Role of Deubiquitinases in Parkinson’s Disease—Therapeutic Perspectives
Abstract
1. Introduction
1.1. Parkinson’s Disease
1.2. Protein Degradation Pathways
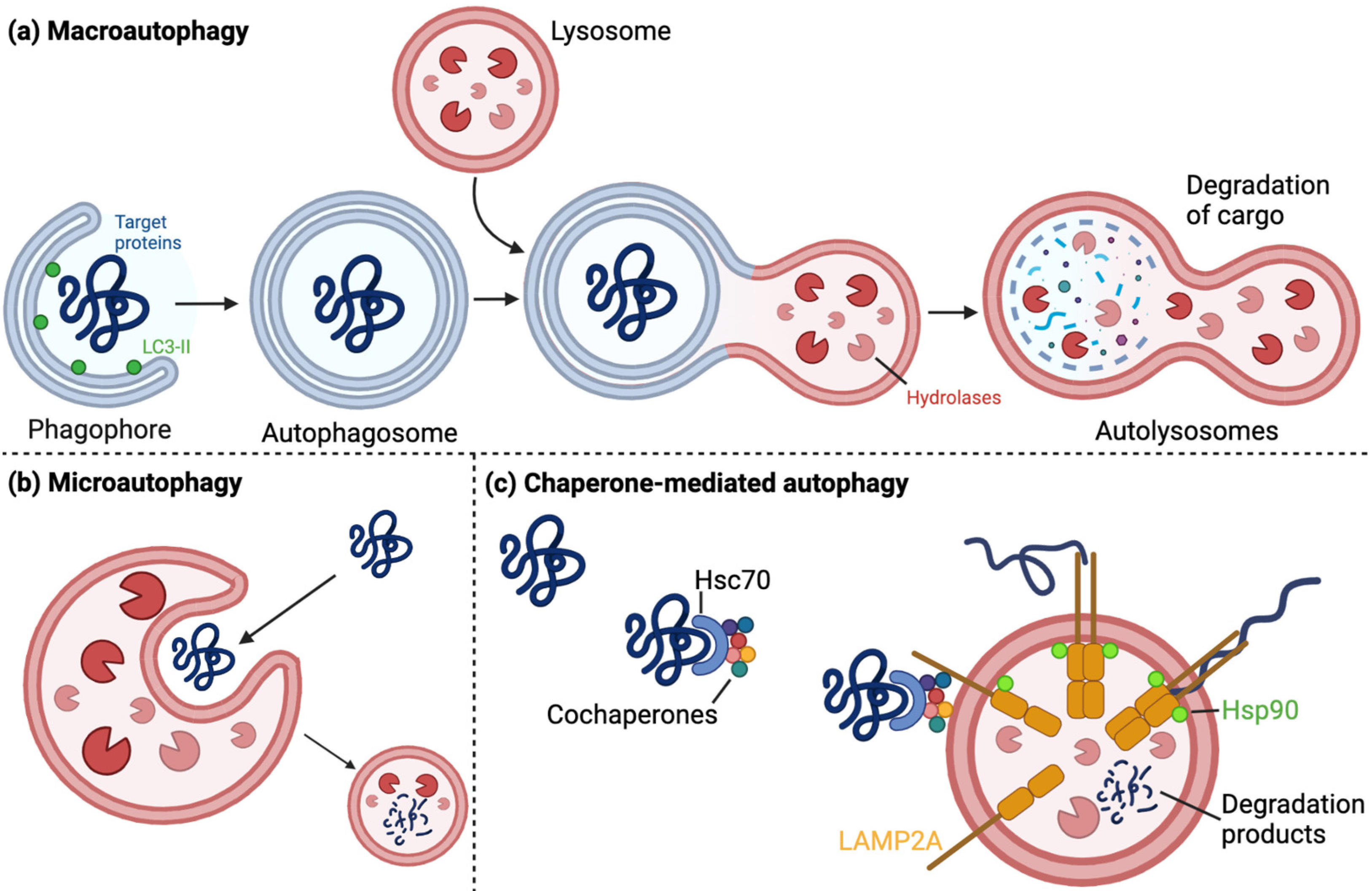
1.2.1. The Autophagy–Lysosomal Pathway
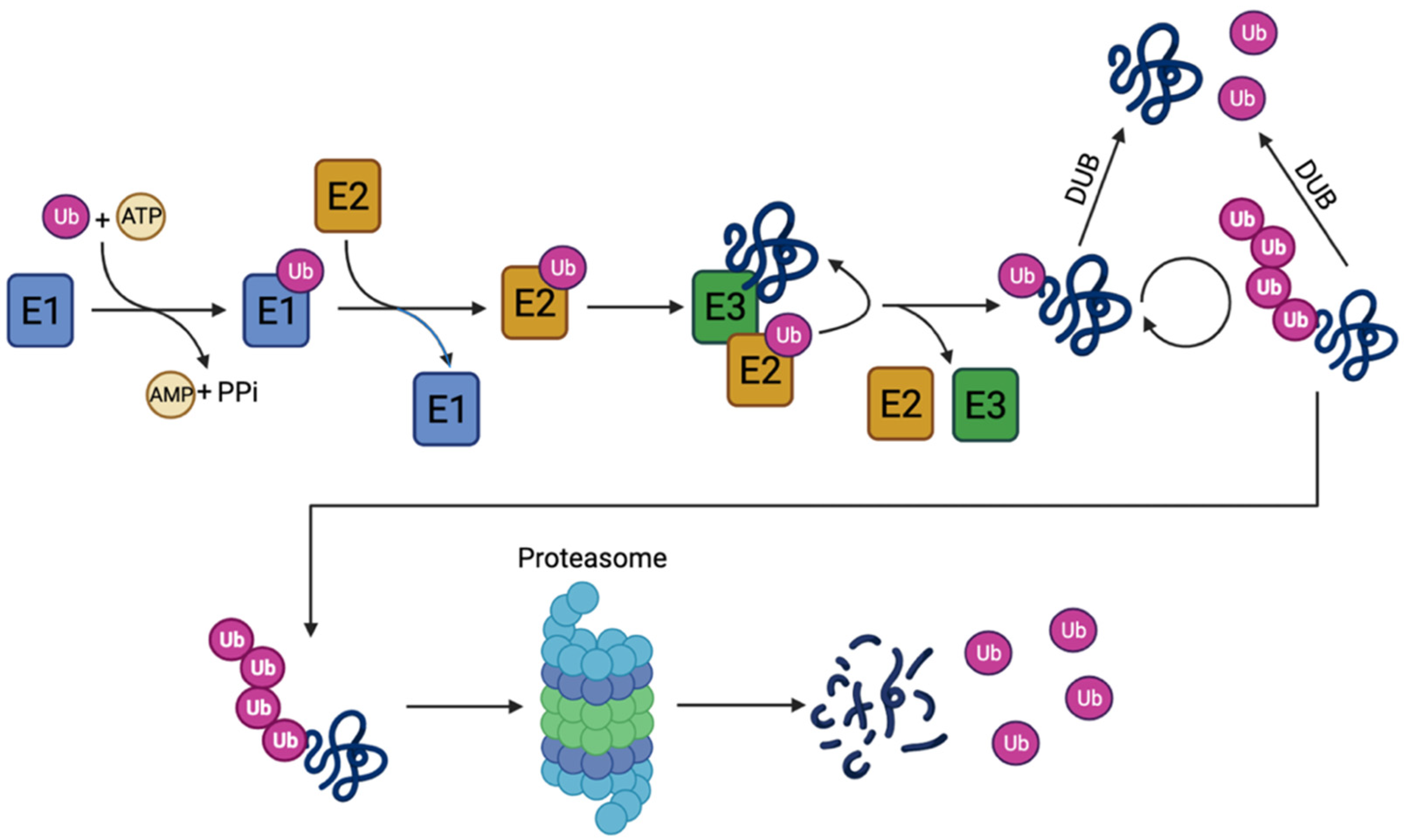
1.2.2. The Ubiquitin–Proteasome Pathway
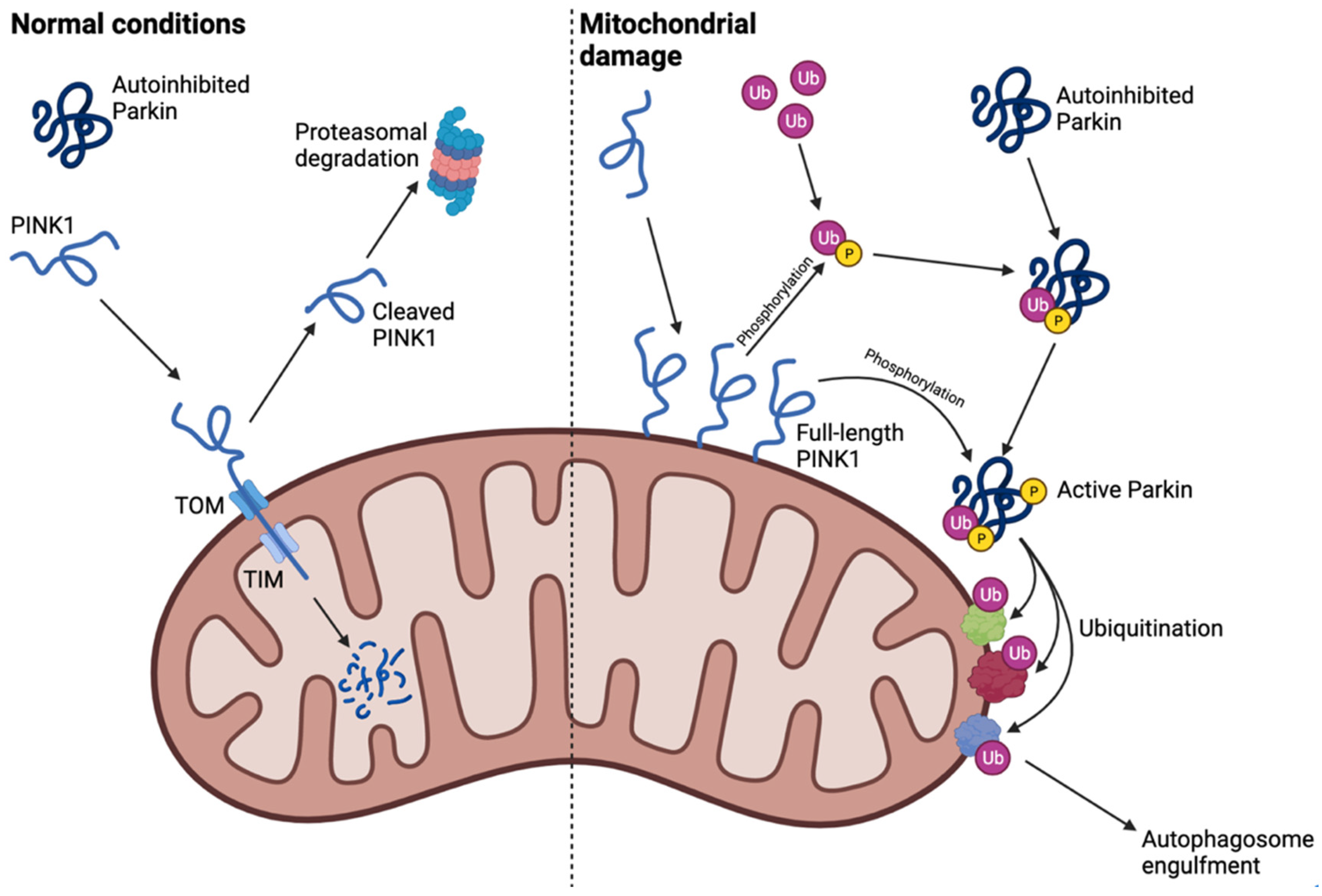
1.2.3. Mitophagy: A Crosstalk between Autophagy and the Ubiquitin–Proteasome System
2. Parkinson’s Disease-Associated Deubiquitinating Enzymes
2.1. USP15 Deubiquitinates Parkin Substrates and Inhibits Mitophagy
2.2. USP30 Inhibits PINK1/Parkin-Dependent Mitophagy by Deubiquitinating Mitochondrial Proteins
2.3. USP33 Deubiquitinates K63-Linked Ubiquitin on Parkin at Lys435
2.4. USP35 Inhibits Parkin-Mediated Mitophagy through Mfn2 Stabilization
2.5. USP8 Regulates Parkin Degradation and Translocation, α-Synuclein Clearance, and Mfn Levels
2.6. Ataxin-3 Is a Machado-Joseph Disease-Associated Protein That Inhibits Parkin Self-Ubiquitination by Diverting Ubiquitin onto Itself
2.7. USP13 KD Protects Dopaminergic Neurons against α-Synuclein-Induced Neuronal Death
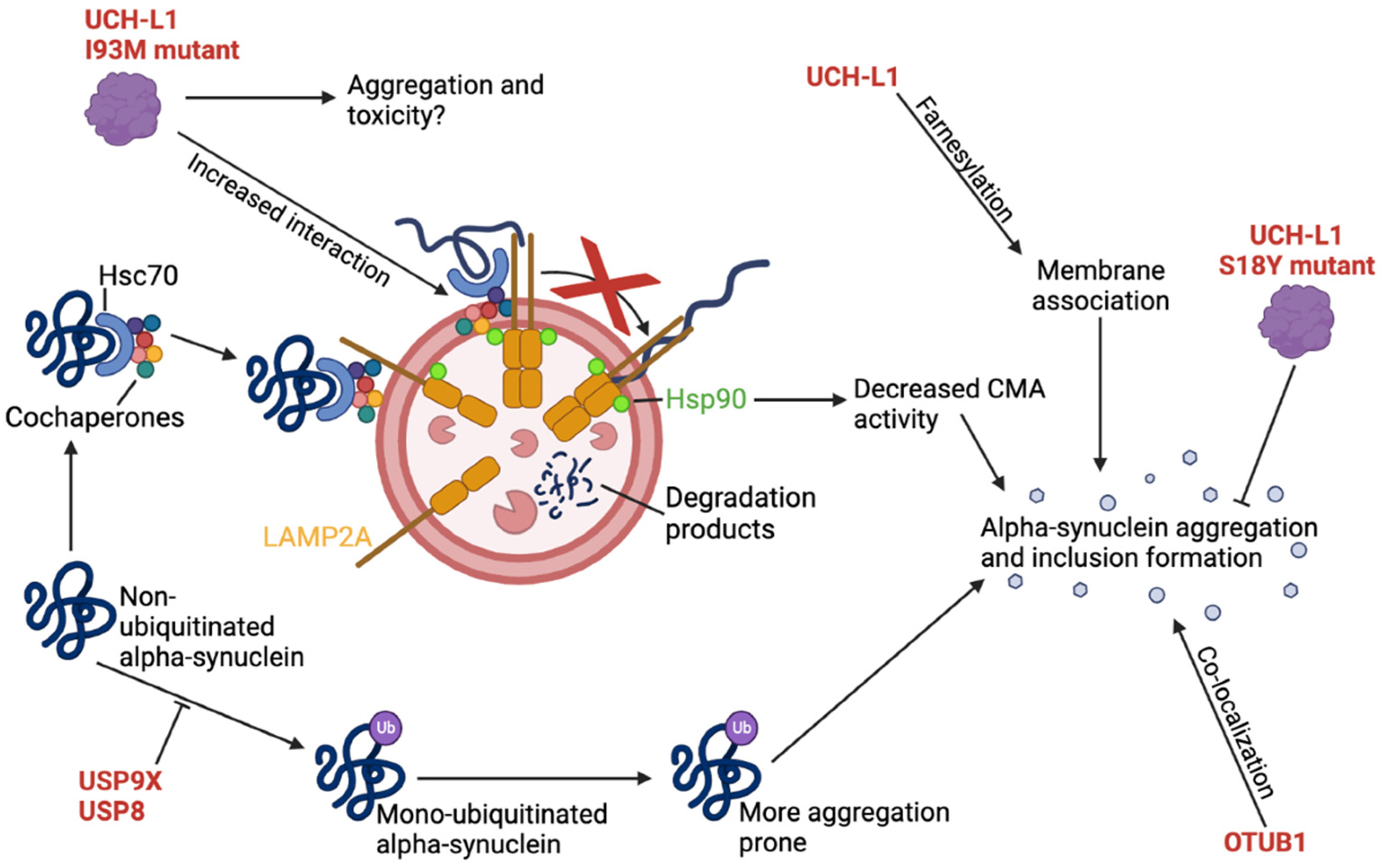
2.8. UCH-L1 I93M Mutant Inhibits CMA-Mediated Degradation of α-Synuclein
2.9. USP9X Deubiquitinates Mono-Ubiquitinated α-Synuclein
2.10. USP14 Is a Negative Regulator of the Proteasome, Autophagy, and Mitophagy
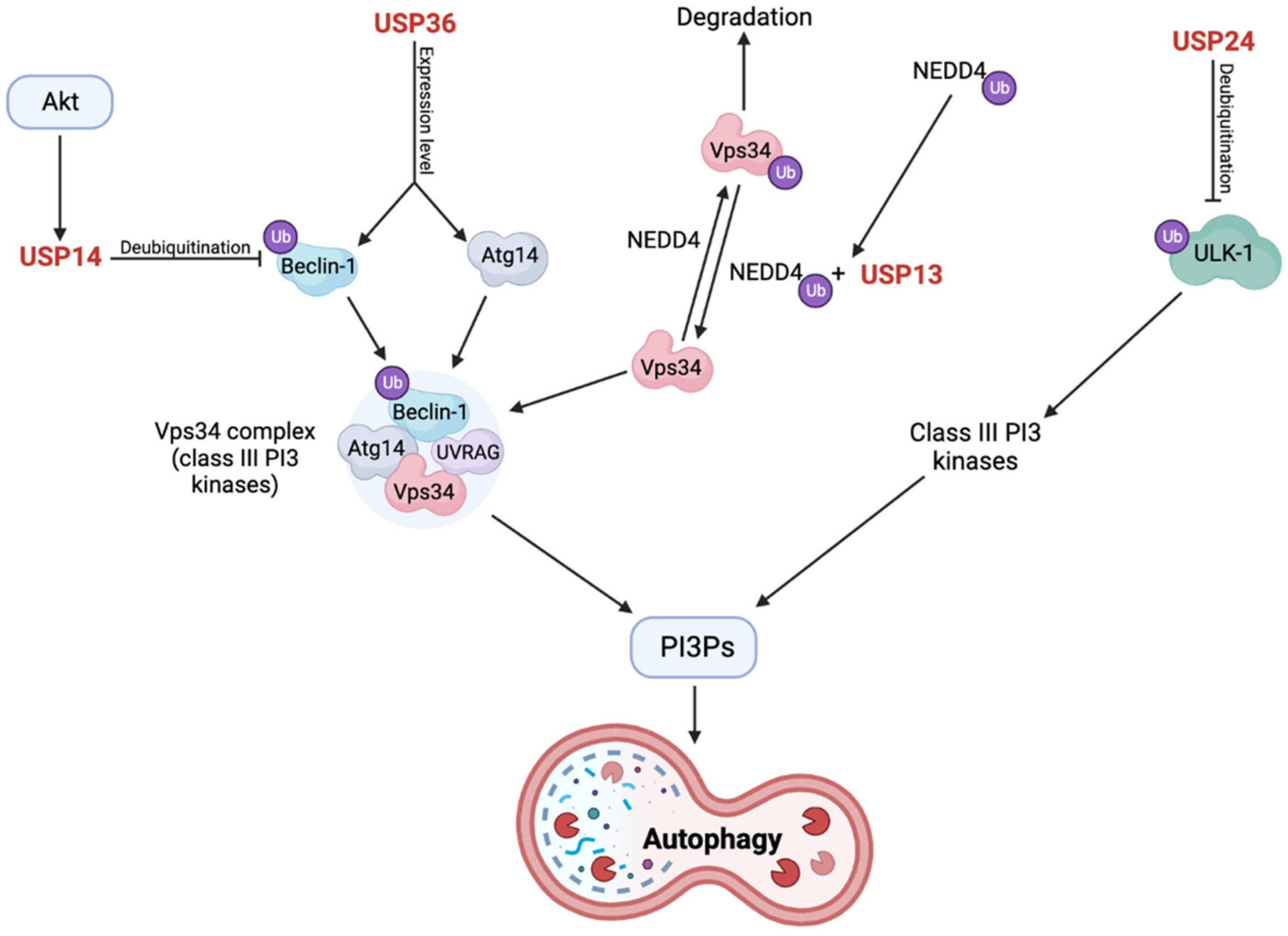
2.11. USP24 Negatively Regulates Autophagy through Destabilization of ULK-1
2.12. Loss of USP36 Causes Impairments in the Mitophagy Process through Altered Expression Levels of Autophagy Complex Proteins
2.13. OTUB1 Has Amyloidogenic Properties and Co-Localizes with α-Synuclein in LBs
2.14. The Role of USP40 in PD Is Still Controversial
3. Discussion, Conclusion, and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Capriotti, T.; Terzakis, K. Parkinson Disease. Home Healthc Now 2016, 34, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Sung, V.W.; Nicholas, A.P. Nonmotor symptoms in Parkinson’s disease: Expanding the view of Parkinson’s disease beyond a pure motor, pure dopaminergic problem. Neurol. Clin. 2013, 31 (3 Suppl.), S1–S16. [Google Scholar] [CrossRef]
- Beitz, J.M. Parkinson’s disease: A review. Front. Biosci. 2014, 6, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Balestrino, R.; Schapira, A.H.V. Parkinson disease. Eur. J. Neurol. 2020, 27, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Fearnley, J.M.; Lees, A.J. Ageing and Parkinson’s disease: Substantia nigra regional selectivity. Brain 1991, 114 Pt 5, 2283–2301. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, S.R.; Chesselet, M.F. Mitochondrial dysfunction and oxidative stress in Parkinson’s disease. Prog. Neurobiol. 2013, 106–107, 17–32. [Google Scholar] [CrossRef]
- Ge, P.; Dawson, V.L.; Dawson, T.M. PINK1 and Parkin mitochondrial quality control: A source of regional vulnerability in Parkinson’s disease. Mol. Neurodegener. 2020, 15, 20. [Google Scholar] [CrossRef]
- Madsen, D.A.; Schmidt, S.I.; Blaabjerg, M.; Meyer, M. Interaction between Parkin and alpha-Synuclein in PARK2-Mediated Parkinson’s Disease. Cells 2021, 10, 283. [Google Scholar] [CrossRef]
- Devine, M.J.; Gwinn, K.; Singleton, A.; Hardy, J. Parkinson’s disease and alpha-synuclein expression. Mov. Disord. 2011, 26, 2160–2168. [Google Scholar] [CrossRef]
- Rocha, E.M.; De Miranda, B.; Sanders, L.H. Alpha-synuclein: Pathology, mitochondrial dysfunction and neuroinflammation in Parkinson’s disease. Neurobiol. Dis. 2018, 109, 249–257. [Google Scholar] [CrossRef]
- Armstrong, M.J.; Okun, M.S. Diagnosis and Treatment of Parkinson Disease: A Review. JAMA 2020, 323, 548–560. [Google Scholar] [CrossRef] [PubMed]
- Cooper, G.M. The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- Gan-Or, Z.; Dion, P.A.; Rouleau, G.A. Genetic perspective on the role of the autophagy-lysosome pathway in Parkinson disease. Autophagy 2015, 11, 1443–1457. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.C.; Bartlett, J.J.; Pulinilkunnil, T. Lysosomal Biology and Function: Modern View of Cellular Debris Bin. Cells 2020, 9, 1131. [Google Scholar] [CrossRef]
- Kocaturk, N.M.; Gozuacik, D. Crosstalk Between Mammalian Autophagy and the Ubiquitin-Proteasome System. Front. Cell Dev. Biol. 2018, 6, 128. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef]
- Mizushima, N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 2018, 20, 521–527. [Google Scholar] [CrossRef]
- Bence, N.F.; Sampat, R.M.; Kopito, R.R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 2001, 292, 1552–1555. [Google Scholar] [CrossRef]
- Ciechanover, A.; Schwartz, A.L. The ubiquitin-proteasome pathway: The complexity and myriad functions of proteins death. Proc. Natl. Acad. Sci. USA 1998, 95, 2727–2730. [Google Scholar] [CrossRef]
- Adams, J. The proteasome: Structure, function, and role in the cell. Cancer Treat Rev. 2003, 29 (Suppl. 1), 3–9. [Google Scholar] [CrossRef]
- Tai, H.C.; Schuman, E.M. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 2008, 9, 826–838. [Google Scholar] [CrossRef]
- Neutzner, M.; Neutzner, A. Enzymes of ubiquitination and deubiquitination. Essays Biochem. 2012, 52, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.; Radulovic, M.; Figueiredo-Pereira, M.E.; Cardozo, C. The Ubiquitin-Proteasome System: Potential Therapeutic Targets for Alzheimer’s Disease and Spinal Cord Injury. Front. Mol. Neurosci. 2016, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef] [PubMed]
- Grice, G.L.; Lobb, I.T.; Weekes, M.P.; Gygi, S.P.; Antrobus, R.; Nathan, J.A. The Proteasome Distinguishes between Heterotypic and Homotypic Lysine-11-Linked Polyubiquitin Chains. Cell Rep. 2015, 12, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef]
- Chen, R.H.; Chen, Y.H.; Huang, T.Y. Ubiquitin-mediated regulation of autophagy. J. Biomed. Sci. 2019, 26, 80. [Google Scholar] [CrossRef]
- Korolchuk, V.I.; Menzies, F.M.; Rubinsztein, D.C. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010, 584, 1393–1398. [Google Scholar] [CrossRef]
- Liu, J.; Liu, W.; Li, R.; Yang, H. Mitophagy in Parkinson’s Disease: From Pathogenesis to Treatment. Cells 2019, 8, 712. [Google Scholar] [CrossRef]
- Narendra, D.; Tanaka, A.; Suen, D.F.; Youle, R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell. Biol. 2008, 183, 795–803. [Google Scholar] [CrossRef]
- Gladkova, C.; Maslen, S.L.; Skehel, J.M.; Komander, D. Mechanism of parkin activation by PINK1. Nature 2018, 559, 410–414. [Google Scholar] [CrossRef]
- Jin, S.M.; Lazarou, M.; Wang, C.; Kane, L.A.; Narendra, D.P.; Youle, R.J. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell. Biol. 2010, 191, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Narendra, D.P.; Jin, S.M.; Tanaka, A.; Suen, D.F.; Gautier, C.A.; Shen, J.; Cookson, M.R.; Youle, R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010, 8, e1000298. [Google Scholar] [CrossRef] [PubMed]
- Kane, L.A.; Lazarou, M.; Fogel, A.I.; Li, Y.; Yamano, K.; Sarraf, S.A.; Banerjee, S.; Youle, R.J. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 2014, 205, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Kondapalli, C.; Kazlauskaite, A.; Zhang, N.; Woodroof, H.I.; Campbell, D.G.; Gourlay, R.; Burchell, L.; Walden, H.; Macartney, T.J.; Deak, M.; et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012, 2, 120080. [Google Scholar] [CrossRef]
- Ziviani, E.; Tao, R.N.; Whitworth, A.J. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl. Acad. Sci. USA 2010, 107, 5018–5023. [Google Scholar] [CrossRef]
- Geisler, S.; Holmstrom, K.M.; Skujat, D.; Fiesel, F.C.; Rothfuss, O.C.; Kahle, P.J.; Springer, W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010, 12, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Grenier, K.; McLelland, G.L.; Fon, E.A. Parkin- and PINK1-Dependent Mitophagy in Neurons: Will the Real Pathway Please Stand Up? Front. Neurol. 2013, 4, 100. [Google Scholar] [CrossRef]
- Park, G.H.; Park, J.H.; Chung, K.C. Precise control of mitophagy through ubiquitin proteasome system and deubiquitin proteases and their dysfunction in Parkinson’s disease. BMB Rep. 2021, 54, 592–600. [Google Scholar] [CrossRef]
- Komander, D.; Clague, M.J.; Urbe, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef]
- Clague, M.J.; Urbe, S.; Komander, D. Breaking the chains: Deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 2019, 20, 338–352. [Google Scholar] [CrossRef]
- Nijman, S.M.; Luna-Vargas, M.P.; Velds, A.; Brummelkamp, T.R.; Dirac, A.M.; Sixma, T.K.; Bernards, R. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005, 123, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, M.; Jolly, L.A.; Gecz, J.; Wood, S.A. La FAM fatale: USP9X in development and disease. Cell. Mol. Life. Sci. 2015, 72, 2075–2089. [Google Scholar] [CrossRef]
- Bingol, B.; Sheng, M. Mechanisms of mitophagy: PINK1, Parkin, USP30 and beyond. Free Radic. Biol. Med. 2016, 100, 210–222. [Google Scholar] [CrossRef] [PubMed]
- Cornelissen, T.; Haddad, D.; Wauters, F.; Van Humbeeck, C.; Mandemakers, W.; Koentjoro, B.; Sue, C.; Gevaert, K.; De Strooper, B.; Verstreken, P.; et al. The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum. Mol. Genet. 2014, 23, 5227–5242. [Google Scholar] [CrossRef] [PubMed]
- Bingol, B.; Tea, J.S.; Phu, L.; Reichelt, M.; Bakalarski, C.E.; Song, Q.; Foreman, O.; Kirkpatrick, D.S.; Sheng, M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 2014, 510, 370–375. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Serricchio, M.; Jauregui, M.; Shanbhag, R.; Stoltz, T.; Di Paolo, C.T.; Kim, P.K.; McQuibban, G.A. Deubiquitinating enzymes regulate PARK2-mediated mitophagy. Autophagy 2015, 11, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Gersch, M.; Gladkova, C.; Schubert, A.F.; Michel, M.A.; Maslen, S.; Komander, D. Mechanism and regulation of the Lys6-selective deubiquitinase USP30. Nat. Struct. Mol. Biol. 2017, 24, 920–930. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, C.N.; Baughman, J.M.; Phu, L.; Tea, J.S.; Yu, C.; Coons, M.; Kirkpatrick, D.S.; Bingol, B.; Corn, J.E. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat. Cell Biol. 2015, 17, 160–169. [Google Scholar] [CrossRef]
- Durcan, T.M.; Tang, M.Y.; Perusse, J.R.; Dashti, E.A.; Aguileta, M.A.; McLelland, G.L.; Gros, P.; Shaler, T.A.; Faubert, D.; Coulombe, B.; et al. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J. 2014, 33, 2473–2491. [Google Scholar] [CrossRef]
- Kluge, A.F.; Lagu, B.R.; Maiti, P.; Jaleel, M.; Webb, M.; Malhotra, J.; Mallat, A.; Srinivas, P.A.; Thompson, J.E. Novel highly selective inhibitors of ubiquitin specific protease 30 (USP30) accelerate mitophagy. Bioorg. Med. Chem. Lett. 2018, 28, 2655–2659. [Google Scholar] [CrossRef]
- Mandal, S.; Kumar BR, P.; Alam, M.T.; Tripathi, P.P.; Channappa, B. Novel Imidazole Phenoxyacetic Acids as Inhibitors of USP30 for Neuroprotection Implication via the Ubiquitin-Rho-110 Fluorometric Assay: Design, Synthesis, and In Silico and Biochemical Assays. ACS Chem. Neurosci. 2022, 13, 1433–1445. [Google Scholar] [CrossRef]
- Rusilowicz-Jones, E.V.; Barone, F.G.; Lopes, F.M.; Stephen, E.; Mortiboys, H.; Urbe, S.; Clague, M.J. Benchmarking a highly selective USP30 inhibitor for enhancement of mitophagy and pexophagy. Life Sci. Alliance 2022, 5. [Google Scholar] [CrossRef] [PubMed]
- Yue, W.; Chen, Z.; Liu, H.; Yan, C.; Chen, M.; Feng, D.; Yan, C.; Wu, H.; Du, L.; Wang, Y.; et al. A small natural molecule promotes mitochondrial fusion through inhibition of the deubiquitinase USP30. Cell Res. 2014, 24, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Tsefou, E.; Walker, A.S.; Clark, E.H.; Hicks, A.R.; Luft, C.; Takeda, K.; Watanabe, T.; Ramazio, B.; Staddon, J.M.; Briston, T.; et al. Investigation of USP30 inhibition to enhance Parkin-mediated mitophagy: Tools and approaches. Biochem. J. 2021, 478, 4099–4118. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Krigman, J.; Zhang, R.; Yang, M.; Sun, N. Pharmacological inhibition of USP30 activates tissue-specific mitophagy. Acta Physiol. (Oxf.) 2021, 232, e13666. [Google Scholar] [CrossRef] [PubMed]
- Niu, K.; Fang, H.; Chen, Z.; Zhu, Y.; Tan, Q.; Wei, D.; Li, Y.; Balajee, A.S.; Zhao, Y. USP33 deubiquitinates PRKN/parkin and antagonizes its role in mitophagy. Autophagy 2020, 16, 724–734. [Google Scholar] [CrossRef]
- Alexopoulou, Z.; Lang, J.; Perrett, R.M.; Elschami, M.; Hurry, M.E.; Kim, H.T.; Mazaraki, D.; Szabo, A.; Kessler, B.M.; Goldberg, A.L.; et al. Deubiquitinase Usp8 regulates alpha-synuclein clearance and modifies its toxicity in Lewy body disease. Proc. Natl. Acad. Sci. USA 2016, 113, E4688–E4697. [Google Scholar] [CrossRef]
- von Stockum, S.; Sanchez-Martinez, A.; Corra, S.; Chakraborty, J.; Marchesan, E.; Locatello, L.; Da Re, C.; Cusumano, P.; Caicci, F.; Ferrari, V.; et al. Inhibition of the deubiquitinase USP8 corrects a Drosophila PINK1 model of mitochondria dysfunction. Life Sci. Alliance 2019, 2. [Google Scholar] [CrossRef] [PubMed]
- Durcan, T.M.; Kontogiannea, M.; Bedard, N.; Wing, S.S.; Fon, E.A. Ataxin-3 deubiquitination is coupled to Parkin ubiquitination via E2 ubiquitin-conjugating enzyme. J. Biol. Chem. 2012, 287, 531–541. [Google Scholar] [CrossRef]
- Durcan, T.M.; Kontogiannea, M.; Thorarinsdottir, T.; Fallon, L.; Williams, A.J.; Djarmati, A.; Fantaneanu, T.; Paulson, H.L.; Fon, E.A. The Machado-Joseph disease-associated mutant form of ataxin-3 regulates parkin ubiquitination and stability. Hum. Mol. Genet. 2011, 20, 141–154. [Google Scholar] [CrossRef]
- Durcan, T.M.; Fon, E.A. Mutant ataxin-3 promotes the autophagic degradation of parkin. Autophagy 2011, 7, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Noronha, C.; Perfeito, R.; Laco, M.; Wullner, U.; Rego, A.C. Expanded and Wild-type Ataxin-3 Modify the Redox Status of SH-SY5Y Cells Overexpressing alpha-Synuclein. Neurochem. Res. 2017, 42, 1430–1437. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hebron, M.; Shi, W.; Lonskaya, I.; Moussa, C.E. Ubiquitin specific protease-13 independently regulates parkin ubiquitination and alpha-synuclein clearance in alpha-synucleinopathies. Hum. Mol. Genet. 2019, 28, 548–560. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, T.; Nihira, T.; Ren, Y.R.; Cao, X.Q.; Wada, K.; Setsuie, R.; Kabuta, T.; Wada, K.; Hattori, N.; Mizuno, Y.; et al. Effects of UCH-L1 on alpha-synuclein over-expression mouse model of Parkinson’s disease. J. Neurochem. 2009, 108, 932–944. [Google Scholar] [CrossRef]
- Kabuta, T.; Furuta, A.; Aoki, S.; Furuta, K.; Wada, K. Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J. Biol. Chem. 2008, 283, 23731–23738. [Google Scholar] [CrossRef]
- Carmine Belin, A.; Westerlund, M.; Bergman, O.; Nissbrandt, H.; Lind, C.; Sydow, O.; Galter, D. S18Y in ubiquitin carboxy-terminal hydrolase L1 (UCH-L1) associated with decreased risk of Parkinson’s disease in Sweden. Parkinsonism Relat. Disord. 2007, 13, 295–298. [Google Scholar] [CrossRef]
- Elbaz, A.; Levecque, C.; Clavel, J.; Vidal, J.S.; Richard, F.; Correze, J.R.; Delemotte, B.; Amouyel, P.; Alperovitch, A.; Chartier-Harlin, M.C.; et al. S18Y polymorphism in the UCH-L1 gene and Parkinson’s disease: Evidence for an age-dependent relationship. Mov. Disord. 2003, 18, 130–137. [Google Scholar] [CrossRef]
- Andersson, F.I.; Werrell, E.F.; McMorran, L.; Crone, W.J.; Das, C.; Hsu, S.T.; Jackson, S.E. The effect of Parkinson’s-disease-associated mutations on the deubiquitinating enzyme UCH-L1. J. Mol. Biol. 2011, 407, 261–272. [Google Scholar] [CrossRef]
- Ardley, H.C.; Scott, G.B.; Rose, S.A.; Tan, N.G.; Robinson, P.A. UCH-L1 aggresome formation in response to proteasome impairment indicates a role in inclusion formation in Parkinson’s disease. J. Neurochem. 2004, 90, 379–391. [Google Scholar] [CrossRef]
- Nishikawa, K.; Li, H.; Kawamura, R.; Osaka, H.; Wang, Y.L.; Hara, Y.; Hirokawa, T.; Manago, Y.; Amano, T.; Noda, M.; et al. Alterations of structure and hydrolase activity of parkinsonism-associated human ubiquitin carboxyl-terminal hydrolase L1 variants. Biochem. Biophys. Res. Commun. 2003, 304, 176–183. [Google Scholar] [CrossRef]
- Shimshek, D.R.; Schweizer, T.; Schmid, P.; van der Putten, P.H. Excess alpha-synuclein worsens disease in mice lacking ubiquitin carboxy-terminal hydrolase L1. Sci. Rep. 2012, 2, 262. [Google Scholar] [CrossRef] [PubMed]
- Cartier, A.E.; Ubhi, K.; Spencer, B.; Vazquez-Roque, R.A.; Kosberg, K.A.; Fourgeaud, L.; Kanayson, P.; Patrick, C.; Rockenstein, E.; Patrick, G.N.; et al. Differential effects of UCHL1 modulation on alpha-synuclein in PD-like models of alpha-synucleinopathy. PLoS ONE 2012, 7, e34713. [Google Scholar] [CrossRef]
- Liu, Z.; Meray, R.K.; Grammatopoulos, T.N.; Fredenburg, R.A.; Cookson, M.R.; Liu, Y.; Logan, T.; Lansbury, P.T., Jr. Membrane-associated farnesylated UCH-L1 promotes alpha-synuclein neurotoxicity and is a therapeutic target for Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2009, 106, 4635–4640. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, F.M.; von Stockum, S.; Giacomello, M.; Goliand, I.; Kakimoto, P.; Marchesan, E.; De Stefani, D.; Kowaltowski, A.J.; Ziviani, E.; Shirihai, O.S. A new target for an old DUB: UCH-L1 regulates mitofusin-2 levels, altering mitochondrial morphology, function and calcium uptake. Redox. Biol. 2020, 37, 101676. [Google Scholar] [CrossRef]
- Contu, V.R.; Kotake, Y.; Toyama, T.; Okuda, K.; Miyara, M.; Sakamoto, S.; Samizo, S.; Sanoh, S.; Kumagai, Y.; Ohta, S. Endogenous neurotoxic dopamine derivative covalently binds to Parkinson’s disease-associated ubiquitin C-terminal hydrolase L1 and alters its structure and function. J. Neurochem. 2014, 130, 826–838. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, H.J.; Jeong, J.E.; Baek, J.Y.; Jeong, J.; Kim, S.; Kim, Y.M.; Kim, Y.; Nam, J.H.; Huh, S.H.; et al. N-terminal truncated UCH-L1 prevents Parkinson’s disease associated damage. PLoS ONE 2014, 9, e99654. [Google Scholar] [CrossRef]
- Wang, K.K.; Yang, Z.; Sarkis, G.; Torres, I.; Raghavan, V. Ubiquitin C-terminal hydrolase-L1 (UCH-L1) as a therapeutic and diagnostic target in neurodegeneration, neurotrauma and neuro-injuries. Expert. Opin. Ther. Targets. 2017, 21, 627–638. [Google Scholar] [CrossRef]
- Rott, R.; Szargel, R.; Haskin, J.; Bandopadhyay, R.; Lees, A.J.; Shani, V.; Engelender, S. alpha-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc. Natl. Acad. Sci. USA 2011, 108, 18666–18671. [Google Scholar] [CrossRef]
- Hu, M.; Li, P.; Song, L.; Jeffrey, P.D.; Chenova, T.A.; Wilkinson, K.D.; Cohen, R.E.; Shi, Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005, 24, 3747–3756. [Google Scholar] [CrossRef]
- Lee, B.H.; Lee, M.J.; Park, S.; Oh, D.C.; Elsasser, S.; Chen, P.C.; Gartner, C.; Dimova, N.; Hanna, J.; Gygi, S.P.; et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 2010, 467, 179–184. [Google Scholar] [CrossRef]
- Xu, D.; Shan, B.; Sun, H.; Xiao, J.; Zhu, K.; Xie, X.; Li, X.; Liang, W.; Lu, X.; Qian, L.; et al. USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1. Genes. Dev. 2016, 30, 1718–1730. [Google Scholar] [CrossRef] [PubMed]
- Peth, A.; Besche, H.C.; Goldberg, A.L. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol. Cell 2009, 36, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, J.; von Stockum, S.; Marchesan, E.; Caicci, F.; Ferrari, V.; Rakovic, A.; Klein, C.; Antonini, A.; Bubacco, L.; Ziviani, E. USP14 inhibition corrects an in vivo model of impaired mitophagy. EMBO Mol. Med. 2018, 10, e9014. [Google Scholar] [CrossRef] [PubMed]
- Thayer, J.A.; Awad, O.; Hegdekar, N.; Sarkar, C.; Tesfay, H.; Burt, C.; Zeng, X.; Feldman, R.A.; Lipinski, M.M. The PARK10 gene USP24 is a negative regulator of autophagy and ULK1 protein stability. Autophagy 2020, 16, 140–153. [Google Scholar] [CrossRef]
- Wu, Y.R.; Chen, C.M.; Chen, Y.C.; Chao, C.Y.; Ro, L.S.; Fung, H.C.; Hsiao, Y.C.; Hu, F.J.; Lee-Chen, G.J. Ubiquitin specific proteases USP24 and USP40 and ubiquitin thiolesterase UCHL1 polymorphisms have synergic effect on the risk of Parkinson’s disease among Taiwanese. Clin. Chim. Acta. 2010, 411, 955–958. [Google Scholar] [CrossRef]
- Haugarvoll, K.; Toft, M.; Skipper, L.; Heckman, M.G.; Crook, J.E.; Soto, A.; Ross, O.A.; Hulihan, M.M.; Kachergus, J.M.; Sando, S.B.; et al. Fine-mapping and candidate gene investigation within the PARK10 locus. Eur. J. Hum. Genet. 2009, 17, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Schrodi, S.; Rowland, C.; Tacey, K.; Catanese, J.; Grupe, A. Genetic evidence for ubiquitin-specific proteases USP24 and USP40 as candidate genes for late-onset Parkinson disease. Hum. Mutat. 2006, 27, 1017–1023. [Google Scholar] [CrossRef]
- Wan, J.Y.; Edwards, K.L.; Hutter, C.M.; Mata, I.F.; Samii, A.; Roberts, J.W.; Agarwal, P.; Checkoway, H.; Farin, F.M.; Yearout, D.; et al. Association mapping of the PARK10 region for Parkinson’s disease susceptibility genes. Parkinsonism. Relat. Disord. 2014, 20, 93–98. [Google Scholar] [CrossRef]
- Zhao, B.; Song, W.; Chen, Y.P.; Huang, R.; Chen, K.; Cao, B.; Yang, Y.; Shang, H.F. Association analysis of single-nucleotide polymorphisms of USP24 and USP40 with Parkinson’s disease in the Han Chinese population. Eur. Neurol. 2012, 68, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Geisler, S.; Jager, L.; Golombek, S.; Nakanishi, E.; Hans, F.; Casadei, N.; Terradas, A.L.; Linnemann, C.; Kahle, P.J. Ubiquitin-specific protease USP36 knockdown impairs Parkin-dependent mitophagy via downregulation of Beclin-1-associated autophagy-related ATG14L. Exp. Cell Res. 2019, 384, 111641. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Kumar, S.; Singh, A.K.; Hanpude, P.; Jangir, D.; Maiti, T.K. Amyloid aggregates of the deubiquitinase OTUB1 are neurotoxic, suggesting that they contribute to the development of Parkinson’s disease. J. Biol. Chem. 2020, 295, 3466–3484. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Kumar, R.; Dey, A.K.; Saha, S.; Maiti, T.K. S-Nitrosylation of OTUB1 Alters Its Stability and Ubc13 Binding. ACS Chem. Neurosci. 2022, 13, 1517–1525. [Google Scholar] [CrossRef]
- Chou, C.K.; Chang, Y.T.; Korinek, M.; Chen, Y.T.; Yang, Y.T.; Leu, S.; Lin, I.L.; Tang, C.J.; Chiu, C.C. The Regulations of Deubiquitinase USP15 and Its Pathophysiological Mechanisms in Diseases. Int. J. Mol. Sci. 2017, 18, 483. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, N.; Kitami, T.; Suzuki, T.; Mizuno, Y.; Hattori, N.; Tanaka, K. Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J. Biol. Chem. 2006, 281, 3204–3209. [Google Scholar] [CrossRef]
- Hampe, C.; Ardila-Osorio, H.; Fournier, M.; Brice, A.; Corti, O. Biochemical analysis of Parkinson’s disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum. Mol. Genet. 2006, 15, 2059–2075. [Google Scholar] [CrossRef]
- Cornelissen, T.; Vilain, S.; Vints, K.; Gounko, N.; Verstreken, P.; Vandenberghe, W. Deficiency of parkin and PINK1 impairs age-dependent mitophagy in Drosophila. Elife 2018, 7, e35878. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, N.; Hirose, S. Regulation of mitochondrial morphology by USP30, a deubiquitinating enzyme present in the mitochondrial outer membrane. Mol. Biol. Cell 2008, 19, 1903–1911. [Google Scholar] [CrossRef] [PubMed]
- Ordureau, A.; Sarraf, S.A.; Duda, D.M.; Heo, J.M.; Jedrychowski, M.P.; Sviderskiy, V.O.; Olszewski, J.L.; Koerber, J.T.; Xie, T.; Beausoleil, S.A.; et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 2014, 56, 360–375. [Google Scholar] [CrossRef] [PubMed]
- Wauer, T.; Swatek, K.N.; Wagstaff, J.L.; Gladkova, C.; Pruneda, J.N.; Michel, M.A.; Gersch, M.; Johnson, C.M.; Freund, S.M.; Komander, D. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 2015, 34, 307–325. [Google Scholar] [CrossRef] [PubMed]
- Okarmus, J.; Agergaard, J.B.; Stummann, T.C.; Haukedal, H.; Ambjørn, M.; Freude, K.K.; Fog, K.; Meyer, M. USP30 inhibition induces mitophagy and reduces oxidative stress in parkin-deficient and CCCP-stressed human iPSC-derived neurons. bioRxiv 2022. [Google Scholar] [CrossRef]
- Allen, M.D.; Bycroft, M. The solution structure of the ZnF UBP domain of USP33/VDU1. Protein Sci. 2007, 16, 2072–2075. [Google Scholar] [CrossRef]
- Li, Z.; Na, X.; Wang, D.; Schoen, S.R.; Messing, E.M.; Wu, G. Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 2002, 277, 4656–4662. [Google Scholar] [CrossRef] [PubMed]
- Fekete, C.; Freitas, B.C.; Zeold, A.; Wittmann, G.; Kadar, A.; Liposits, Z.; Christoffolete, M.A.; Singru, P.; Lechan, R.M.; Bianco, A.C.; et al. Expression patterns of WSB-1 and USP-33 underlie cell-specific posttranslational control of type 2 deiodinase in the rat brain. Endocrinology 2007, 148, 4865–4874. [Google Scholar] [CrossRef] [PubMed]
- Yuasa-Kawada, J.; Kinoshita-Kawada, M.; Wu, G.; Rao, Y.; Wu, J.Y. Midline crossing and Slit responsiveness of commissural axons require USP33. Nat. Neurosci. 2009, 12, 1087–1089. [Google Scholar] [CrossRef] [PubMed]
- Thorne, C.; Eccles, R.L.; Coulson, J.M.; Urbe, S.; Clague, M.J. Isoform-specific localization of the deubiquitinase USP33 to the Golgi apparatus. Traffic 2011, 12, 1563–1574. [Google Scholar] [CrossRef] [PubMed]
- Bruzzone, F.; Vallarino, M.; Berruti, G.; Angelini, C. Expression of the deubiquitinating enzyme mUBPy in the mouse brain. Brain. Res. 2008, 1195, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Celardo, I.; Costa, A.C.; Lehmann, S.; Jones, C.; Wood, N.; Mencacci, N.E.; Mallucci, G.R.; Loh, S.H.Y.; Martins, L.M. Mitofusin-mediated ER stress triggers neurodegeneration in pink1/parkin models of Parkinson’s disease. Cell Death Dis. 2016, 7, e2271. [Google Scholar] [CrossRef]
- Deng, H.; Dodson, M.W.; Huang, H.; Guo, M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl. Acad. Sci. USA 2008, 105, 14503–14508. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Okamoto, T.; Taniwaki, M.; Aizawa, M.; Inoue, M.; Katayama, S.; Kawakami, H.; Nakamura, S.; Nishimura, M.; Akiguchi, I.; et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 1994, 8, 221–228. [Google Scholar] [CrossRef]
- Klein, C.; Schneider, S.A.; Lang, A.E. Hereditary parkinsonism: Parkinson disease look-alikes--an algorithm for clinicians to “PARK” genes and beyond. Mov. Disord. 2009, 24, 2042–2058. [Google Scholar] [CrossRef]
- Wang, J.L.; Xiao, B.; Cui, X.X.; Guo, J.F.; Lei, L.F.; Song, X.W.; Shen, L.; Jiang, H.; Yan, X.X.; Pan, Q.; et al. Analysis of SCA2 and SCA3/MJD repeats in Parkinson’s disease in mainland China: Genetic, clinical, and positron emission tomography findings. Mov. Disord. 2009, 24, 2007–2011. [Google Scholar] [CrossRef]
- Gwinn-Hardy, K.; Singleton, A.; O’Suilleabhain, P.; Boss, M.; Nicholl, D.; Adam, A.; Hussey, J.; Critchley, P.; Hardy, J.; Farrer, M. Spinocerebellar ataxia type 3 phenotypically resembling parkinson disease in a black family. Arch. Neurol. 2001, 58, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Socal, M.P.; Emmel, V.E.; Rieder, C.R.; Hilbig, A.; Saraiva-Pereira, M.L.; Jardim, L.B. Intrafamilial variability of Parkinson phenotype in SCAs: Novel cases due to SCA2 and SCA3 expansions. Park. Relat. Disord. 2009, 15, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.C.; Fishman, P.S.; Thakor, N.V.; Oyler, G.A. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J. Biol. Chem. 2003, 278, 22044–22055. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Balaraman, K.; Lynch, C.C.; Hebron, M.; Wolf, C.; Moussa, C. Novel Ubiquitin Specific Protease-13 Inhibitors Alleviate Neurodegenerative Pathology. Metabolites 2021, 11, 622. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Balaraman, K.; Lynch, C.C.; Hebron, M.; Shah, P.K.; Hu, S.; Stevenson, M.; Wolf, C.; Moussa, C. Inhibition of Ubiquitin-Specific Protease-13 Improves Behavioral Performance in Alpha-Synuclein Expressing Mice. Int. J. Mol. Sci. 2022, 23, 8131. [Google Scholar] [CrossRef]
- Vajhoj, C.; Schmid, B.; Alik, A.; Melki, R.; Fog, K.; Holst, B.; Stummann, T.C. Establishment of a human induced pluripotent stem cell neuronal model for identification of modulators of A53T alpha-synuclein levels and aggregation. PLoS ONE 2021, 16, e0261536. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Jin, S.; Wu, Y.; Xian, H.; Tian, S.; Liu, D.A.; Guo, Z.; Cui, J. Auto-ubiquitination of NEDD4-1 Recruits USP13 to Facilitate Autophagy through Deubiquitinating VPS34. Cell Rep. 2020, 30, 2807–2819. [Google Scholar] [CrossRef]
- Choi, J.; Levey, A.I.; Weintraub, S.T.; Rees, H.D.; Gearing, M.; Chin, L.S.; Li, L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J. Biol. Chem. 2004, 279, 13256–13264. [Google Scholar] [CrossRef] [PubMed]
- Solano, S.M.; Miller, D.W.; Augood, S.J.; Young, A.B.; Penney, J.B., Jr. Expression of alpha-synuclein, parkin, and ubiquitin carboxy-terminal hydrolase L1 mRNA in human brain: Genes associated with familial Parkinson’s disease. Ann. Neurol. 2000, 47, 201–210. [Google Scholar] [CrossRef]
- Mondello, S.; Constantinescu, R.; Zetterberg, H.; Andreasson, U.; Holmberg, B.; Jeromin, A. CSF alpha-synuclein and UCH-L1 levels in Parkinson’s disease and atypical parkinsonian disorders. Park. Relat. Disord. 2014, 20, 382–387. [Google Scholar] [CrossRef]
- Liu, Y.; Fallon, L.; Lashuel, H.A.; Liu, Z.; Lansbury, P.T., Jr. The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell 2002, 111, 209–218. [Google Scholar] [CrossRef]
- Osaka, H.; Wang, Y.L.; Takada, K.; Takizawa, S.; Setsuie, R.; Li, H.; Sato, Y.; Nishikawa, K.; Sun, Y.J.; Sakurai, M.; et al. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum. Mol. Genet. 2003, 12, 1945–1958. [Google Scholar] [CrossRef]
- Cartier, A.E.; Djakovic, S.N.; Salehi, A.; Wilson, S.M.; Masliah, E.; Patrick, G.N. Regulation of synaptic structure by ubiquitin C-terminal hydrolase L1. J. Neurosci. 2009, 29, 7857–7868. [Google Scholar] [CrossRef] [PubMed]
- Barrachina, M.; Castano, E.; Dalfo, E.; Maes, T.; Buesa, C.; Ferrer, I. Reduced ubiquitin C-terminal hydrolase-1 expression levels in dementia with Lewy bodies. Neurobiol. Dis. 2006, 22, 265–273. [Google Scholar] [CrossRef]
- Chakraborty, J.; Ziviani, E. Deubiquitinating Enzymes in Parkinson’s Disease. Front. Physiol. 2020, 11, 535. [Google Scholar] [CrossRef] [PubMed]
- Saini, P.; Rudakou, U.; Yu, E.; Ruskey, J.A.; Asayesh, F.; Laurent, S.B.; Spiegelman, D.; Fahn, S.; Waters, C.; Monchi, O.; et al. Association study of DNAJC13, UCHL1, HTRA2, GIGYF2, and EIF4G1 with Parkinson’s disease. Neurobiol. Aging. 2021, 100, 119.e113–119.e117. [Google Scholar] [CrossRef] [PubMed]
- Lowe, J.; McDermott, H.; Landon, M.; Mayer, R.J.; Wilkinson, K.D. Ubiquitin carboxyl-terminal hydrolase (PGP 9.5) is selectively present in ubiquitinated inclusion bodies characteristic of human neurodegenerative diseases. J. Pathol. 1990, 161, 153–160. [Google Scholar] [CrossRef]
- Leroy, E.; Boyer, R.; Auburger, G.; Leube, B.; Ulm, G.; Mezey, E.; Harta, G.; Brownstein, M.J.; Jonnalagada, S.; Chernova, T.; et al. The ubiquitin pathway in Parkinson’s disease. Nature 1998, 395, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Setsuie, R.; Wang, Y.L.; Mochizuki, H.; Osaka, H.; Hayakawa, H.; Ichihara, N.; Li, H.; Furuta, A.; Sano, Y.; Sun, Y.J.; et al. Dopaminergic neuronal loss in transgenic mice expressing the Parkinson’s disease-associated UCH-L1 I93M mutant. Neurochem. Int. 2007, 50, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Maraganore, D.M.; Farrer, M.J.; Hardy, J.A.; Lincoln, S.J.; McDonnell, S.K.; Rocca, W.A. Case-control study of the ubiquitin carboxy-terminal hydrolase L1 gene in Parkinson’s disease. Neurology 1999, 53, 1858–1860. [Google Scholar] [CrossRef] [PubMed]
- Wintermeyer, P.; Kruger, R.; Kuhn, W.; Muller, T.; Woitalla, D.; Berg, D.; Becker, G.; Leroy, E.; Polymeropoulos, M.; Berger, K.; et al. Mutation analysis and association studies of the UCHL1 gene in German Parkinson’s disease patients. Neuroreport 2000, 11, 2079–2082. [Google Scholar] [CrossRef] [PubMed]
- Levecque, C.; Destee, A.; Mouroux, V.; Becquet, E.; Defebvre, L.; Amouyel, P.; Chartier-Harlin, M.C. No genetic association of the ubiquitin carboxy-terminal hydrolase-L1 gene S18Y polymorphism with familial Parkinson’s disease. J. Neural. Transm. (Vienna) 2001, 108, 979–984. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, C.Y.; Si, Y.M.; Liu, Z.L.; Chen, B.; Yu, L. ACT and UCH-L1 polymorphisms in Parkinson’s disease and age of onset. Mov. Disord. 2002, 17, 767–771. [Google Scholar] [CrossRef]
- Maraganore, D.M.; Lesnick, T.G.; Elbaz, A.; Chartier-Harlin, M.C.; Gasser, T.; Kruger, R.; Hattori, N.; Mellick, G.D.; Quattrone, A.; Satoh, J.; et al. UCHL1 is a Parkinson’s disease susceptibility gene. Ann. Neurol. 2004, 55, 512–521. [Google Scholar] [CrossRef]
- Tan, E.K.; Puong, K.Y.; Fook-Chong, S.; Chua, E.; Shen, H.; Yuen, Y.; Pavanni, R.; Wong, M.C.; Puvan, K.; Zhao, Y. Case-control study of UCHL1 S18Y variant in Parkinson’s disease. Mov. Disord. 2006, 21, 1765–1768. [Google Scholar] [CrossRef]
- Facheris, M.; Strain, K.J.; Lesnick, T.G.; de Andrade, M.; Bower, J.H.; Ahlskog, J.E.; Cunningham, J.M.; Lincoln, S.; Farrer, M.J.; Rocca, W.A.; et al. UCHL1 is associated with Parkinson’s disease: A case-unaffected sibling and case-unrelated control study. Neurosci. Lett. 2005, 381, 131–134. [Google Scholar] [CrossRef]
- Zhang, J.; Hattori, N.; Leroy, E.; Morris, H.R.; Kubo, S.; Kobayashi, T.; Wood, N.W.; Polymeropoulos, M.H.; Mizuno, Y. Association between a polymorphism of ubiquitin carboxy-terminal hydrolase L1 (UCH-L1) gene and sporadic Parkinson’s disease. Parkinsonism. Relat. Disord. 2000, 6, 195–197. [Google Scholar] [CrossRef]
- Satoh, J.; Kuroda, Y. A polymorphic variation of serine to tyrosine at codon 18 in the ubiquitin C-terminal hydrolase-L1 gene is associated with a reduced risk of sporadic Parkinson’s disease in a Japanese population. J. Neurol. Sci. 2001, 189, 113–117. [Google Scholar] [CrossRef]
- Mellick, G.D.; Silburn, P.A. The ubiquitin carboxy-terminal hydrolase-L1 gene S18Y polymorphism does not confer protection against idiopathic Parkinson’s disease. Neurosci. Lett. 2000, 293, 127–130. [Google Scholar] [CrossRef]
- Zhu, R.; Zhu, Y.; Liu, X.; He, Z. UCH-L1 S18Y variant and risk of Parkinson’s disease in Asian populations: An updated meta-analysis. Neurodegener Dis. 2014, 14, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Guo, J.F.; Nie, L.L.; Luo, L.; Zuo, X.; Shen, L.; Jiang, H.; Yan, X.X.; Xia, K.; Pan, Q.; et al. Case-control study of the UCH-L1 S18Y variant in sporadic Parkinson’s disease in the Chinese population. J. Clin. Neurosci. 2011, 18, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.J.; Burgunder, J.M.; An, X.K.; Wu, Y.; Chen, W.J.; Zhang, J.H.; Wang, Y.C.; Xu, Y.M.; Gou, Y.R.; Yuan, G.G.; et al. Lack of evidence for association of a UCH-L1 S18Y polymorphism with Parkinson’s disease in a Han-Chinese population. Neurosci. Lett. 2008, 442, 200–202. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Tao, E. An Ile93Met substitution in the UCH-L1 gene is not a disease-causing mutation for idiopathic Parkinson’s disease. Chin. Med. J. 2003, 116, 312–313. [Google Scholar]
- Harhangi, B.S.; Farrer, M.J.; Lincoln, S.; Bonifati, V.; Meco, G.; De Michele, G.; Brice, A.; Durr, A.; Martinez, M.; Gasser, T.; et al. The Ile93Met mutation in the ubiquitin carboxy-terminal-hydrolase-L1 gene is not observed in European cases with familial Parkinson’s disease. Neurosci. Lett. 1999, 270, 1–4. [Google Scholar] [CrossRef]
- Zhang, J.; Hattori, N.; Giladi, N.; Mizuno, Y. Failure to find mutations in ubiquitin carboxy-terminal hydrolase L1 gene in familial Parkinson’s disease. Park. Relat. Disord 2000, 6, 199–200. [Google Scholar] [CrossRef]
- Kyratzi, E.; Pavlaki, M.; Stefanis, L. The S18Y polymorphic variant of UCH-L1 confers an antioxidant function to neuronal cells. Hum. Mol. Genet. 2008, 17, 2160–2171. [Google Scholar] [CrossRef]
- Xilouri, M.; Kyratzi, E.; Pitychoutis, P.M.; Papadopoulou-Daifoti, Z.; Perier, C.; Vila, M.; Maniati, M.; Ulusoy, A.; Kirik, D.; Park, D.S.; et al. Selective neuroprotective effects of the S18Y polymorphic variant of UCH-L1 in the dopaminergic system. Hum. Mol. Genet. 2012, 21, 874–889. [Google Scholar] [CrossRef]
- Lee, Y.C.; Hsu, S.D. Familial Mutations and Post-translational Modifications of UCH-L1 in Parkinson’s Disease and Neurodegenerative Disorders. Curr. Protein. Pept. Sci. 2017, 18, 733–745. [Google Scholar] [CrossRef]
- Bishop, P.; Rubin, P.; Thomson, A.R.; Rocca, D.; Henley, J.M. The ubiquitin C-terminal hydrolase L1 (UCH-L1) C terminus plays a key role in protein stability, but its farnesylation is not required for membrane association in primary neurons. J. Biol. Chem. 2014, 289, 36140–36149. [Google Scholar] [CrossRef]
- Khut, P.Y.; Tucker, B.; Lardelli, M.; Wood, S.A. Evolutionary and expression analysis of the zebrafish deubiquitylating enzyme, usp9. Zebrafish 2007, 4, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Homan, C.C.; Kumar, R.; Nguyen, L.S.; Haan, E.; Raymond, F.L.; Abidi, F.; Raynaud, M.; Schwartz, C.E.; Wood, S.A.; Gecz, J.; et al. Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am. J. Hum. Genet. 2014, 94, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Jolly, L.A.; Taylor, V.; Wood, S.A. USP9X enhances the polarity and self-renewal of embryonic stem cell-derived neural progenitors. Mol. Biol. Cell 2009, 20, 2015–2029. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.A.; Pascoe, W.S.; Ru, K.; Yamada, T.; Hirchenhain, J.; Kemler, R.; Mattick, J.S. Cloning and expression analysis of a novel mouse gene with sequence similarity to the Drosophila fat facets gene. Mech. Dev. 1997, 63, 29–38. [Google Scholar] [CrossRef]
- Murray, R.Z.; Jolly, L.A.; Wood, S.A. The FAM deubiquitylating enzyme localizes to multiple points of protein trafficking in epithelia, where it associates with E-cadherin and beta-catenin. Mol. Biol. Cell 2004, 15, 1591–1599. [Google Scholar] [CrossRef]
- Schwickart, M.; Huang, X.; Lill, J.R.; Liu, J.; Ferrando, R.; French, D.M.; Maecker, H.; O’Rourke, K.; Bazan, F.; Eastham-Anderson, J.; et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 2010, 463, 103–107. [Google Scholar] [CrossRef]
- Trinkle-Mulcahy, L.; Boulon, S.; Lam, Y.W.; Urcia, R.; Boisvert, F.M.; Vandermoere, F.; Morrice, N.A.; Swift, S.; Rothbauer, U.; Leonhardt, H.; et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 2008, 183, 223–239. [Google Scholar] [CrossRef]
- Jesko, H.; Lenkiewicz, A.M.; Wilkaniec, A.; Adamczyk, A. The interplay between parkin and alpha-synuclein; possible implications for the pathogenesis of Parkinson’s disease. Acta Neurobiol. Exp. 2019, 79, 276–289. [Google Scholar]
- Engelender, S. alpha-Synuclein fate: Proteasome or autophagy? Autophagy. 2012, 8, 418–420. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Hathaway, N.A.; Tone, Y.; Crosas, B.; Elsasser, S.; Kirkpatrick, D.S.; Leggett, D.S.; Gygi, S.P.; King, R.W.; Finley, D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 2006, 127, 99–111. [Google Scholar] [CrossRef]
- Jaber, N.; Zong, W.X. Class III PI3K Vps34: Essential roles in autophagy, endocytosis, and heart and liver function. Ann. N.Y. Acad. Sci. 2013, 1280, 48–51. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Park, S.; Lee, J.H.; Mun, J.Y.; Choi, W.H.; Yun, Y.; Lee, J.; Kim, J.H.; Kang, M.J.; Lee, M.J. Dual Function of USP14 Deubiquitinase in Cellular Proteasomal Activity and Autophagic Flux. Cell Rep. 2018, 24, 732–743. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, A.; Ohnuma, S.; Kodani, Y.; Kaneko, Y.S.; Nagasaki, H.; Nagatsu, T.; Ota, A. Inhibition of deubiquitinating activity of USP14 decreases tyrosine hydroxylase phosphorylated at Ser19 in PC12D cells. Biochem. Biophys. Res. Commun. 2016, 472, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, C.; Roy, M.; Mondal, R.; Chakraborty, J. USP14 as a Therapeutic Target Against Neurodegeneration: A Rat Brain Perspective. Front. Cell Dev. Biol. 2020, 8, 727. [Google Scholar] [CrossRef]
- Ortuno, D.; Carlisle, H.J.; Miller, S. Does inactivation of USP14 enhance degradation of proteasomal substrates that are associated with neurodegenerative diseases? F1000Res 2016, 5, 137. [Google Scholar] [CrossRef]
- Wang, K.; Liu, S.; Wang, J.; Wu, Y.; Cai, F.; Song, W. Transcriptional regulation of human USP24 gene expression by NF-kappa B. J. Neurochem. 2014, 128, 818–828. [Google Scholar] [CrossRef]
- Lipinski, M.M.; Zheng, B.; Lu, T.; Yan, Z.; Py, B.F.; Ng, A.; Xavier, R.J.; Li, C.; Yankner, B.A.; Scherzer, C.R.; et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2010, 107, 14164–14169. [Google Scholar] [CrossRef]
- Lipinski, M.M.; Hoffman, G.; Ng, A.; Zhou, W.; Py, B.F.; Hsu, E.; Liu, X.; Eisenberg, J.; Liu, J.; Blenis, J.; et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev. Cell. 2010, 18, 1041–1052. [Google Scholar] [CrossRef]
- Balakirev, M.Y.; Tcherniuk, S.O.; Jaquinod, M.; Chroboczek, J. Otubains: A new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 2003, 4, 517–522. [Google Scholar] [CrossRef]
- Xia, Q.; Liao, L.; Cheng, D.; Duong, D.M.; Gearing, M.; Lah, J.J.; Levey, A.I.; Peng, J. Proteomic identification of novel proteins associated with Lewy bodies. Front. Biosci. 2008, 13, 3850–3856. [Google Scholar] [CrossRef]
- Nakamura, T.; Cho, D.H.; Lipton, S.A. Redox regulation of protein misfolding, mitochondrial dysfunction, synaptic damage, and cell death in neurodegenerative diseases. Exp. Neurol. 2012, 238, 12–21. [Google Scholar] [CrossRef] [PubMed]
- An, W.; Yao, S.; Sun, X.; Hou, Z.; Lin, Y.; Su, L.; Liu, X. Glucocorticoid modulatory element-binding protein 1 (GMEB1) interacts with the de-ubiquitinase USP40 to stabilize CFLARL and inhibit apoptosis in human non-small cell lung cancer cells. J. Exp. Clin. Cancer. Res. 2019, 38, 181. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.H.; Wang, H.; Ji, X.Y.; Zhang, F.X.; Gao, B.L.; Hu, J.A.; Zheng, J. A detrimental mutation on USP40 unlocks the tumorigenesis in a rare case of lung cancer. Int. J. Clin. Exp. Pathol. 2019, 12, 740–749. [Google Scholar] [PubMed]
- Cleynen, I.; Vazeille, E.; Artieda, M.; Verspaget, H.W.; Szczypiorska, M.; Bringer, M.A.; Lakatos, P.L.; Seibold, F.; Parnell, K.; Weersma, R.K.; et al. Genetic and microbial factors modulating the ubiquitin proteasome system in inflammatory bowel disease. Gut 2014, 63, 1265–1274. [Google Scholar] [CrossRef]
- Wang, X.; Michaelis, E.K. Selective neuronal vulnerability to oxidative stress in the brain. Front. Aging Neurosci. 2010, 2, 12. [Google Scholar] [CrossRef]

| Protein Name | Chromosomal Location | PD-Related Function |
|---|---|---|
| USP15 | 12q14.1 |
|
| USP30 | 12q24.11 | |
| USP33 (VDU1) | 1p31.1 |
|
| USP35 | 11q14.1 |
|
| USP8 | 15q11.2-q21 |
|
| Ataxin-3 | 14q32.1 |
|
| USP13 | 3q26.33 | |
| UCH-L1 | 4p14 |
|
| USP9X | Xp11.4 | |
| USP14 | 18p11.32 | |
| USP24 | 1p31.1 |
|
| USP36 | 17q25.3 | |
| OTUB1 | 11q13.1 |
|
| USP40 | 2q37.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nielsen, P.Y.Ø.; Okarmus, J.; Meyer, M. Role of Deubiquitinases in Parkinson’s Disease—Therapeutic Perspectives. Cells 2023, 12, 651. https://doi.org/10.3390/cells12040651
Nielsen PYØ, Okarmus J, Meyer M. Role of Deubiquitinases in Parkinson’s Disease—Therapeutic Perspectives. Cells. 2023; 12(4):651. https://doi.org/10.3390/cells12040651
Chicago/Turabian StyleNielsen, Pernille Y. Ø., Justyna Okarmus, and Morten Meyer. 2023. "Role of Deubiquitinases in Parkinson’s Disease—Therapeutic Perspectives" Cells 12, no. 4: 651. https://doi.org/10.3390/cells12040651
APA StyleNielsen, P. Y. Ø., Okarmus, J., & Meyer, M. (2023). Role of Deubiquitinases in Parkinson’s Disease—Therapeutic Perspectives. Cells, 12(4), 651. https://doi.org/10.3390/cells12040651







