef]
- Mali, P.; Ye, Z.; Hommond, H.H.; Yu, X.; Lin, J.; Chen, G.; Zou, J.; Cheng, L. Improved Efficiency and Pace of Generating Induced Pluripotent Stem Cells from Human Adult and Fetal Fibroblasts. Stem Cells Dayt. Ohio 2008, 26, 1998–2005. [Google Scholar] [CrossRef]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Wu, Z.; Wang, Y.; Cheng, L.; Cui, C.; Gao, Y.; Chen, T.; Rao, L.; Chen, S.; Jia, N.; et al. Enhanced Efficiency of Generating Induced Pluripotent Stem (IPS) Cells from Human Somatic Cells by a Combination of Six Transcription Factors. Cell Res. 2008, 18, 600–603. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Li, Y.; Jin, H.; Xie, L.; Liu, C.; Jiang, F.; Luo, Y.; Yin, G.; Li, Y.; Wang, J.; et al. Rapid and Efficient Reprogramming of Human Amnion-Derived Cells into Pluripotency by Three Factors OCT4/SOX2/NANOG. Differ. Res. Biol. Divers. 2010, 80, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Li, W.; Zhou, H.; Wei, W.; Ambasudhan, R.; Lin, T.; Kim, J.; Zhang, K.; Ding, S. Reprogramming of Human Primary Somatic Cells by OCT4 and Chemical Compounds. Cell Stem Cell 2010, 7, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Chang, J.C.; Lin, C.; Qi, Z.; Yu, J.; Kan, Y.W. Generation of Induced Pluripotent Stem Cells Using Site-Specific Integration with Phage Integrase. Proc. Natl. Acad. Sci. USA 2010, 107, 19467–19472. [Google Scholar] [CrossRef]
- Zhou, W.; Freed, C.R. Adenoviral Gene Delivery Can Reprogram Human Fibroblasts to Induced Pluripotent Stem Cells. Stem Cells Dayt. Ohio 2009, 27, 2667–2674. [Google Scholar] [CrossRef]
- Ban, H.; Nishishita, N.; Fusaki, N.; Tabata, T.; Saeki, K.; Shikamura, M.; Takada, N.; Inoue, M.; Hasegawa, M.; Kawamata, S.; et al. Efficient Generation of Transgene-Free Human Induced Pluripotent Stem Cells (IPSCs) by Temperature-Sensitive Sendai Virus Vectors. Proc. Natl. Acad. Sci. USA 2011, 108, 14234–14239. [Google Scholar] [CrossRef]
- Fusaki, N.; Ban, H.; Nishiyama, A.; Saeki, K.; Hasegawa, M. Efficient Induction of Transgene-Free Human Pluripotent Stem Cells Using a Vector Based on Sendai Virus, an RNA Virus That Does Not Integrate into the Host Genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 348–362. [Google Scholar] [CrossRef]
- Kaji, K.; Norrby, K.; Paca, A.; Mileikovsky, M.; Mohseni, P.; Woltjen, K. Virus-Free Induction of Pluripotency and Subsequent Excision of Reprogramming Factors. Nature 2009, 458, 771–775. [Google Scholar] [CrossRef]
- Woltjen, K.; Michael, I.P.; Mohseni, P.; Desai, R.; Mileikovsky, M.; Hämäläinen, R.; Cowling, R.; Wang, W.; Liu, P.; Gertsenstein, M.; et al. PiggyBac Transposition Reprograms Fibroblasts to Induced Pluripotent Stem Cells. Nature 2009, 458, 766–770. [Google Scholar] [CrossRef]
- Si-Tayeb, K.; Noto, F.K.; Sepac, A.; Sedlic, F.; Bosnjak, Z.J.; Lough, J.W.; Duncan, S.A. Generation of Human Induced Pluripotent Stem Cells by Simple Transient Transfection of Plasmid DNA Encoding Reprogramming Factors. BMC Dev. Biol. 2010, 10, 81. [Google Scholar] [CrossRef]
- Yu, J.; Hu, K.; Smuga-Otto, K.; Tian, S.; Stewart, R.; Slukvin, I.I.; Thomson, J.A. Human Induced Pluripotent Stem Cells Free of Vector and Transgene Sequences. Science 2009, 324, 797–801. [Google Scholar] [CrossRef]
- Yu, J.; Chau, K.F.; Vodyanik, M.A.; Jiang, J.; Jiang, Y. Efficient Feeder-Free Episomal Reprogramming with Small Molecules. PLoS ONE 2011, 6, e17557. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Matsumura, Y.; Sato, Y.; Okada, A.; Morizane, A.; Okamoto, S.; Hong, H.; Nakagawa, M.; Tanabe, K.; Tezuka, K.; et al. A More Efficient Method to Generate Integration-Free Human IPS Cells. Nat. Methods 2011, 8, 409–412. [Google Scholar] [CrossRef]
- Jia, F.; Wilson, K.D.; Sun, N.; Gupta, D.M.; Huang, M.; Li, Z.; Panetta, N.J.; Chen, Z.Y.; Robbins, R.C.; Kay, M.A.; et al. A Nonviral Minicircle Vector for Deriving Human IPS Cells. Nat. Methods 2010, 7, 197–199. [Google Scholar] [CrossRef]
- Kim, D.; Kim, C.-H.; Moon, J.-I.; Chung, Y.-G.; Chang, M.-Y.; Han, B.-S.; Ko, S.; Yang, E.; Cha, K.Y.; Lanza, R.; et al. Generation of Human Induced Pluripotent Stem Cells by Direct Delivery of Reprogramming Proteins. Cell Stem Cell 2009, 4, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Yakubov, E.; Rechavi, G.; Rozenblatt, S.; Givol, D. Reprogramming of Human Fibroblasts to Pluripotent Stem Cells Using MRNA of Four Transcription Factors. Biochem. Biophys. Res. Commun. 2010, 394, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.-H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified MRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, N.; Ishii, H.; Nagano, H.; Haraguchi, N.; Dewi, D.L.; Kano, Y.; Nishikawa, S.; Tanemura, M.; Mimori, K.; Tanaka, F.; et al. Reprogramming of Mouse and Human Cells to Pluripotency Using Mature MicroRNAs. Cell Stem Cell 2011, 8, 633–638. [Google Scholar] [CrossRef]
- Trokovic, R.; Weltner, J.; Nishimura, K.; Ohtaka, M.; Nakanishi, M.; Salomaa, V.; Jalanko, A.; Otonkoski, T.; Kyttälä, A. Advanced Feeder-Free Generation of Induced Pluripotent Stem Cells Directly From Blood Cells. Stem Cells Transl. Med. 2014, 3, 1402–1409. [Google Scholar] [CrossRef]
- Schlaeger, T.M.; Daheron, L.; Brickler, T.R.; Entwisle, S.; Chan, K.; Cianci, A.; DeVine, A.; Ettenger, A.; Fitzgerald, K.; Godfrey, M.; et al. A Comparison of Non-Integrating Reprogramming Methods. Nat. Biotechnol. 2015, 33, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Kogut, I.; McCarthy, S.M.; Pavlova, M.; Astling, D.P.; Chen, X.; Jakimenko, A.; Jones, K.L.; Getahun, A.; Cambier, J.C.; Pasmooij, A.M.G.; et al. High-Efficiency RNA-Based Reprogramming of Human Primary Fibroblasts. Nat. Commun. 2018, 9, 745. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Y.L. Application of Modified MRNA in Somatic Reprogramming to Pluripotency and Directed Conversion of Cell Fate. Int. J. Mol. Sci. 2021, 22, 8148. [Google Scholar] [CrossRef]
- Cabrera, S.; Ji, A.-R.; Frejo, L.; Ramos-Mejia, V.; Romero, T.; Real, P.; Lopez-Escamez, J.A. Generation of Human IPSC Line GRX-MCiPS4F-A2 from Adult Peripheral Blood Mononuclear Cells (PBMCs) with Spanish Genetic Background. Stem Cell Res. 2015, 15, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Lamolda, M.; Montes, R.; Simón, I.; Perales, S.; Martínez-Navajas, G.; Lopez-Onieva, L.; Ríos-Pelegrina, R.; del Moral, R.G.; Griñan-Lison, C.; Marchal, J.A.; et al. GENYOi005-A: An Induced Pluripotent Stem Cells (IPSCs) Line Generated from a Patient with Familial Platelet Disorder with Associated Myeloid Malignancy (FPDMM) Carrying a p.Thr196Ala Variant. Stem Cell Res. 2019, 41, 101603. [Google Scholar] [CrossRef]
- Cimmino, L.; Neel, B.G.; Aifantis, I. Vitamin C in Stem Cell Reprogramming and Cancer. Trends Cell Biol. 2018, 28, 698–708. [Google Scholar] [CrossRef]
- Steinle, H.; Weber, M.; Behring, A.; Mau-Holzmann, U.; Schlensak, C.; Wendel, H.P.; Avci-Adali, M. Generation of IPSCs by Nonintegrative RNA-Based Reprogramming Techniques: Benefits of Self-Replicating RNA versus Synthetic MRNA. Stem Cells Int. 2019, 2019, 7641767. [Google Scholar] [CrossRef]
- Park, I.-H.; Arora, N.; Huo, H.; Maherali, N.; Ahfeldt, T.; Shimamura, A.; Lensch, M.W.; Cowan, C.; Hochedlinger, K.; Daley, G.Q. Disease-Specific Induced Pluripotent Stem Cells. Cell 2008, 134, 877–886. [Google Scholar] [CrossRef]
- Francis, K.R.; Wei, L. Human Embryonic Stem Cell Neural Differentiation and Enhanced Cell Survival Promoted by Hypoxic Preconditioning. Cell Death Dis. 2010, 1, e22. [Google Scholar] [CrossRef]
- Hu, B.Y.; Weick, J.P.; Yu, J.; Ma, L.X.; Zhang, X.Q.; Thomson, J.A.; Zhang, S.C. Neural Differentiation of Human Induced Pluripotent Stem Cells Follows Developmental Principles but with Variable Potency. Proc. Natl. Acad. Sci. USA 2010, 107, 4335–4340. [Google Scholar] [CrossRef]
- Kuruş, M.; Akbari, S.; Eskier, D.; Bursalı, A.; Ergin, K.; Erdal, E.; Karakülah, G. Transcriptome Dynamics of Human Neuronal Differentiation From IPSC. Front. Cell Dev. Biol. 2021, 9, 3477. [Google Scholar] [CrossRef] [PubMed]
- Israel, M.A.; Yuan, S.H.; Bardy, C.; Reyna, S.M.; Mu, Y.; Herrera, C.; Hefferan, M.P.; Van Gorp, S.; Nazor, K.L.; Boscolo, F.S.; et al. Probing Sporadic and Familial Alzheimer’s Disease Using Induced Pluripotent Stem Cells. Nature 2012, 482, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Juopperi, T.A.; Kim, W.R.; Chiang, C.-H.; Yu, H.; Margolis, R.L.; Ross, C.A.; Ming, G.; Song, H. Astrocytes Generated from Patient Induced Pluripotent Stem Cells Recapitulate Features of Huntington’s Disease Patient Cells. Mol. Brain 2012, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Song, J.; Huang, H.; Chen, H.; Qian, K. Modeling Hallmark Pathology Using Motor Neurons Derived from the Family and Sporadic Amyotrophic Lateral Sclerosis Patient-Specific IPS Cells. Stem Cell Res. Ther. 2018, 9, 315. [Google Scholar] [CrossRef]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed Differentiation of Human Pluripotent Stem Cells into Intestinal Tissue in Vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef]
- Nakano, T.; Ando, S.; Takata, N.; Kawada, M.; Muguruma, K.; Sekiguchi, K.; Saito, K.; Yonemura, S.; Eiraku, M.; Sasai, Y. Self-Formation of Optic Cups and Storable Stratified Neural Retina from Human ESCs. Cell Stem Cell 2012, 10, 771–785. [Google Scholar] [CrossRef]
- Fligor, C.M.; Huang, K.-C.; Lavekar, S.S.; VanderWall, K.B.; Meyer, J.S. Differentiation of Retinal Organoids from Human Pluripotent Stem Cells. Methods Cell Biol. 2020, 159, 279–302. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.-A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral Organoids Model Human Brain Development and Microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Chari, D.A.; Chan, D.K. Diagnosis and Treatment of Congenital Sensorineural Hearing Loss. Curr. Otorhinolaryngol. Rep. 2017, 5, 251–258. [Google Scholar] [CrossRef]
- Renauld, J.M.; Basch, M.L. Congenital Deafness and Recent Advances Towards Restoring Hearing Loss. Curr. Protoc. 2021, 1, e76. [Google Scholar] [CrossRef]
- Sheikh, A.; Bint-e-Zainab; Shabbir, K.; Imtiaz, A. Structure and Physiology of Human Ear Involved in Hearing; IntechOpen: London, UK, 2022; ISBN 978-1-80355-190-6. [Google Scholar] [CrossRef]
- Zhang, W.; Kim, S.M.; Wang, W.; Cai, C.; Feng, Y.; Kong, W.; Lin, X. Cochlear Gene Therapy for Sensorineural Hearing Loss: Current Status and Major Remaining Hurdles for Translational Success. Front. Mol. Neurosci. 2018, 11, 221. [Google Scholar] [CrossRef]
- Lopez-Escamez, J.A.; Carey, J.; Chung, W.-H.; Goebel, J.A.; Magnusson, M.; Mandalà, M.; Newman-Toker, D.E.; Strupp, M.; Suzuki, M.; Trabalzini, F.; et al. Diagnostic Criteria for Menière’s Disease. J. Vestib. Res. Equilib. Orientat. 2015, 25, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Escamez, J.A.; Batuecas-Caletrio, A.; Bisdorff, A. Towards Personalized Medicine in Ménière’s Disease. F1000Research 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Gibson, W.P.R. Meniere’s Disease. Adv. Otorhinolaryngol. 2019, 82, 77–86. [Google Scholar] [CrossRef]
- Perez-Carpena, P.; Lopez-Escamez, J.A. Current Understanding and Clinical Management of Meniere’s Disease: A Systematic Review. Semin. Neurol. 2020, 40, 138–150. [Google Scholar] [CrossRef]
- Frejo, L.; Martin-Sanz, E.; Teggi, R.; Trinidad, G.; Soto-Varela, A.; Santos-Perez, S.; Manrique, R.; Perez, N.; Aran, I.; Almeida-Branco, M.S.; et al. Extended Phenotype and Clinical Subgroups in Unilateral Meniere Disease: A Cross-Sectional Study with Cluster Analysis. Clin. Otolaryngol. 2017, 42, 1172–1180. [Google Scholar] [CrossRef]
- Martín-Sierra, C.; Gallego-Martinez, A.; Requena, T.; Frejo, L.; Batuecas-Caletrío, A.; Lopez-Escamez, J.A. Variable Expressivity and Genetic Heterogeneity Involving DPT and SEMA3D Genes in Autosomal Dominant Familial Meniere’s Disease. Eur. J. Hum. Genet. EJHG 2017, 25, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Roman-Naranjo, P.; Gallego-Martinez, A.; Soto-Varela, A.; Aran, I.; Moleon, M.D.C.; Espinosa-Sanchez, J.M.; Amor-Dorado, J.C.; Batuecas-Caletrio, A.; Perez-Vazquez, P.; Lopez-Escamez, J.A. Burden of Rare Variants in the OTOG Gene in Familial Meniere’s Disease. Ear Hear. 2020, 41, 1598–1605. [Google Scholar] [CrossRef]
- Roman-Naranjo, P.; Moleon, M.D.C.; Aran, I.; Escalera-Balsera, A.; Soto-Varela, A.; Bächinger, D.; Gomez-Fiñana, M.; Eckhard, A.H.; Lopez-Escamez, J.A. Rare Coding Variants Involving MYO7A and Other Genes Encoding Stereocilia Link Proteins in Familial Meniere Disease. Hear. Res. 2021, 409, 108329. [Google Scholar] [CrossRef]
- Roman-Naranjo, P.; Parra-Perez, A.M.; Escalera-Balsera, A.; Soto-Varela, A.; Gallego-Martinez, A.; Aran, I.; Perez-Fernandez, N.; Bächinger, D.; Eckhard, A.H.; Gonzalez-Aguado, R.; et al. Defective α-Tectorin May Involve Tectorial Membrane in Familial Meniere Disease. Clin. Transl. Med. 2022, 12, e829. [Google Scholar] [CrossRef]
- Requena, T.; Cabrera, S.; Martín-Sierra, C.; Price, S.D.; Lysakowski, A.; Lopez-Escamez, J.A. Identification of Two Novel Mutations in FAM136A and DTNA Genes in Autosomal-Dominant Familial Meniere’s Disease. Hum. Mol. Genet. 2015, 24, 1119–1126. [Google Scholar] [CrossRef]
- Hoffman, H.J.; Dobie, R.A.; Losonczy, K.G.; Themann, C.L.; Flamme, G.A. Declining Prevalence of Hearing Loss in US Adults Aged 20 to 69 Years. JAMA Otolaryngol. Neck Surg. 2017, 143, 274. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Liu, Y.; He, J.; Li, S.; Zhang, Q.; Duan, M.; Yang, J.; Jin, Y. A Comparison of Local Endolymphatic Sac Decompression, Endolymphatic Mastoid Shunt, and Wide Endolymphatic Sac Decompression in the Treatment of Intractable Meniere’s Disease: A Short-Term Follow-Up Investigation. Front. Neurol. 2022, 13, 810352. [Google Scholar] [CrossRef]
- De Luca, P.; Cassandro, C.; Ralli, M.; Gioacchini, F.M.; Turchetta, R.; Orlando, M.P.; Iaccarino, I.; Cavaliere, M.; Cassandro, E.; Scarpa, A. Dietary Restriction for The Treatment of Meniere’s Disease. Transl. Med. UniSa 2020, 22, 5–9. [Google Scholar] [PubMed]
- Castiglione, A.; Benatti, A.; Velardita, C.; Favaro, D.; Padoan, E.; Severi, D.; Pagliaro, M.; Bovo, R.; Vallesi, A.; Gabelli, C.; et al. Aging, Cognitive Decline and Hearing Loss: Effects of Auditory Rehabilitation and Training with Hearing Aids and Cochlear Implants on Cognitive Function and Depression among Older Adults. Audiol. Neurotol. 2016, 21, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Grayden, D.; McDonnell, M. Unifying Information Theory and Machine Learning in a Model of Electrode Discrimination in Cochlear Implants. PLoS ONE 2021, 16, e0257568. [Google Scholar] [CrossRef]
- de Cates, C.; Winters, R. Intratympanic Steroid Injection. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Shin, S.-H.; Byun, S.W.; Park, S.; Kim, E.H.; Kim, M.W.; Lee, H.Y. Optimal First-Line Therapy for Acute Low-Tone Sensorineural Hearing Loss. J. Audiol. Otol. 2021, 25, 209–216. [Google Scholar] [CrossRef]
- Goudey, B.; Plant, K.; Kiral, I.; Jimeno-Yepes, A.; Swan, A.; Gambhir, M.; Büchner, A.; Kludt, E.; Eikelboom, R.H.; Sucher, C.; et al. A MultiCenter Analysis of Factors Associated with Hearing Outcome for 2,735 Adults with Cochlear Implants. Trends Hear. 2021, 25, 233121652110375. [Google Scholar] [CrossRef]
- Roccio, M.; Senn, P.; Heller, S. Novel Insights into Inner Ear Development and Regeneration for Targeted Hearing Loss Therapies. Hear. Res. 2020, 397, 107859. [Google Scholar] [CrossRef]
- He, Z.; Ding, Y.; Mu, Y.; Xu, X.; Kong, W.; Chai, R.; Chen, X. Stem Cell-Based Therapies in Hearing Loss. Front. Cell Dev. Biol. 2021, 9, 730042. [Google Scholar] [CrossRef]
- Nacher-Soler, G.; Garrido, J.M.; Rodríguez-Serrano, F. Hearing Regeneration and Regenerative Medicine: Present and Future Approaches. Arch. Med. Sci. AMS 2019, 15, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Johnson Chacko, L.; Wertjanz, D.; Sergi, C.; Dudas, J.; Fischer, N.; Eberharter, T.; Hoermann, R.; Glueckert, R.; Fritsch, H.; Rask-Andersen, H.; et al. Growth and Cellular Patterning during Fetal Human Inner Ear Development Studied by a Correlative Imaging Approach. BMC Dev. Biol. 2019, 19, 11. [Google Scholar] [CrossRef]
- Filova, I.; Bohuslavova, R.; Tavakoli, M.; Yamoah, E.N.; Fritzsch, B.; Pavlinkova, G. Early Deletion of Neurod1 Alters Neuronal Lineage Potential and Diminishes Neurogenesis in the Inner Ear. Front. Cell Dev. Biol. 2022, 10, 845461. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Jadali, A.; Fritzsch, B.; Kwan, K.Y. NEUROG1 Regulates CDK2 to Promote Proliferation in Otic Progenitors. Stem Cell Rep. 2017, 9, 1516–1529. [Google Scholar] [CrossRef] [PubMed]
- Filova, I.; Dvorakova, M.; Bohuslavova, R.; Pavlinek, A.; Elliott, K.L.; Vochyanova, S.; Fritzsch, B.; Pavlinkova, G. Combined Atoh1 and Neurod1 Deletion Reveals Autonomous Growth of Auditory Nerve Fibers. Mol. Neurobiol. 2020, 57, 5307–5323. [Google Scholar] [CrossRef]
- Sherrill, H.E.; Jean, P.; Driver, E.C.; Sanders, T.R.; Fitzgerald, T.S.; Moser, T.; Kelley, M.W. Pou4f1 Defines a Subgroup of Type I Spiral Ganglion Neurons and Is Necessary for Normal Inner Hair Cell Presynaptic Ca2+ Signaling. J. Neurosci. Off. J. Soc. Neurosci. 2019, 39, 5284–5298. [Google Scholar] [CrossRef]
- Lahlou, H.; Nivet, E.; Lopez-Juarez, A.; Fontbonne, A.; Assou, S.; Zine, A. Enriched Differentiation of Human Otic Sensory Progenitor Cells Derived From Induced Pluripotent Stem Cells. Front. Mol. Neurosci. 2018, 11, 452. [Google Scholar] [CrossRef]
- Kurihara, S.; Fujioka, M.; Hirabayashi, M.; Yoshida, T.; Hosoya, M.; Nagase, M.; Kato, F.; Ogawa, K.; Okano, H.; Kojima, H.; et al. Otic Organoids Containing Spiral Ganglion Neuron-like Cells Derived from Human-Induced Pluripotent Stem Cells as a Model of Drug-Induced Neuropathy. Stem Cells Transl. Med. 2022, 11, 282–296. [Google Scholar] [CrossRef]
- Boddy, S.L.; Romero-Guevara, R.; Ji, A.-R.; Unger, C.; Corns, L.; Marcotti, W.; Rivolta, M.N. Generation of Otic Lineages from Integration-Free Human-Induced Pluripotent Stem Cells Reprogrammed by MRNAs. Stem Cells Int. 2020, 2020, 3692937. [Google Scholar] [CrossRef]
- Chen, J.-R.; Tang, Z.-H.; Zheng, J.; Shi, H.-S.; Ding, J.; Qian, X.-D.; Zhang, C.; Chen, J.-L.; Wang, C.-C.; Li, L.; et al. Effects of Genetic Correction on the Differentiation of Hair Cell-like Cells from IPSCs with MYO15A Mutation. Cell Death Differ. 2016, 23, 1347–1357. [Google Scholar] [CrossRef]
- Chen, W.; Jongkamonwiwat, N.; Abbas, L.; Eshtan, S.J.; Johnson, S.L.; Kuhn, S.; Milo, M.; Thurlow, J.K.; Andrews, P.W.; Marcotti, W.; et al. Restoration of Auditory Evoked Responses by Human ES-Cell-Derived Otic Progenitors. Nature 2012, 490, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Ronaghi, M.; Nasr, M.; Ealy, M.; Durruthy-Durruthy, R.; Waldhaus, J.; Diaz, G.H.; Joubert, L.-M.; Oshima, K.; Heller, S. Inner Ear Hair Cell-like Cells from Human Embryonic Stem Cells. Stem Cells Dev. 2014, 23, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- Kenna, M.A.; Feldman, H.A.; Neault, M.W.; Frangulov, A.; Wu, B.-L.; Fligor, B.; Rehm, H.L. Audiologic Phenotype and Progression in GJB2 (Connexin 26) Hearing Loss. Arch. Otolaryngol. Head Neck Surg. 2010, 136, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, I.; Oe, Y.; Danzaki, K.; Ohta, S.; Chen, C.; Shirai, K.; Kawano, A.; Ikeda, K.; Kamiya, K. Modeling Gap Junction Beta 2 Gene-Related Deafness with Human IPSC. Hum. Mol. Genet. 2021, 30, 1429–1442. [Google Scholar] [CrossRef]
- Frejo, L.; Cara, F.; Gallego-Martin, A.; Lopez-Escamez, A. An Inner Ear Organoid Model of Meniere Disease; Trieste, Italy, September 2022. (P3.15.01). Available online: http://www.innerearbiology.eu/abstracts/ieb2022_abstractbook.pdf (accessed on 15 March 2023).
- Frejo, L.; Cara, F.; Gallego-Martin, A.; Lopez-Escamez, A. DTNA and FAM136A Expression in a 3D Inner Ear Organoid Model of Meniere Disease; Granada, Spain, May 2022. (ST0128). Available online: https://content.iospress.com/articles/journal-of-vestibular-research/ves220211 (accessed on 15 March 2023).
- Nie, J.; Hashino, E. Generation of Inner Ear Organoids from Human Pluripotent Stem Cells. Methods Cell Biol. 2020, 159, 303–321. [Google Scholar] [CrossRef]
- Kawasaki, H.; Mizuseki, K.; Nishikawa, S.; Kaneko, S.; Kuwana, Y.; Nakanishi, S.; Nishikawa, S.I.; Sasai, Y. Induction of Midbrain Dopaminergic Neurons from ES Cells by Stromal Cell-Derived Inducing Activity. Neuron 2000, 28, 31–40. [Google Scholar] [CrossRef]
- Chen, Y.; Xiong, M.; Dong, Y.; Haberman, A.; Cao, J.; Liu, H.; Zhou, W.; Zhang, S.-C. Chemical Control of Grafted Human PSC-Derived Neurons in a Mouse Model of Parkinson’s Disease. Cell Stem Cell 2016, 18, 817–826. [Google Scholar] [CrossRef]
- Oestreicher, E.; Wolfgang, A.; Felix, D. Neurotransmission of the Cochlear Inner Hair Cell Synapse—Implications for Inner Ear Therapy. Adv. Otorhinolaryngol. 2002, 59, 131–139. [Google Scholar] [CrossRef]
- Gunewardene, N.; Bergen, N.V.; Crombie, D.; Needham, K.; Dottori, M.; Nayagam, B.A. Directing Human Induced Pluripotent Stem Cells into a Neurosensory Lineage for Auditory Neuron Replacement. BioRes. Open Access 2014, 3, 162–175. [Google Scholar] [CrossRef]
- Matsuoka, A.J.; Morrissey, Z.D.; Zhang, C.; Homma, K.; Belmadani, A.; Miller, C.A.; Chadly, D.M.; Kobayashi, S.; Edelbrock, A.N.; Tanaka-Matakatsu, M.; et al. Directed Differentiation of Human Embryonic Stem Cells Toward Placode-Derived Spiral Ganglion-Like Sensory Neurons. Stem Cells Transl. Med. 2017, 6, 923–936. [Google Scholar] [CrossRef]
- Needham, K.; Hyakumura, T.; Gunewardene, N.; Dottori, M.; Nayagam, B.A. Electrophysiological Properties of Neurosensory Progenitors Derived from Human Embryonic Stem Cells. Stem Cell Res. 2014, 12, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hong, F.; Zhang, C.; Li, L.; Wang, C.; Shi, H.; Fu, Y.; Wang, J. Differentiation and Transplantation of Human Induced Pluripotent Stem Cell-Derived Otic Epithelial Progenitors in Mouse Cochlea. Stem Cell Res. Ther. 2018, 9, 230. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.-C.; Alex, A.L.; Nie, J.; Lee, J.; Roth, A.A.; Booth, K.T.; Koehler, K.R.; Hashino, E.; Nelson, R.F. Defective Tmprss3-Associated Hair Cell Degeneration in Inner Ear Organoids. Stem Cell Rep. 2019, 13, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, J. Safety of Autologous Stem Cell Infusion for Children With Acquired Hearing Loss; 2018. Available online: clinicaltrials.gov (accessed on 1 October 2022).
- Kanzaki, S.; Toyoda, M.; Umezawa, A.; Ogawa, K. Application of Mesenchymal Stem Cell Therapy and Inner Ear Regeneration for Hearing Loss: A Review. Int. J. Mol. Sci. 2020, 21, 5764. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.-H.; Chen, J.-R.; Zheng, J.; Shi, H.-S.; Ding, J.; Qian, X.-D.; Zhang, C.; Chen, J.-L.; Wang, C.-C.; Li, L.; et al. Genetic Correction of Induced Pluripotent Stem Cells From a Deaf Patient With MYO7A Mutation Results in Morphologic and Functional Recovery of the Derived Hair Cell-Like Cells. Stem Cells Transl. Med. 2016, 5, 561–571. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Tsai, C.-L.; Wei, Y.-H.; Wu, Y.-T.; Hsu, W.-T.; Lin, H.-C.; Hsu, Y.-C. ATOH1/RFX1/RFX3 Transcription Factors Facilitate the Differentiation and Characterisation of Inner Ear Hair Cell-like Cells from Patient-Specific Induced Pluripotent Stem Cells Harbouring A8344G Mutation of Mitochondrial DNA. Cell Death Dis. 2018, 9, 437. [Google Scholar] [CrossRef]
- Fukunaga, I.; Fujimoto, A.; Hatakeyama, K.; Aoki, T.; Nishikawa, A.; Noda, T.; Minowa, O.; Kurebayashi, N.; Ikeda, K.; Kamiya, K. In Vitro Models of GJB2-Related Hearing Loss Recapitulate Ca2+ Transients via a Gap Junction Characteristic of Developing Cochlea. Stem Cell Rep. 2016, 7, 1023–1036. [Google Scholar] [CrossRef]
- Hosoya, M.; Fujioka, M.; Sone, T.; Okamoto, S.; Akamatsu, W.; Ukai, H.; Ueda, H.R.; Ogawa, K.; Matsunaga, T.; Okano, H. Cochlear Cell Modeling Using Disease-Specific IPSCs Unveils a Degenerative Phenotype and Suggests Treatments for Congenital Progressive Hearing Loss. Cell Rep. 2017, 18, 68–81. [Google Scholar] [CrossRef]
- Hosoya, M.; Saeki, T.; Saegusa, C.; Matsunaga, T.; Okano, H.; Fujioka, M.; Ogawa, K. Estimating the Concentration of Therapeutic Range Using Disease-Specific IPS Cells: Low-Dose Rapamycin Therapy for Pendred Syndrome. Regen. Ther. 2019, 10, 54–63. [Google Scholar] [CrossRef]
- Fujioka, M.; Akiyama, T.; Hosoya, M.; Kikuchi, K.; Fujiki, Y.; Saito, Y.; Yoshihama, K.; Ozawa, H.; Tsukada, K.; Nishio, S.-Y.; et al. A Phase I/IIa Double Blind Single Institute Trial of Low Dose Sirolimus for Pendred Syndrome/DFNB4. Medicine 2020, 99, e19763. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).