Glucagon Promotes Gluconeogenesis through the GCGR/PKA/CREB/PGC-1α Pathway in Hepatocytes of the Japanese Flounder Paralichthys olivaceus
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Cloning and Tissue Distribution Analysis of GCG
2.3. Sequence Analysis and 3D Structure of GCG
2.4. Synthesis of Glucagon
2.5. Treatments with Glucagon and Inhibitors in Hepatocytes
2.6. Overexpression of gcgr
2.7. Measurements of Metabolites
2.8. Real-Time Quantitative RT-PCR Analysis
2.9. Western Blot Analysis
2.10. Statistics Analysis
3. Results
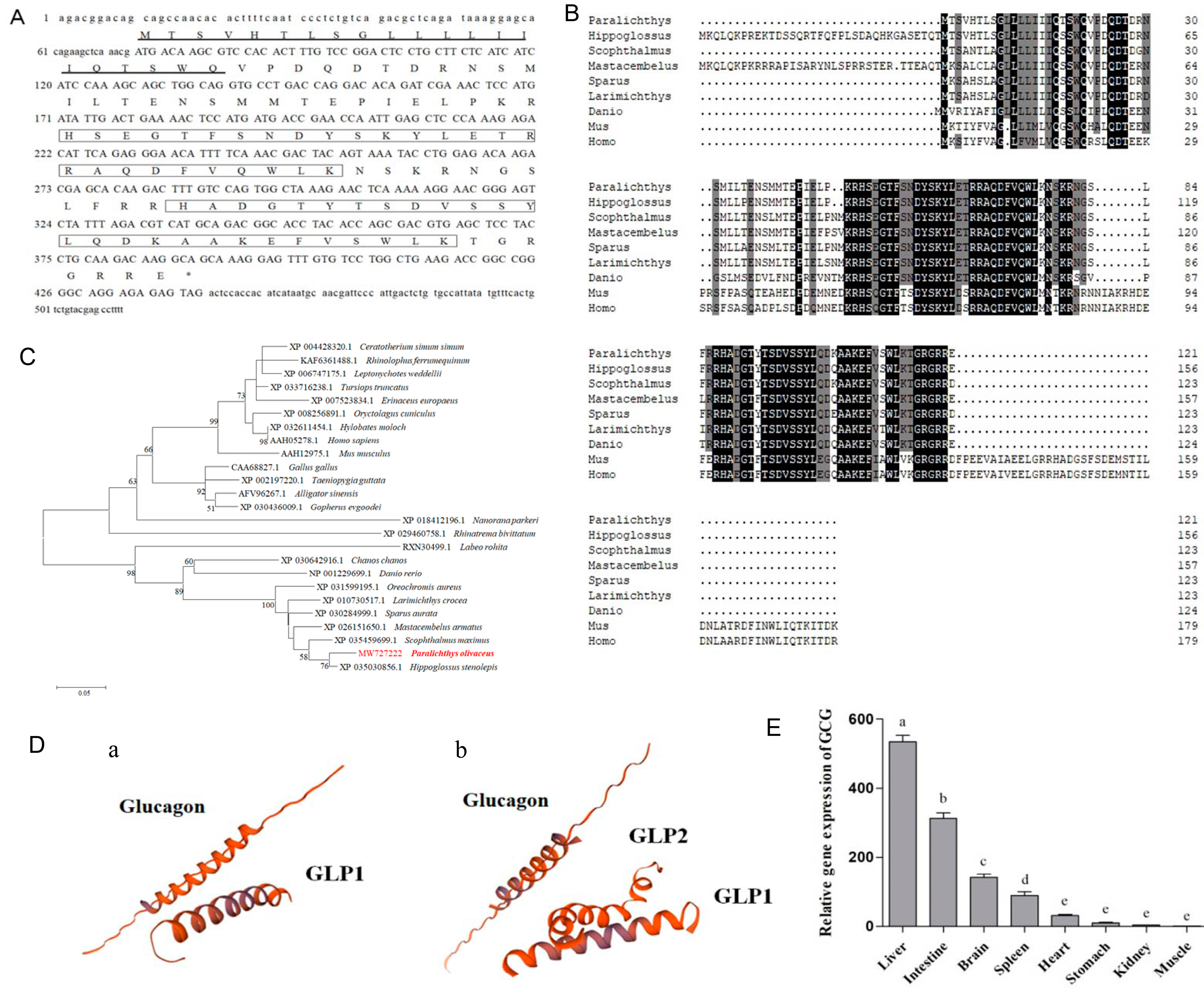
3.1. Gene Cloning, Functional Analysis, and Tissue Distribution of GCG and Synthesis of Glucagon
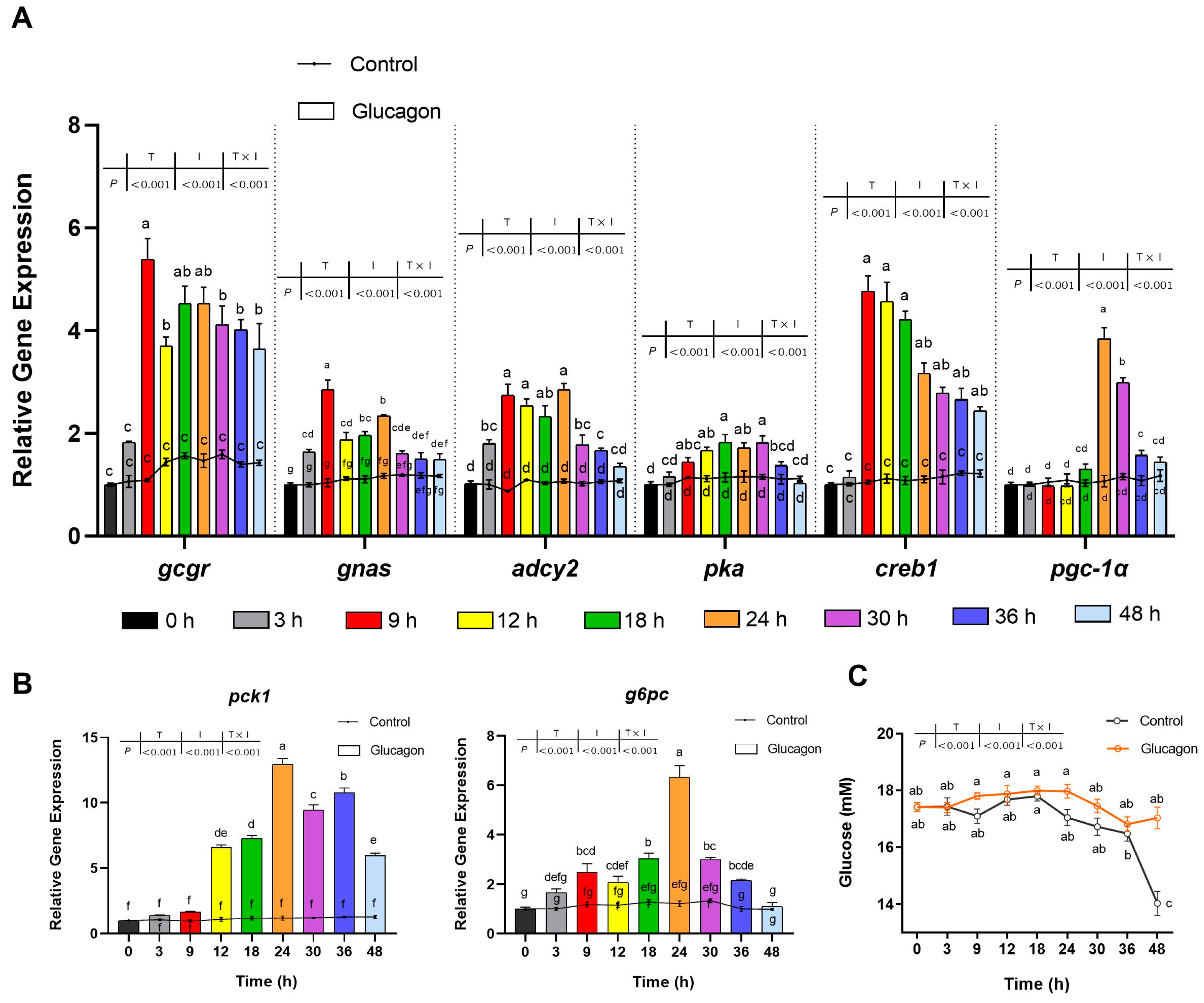
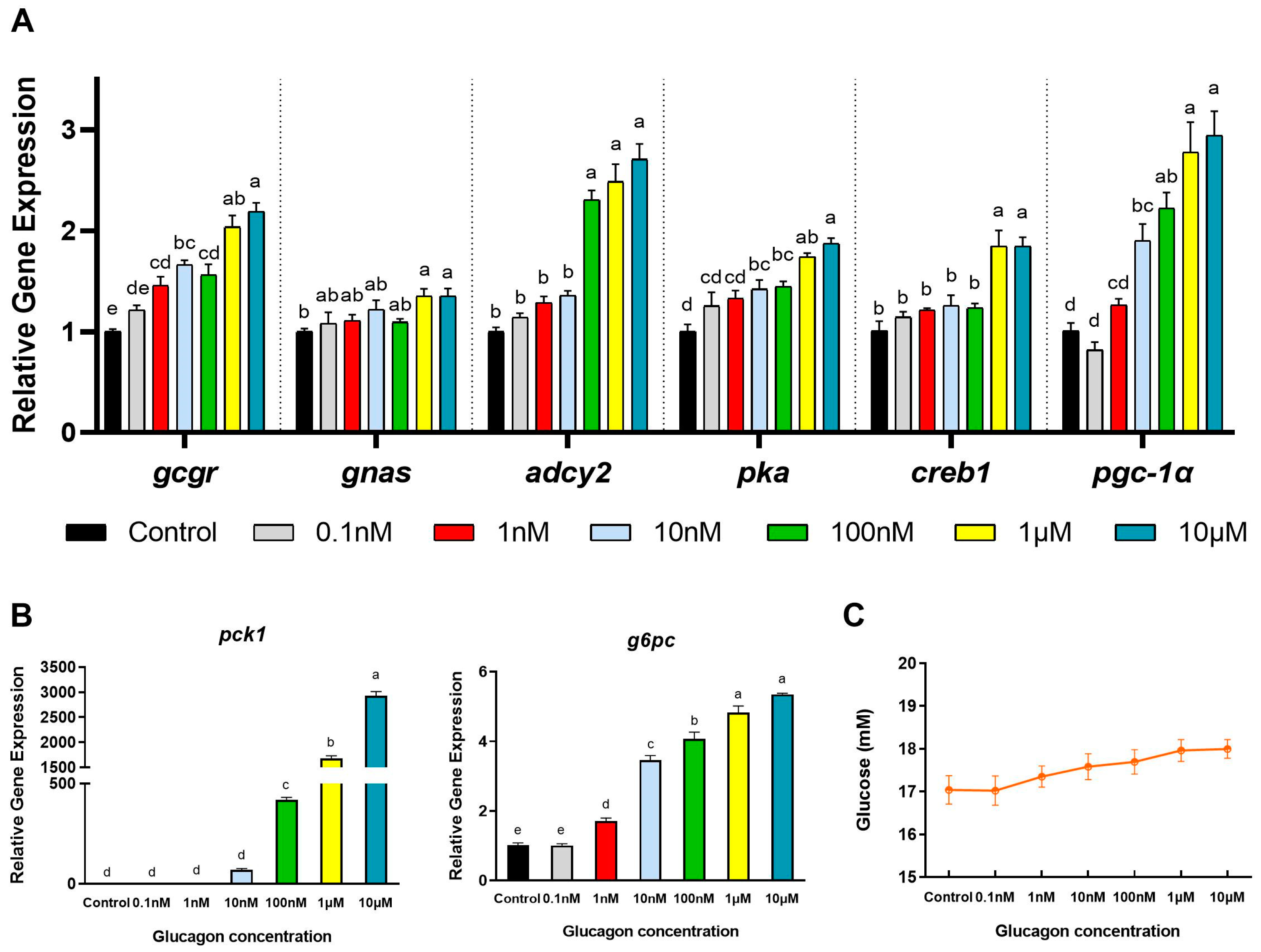
3.2. Glucagon Induces Gene Expression in Hepatocytes and Increases the Glucose Concentration in the Medium
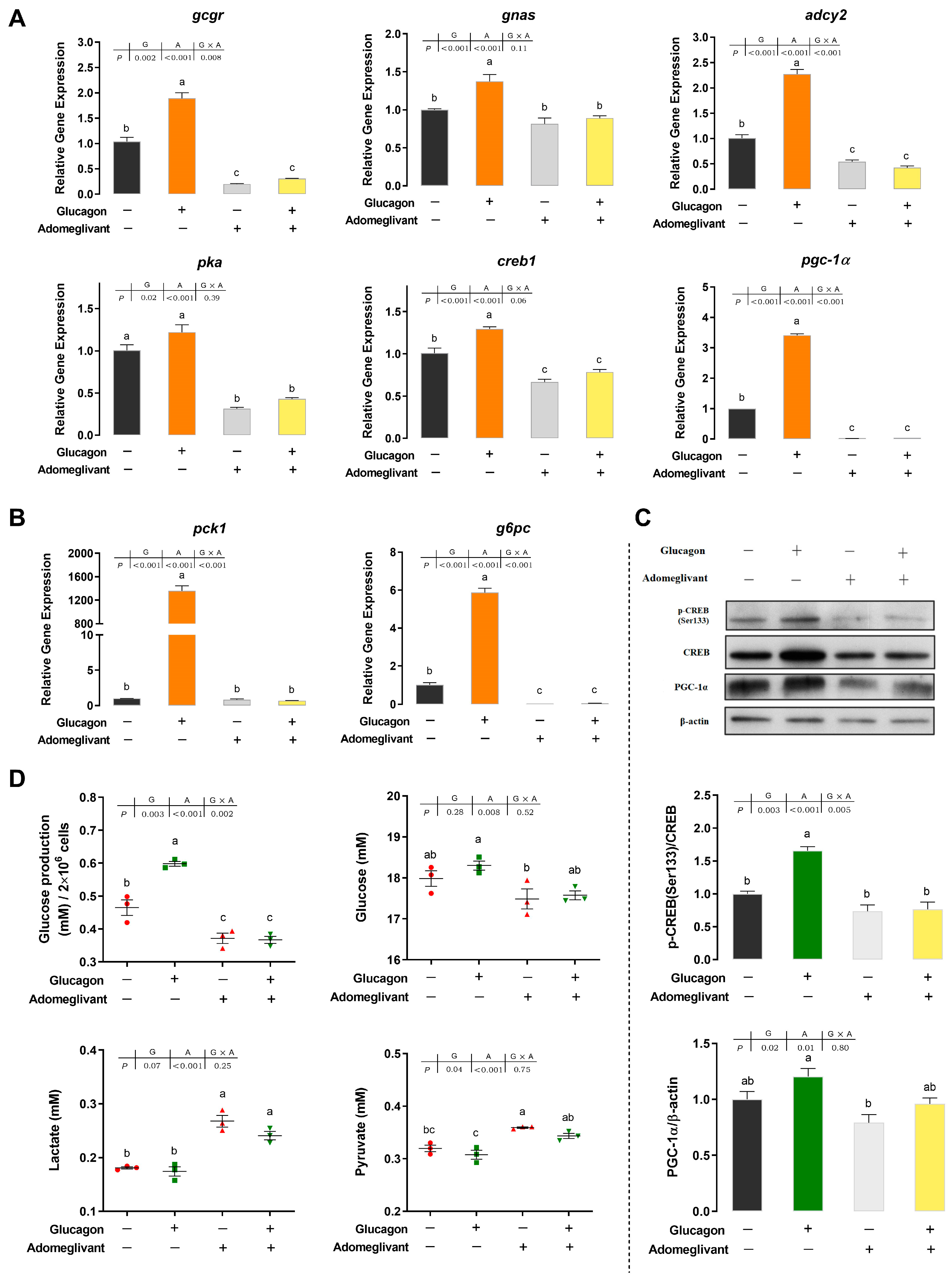
3.3. Adomeglivant Inhibits the Glucagon Pathway in Hepatocytes by Targeting GCGR
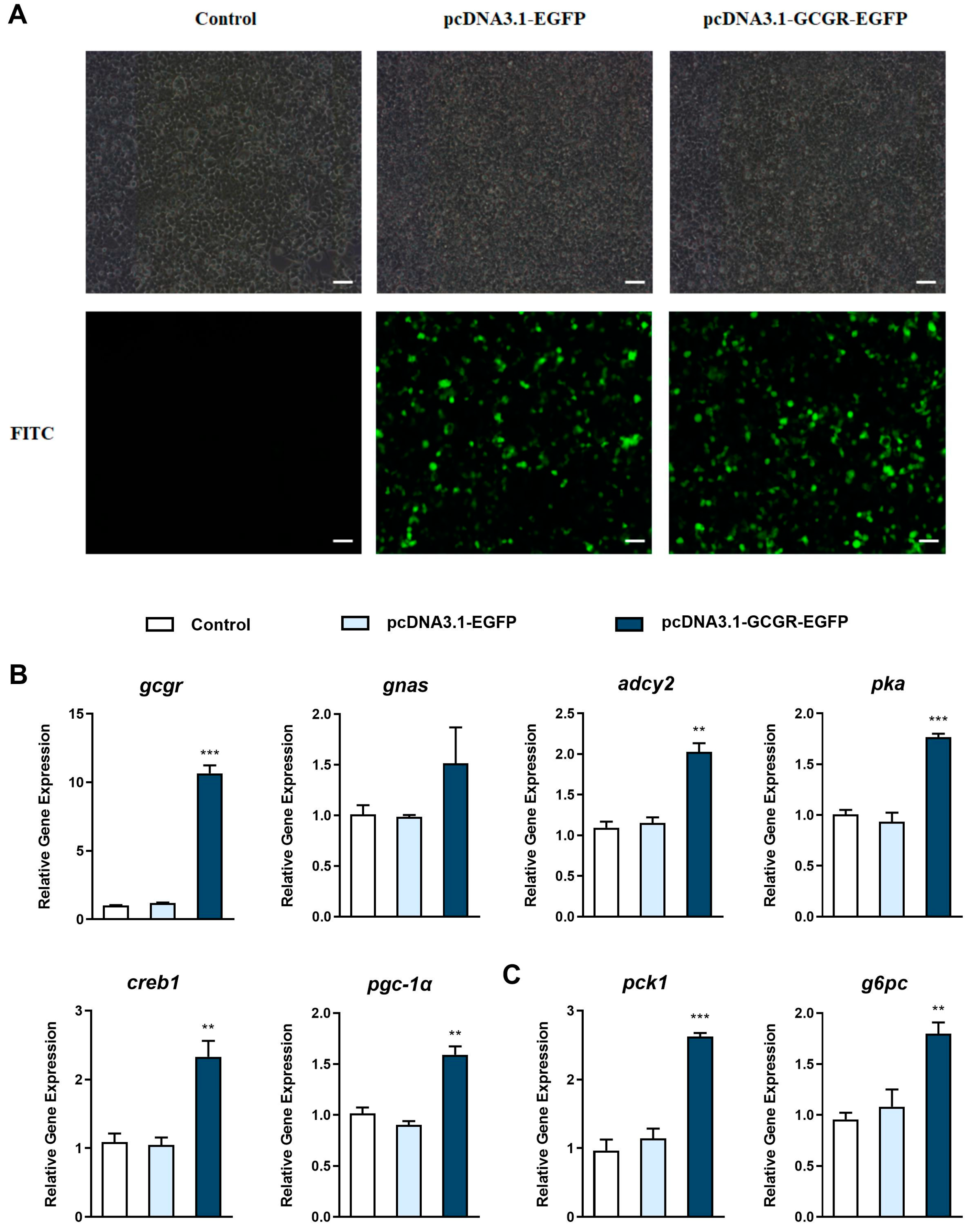
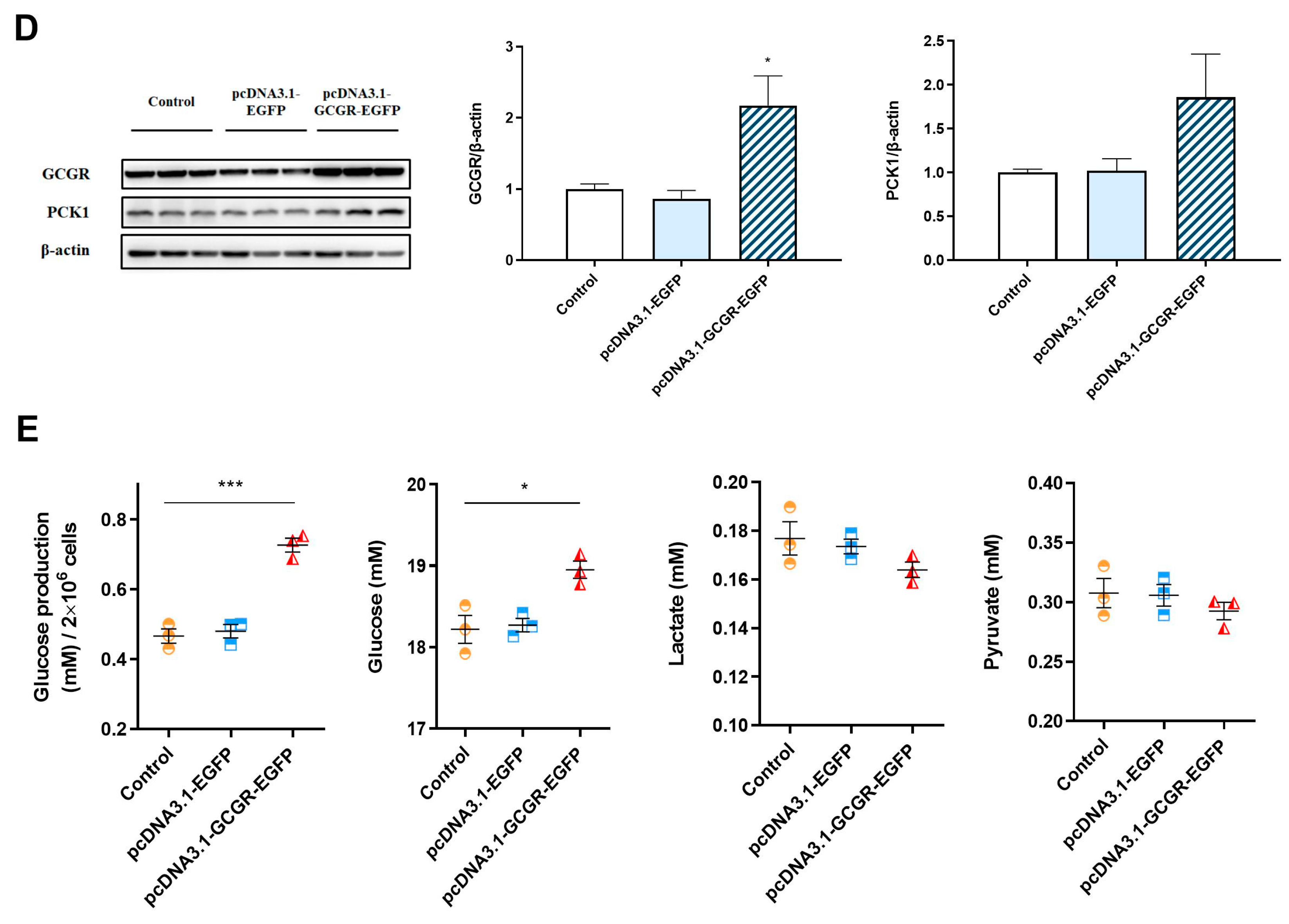
3.4. Overexpression of Gcgr Activates the Glucagon Pathway in Hepatocytes
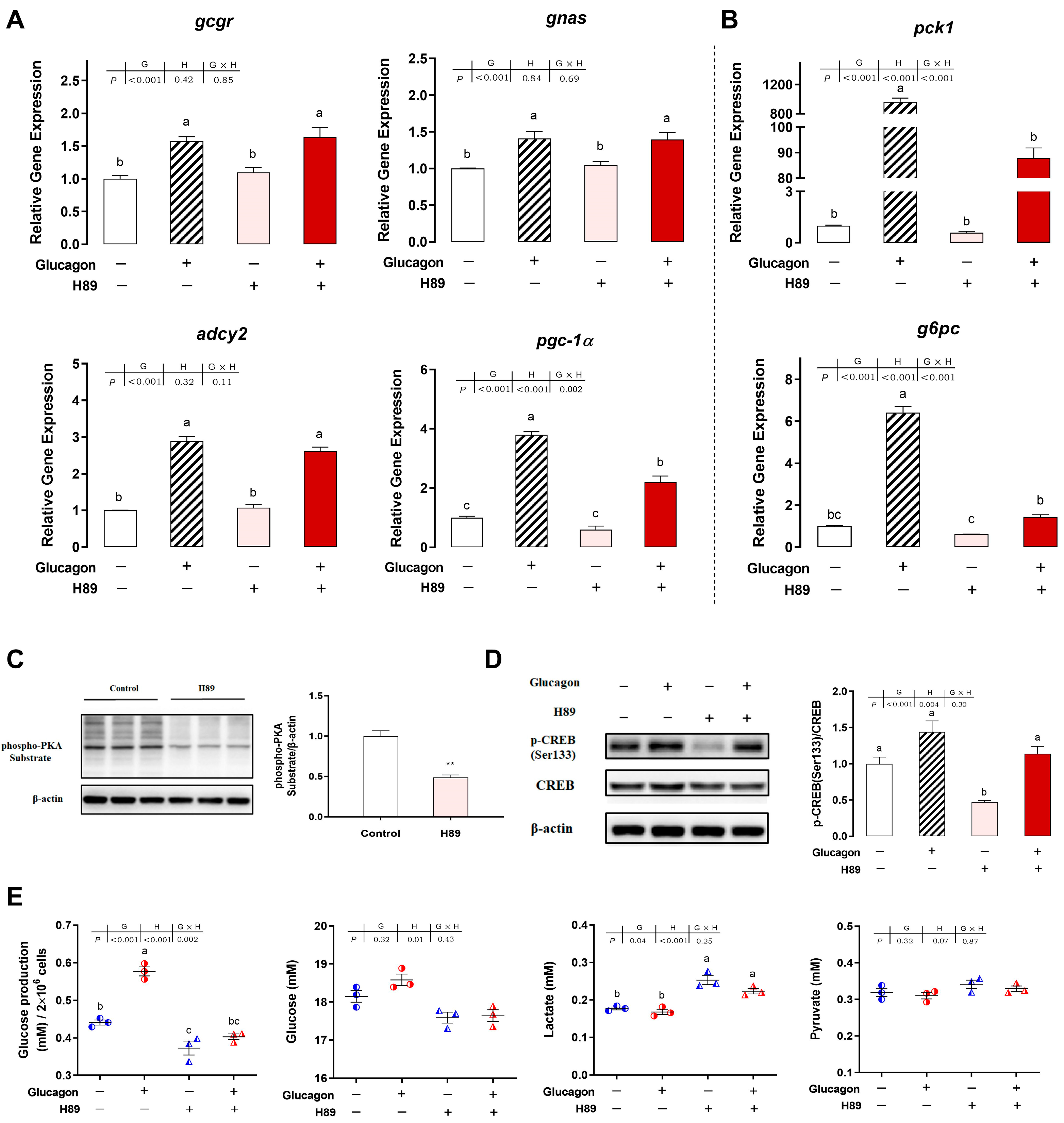
3.5. H89 Inhibits the Glucagon Pathway in Hepatocytes by Targeting PKA
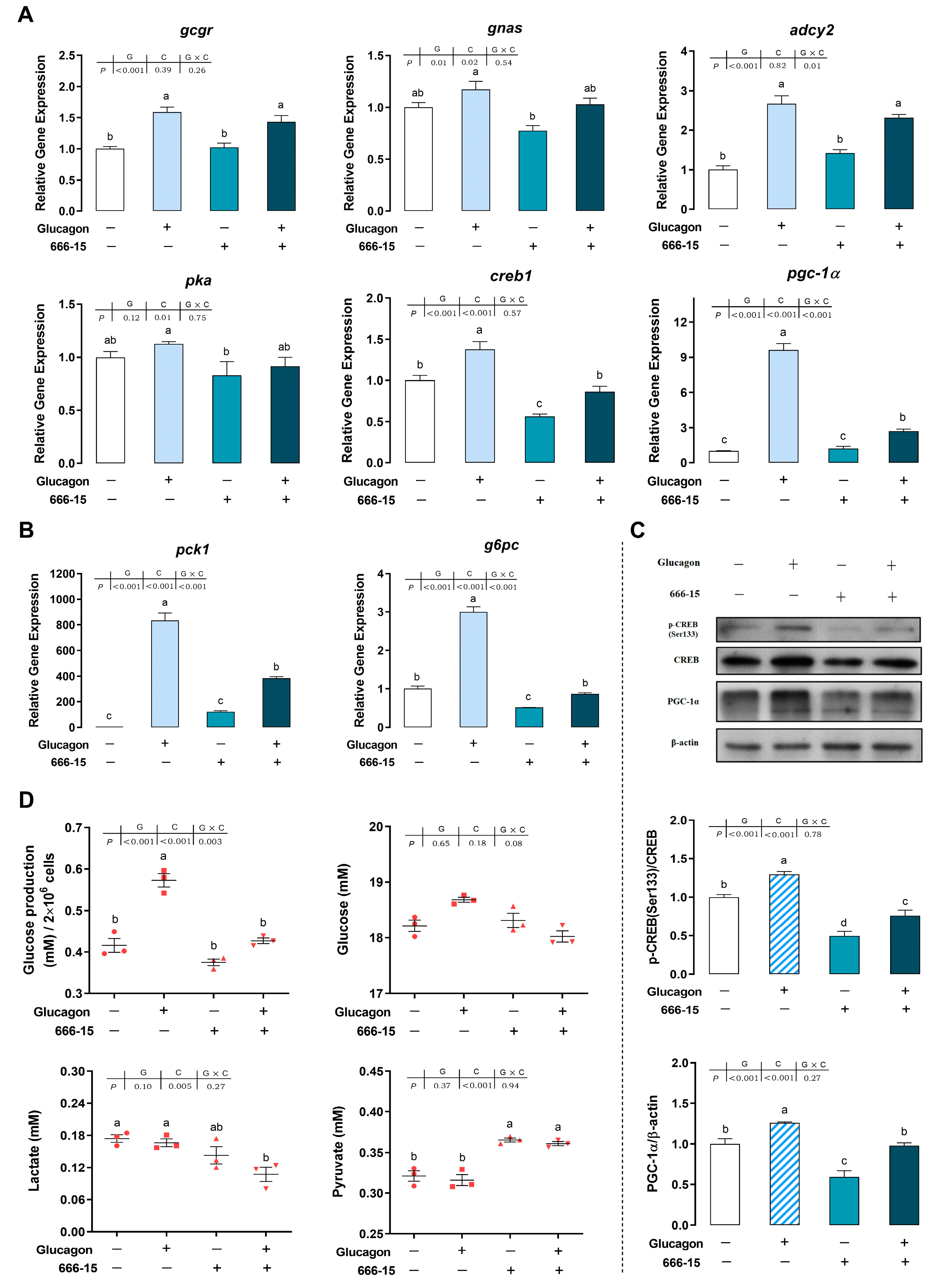
3.6. The Molecule 666-15 Inhibits the Glucagon Pathway in Hepatocytes by Targeting CREB
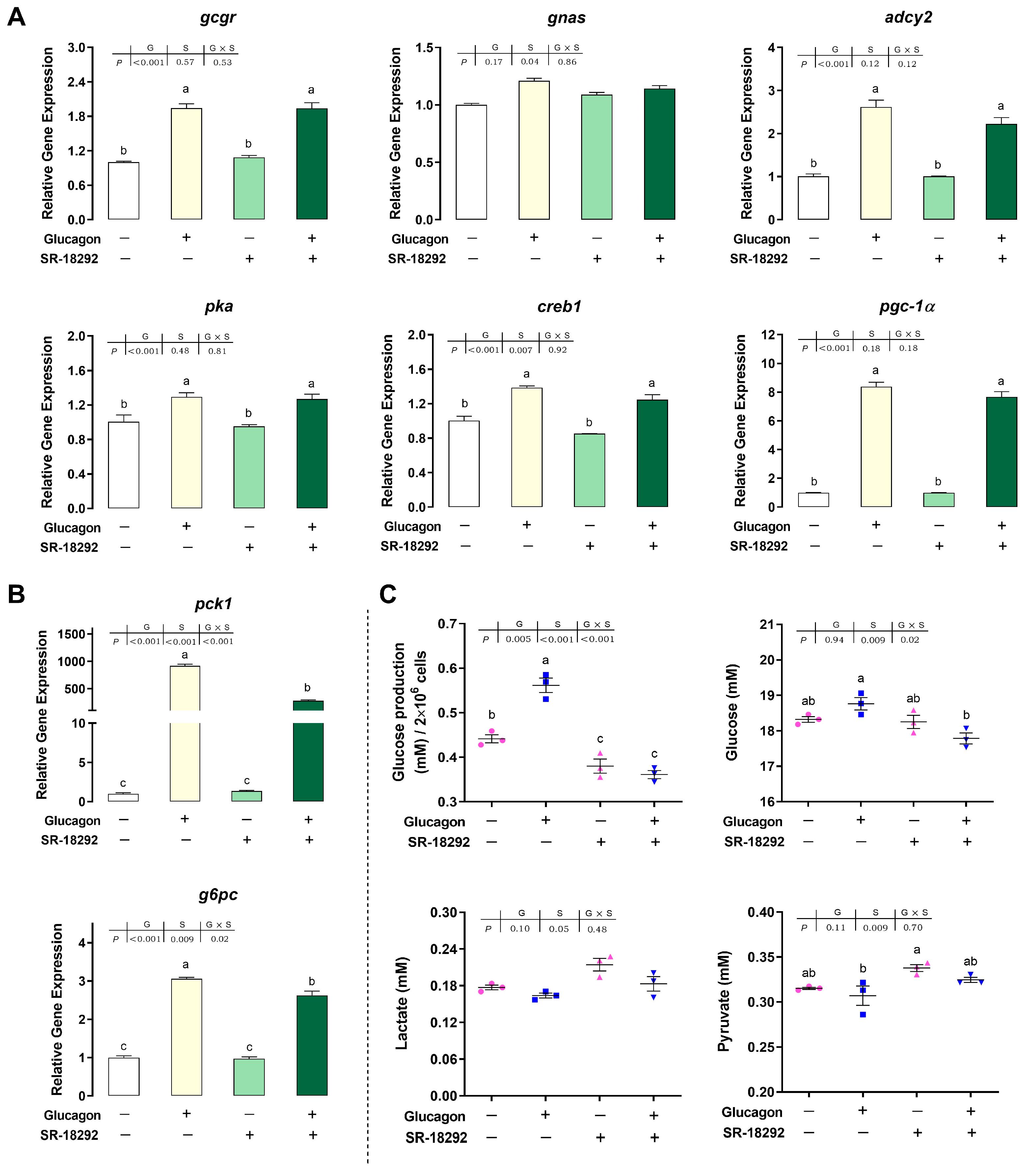
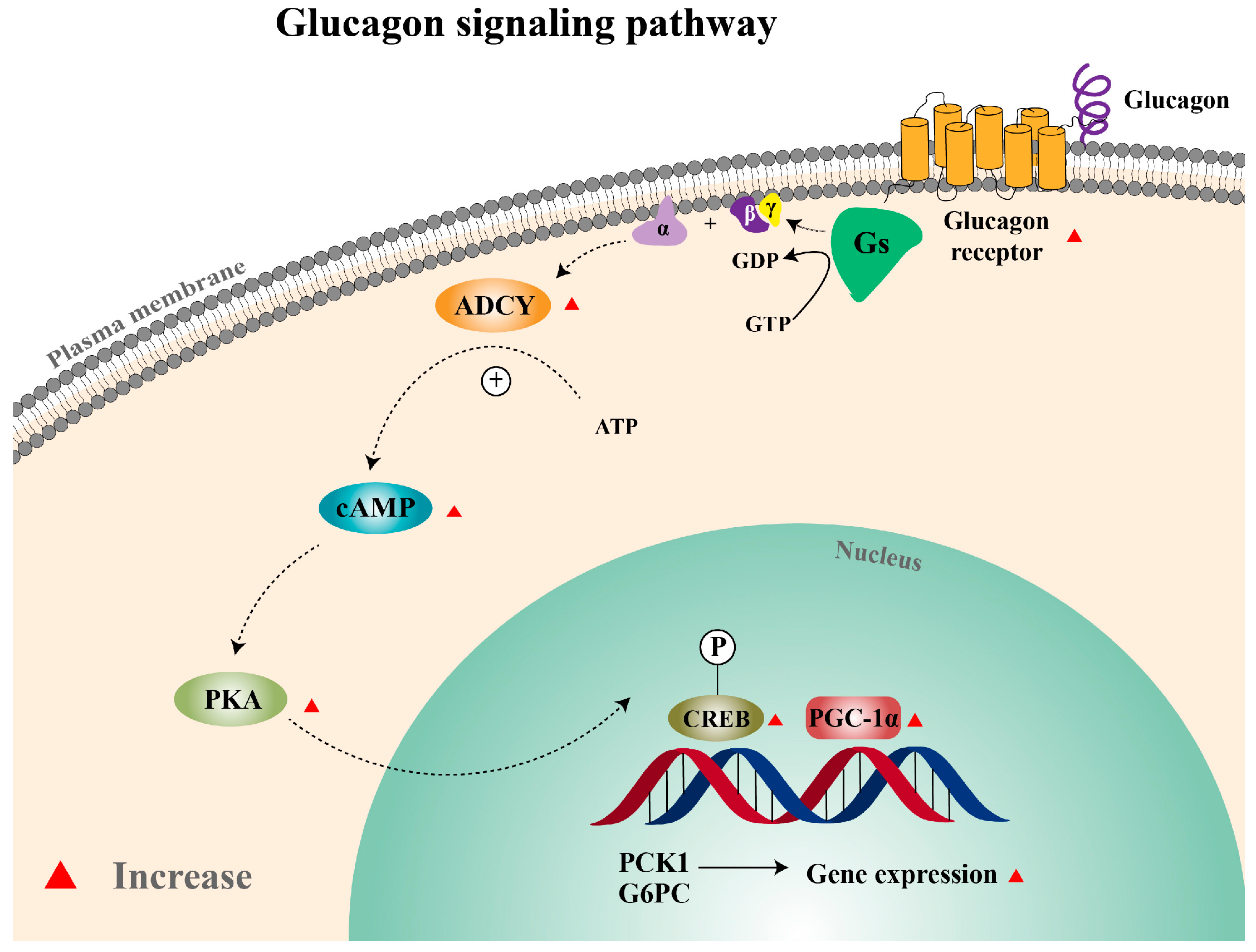
3.7. SR-18292 Inhibits the Glucagon Pathway in Hepatocytes by Targeting PGC-1α
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Park, M.K. Glucagon. In Handbook of Hormones; Takei, Y., Ando, H., Tsutsui, K., Eds.; Academic Press: Oxford, UK, 2016; pp. 129–131. [Google Scholar]
- Herzig, S.; Long, F.X.; Jhala, U.S.; Hedrick, S.; Quinn, R.; Bauer, A.; Rudolph, D.; Schutz, G.; Yoon, C.; Puigserver, P.; et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 2001, 413, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Zhang, B.B. Glucagon and regulation of glucose metabolism. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E671–E678. [Google Scholar] [CrossRef] [PubMed]
- Forbes, J.M.; Cooper, M.E. Mechanisms of diabetic complications. Physiol. Rev. 2013, 93, 137–188. [Google Scholar] [CrossRef] [PubMed]
- Richardson, L.C.; Pollack, L.A. Therapy insight: Influence of type 2 diabetes on the development, treatment and outcomes of cancer. Nat. Clin. Pract. Oncol. 2005, 2, 48–53. [Google Scholar] [CrossRef]
- Lund, A.; Bagger, J.I.; Christensen, M.; Knop, F.K.; Vilsbøll, T. Glucagon and type 2 diabetes: The return of the alpha cell. Curr. Diabetes Rep. 2014, 14, 1–7. [Google Scholar] [CrossRef]
- D’alessio, D. The role of dysregulated glucagon secretion in type 2 diabetes. Diabetes Obes. Metab. 2011, 13, 126–132. [Google Scholar] [CrossRef]
- Godoy-Matos, A.F. The role of glucagon on type 2 diabetes at a glance. Diabetol. Metab. Syndr. 2014, 6, 1–5. [Google Scholar] [CrossRef]
- Yan, H.; Gu, W.; Yang, J.; Bi, V.; Shen, Y.; Lee, E.; Winters, K.A.; Komorowski, R.; Zhang, C.; Patel, J.J.; et al. Fully human monoclonal antibodies antagonizing the glucagon receptor improve glucose homeostasis in mice and monkeys. J. Pharmacol. Exp. Ther. 2009, 329, 102–111. [Google Scholar] [CrossRef]
- Hare, K.J.; Vilsboll, T.; Asmar, M.; Deacon, C.F.; Knop, F.K.; Holst, J.J. The glucagonostatic and insulinotropic effects of glucagon-like peptide 1 contribute equally to its glucose-lowering action. Diabetes 2010, 59, 1765–1770. [Google Scholar] [CrossRef]
- Liang, Y.; Osborne, M.C.; Monia, B.P.; Bhanot, S.; Gaarde, W.A.; Reed, C.; She, P.X.; Jetton, T.L.; Demarest, K.T. Reduction in glucagon receptor expression by an antisense oligonucleotide ameliorates diabetic syndrome in db/db mice. Diabetes 2004, 53, 410–417. [Google Scholar] [CrossRef]
- Yang, H.; Yang, L. Targeting cAMP/PKA signaling pathway for glycemic control and type 2 diabetes therapy. J. Mol. Endocrinol. 2016, 57, R93–R108. [Google Scholar] [CrossRef]
- Furuichi, M.; Yone, Y. Change of blood sugar and plasma insulin levels of fishes in glucose tolerance test. Nippon. Suisan Gakkaishi 1981, 47, 761–764. [Google Scholar] [CrossRef]
- Malik, M.A.; Komarewar, S.; Dar, S.A. Why fish is a natural diabetic animal? World J. Aquac Res. Dev. 2020, 2, 016–017. [Google Scholar]
- Kamalam, B.S.; Panserat, S. Carbohydrates in fish nutrition. Int. Aquafeed. 2016, 20–23. [Google Scholar] [CrossRef]
- Su, J.Z.; Gong, Y.L.; Mei, L.Y.; Xi, L.W.; Chi, S.Y.; Yang, Y.X.; Jin, J.Y.; Liu, H.K.; Zhu, X.M.; Xie, S.Q.; et al. The characteristics of glucose homoeostasis in grass carp and Chinese longsnout catfish after oral starch administration: A comparative study between herbivorous and carnivorous species of fish. Br. J. Nutr. 2020, 123, 627–641. [Google Scholar] [CrossRef]
- Viegas, I.; Mendes, V.M.; Leston, S.; Jarak, I.; Carvalho, R.A.; Pardal, M.A.; Manadas, B.; Jones, J.G. Analysis of glucose metabolism in farmed European sea bass (Dicentrarchus labrax L.) using deuterated water. Comp. Biochem. Physiol. A-Mol. Integr. Physiol. 2011, 160, 341–347. [Google Scholar] [CrossRef]
- Higuera, M.; Cardenas, P. Hormonal effects on gluconeogenesis from (U-14C) glutamate in rainbow trout (Salmo gairdneri). Comp. Biochem. Physiol. B-Biochem. Mol. Biol. 1986, 85, 517–521. [Google Scholar] [CrossRef]
- Ottolenghi, C.; Puviani, A.C.; Baruffaldi, A.; Gavioli, M.E.; Brighenti, L. Glucagon control of glycogenolysis in catfish tissues. Comp. Biochem. Physiol. B-Biochem. Mol. Biol. 1988, 90, 285–290. [Google Scholar] [CrossRef]
- Deng, K.; Pan, M.Z.; Liu, J.H.; Yang, M.X.; Gu, Z.X.; Zhang, Y.; Liu, G.X.; Liu, D.; Zhang, W.B.; Mai, K.S. Chronic stress of high dietary carbohydrate level causes inflammation and influences glucose transport through SOCS3 in Japanese flounder Paralichthys olivaceus. Sci. Rep. 2018, 8, 7415. [Google Scholar] [CrossRef]
- Liu, D.; Deng, K.Y.; Sampath, W.W.H.A.; Gu, Z.X.; Pan, M.Z.; Zhang, Y.; Zhang, W.B.; Mai, K.S. Responses of glucosensing system to glucose in Japanese flounder Paralichthys olivaceus fed diets with different carbohydrate content. Comp. Biochem. Physiol. B-Biochem. Mol. Biol. 2019, 232, 72–78. [Google Scholar] [CrossRef]
- Liu, J.H.; Deng, K.Y.; Pan, M.Z.; Liu, G.X.; Wu, J.; Yang, M.X.; Huang, D.; Zhang, W.B.; Mai, K.S. Dietary carbohydrates influence muscle texture of olive flounder Paralichthys olivaceus through impacting mitochondria function and metabolism of glycogen and protein. Sci. Rep. 2020, 10, 21811. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.X.; Deng, K.Y.; Pan, M.Z.; Gu, Z.X.; Liu, D.; Zhang, Y.; Zhang, W.B.; Mai, K.S. Glucose and lipid metabolic adaptations during postprandial starvation of Japanese flounder Paralichthys olivaceus previously fed different levels of dietary carbohydrates. Aquaculture 2019, 501, 416–429. [Google Scholar] [CrossRef]
- Yang, M.X.; Deng, K.Y.; Pan, M.Z.; Zhang, Y.; Sampath, W.W.H.A.; Zhang, W.B.; Mai, K.S. Molecular adaptations of glucose and lipid metabolism to different levels of dietary carbohydrates in juvenile Japanese flounder Paralichthys olivaceus. Aquac. Nutr. 2019, 26, 516–527. [Google Scholar] [CrossRef]
- Pan, M.Z.; Zhang, Y.; Deng, K.Y.; Liu, G.X.; Gu, Z.X.; Liu, J.H.; Luo, K.; Zhang, W.B.; Mai, K.S. Forkhead box O1 in turbot Scophthalmus maximus: Molecular characterization, gene structure, tissue distribution and the role in glucose metabolism. Gene 2019, 708, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Pan, M.; Liu, J.; Yang, M.; Zhang, W.; Mai, K. Molecular cloning of AKT1 and AKT2 and their divergent responses to insulin and glucose at transcriptional level in the liver of Japanese flounder Paralichthys olivaceus. Aquac. Rep. 2022, 23, 101066. [Google Scholar] [CrossRef]
- Peng, Y.H.; Wang, P.; He, X.Q.; Hong, M.Z.; Liu, F. Micro ribonucleic acid-363 regulates the phosphatidylinositol 3-kinase/threonine protein kinase axis by targeting NOTCH1 and forkhead box C2, leading to hepatic glucose and lipids metabolism disorder in type 2 diabetes mellitus. J. Diabetes Investig. 2022, 13, 236–248. [Google Scholar] [CrossRef]
- MacQueen, J.; Plaut, D. Colorimetric microdetermination of plasma lactate. Am. J. Med. Technol. 1979, 45, 34–37. [Google Scholar]
- Xie, H.L.; Zhu, S.; Zhang, J.; Wen, J.; Yuan, H.J.; Pan, L.Z.; Luo, M.J.; Tan, J.H. Glucose metabolism during in vitro maturation of mouse oocytes: An study using RNA interference. J. Cell. Physiol. 2018, 233, 6952–6964. [Google Scholar] [CrossRef]
- Sun, C.; Wang, M.H.; Liu, X.Y.; Luo, L.; Li, K.X.; Zhang, S.Q.; Wang, Y.J.; Yang, Y.M.; Ding, F.; Gu, X.S.; et al. PCAF improves glucose homeostasis by suppressing the gluconeogenic activity of PGC-1α. Cell Rep. 2014, 9, 2250–2262. [Google Scholar] [CrossRef]
- Liu, J.H.; Pan, M.Z.; Huang, D.; Guo, Y.L.; Yang, M.X.; Zhang, W.B.; Mai, K.S. Myostatin-1 inhibits cell proliferation by inhibiting the mTOR signal pathway and MRFs, and activating the ubiquitin-proteasomal system in skeletal muscle cells of Japanese flounder Paralichthys olivaceus. Cells 2020, 9, 2376. [Google Scholar] [CrossRef]
- Irwin, D.M. Ancient duplications of the human proglucagon gene. Genomics 2002, 79, 741–746. [Google Scholar] [CrossRef]
- Cardoso, J.; Félix, R.; Costa, C.; Palma, P.; Canário, A.; Power, D.M. Evolution of the glucagon-like system across fish. Gen. Comp. Endocrinol. 2017, 264, 113–130. [Google Scholar] [CrossRef]
- Plisetskaya, E.M.; Mommsen, T.P. Glucagon and glucagon-like peptides in fishes. Int. Rev. Cytol. 1996, 168, 187–257. [Google Scholar]
- Zhou, L.; Irwin, D.M. Fish proglucagon genes have differing coding potential. Comp. Biochem. Physiol. B-Biochem. Mol. Biol. 2004, 137, 255–264. [Google Scholar] [CrossRef]
- Forbes, J.L.I.; Kostyniuk, D.J.; Mennigen, J.A.; Weber, J.M. Glucagon regulation of carbohydrate metabolism in rainbow trout: In vivo glucose fluxes and gene expression. J. Exp. Biol. 2019, 222, jeb211730. [Google Scholar] [CrossRef]
- Shi, H.J.; Liu, W.B.; Xu, C.; Zhang, L.; Liu, J.; Zhang, D.D.; Zhang, L.; Li, X.F. Transcriptional regulation of the AMP-activated protein kinase and glycolipid metabolism-related genes by insulin and glucagon in blunt snout bream (Megalobrama amblycephala): A comparative study. Aquaculture 2020, 515, 734553. [Google Scholar] [CrossRef]
- Moon, T.W.; Gambarotta, A.; Capuzzo, A.; Fabbri, E. Glucagon and glucagon-like peptide signaling pathways in the liver of two fish species, the American eel and the black bullhead. J. Exp. Zool. 1997, 279, 62–70. [Google Scholar] [CrossRef]
- Navarro, I.; Moon, T.W. Glucagon binding to hepatocytes isolated from two teleost fishes, the American eel and the brown bullhead. J. Endocrinol. 1994, 140, 217–227. [Google Scholar] [CrossRef]
- Kazda, C.M.; Ying, D.; Kelly, R.P.; Garhyan, P.; Hardy, T.A. Evaluation of efficacy and safety of the glucagon receptor antagonist LY2409021 in patients with type 2 diabetes: 12- and 24-week phase 2 studies. Diabetes Care 2016, 39, e199. [Google Scholar] [CrossRef]
- Gonzalez, G.A.; Montminy, M.R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 1989, 59, 675–680. [Google Scholar] [CrossRef]
- Oh, K.J.; Han, H.S.; Kim, M.J.; Koo, S.H. CREB and FoxO1: Two transcription factors for the regulation of hepatic gluconeogenesis. Bmb Rep. 2013, 46, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Jiang, X.; He, M.; Chan, B.; Wong, A. Novel mechanisms for IGF-I regulation by glucagon in carp hepatocytes: Up-regulation of HNF1α and CREB expression via signaling crosstalk for IGF-I gene transcription. Front. Endocrinol. 2019, 10, 605. [Google Scholar] [CrossRef] [PubMed]
- Yan, A.F.; Chen, T.; Shuang, C.; Tang, D.S.; Hu, C.Q. Signal transduction mechanism for glucagon-induced leptin gene expression in goldfish liver. Int. J. Biol. Sci. 2016, 12, 1544–1554. [Google Scholar] [CrossRef] [PubMed]
- Corona, J.C.; Duchen, M.R. PPARγ and PGC-1α as therapeutic targets in Parkinson’s. Neurochem. Res. 2015, 40, 308–316. [Google Scholar] [CrossRef]
- Rhee, J.; Inoue, Y.; Yoon, J.C.; Puigserver, P.; Fan, M.; Gonzalez, F.J.; Spiegelman, B.M. Regulation of hepatic fasting response by PPARγ coactivator-1α (PGC-1): Requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 4012–4017. [Google Scholar] [CrossRef]
- Wu, H.; Deng, X.; Shi, Y.; Ye, S.; Duan, H. PGC-1α, glucose metabolism and type 2 diabetes mellitus. J. Endocrinol. 2016, 229, R99–R115. [Google Scholar] [CrossRef]
- Yoon, J.C.; Pulgserver, P.; Chen, G.; Donovan, J.; Wu, Z.D.; Rhee, J.; Adelmant, G.; Stafford, J.; Kahn, C.R.; Granner, D.K.; et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 2001, 413, 131–138. [Google Scholar] [CrossRef]
- Sharabi, K.; Hua, L.; Tavares, C.; Dominy, J.E.; Puigserver, P. Selective chemical inhibition of PGC-1α gluconeogenic activity ameliorates type 2 diabetes. Cell 2017, 169, 148–160. [Google Scholar] [CrossRef]
| Primer | Sequences (5′-3′) | Accession Number |
|---|---|---|
| Primers for partial cDNA | ||
| a GCG-F | YGGACTCCTGCTCYTCAT | MW727222 |
| GCG-R | AACTCCTTGGCKGCCTGG | |
| Primers for RACE PCR | ||
| 5′-GCG-R1 GGTTCGGTCATCATGGAGT | GGTTCGGTCATCATGGAGT | MW727222 |
| 5′-GCG-R2 | GATGAGAAGCAGGAGTCCG | |
| 3′-GCG-F1 | CGAGCACAAGACTTTGTCCA | |
| 3′-GCG-F2 | ATGCAGACGGCACCTACAC | |
| UPM (long) | ACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | |
| UPM (short) | CTAATACGACTACTATAGGGC | |
| NUP | AAGCAGTGGTATCAACGCAGAGT | |
| Primers for GCGR cds | ||
| b GCGR-F | GAGCAGCGGTGAAGAGCAG | KY742714 |
| GCGR-R | ATGTCAGGGCCAGACGAGA | |
| Primers with homology arms | ||
| GCGR-h-F | 5′-cttggtaccgagctcggatccATGT CACGGCTGTTCCTCCTT-3′ | KY742714 |
| GCGR-h-R | 5′-atggtggcgaccggtggatccCAGGTG GCTCTCTGCGTTCT-3′ | |
| Primers for qPCR | ||
| a gcg-q-F | CGGACTCCTGCTTCTCAT | MW727222 |
| gcg-q-R | TTTGCTGCCTTGTCTTGC | |
| b gcgr-q-F | AGGACGTTTGACAATTATGC | KY742714 |
| gcgr-q-R | TCTTCGTAGCCACCCACT | |
| c gnas-q-F | TCCGGCAGACTGACTACAC | KY742715 |
| gnas-q-R | GACCTCCAACATCAAACATA | |
| d adcy2-q-F | AGAGTGATGAAAGCCGAAAT | KY742716 |
| adcy2-q-R | TGGTCAAACTTGCCGAAC | |
| e pka-q-F | GCATTGGACCACTTTGAG | KY742718 |
| pka-q-R | GACAGCTTGCAGGATACG | |
| f creb1-q-F | CGCAGGAAGAAGAAGGAGT | MT215340 |
| creb1-q-R | GGTCTTTAAGGGCTTTGAGT | |
| g pgc-1α-q-F | GGATTGCCTTCGTTTGAGG | MT215340 |
| pgc-1α-q-R | GCTGGCTGCATGGTTCTG | |
| h g6pc-q-F | TACCCCGTGACCTGTGAGAC | KY742717 |
| g6pc-q-R | TTGGTGGATTTCTTGCTTCC | |
| i pck1-q-F | ACCAGGGACCAGAAGGACA | MN173838 |
| pck1-q-R | TGGCGACCACGTAGGGAGA | |
| β-actin-q-F | GGAAATCGTGCGTGACATTAAG | HQ386788 |
| β-actin-q-R | CCTCTGGACAACGGAACCTCT | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Pan, M.; Huang, D.; Liu, J.; Guo, Y.; Liu, Y.; Zhang, W. Glucagon Promotes Gluconeogenesis through the GCGR/PKA/CREB/PGC-1α Pathway in Hepatocytes of the Japanese Flounder Paralichthys olivaceus. Cells 2023, 12, 1098. https://doi.org/10.3390/cells12071098
Yang M, Pan M, Huang D, Liu J, Guo Y, Liu Y, Zhang W. Glucagon Promotes Gluconeogenesis through the GCGR/PKA/CREB/PGC-1α Pathway in Hepatocytes of the Japanese Flounder Paralichthys olivaceus. Cells. 2023; 12(7):1098. https://doi.org/10.3390/cells12071098
Chicago/Turabian StyleYang, Mengxi, Mingzhu Pan, Dong Huang, Jiahuan Liu, Yanlin Guo, Yue Liu, and Wenbing Zhang. 2023. "Glucagon Promotes Gluconeogenesis through the GCGR/PKA/CREB/PGC-1α Pathway in Hepatocytes of the Japanese Flounder Paralichthys olivaceus" Cells 12, no. 7: 1098. https://doi.org/10.3390/cells12071098
APA StyleYang, M., Pan, M., Huang, D., Liu, J., Guo, Y., Liu, Y., & Zhang, W. (2023). Glucagon Promotes Gluconeogenesis through the GCGR/PKA/CREB/PGC-1α Pathway in Hepatocytes of the Japanese Flounder Paralichthys olivaceus. Cells, 12(7), 1098. https://doi.org/10.3390/cells12071098






