Novel Components of the Stress Assembly Sec Body Identified by Proximity Labeling
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Amino Acid Starvation
2.2. Molecular Cloning and Transfection
- GFP-APEX2 forward: 5′-CATGTTCGAACTATGGTGAGCAAGGGCG-3′,
- GFP-APEX2 reverse: 5′-CATGGTTTAAACTTAGGCATCAGCAAACCC-3′
- Sec24AB forward: 5′-CATGGAATTCCACCATGTCGACTTACAATCCGAACTTC-3′
- Sec24AB reverse: 5′-CATGGGGCCCTTTAACCTGAGCCCGAATGT-3′
- Rox8 forward: 5′-GGGATCTAGATCGGGGTACCATGGACGAGTCGCAACCG-3’,
- Rox8 reverse: 5′-GCCACTGTGCTGGATATCTTGGGTCTGGTATTGTGGCATCG-3’.
2.3. Stable Cell Line
2.4. Antibodies
2.5. Immunofluorescence
2.6. Western Blotting
2.7. APEX Proximity Labeling and Analysis of Biotinylated Proteins
2.8. Mass Spectrometry Proteome Analysis
2.9. Mass Spectrometry Database Search
3. Results and Discussion:
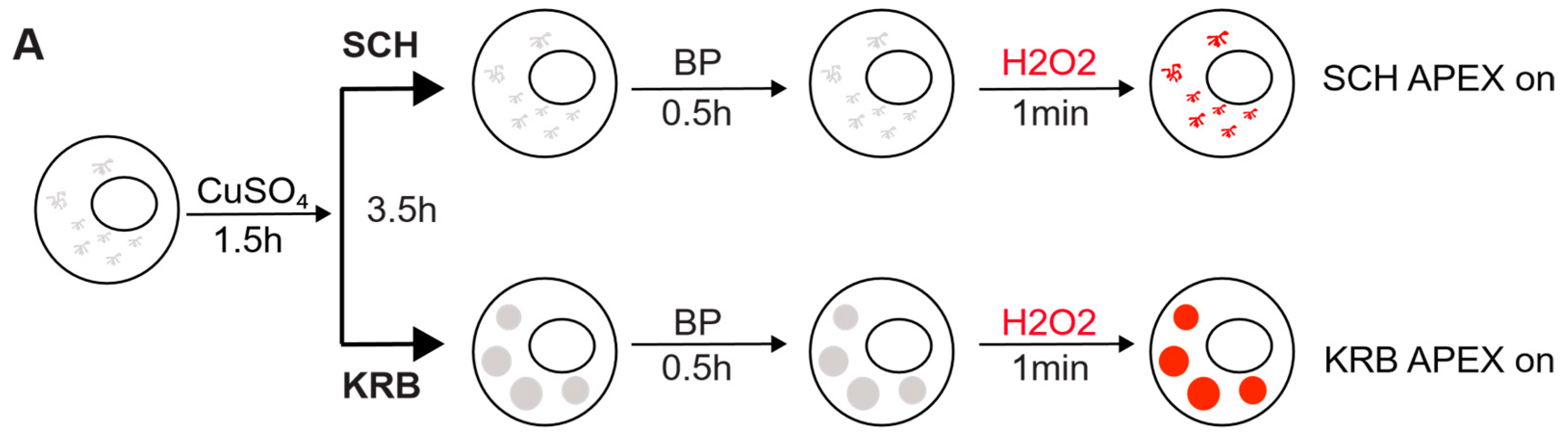
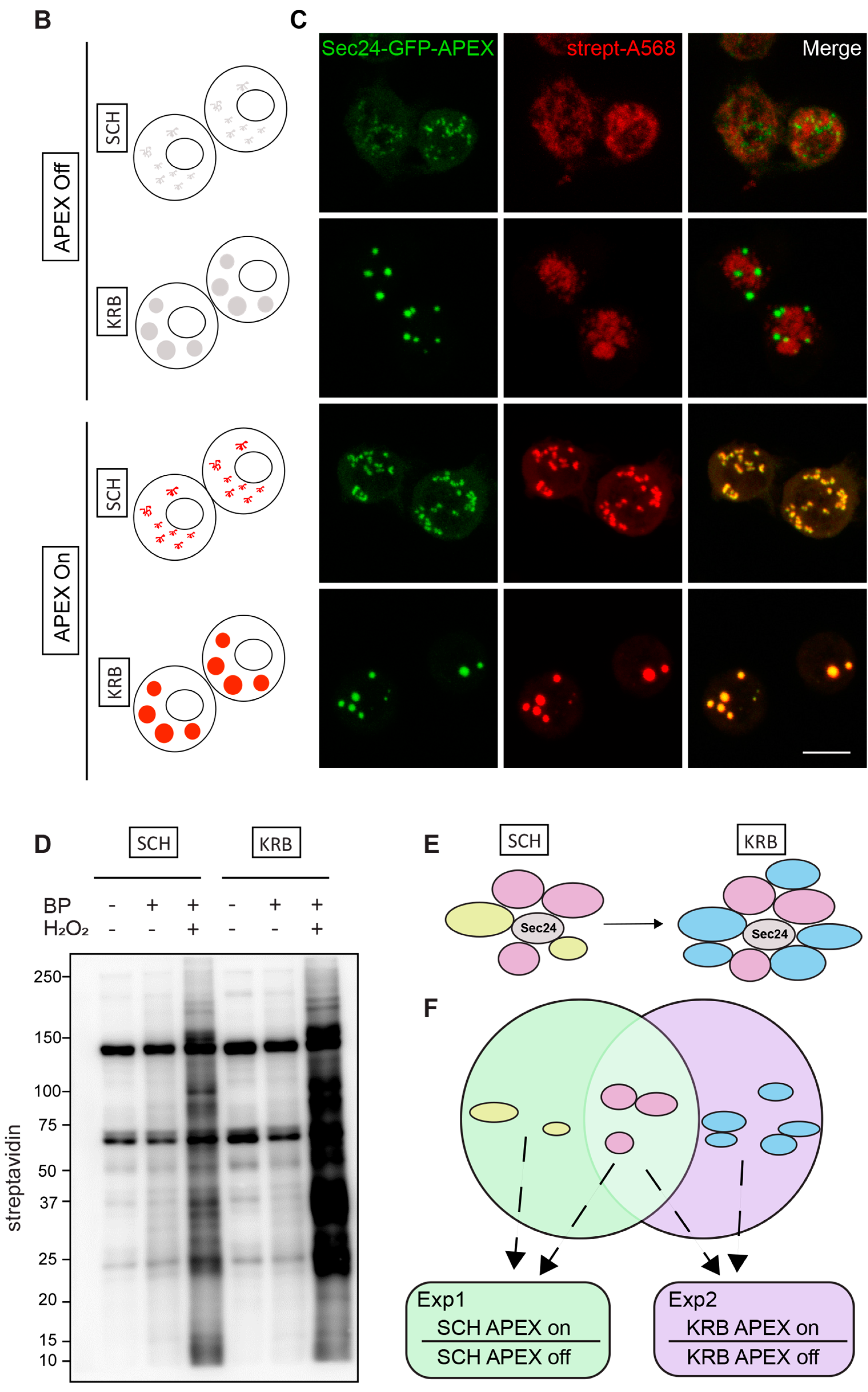
3.1. Sec24-APEX2 Mediates Biotinylation of Sec Body Proteins
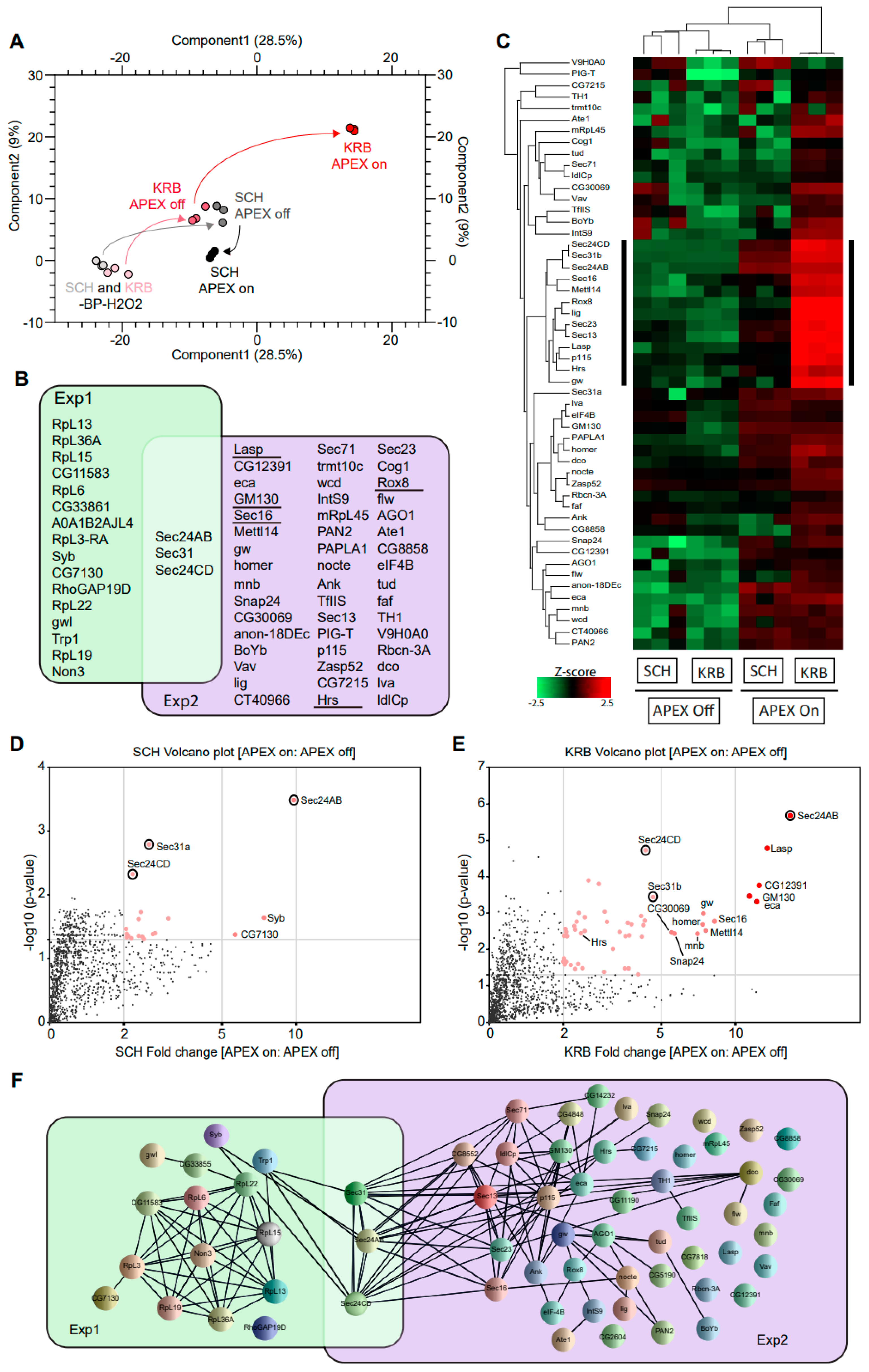
3.2. The Sec24 Spatial Interactome
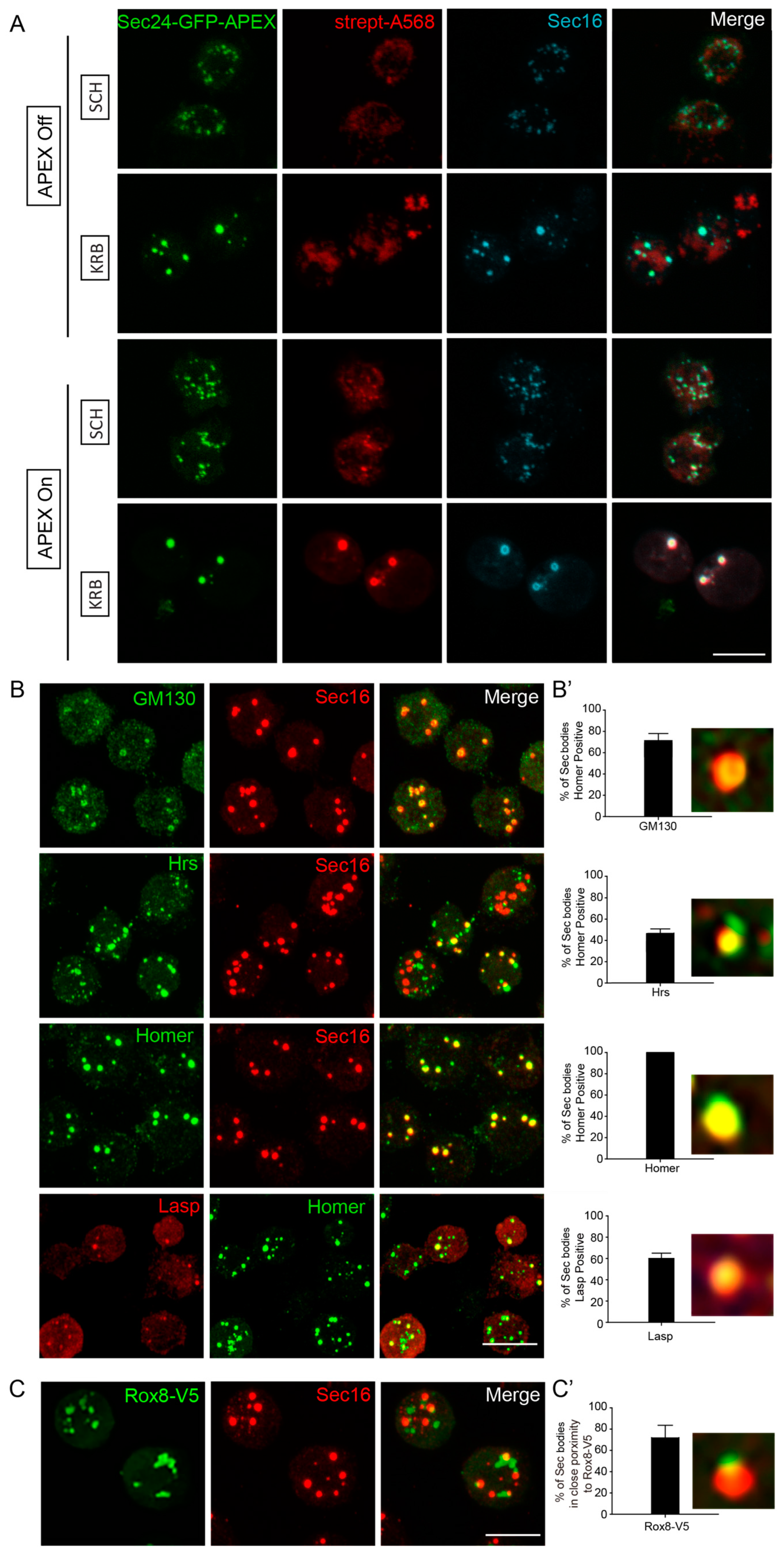
3.3. Validation of Sec Body Components Identified by APEX
3.4. Sec Bodies Form from the Re-Localization of Existing Intracellular Proteins
3.5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mellman, I.; Warren, G. The Road Taken. Cell 2000, 100, 99–112. [Google Scholar] [PubMed]
- Hyman, A.A.; Weber, C.A.; Jülicher, F. Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, W.; Rabouille, C. Cellular stress leads to the formation of membraneless stress assemblies in eukaryotic cells. Traffic 2019, 20, 623–638. [Google Scholar] [PubMed]
- Zhang, C.; Rabouille, C. Membrane-Bound Meet Membraneless in Health and Disease. Cells 2019, 8, 1000. [Google Scholar] [CrossRef] [PubMed]
- Berchtold, D.; Battich, N.; Pelkmans, L. A Systems-Level Study Reveals Regulators of Membrane-less Organelles in Human Cells. Mol. Cell 2018, 72, 1035–1049.e5. [Google Scholar] [CrossRef]
- Aulas, A.; Lyons, S.M.; Fay, M.M.; Anderson, P.; Ivanov, P. Nitric oxide triggers the assembly of “type II” stress granules linked to decreased cell viability. Cell Death Dis. 2018, 9, 1129. [Google Scholar] [CrossRef]
- Aulas, A.; Fay, M.M.; Lyons, S.M.; Achorn, C.A.; Kedersha, N.; Anderson, P.; Ivanov, P. Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci. 2017, 130, 927–937. [Google Scholar] [CrossRef]
- Jevtov, I.; Zacharogianni, M.; van Oorschot, M.M.; van Zadelhoff, G.; Aguilera-Gomez, A.; Vuillez, I.; Braakman, I.; Hafen, E.; Stocker, H.; Rabouille, C. TORC2 mediates the heat stress response in Drosophila by promoting the formation of stress granules. J. Cell Sci. 2015, 128, 2497–2508. [Google Scholar] [CrossRef]
- Khong, A.; Matheny, T.; Jain, S.; Mitchell, S.F.; Wheeler, J.R.; Parker, R. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol. Cell 2017, 68, 808–820.e5. [Google Scholar] [CrossRef] [PubMed]
- Protter, D.S.; Parker, R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016, 26, 668–679. [Google Scholar] [CrossRef]
- Aguilera-Gomez, A.; Zacharogianni, M.; van Oorschot, M.M.; Genau, H.; Grond, R.; Veenendaal, T.; Sinsimer, K.S.; Gavis, E.R.; Behrends, C.; Rabouille, C. Phospho-Rasputin Stabilization by Sec16 Is Required for Stress Granule Formation upon Amino Acid Starvation. Cell Rep. 2017, 20, 935–948. [Google Scholar] [PubMed]
- Zacharogianni, M.; Aguilera-Gomez, A.; Veenendaal, T.; Smout, J.; Rabouille, C. A stress assembly that confers cell viability by preserving ERES components during amino-acid starvation. Elife 2014, 3, e04132. [Google Scholar] [PubMed]
- Zhang, C.; van Leeuwen, W.; Blotenburg, M.; Aguilera-Gomez, A.; Brussee, S.; Grond, R.; Kampinga, H.H.; Rabouille, C. Activation of IRE1, PERK and salt-inducible kinases leads to Sec body formation in Drosophila S2 cells. J. Cell Sci. 2021, 134, jcs258685. [Google Scholar]
- Barlowe, C.; Schekman, R. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature 1993, 365, 347–349. [Google Scholar] [CrossRef] [PubMed]
- Barlowe, C. COPII: A membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 1994, 77, 895–907. [Google Scholar] [CrossRef]
- Ivan, V.; de Voer, G.; Xanthakis, D.; Spoorendonk, K.M.; Kondylis, V.; Rabouille, C. Drosophila Sec16 Mediates the Biogenesis of tER Sites Upstream of Sar1 through an Arginine-Rich Motif. Mol. Biol. Cell 2008, 19, 4352–4365. [Google Scholar] [CrossRef] [PubMed]
- Kondylis, V.; Spoorendonk, K.M.; Rabouille, C. dGRASP Localization and Function in the Early Exocytic Pathway in Drosophila S2 Cells. Mol. Biol. Cell 2005, 16, 4061–4072. [Google Scholar] [CrossRef]
- Stagg, S.M.; Gürkan, C.; Fowler, D.M.; LaPointe, P.; Foss, T.R.; Potter, C.S.; Carragher, B.; Balch, W.E. Structure of the Sec13/31 COPII coat cage. Nature 2006, 439, 234–238. [Google Scholar]
- Gomez-Navarro, N.; Miller, E. Protein sorting at the ER–Golgi interface. J. Cell Biol. 2016, 215, 769–778. [Google Scholar]
- Sprangers, J.; Rabouille, C. SEC16 in COPII coat dynamics at ER exit sites. Biochem. Soc. Trans. 2015, 43, 97–103. [Google Scholar] [CrossRef]
- Connerly, P.L.; Esaki, M.; Montegna, E.A.; Strongin, D.E.; Levi, S.; Soderholm, J.; Glick, B.S. Sec16 is a Determinant of Transitional ER Organization. Curr. Biol. 2005, 15, 1439–1447. [Google Scholar] [CrossRef] [PubMed]
- Farhan, H.; Wendeler, M.W.; Mitrovic, S.; Fava, E.; Silberberg, Y.; Sharan, R.; Zerial, M.; Hauri, H.-P. MAPK signaling to the early secretory pathway revealed by kinase/phosphatase functional screening. J. Cell Biol. 2010, 189, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Hughes, H.; Budnik, A.; Schmidt, K.; Palmer, K.J.; Mantell, J.; Noakes, C.; Johnson, A.; Carter, D.A.; Verkade, P.; Watson, P.; et al. Organisation of human ER-exit sites: Requirements for the localisation of Sec16 to transitional ER. J. Cell Sci. 2009, 122, 2924–2934. [Google Scholar] [CrossRef]
- Joo, J.H.; Wang, B.; Frankel, E.; Ge, L.; Xu, L.; Iyengar, R.; Li-Harms, X.; Wright, C.; Shaw, T.I.; Lindsten, T.; et al. The Noncanonical Role of ULK/ATG1 in ER-to-Golgi Trafficking Is Essential for Cellular Homeostasis. Mol. Cell 2016, 62, 491–506. [Google Scholar] [PubMed]
- Kaiser, C.A.; Schekman, R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 1990, 61, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Madden, D.T.; Hamamoto, S.; Orci, L.; Schekman, R. Sec16p potentiates the action of COPII proteins to bud transport vesicles. J. Cell Biol. 2002, 158, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Wilhelmi, I.; Kanski, R.; Neumann, A.; Herdt, O.; Hoff, F.; Jacob, R.; Preußner, M.; Heyd, F. Sec16 alternative splicing dynamically controls COPII transport efficiency. Nat. Commun. 2016, 7, 12347. [Google Scholar] [CrossRef]
- Weigel, A.V.; Chang, C.L.; Shtengel, G.; Xu, C.S.; Hoffman, D.P.; Freeman, M.; Iyer, N.; Aaron, J.; Khuon, S.; Bogovic, J.; et al. ER-to-Golgi protein delivery through an interwoven, tubular network extending from ER. Cell 2021, 184, 2412–2429.e16. [Google Scholar]
- Shomron, O.; Nevo-Yassaf, I.; Aviad, T.; Yaffe, Y.; Zahavi, E.E.; Dukhovny, A.; Perlson, E.; Brodsky, I.; Yeheskel, A.; Pasmanik-Chor, M.; et al. COPII collar defines the boundary between ER and ER exit site and does not coat cargo containers. J. Cell Biol. 2021, 220, e201907224. [Google Scholar] [CrossRef]
- Aguilera-Gomez, A.; van Oorschot, M.M.; Veenendaal, T.; Rabouille, C. In vivo vizualisation of mono-ADP-ribosylation by dPARP16 upon amino-acid starvation. Elife 2016, 5, e21475. [Google Scholar] [CrossRef]
- Banani, S.F.; Rice, A.M.; Peeples, W.B.; Lin, Y.; Jain, S.; Parker, R.; Rosen, M.K. Compositional Control of Phase-Separated Cellular Bodies. Cell 2016, 166, 651–663. [Google Scholar] [CrossRef]
- Hung, V.; Udeshi, N.D.; Lam, S.S.-M.; Loh, K.H.; Cox, K.J.; Pedram, K.; Carr, S.A.; Ting, V.H.S.S.L.K.H.L.K.J.C.K.P.A.Y. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 2016, 11, 456–475. [Google Scholar] [CrossRef] [PubMed]
- Markmiller, S.; Soltanieh, S.; Server, K.L.; Mak, R.; Jin, W.; Fang, M.Y.; Luo, E.-C.; Krach, F.; Yang, D.; Sen, A.; et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 2018, 172, 590–604.e13. [Google Scholar] [CrossRef] [PubMed]
- Marmor-Kollet, H.; Siany, A.; Kedersha, N.; Knafo, N.; Rivkin, N.; Danino, Y.M.; Moens, T.G.; Olender, T.; Sheban, D.; Cohen, N.; et al. Spatiotemporal Proteomic Analysis of Stress Granule Disassembly Using APEX Reveals Regulation by SUMOylation and Links to ALS Pathogenesis. Mol. Cell 2020, 80, 876–891.e6. [Google Scholar] [CrossRef]
- Babu, M.M.; Kriwacki, R.W.; Pappu, R.V. Versatility from Protein Disorder. Science 2012, 337, 1460–1461. [Google Scholar] [CrossRef] [PubMed]
- Guillén-Boixet, J.; Kopach, A.; Holehouse, A.S.; Wittmann, S.; Jahnel, M.; Schlüßler, R.; Kim, K.; Trussina, I.R.; Wang, J.; Mateju, D.; et al. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 2020, 181, 346–361.e17. [Google Scholar] [CrossRef] [PubMed]
- Zacharogianni, M.; Rabouille, C. Trafficking Along the Secretory Pathway in Drosophila Cell Line and Tissues. In Methods in Cell Biology; Academic Press: Cambridge, MA, USA, 2013; Volume 118, pp. 35–49. [Google Scholar] [CrossRef]
- Seemann, J.; Pypaert, M.; Taguchi, T.; Malsam, J.; Warren, G. Partitioning of the Matrix Fraction of the Golgi Apparatus During Mitosis in Animal Cells. Science 2002, 295, 848–851. [Google Scholar] [CrossRef]
- Riedel, F.; Gillingham, A.K.; Rosa-Ferreira, C.; Galindo, A.; Munro, S. An antibody toolkit for the study of membrane traffic in Drosophila melanogaster. Biol. Open 2016, 5, 987–992. [Google Scholar] [CrossRef] [PubMed]
- Diagana, T.T.; Thomas, U.; Prokopenko, S.N.; Xiao, B.; Worley, P.F.; Thomas, J.B. Mutation of Drosophila homer Disrupts Control of Locomotor Activity and Behavioral Plasticity. J. Neurosci. 2002, 22, 428–436. [Google Scholar] [CrossRef]
- Suyama, R.; Jenny, A.; Curado, S.; Berkel, W.P.-V.; Ephrussi, A. The actin-binding protein Lasp promotes Oskar accumulation at the posterior pole of the Drosophila embryo. Development 2009, 136, 95–105. [Google Scholar] [CrossRef]
- Darbyson, A.; Ngsee, J.K. Oxysterol-binding protein ORP3 rescues the Amyotrophic Lateral Sclerosis-linked mutant VAPB phenotype. Exp. Cell Res. 2016, 341, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.; Chalhoub, A.; Schooley, A.; Zhang, W.; Ngsee, J.K. A mutation in VAPB that causes amyotrophic lateral sclerosis also causes a nuclear envelope defect. J. Cell Sci. 2012, 125, 2831–2836. [Google Scholar] [PubMed]
- Nakamura, N. Emerging New Roles of GM130, a cis-Golgi Matrix Protein, in Higher Order Cell Functions. J. Pharmacol. Sci. 2010, 112, 255–264. [Google Scholar] [CrossRef]
- Pullan, L.; Mullapudi, S.; Huang, Z.; Baldwin, P.R.; Chin, C.; Sun, W.; Tsujimoto, S.; Kolodziej, S.J.; Stoops, J.K.; Lee, J.C.; et al. The Endosome-Associated Protein Hrs Is Hexameric and Controls Cargo Sorting as a “Master Molecule”. Structure 2006, 14, 661–671. [Google Scholar] [CrossRef]
- Luo, P.; Li, X.; Fei, Z.; Poon, W. Scaffold protein Homer 1: Implications for neurological diseases. Neurochem. Int. 2012, 61, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Lepa, C.; Möller-Kerutt, A.; Stölting, M.; Picciotto, C.; Eddy, M.; Butt, E.; Kerjaschki, D.; Korb-Pap, A.; Vollenbröker, B.; Weide, T.; et al. LIM and SH3 protein 1 (LASP-1): A novel link between the slit membrane and actin cytoskeleton dynamics in podocytes. FASEB J. 2020, 34, 5453–5464. [Google Scholar]
- Waris, S.; Wilce, M.C.J.; Wilce, J.A. RNA Recognition and Stress Granule Formation by TIA Proteins. Int. J. Mol. Sci. 2014, 15, 23377–23388. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, C.J.; Dunker, A.K. Intrinsically Disordered Proteins and Intrinsically Disordered Protein Regions. Annu. Rev. Biochem. 2014, 83, 553–584. [Google Scholar] [CrossRef]
- Jain, S.; Wheeler, J.R.; Walters, R.W.; Agrawal, A.; Barsic, A.; Parker, R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 2016, 164, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Han, T.W.; Xie, S.; Shi, K.; Du, X.; Wu, L.C.; Mirzaei, H.; Goldsmith, E.J.; Longgood, J.; Pei, J.; et al. Cell-free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers within Hydrogels. Cell 2012, 149, 753–767. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Kalaitsidou, E.; Damen, J.M.A.; Grond, R.; Rabouille, C.; Wu, W. Novel Components of the Stress Assembly Sec Body Identified by Proximity Labeling. Cells 2023, 12, 1055. https://doi.org/10.3390/cells12071055
Zhang C, Kalaitsidou E, Damen JMA, Grond R, Rabouille C, Wu W. Novel Components of the Stress Assembly Sec Body Identified by Proximity Labeling. Cells. 2023; 12(7):1055. https://doi.org/10.3390/cells12071055
Chicago/Turabian StyleZhang, Chujun, Elisavet Kalaitsidou, J. Mirjam A. Damen, Rianne Grond, Catherine Rabouille, and Wei Wu. 2023. "Novel Components of the Stress Assembly Sec Body Identified by Proximity Labeling" Cells 12, no. 7: 1055. https://doi.org/10.3390/cells12071055
APA StyleZhang, C., Kalaitsidou, E., Damen, J. M. A., Grond, R., Rabouille, C., & Wu, W. (2023). Novel Components of the Stress Assembly Sec Body Identified by Proximity Labeling. Cells, 12(7), 1055. https://doi.org/10.3390/cells12071055




