Application of Digital Holographic Microscopy to Analyze Changes in T-Cell Morphology in Response to Bacterial Challenge
Abstract
1. Introduction
2. Materials and Methods
2.1. T-Cell Isolation
2.2. Bacterial Strains and Culture Conditions
2.3. Preparation of Bacterial MVs
2.4. Preparation of Sterile Culture Supernatants
2.5. In Vitro Infection
2.6. Digital Holographic Microscopy (DHM)
3. Results
3.1. Strain- and Species-Dependent Morphological Changes in Reaction to Bacterial Culture Supernatants
3.2. T-Cell Response to S. aureus MVs Depends on the Strain
3.3. Cellular Changes in Response to Bacterial Stress Are Concentration-Dependent
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singer, M.; Deutschman, C.S.; Seymour, C.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA—J. Am. Med. Assoc. 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, Regional, and National Sepsis Incidence and Mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef]
- Barichello, T.; Generoso, J.S.; Singer, M.; Dal-Pizzol, F. Biomarkers for Sepsis: More than Just Fever and Leukocytosis—A Narrative Review. Crit. Care 2022, 26, 14. [Google Scholar] [CrossRef] [PubMed]
- Pierrakos, C.; Velissaris, D.; Bisdorff, M.; Marshall, J.C.; Vincent, J.-L. Biomarkers of Sepsis: Time for a Reappraisal. Crit. Care 2020, 24, 287. [Google Scholar] [CrossRef] [PubMed]
- Mammen, J.; Choudhuri, J.; Paul, J.; Sudarsan, T.I.; Josephine, T.; Mahasampath, G.; Jeyaseelan, V.; Nair, S.C.; Peter, J.V. Cytomorphometric Neutrophil and Monocyte Markers May Strengthen the Diagnosis of Sepsis. J. Intensive Care Med. 2018, 33, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Urrechaga, E.; Bóveda, O.; Aguirre, U. Improvement in Detecting Sepsis Using Leukocyte Cell Population Data (CPD). Clin. Chem. Lab. Med. 2019, 57, 918–926. [Google Scholar] [CrossRef]
- Arora, P.; Gupta, P.K.; Lingaiah, R.; Mukhopadhyay, A.K. Volume, Conductivity, and Scatter Parameters of Leukocytes as Early Markers of Sepsis and Treatment Response. J. Lab. Physicians 2019, 11, 29–33. [Google Scholar] [CrossRef]
- Shalini, P.; Rao, P.S.; Rao, S.; Anil, M.; Benny, A. Diagnostic Utility of Cell Population Data (CPD) in Sepsis Using Automated Hematology Analysers. Ann. Pathol. Lab. Med. 2019, 6, 284. [Google Scholar] [CrossRef]
- Zonneveld, R.; Molema, G.; Plötz, F.B. Analyzing Neutrophil Morphology, Mechanics, and Motility in Sepsis. Crit. Care Med. 2016, 44, 218–228. [Google Scholar] [CrossRef]
- Park, Y.K.; Depeursinge, C.; Popescu, G. Quantitative Phase Imaging in Biomedicine. Nat. Photonics 2018, 12, 578–589. [Google Scholar] [CrossRef]
- Lee, K.; Kim, K.; Jung, J.; Heo, J.; Cho, S.; Lee, S.; Chang, G.; Jo, Y.; Park, H.; Park, Y. Quantitative Phase Imaging Techniques for the Study of Cell Pathophysiology: From Principles to Applications. Sensors 2013, 13, 4170–4191. [Google Scholar] [CrossRef] [PubMed]
- Kemper, B.; Von Bally, G. Digital Holographic Microscopy for Live Cell Applications and Technical Inspection. Appl. Opt. 2008, 47, A52–A61. [Google Scholar] [CrossRef] [PubMed]
- El-Schich, Z.; Janicke, B.; Alm, K.; Dizeyi, N.; Persson, L.J.; Gjörloff Wingren, A. Discrimination between Breast Cancer Cells and White Blood Cells by Non-Invasive Measurements: Implications for a Novel in Vitro-Based Circulating Tumor Cell Model Using Digital Holographic Cytometry. Appl. Sci. 2020, 10, 4854. [Google Scholar] [CrossRef]
- Benzerdjeb, N.; Garbar, C.; Camparo, P.; Sevestre, H. Digital Holographic Microscopy as Screening Tool for Cervical Cancer Preliminary Study. Cancer Cytopathol. 2016, 124, 573–580. [Google Scholar] [CrossRef]
- O’Connor, T.; Anand, A.; Andemariam, B.; Javidi, B. Overview of Cell Motility-Based Sickle Cell Disease Diagnostic System in Shearing Digital Holographic Microscopy. JPhys Photonics 2020, 2, 031002. [Google Scholar] [CrossRef]
- Ugele, M.; Weniger, M.; Leidenberger, M.; Huang, Y.; Bassler, M.; Friedrich, O.; Kappes, B.; Hayden, O.; Richter, L. Label-Free, High-Throughput Detection of P. Falciparum Infection in Sphered Erythrocytes with Digital Holographic Microscopy. Lab Chip 2018, 18, 1704–1712. [Google Scholar] [CrossRef]
- O’Connor, T.; Santaniello, S.; Javidi, B. COVID-19 Detection from Red Blood Cells Using Highly Comparative Time-Series Analysis (HCTSA) in Digital Holographic Microscopy. Opt. Express 2022, 30, 1723–1736. [Google Scholar] [CrossRef]
- Urrechaga, E. Reviewing the Value of Leukocytes Cell Population Data (CPD) in the Management of Sepsis. Ann. Transl. Med. 2020, 8, 953. [Google Scholar] [CrossRef]
- Kunsmann, L.; Rüter, C.; Bauwens, A.; Greune, L.; Glüder, M.; Kemper, B.; Fruth, A.; Wai, S.N.; He, X.; Lloubes, R.; et al. Virulence from Vesicles: Novel Mechanisms of Host Cell Injury by Escherichia Coli O104:H4 Outbreak Strain. Sci. Rep. 2015, 5, 13252. [Google Scholar] [CrossRef]
- Bauwens, A.; Bielaszewska, M.; Kemper, B.; Langehanenberg, P.; Von Bally, G.; Reichelt, R.; Mulac, D.; Humpf, H.U.; Friedrich, A.W.; Kim, K.S.; et al. Differential Cytotoxic Actions of Shiga Toxin 1 and Shiga Toxin 2 on Microvascular and Macrovascular Endothelial Cells. Thromb. Haemost. 2011, 105, 515–528. [Google Scholar] [CrossRef]
- Sela, U.; Euler, C.W.; Correa da Rosa, J.; Fischetti, V.A. Strains of Bacterial Species Induce a Greatly Varied Acute Adaptive Immune Response: The Contribution of the Accessory Genome. PLoS Pathog. 2018, 14, e1006726. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, F.R.; McLaren, J.E. T Cell Immunity to Bacterial Pathogens: Mechanisms of Immune Control and Bacterial Evasion. Int. J. Mol. Sci. 2020, 21, 6144. [Google Scholar] [CrossRef]
- vom Werth, K.L.; Wörmann, T.; Kemper, B.; Kümpers, P.; Kampmeier, S.; Mellmann, A. Investigating Morphological Changes of T-Lymphocytes after Exposure with Bacterial Determinants for Early Detection of Septic Conditions. Microorganisms 2022, 10, 391. [Google Scholar] [CrossRef]
- Sakr, Y.; Jaschinski, U.; Wittebole, X.; Szakmany, T.; Lipman, J.; Ñamendys-Silva, S.A.; Martin-Loeches, I.; Leone, M.; Lupu, M.N.; Vincent, J.-L. Sepsis in Intensive Care Unit Patients: Worldwide Data from the Intensive Care over Nations Audit. Open Forum Infect. Dis. 2018, 5, ofy313. [Google Scholar] [CrossRef] [PubMed]
- GBD 2019 Antimicrobial Resistance Collaborators. Global Mortality Associated with 33 Bacterial Pathogens in 2019: A Systematic Analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 2248. [Google Scholar] [CrossRef]
- Ramachandran, G. Gram-Positive and Gram-Negative Bacterial Toxins in Sepsis: A Brief Review. Virulence 2014, 5, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, C.K.; Meysick, K.C.; O’Brien, A.D. Bacterial Toxins: Friends or Foes? Emerg. Infect. Dis. 1999, 5, 224–234. [Google Scholar] [CrossRef]
- Macion, A.; Wyszyńska, A.; Godlewska, R. Delivery of Toxins and Effectors by Bacterial Membrane Vesicles. Toxins (Basel). 2021, 13, 845. [Google Scholar] [CrossRef]
- Kaparakis-Liaskos, M.; Ferrero, R.L. Immune Modulation by Bacterial Outer Membrane Vesicles. Nat. Rev. Immunol. 2015, 15, 375–387. [Google Scholar] [CrossRef]
- Tulkens, J.; Vergauwen, G.; Van Deun, J.; Geeurickx, E.; Dhondt, B.; Lippens, L.; De Scheerder, M.A.; Miinalainen, I.; Rappu, P.; De Geest, B.G.; et al. Increased Levels of Systemic LPS-Positive Bacterial Extracellular Vesicles in Patients with Intestinal Barrier Dysfunction. Gut 2020, 69, 191–193. [Google Scholar] [CrossRef]
- Peng, Y.; Gao, M.; Liu, Y.; Qiu, X.; Cheng, X.; Yang, X.; Chen, F.; Wang, E. Bacterial Outer Membrane Vesicles Induce Disseminated Intravascular Coagulation through the Caspase-11-Gasdermin D Pathway. Thromb. Res. 2020, 196, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Svennerholm, K.; Park, K.S.; Wikström, J.; Lässer, C.; Crescitelli, R.; Shelke, G.V.; Jang, S.C.; Suzuki, S.; Bandeira, E.; Olofsson, C.S.; et al. Escherichia Coli Outer Membrane Vesicles Can Contribute to Sepsis Induced Cardiac Dysfunction. Sci. Rep. 2017, 7, 17434. [Google Scholar] [CrossRef]
- Vann, J.M.; Proctor, R.A. Ingestion of Staphylococcus Aureus by Bovine Endothelial Cells Results in Time- and Inoculum-Dependent Damage to Endothelial Cell Monolayers. Infect. Immun. 1987, 55, 2155–2163. [Google Scholar] [CrossRef] [PubMed]
- Tenover, F.C.; Goering, R.V. Methicillin-Resistant Staphylococcus Aureus Strain USA300: Origin and Epidemiology. J. Antimicrob. Chemother. 2009, 64, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Planet, P.J. Life after USA300: The Rise and Fall of a Superbug. J. Infect. Dis. 2017, 215, S71–S77. [Google Scholar] [CrossRef] [PubMed]
- Witte, W.; Strommenger, B.; Stanek, C.; Cuny, C. Methicillin-Resistant Staphylococcus Aureus ST398 in Humans and Animals, Central Europe. Emerg. Infect. Dis. 2007, 13, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Berger, H.; Hacker, J.; Juarez, A.; Hughes, C.; Goebel, W. Cloning of the Chromosomal Determinants Encoding Hemolysin Production and Mannose-Resistant Hemagglutination in Escherichia Coli. J. Bacteriol. 1982, 152, 1241–1247. [Google Scholar] [CrossRef]
- Korhonen, T.K.; Valtonen, M.V.; Parkkinen, J.; Väisänen-Rhen, V.; Finne, J.; Orskov, F.; Orskov, I.; Svenson, S.B.; Helena Mäkelä, P. Serotypes, Hemolysin Production, and Receptor Recognition of Escherichia Coli Strains Associated with Neonatal Sepsis and Meningitis. Infect. Immun. 1985, 48, 486–491. [Google Scholar] [CrossRef]
- Bachmann, B.J. Derivations and genotypes of some mutant derivatives of Escherichia coli K12. In Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd ed.; Neidhardt FCCurtiss RIngraham JLLin ECCLow KBMagasanik BReznikoff WSRiley MSchaechter MUmbarger, H.E., Ed.; ASM Press: Washington, DC, USA, 1996; pp. 2460–2488. [Google Scholar]
- Bielaszewska, M.; Rüter, C.; Kunsmann, L.; Greune, L.; Bauwens, A.; Zhang, W.; Kuczius, T.; Kim, K.S.; Mellmann, A.; Schmidt, M.A.; et al. Enterohemorrhagic Escherichia Coli Hemolysin Employs Outer Membrane Vesicles to Target Mitochondria and Cause Endothelial and Epithelial Apoptosis. PLoS Pathog. 2013, 9, e1003797. [Google Scholar] [CrossRef]
- Kemper, B.; Barroso, Á.; Eder, K.; Marzi, A.; Ritz, S.; Ntovas, A.; Schnekenburger, J.; Ketelhut, S. Enhanced Quantitative Phase Imaging in Mach-Zehnder Interferometer-Based Digital Holographic Microscopy by Modulation of the Object Illumination with an Electrically Focus Tunable Lens. Proc. SPIE 2021, 11786, 117860I. [Google Scholar] [CrossRef]
- Kemper, B.; Carl, D.; Schnekenburger, J.; Bredebusch, I.; Schäfer, M.; Domschke, W.; von Bally, G. Investigation of Living Pancreas Tumor Cells by Digital Holographic Microscopy. J. Biomed. Opt. 2006, 11, 034005. [Google Scholar] [CrossRef] [PubMed]
- Min, J.; Yao, B.; Ketelhut, S.; Engwer, C.; Greve, B.; Kemper, B. Simple and Fast Spectral Domain Algorithm for Quantitative Phase Imaging of Living Cells with Digital Holographic Microscopy. Opt. Lett. 2017, 42, 230. [Google Scholar] [CrossRef] [PubMed]
- Kastl, L.; Isbach, M.; Dirksen, D.; Schnekenburger, J.; Kemper, B. Quantitative Phase Imaging for Cell Culture Quality Control. Cytom. Part A 2017, 91, 470–481. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, W.J.; Johnston, E.L.; Zavan, L.; Bitto, N.J.; Kaparakis-Liaskos, M. Immunomodulatory Roles and Novel Applications of Bacterial Membrane Vesicles. Mol. Immunol. 2021, 134, 72–85. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A Reference Database for Bacterial Virulence Factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed]
- van der Poll, T.; Opal, S.M. Host-pathogen interactions in sepsis. In The Lancet Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2008; pp. 32–43. [Google Scholar] [CrossRef]
- Strobel, M.; Pförtner, H.; Tuchscherr, L.; Völker, U.; Schmidt, F.; Kramko, N.; Schnittler, H.J.; Fraunholz, M.J.; Löffler, B.; Peters, G.; et al. Post-Invasion Events after Infection with Staphylococcus Aureus Are Strongly Dependent on Both the Host Cell Type and the Infecting S. Aureus Strain. Clin. Microbiol. Infect. 2016, 22, 799–809. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, X.; Rao, X. Apoptosis Induced by Staphylococcus Aureus Toxins. Microbiol. Res. 2017, 205, 19–24. [Google Scholar] [CrossRef]
- Nygaard, T.K.; Pallister, K.B.; DuMont, A.L.; DeWald, M.; Watkins, R.L.; Pallister, E.Q.; Malone, C.; Griffith, S.; Horswill, A.R.; Torres, V.J.; et al. Alpha-Toxin Induces Programmed Cell Death of Human T Cells, B Cells, and Monocytes during USA 300 Infection. PLoS ONE 2012, 7, e36532. [Google Scholar] [CrossRef]
- King, J.M.; Kulhankova, K.; Stach, C.S.; Vu, B.G.; Salgado-Pabón, W. Phenotypes and Virulence among Staphylococcus Aureus USA100, USA200, USA300, USA400, and USA600 Clonal Lineages. mSphere 2016, 1, e00071-16. [Google Scholar] [CrossRef]
- Jorgensen, I.; Rayamajhi, M.; Miao, E.A. Programmed Cell Death as a Defence against Infection. Nat. Rev. Immunol. 2017, 17, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, U.; Groscurth, P. Morphological Features of Cell Death. Physiology 2004, 19, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Verduijn, J.; Van der Meeren, L.; Krysko, D.V.; Skirtach, A.G. Deep Learning with Digital Holographic Microscopy Discriminates Apoptosis and Necroptosis. Cell Death Discov. 2021, 7, 229. [Google Scholar] [CrossRef] [PubMed]
- Piasecka, B.; Duffy, D.; Urrutia, A.; Quach, H.; Patin, E.; Posseme, C.; Bergstedt, J.; Charbit, B.; Rouilly, V.; MacPherson, C.R.; et al. Distinctive Roles of Age, Sex, and Genetics in Shaping Transcriptional Variation of Human Immune Responses to Microbial Challenges. Proc. Natl. Acad. Sci. USA 2018, 115, E488–E497. [Google Scholar] [CrossRef]
- Macia, L.; Nanan, R.; Hosseini-Beheshti, E.; Grau, G.E. Host-and Microbiota-Derived Extracellular Vesicles, Immune Function, and Disease Development. Int. J. Mol. Sci. 2020, 21, 107. [Google Scholar] [CrossRef]
- Thay, B.; Wai, S.N.; Oscarsson, J. Staphylococcus Aureus α-Toxin-Dependent Induction of Host Cell Death by Membrane-Derived Vesicles. PLoS ONE 2013, 8, e54661. [Google Scholar] [CrossRef]
- Jin, J.S.; Kwon, S.-O.; Moon, D.C.; Gurung, M.; Lee, J.H.; Kim, S.I.; Lee, J.C. Acinetobacter Baumannii Secretes Cytotoxic Outer Membrane Protein A via Outer Membrane Vesicles. PLoS ONE 2011, 6, e17027. [Google Scholar] [CrossRef]
- Jones, E.J.; Booth, C.; Fonseca, S.; Parker, A.; Cross, K.; Miquel-Clopés, A.; Hautefort, I.; Mayer, U.; Wileman, T.; Stentz, R.; et al. The Uptake, Trafficking, and Biodistribution of Bacteroides Thetaiotaomicron Generated Outer Membrane Vesicles. Front. Microbiol. 2020, 11, 57. [Google Scholar] [CrossRef]
- Nagakubo, T.; Nomura, N.; Toyofuku, M. Cracking Open Bacterial Membrane Vesicles. Front. Microbiol. 2020, 10, 3026. [Google Scholar] [CrossRef]
- Jeon, H.; Oh, M.H.; Jun, S.H.; Kim, S.I.; Choi, C.W.; Kwon, H.I.; Na, S.H.; Kim, Y.J.; Nicholas, A.; Selasi, G.N.; et al. Variation among Staphylococcus Aureus Membrane Vesicle Proteomes Affects Cytotoxicity of Host Cells. Microb. Pathog. 2016, 93, 185–193. [Google Scholar] [CrossRef]
- Mehanny, M.; Koch, M.; Lehr, C.-M.; Fuhrmann, G. Streptococcal Extracellular Membrane Vesicles Are Rapidly Internalized by Immune Cells and Alter Their Cytokine Release. Front. Immunol. 2020, 11, 80. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Lee, E.J.; Lee, J.H.; Jun, S.H.; Choi, C.W.; Kim, S.I.; Kang, S.S.; Hyun, S. Klebsiella Pneumoniae Secretes Outer Membrane Vesicles That Induce the Innate Immune Response. FEMS Microbiol. Lett. 2012, 331, 17–24. [Google Scholar] [CrossRef] [PubMed]
- van der Poll, T.; Shankar-Hari, M.; Wiersinga, W.J. The Immunology of Sepsis. Immunity 2021, 54, 2450–2464. [Google Scholar] [CrossRef] [PubMed]
- Caramalho, I.; Lopes-Carvalho, T.; Ostler, D.; Zelenay, S.; Haury, M.; Demengeot, J. Regulatory T Cells Selectively Express Toll-like Receptors and Are Activated by Lipopolysaccharide. J. Exp. Med. 2003, 197, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhang, L.; Zhao, Y. Modulation of Immune Responses through Direct Activation of Toll-like Receptors to T Cells. Clin. Exp. Immunol. 2010, 160, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Piva, E.; Zuin, J.; Pelloso, M.; Tosato, F.; Fogar, P.; Plebani, M. Monocyte Distribution Width (MDW) Parameter as a Sepsis Indicator in Intensive Care Units. Clin. Chem. Lab. Med. 2021, 59, 1307–1314. [Google Scholar] [CrossRef]
- Paoli, C.J.; Reynolds, M.A.; Sinha, M.; Gitlin, M.; Crouser, E. Epidemiology and Costs of Sepsis in the United States-an Analysis Based on Timing of Diagnosis and Severity Level. Crit. Care Med. 2018, 46, 1889–1897. [Google Scholar] [CrossRef]
- Liu, V.X.; Fielding-Singh, V.; Greene, J.D.; Baker, J.M.; Iwashyna, T.J.; Bhattacharya, J.; Escobar, G.J. The Timing of Early Antibiotics and Hospital Mortality in Sepsis. Am. J. Respir. Crit. Care Med. 2017, 196, 856–863. [Google Scholar] [CrossRef]
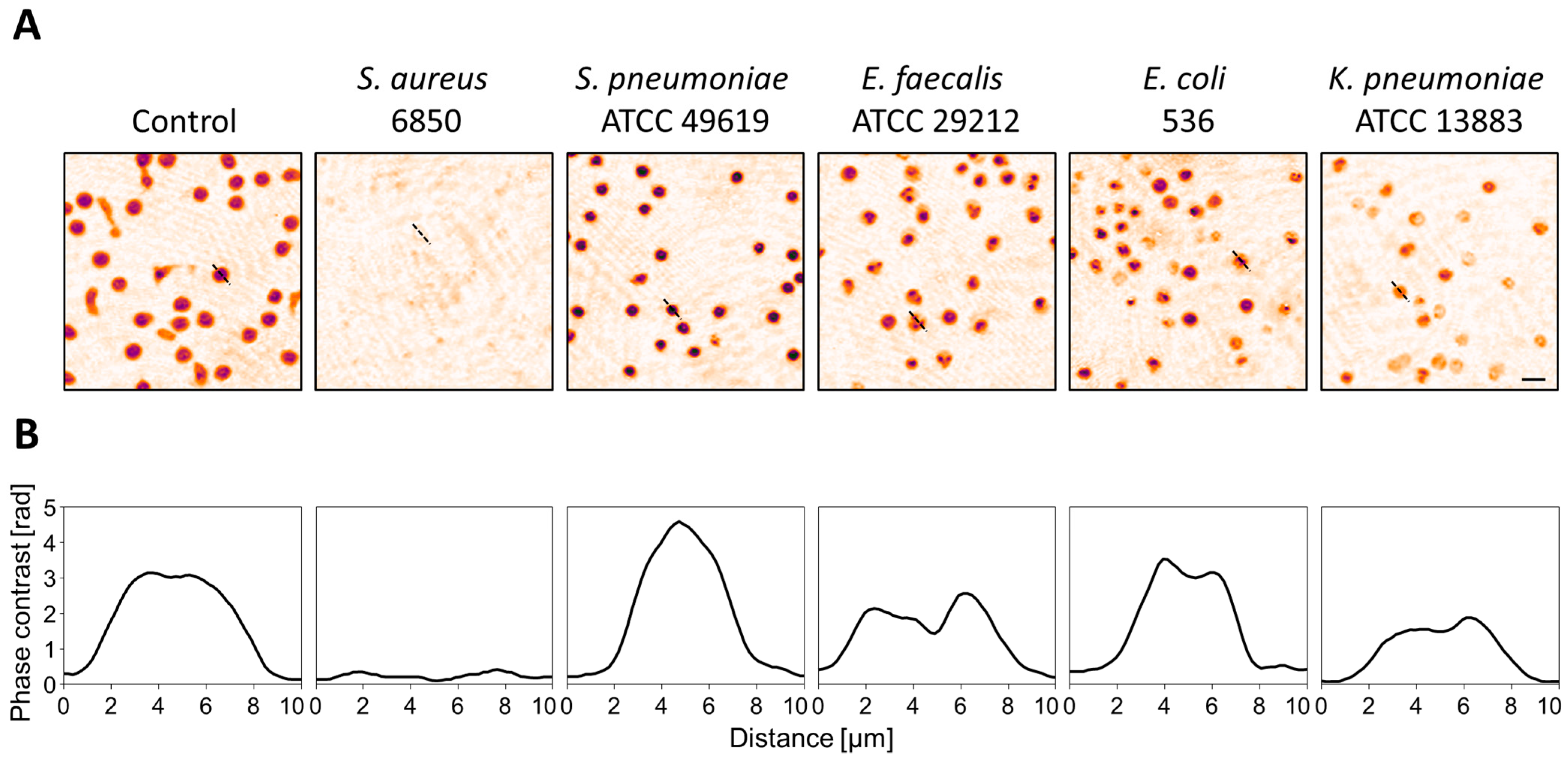
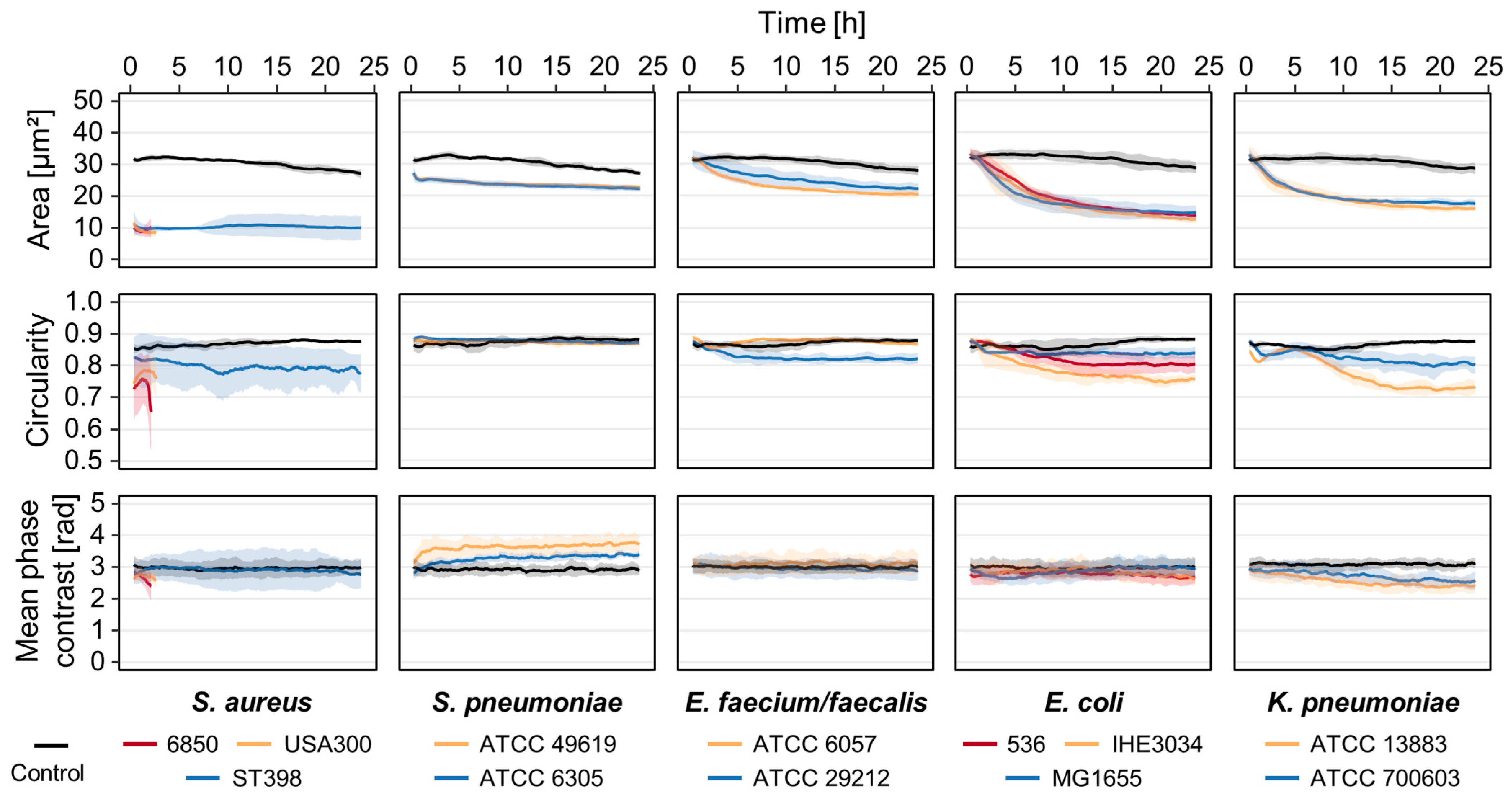
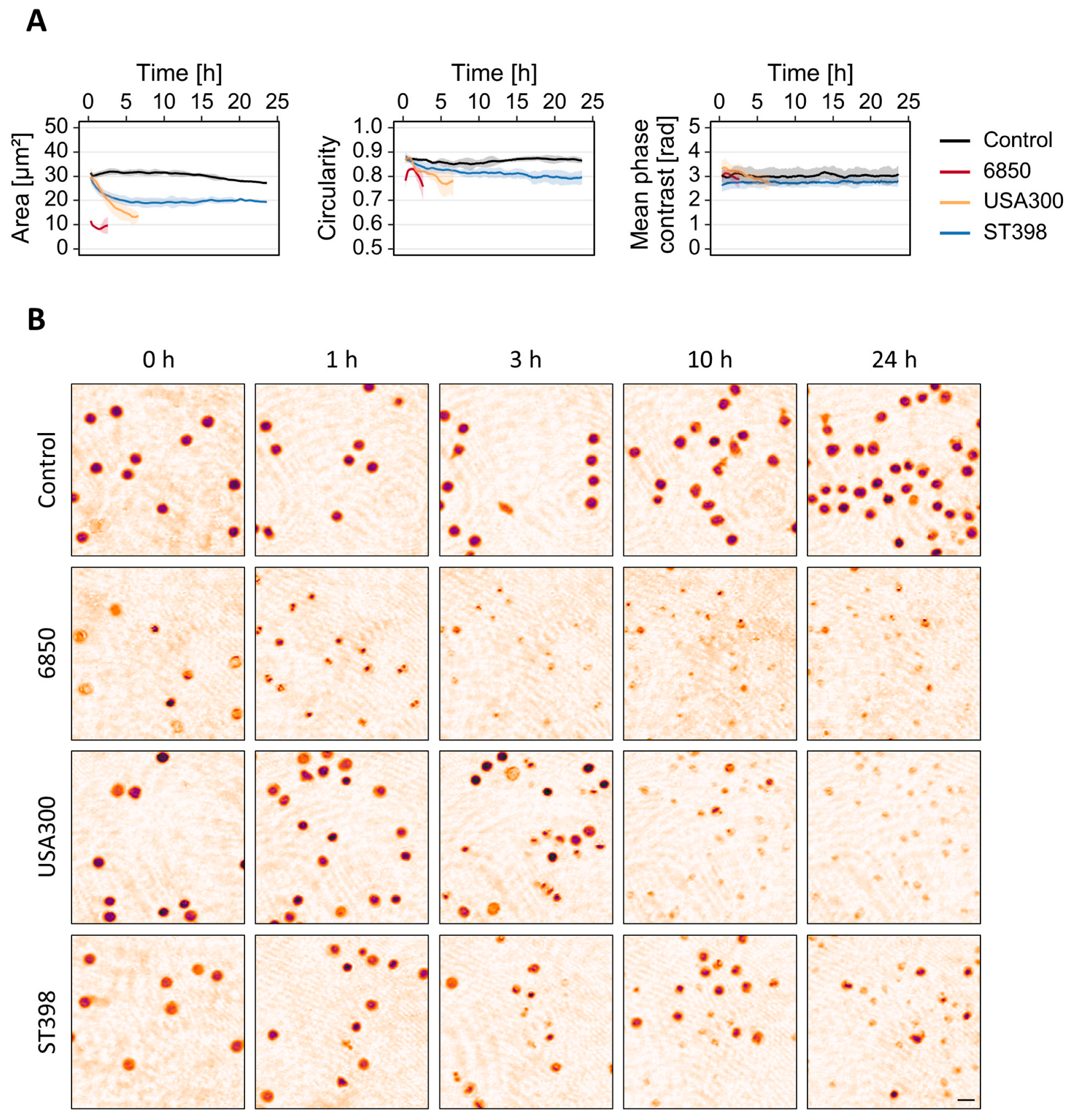

| Species | Strain | Reference |
|---|---|---|
| S. aureus | 6850 | [33] |
| USA300 | [34,35] | |
| ST398 | [36] | |
| S. pneumoniae | ATCC 49619 | ATCC 49619 |
| ATCC 6305 | ATCC 6305 | |
| E. faecium | ATCC 6057 | ATCC 6057 |
| E. faecalis | ATCC 29212 | ATCC 29212 |
| E. coli | 536 | [37] |
| IHE3034 | [38] | |
| MG1655 | [39] | |
| K. pneumoniae | ATCC 13883 | ATCC 13883 |
| ATCC 700603 | ATCC 700603 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
vom Werth, K.L.; Kemper, B.; Kampmeier, S.; Mellmann, A. Application of Digital Holographic Microscopy to Analyze Changes in T-Cell Morphology in Response to Bacterial Challenge. Cells 2023, 12, 762. https://doi.org/10.3390/cells12050762
vom Werth KL, Kemper B, Kampmeier S, Mellmann A. Application of Digital Holographic Microscopy to Analyze Changes in T-Cell Morphology in Response to Bacterial Challenge. Cells. 2023; 12(5):762. https://doi.org/10.3390/cells12050762
Chicago/Turabian Stylevom Werth, Kari Lavinia, Björn Kemper, Stefanie Kampmeier, and Alexander Mellmann. 2023. "Application of Digital Holographic Microscopy to Analyze Changes in T-Cell Morphology in Response to Bacterial Challenge" Cells 12, no. 5: 762. https://doi.org/10.3390/cells12050762
APA Stylevom Werth, K. L., Kemper, B., Kampmeier, S., & Mellmann, A. (2023). Application of Digital Holographic Microscopy to Analyze Changes in T-Cell Morphology in Response to Bacterial Challenge. Cells, 12(5), 762. https://doi.org/10.3390/cells12050762







