The RUNX/CBFβ Complex in Breast Cancer: A Conundrum of Context
Abstract
1. Introduction
2. The RUNX/CBFβ Transcription Complex
3. RUNX1 and CBFβ in Breast Cancer: The Enigmatic Duo
4. RUNX2 in Breast Cancer: Mediator of Metastasis
5. RUNX3 in Breast Cancer: Putative Tumor Suppressor
6. Runx Genes in Mammary Development and Homeostasis
7. Relationship of RUNX/CBFβ with ER Signaling
8. CBFβ as an Emerging Regulator in Breast Cancer
8.1. CBFβ as a Gate Keeper
8.2. CBFβ as a Driver of Tumorigenesis
9. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Dawson, S.-J.; Rueda, O.M.; Aparicio, S.; Caldas, C. A new genome-driven integrated classification of breast cancer and its implications. EMBO J. 2013, 32, 617–628. [Google Scholar] [CrossRef]
- Taneja, P.; Maglic, D.; Kai, F.; Zhu, S.; Kendig, R.D.; Elizabeth, A.F.; Inoue, K. Classical and Novel Prognostic Markers for Breast Cancer and their Clinical Significance. Clin. Med. Insights Oncol. 2010, 4, 15–34. [Google Scholar] [CrossRef]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Chang, M.-S. Tamoxifen Resistance in Breast Cancer. Biomol. Ther. 2012, 20, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto-Ibusuki, M.; Arnedos, M.; André, F. Targeted therapies for ER+/HER2- metastatic breast cancer. BMC Med. 2015, 13, 137. [Google Scholar] [CrossRef] [PubMed]
- Ring, A.; Dowsett, M. Mechanisms of tamoxifen resistance. Endocr. Relat. Cancer 2004, 11, 643–658. [Google Scholar] [CrossRef]
- Hultsch, S.; Kankainen, M.; Paavolainen, L.; Kovanen, R.-M.; Ikonen, E.; Kangaspeska, S.; Pietiäinen, V.; Kallioniemi, O. Association of tamoxifen resistance and lipid reprogramming in breast cancer. BMC Cancer 2018, 18, 850. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K.W. Cancer genes and the pathways they control. Nat. Med. 2004, 10, 789–799. [Google Scholar] [CrossRef]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015, 163, 506–519. [Google Scholar] [CrossRef]
- Nik-Zainal, S.; Davies, H.; Staaf, J.; Ramakrishna, M.; Glodzik, D.; Zou, X.; Martincorena, I.; Alexandrov, L.B.; Martin, S.; Wedge, D.C.; et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016, 534, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304.e6. [Google Scholar] [CrossRef] [PubMed]
- Ellrott, K.; Bailey, M.H.; Saksena, G.; Covington, K.R.; Kandoth, C.; Stewart, C.; Hess, J.; Ma, S.; Chiotti, K.E.; McLellan, M.; et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst. 2018, 6, 271–281.e7. [Google Scholar] [CrossRef]
- Taylor, A.M.; Shih, J.; Ha, G.; Gao, G.F.; Zhang, X.; Berger, A.C.; Schumacher, S.E.; Wang, C.; Hu, H.; Liu, J.; et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018, 33, 676–689.e3. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.M.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e11. [Google Scholar] [CrossRef]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e10. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Liang, W.-W.; Foltz, S.M.; Mutharasu, G.; Jayasinghe, R.G.; Cao, S.; Liao, W.-W.; Reynolds, S.M.; Wyczalkowski, M.A.; Yao, L.; et al. Driver Fusions and Their Implications in the Development and Treatment of Human Cancers. Cell Rep. 2018, 23, 227–238.e3. [Google Scholar] [CrossRef]
- Bhandari, V.; Hoey, C.; Liu, L.Y.; Lalonde, E.; Ray, J.; Livingstone, J.; Lesurf, R.; Shiah, Y.-J.; Vujcic, T.; Huang, X.; et al. Molecular landmarks of tumor hypoxia across cancer types. Nat. Genet. 2019, 51, 308–318. [Google Scholar] [CrossRef]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef]
- Ding, L.; Bailey, M.H.; Porta-Pardo, E.; Thorsson, V.; Colaprico, A.; Bertrand, D.; Gibbs, D.L.; Weerasinghe, A.; Huang, K.-L.; Tokheim, C.; et al. Perspective on Oncogenic Processes at the End of the Beginning of Cancer Genomics. Cell 2018, 173, 305–320.e10. [Google Scholar] [CrossRef]
- Bonneville, R.; Krook, M.A.; Kautto, E.A.; Miya, J.; Wing, M.R.; Chen, H.-Z.; Reeser, J.W.; Yu, L.; Roychowdhury, S. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis. Oncol. 2017, 2017, 1–15. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.E.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Pereira, B.; Chin, S.-F.; Rueda, O.M.; Vollan, H.-K.M.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.; Russell, R.; Sammut, S.-J.; et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat. Commun. 2016, 7, 11479. [Google Scholar] [CrossRef]
- van Wijnen, A.J.; Stein, G.S.; Gergen, J.P.; Groner, Y.; Hiebert, S.W.; Ito, Y.; Liu, P.; Neil, J.C.; Ohki, M.; Speck, N. Nomenclature for Runt-related (RUNX) proteins. Oncogene 2004, 23, 4209–4210. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, Q.; E Crute, B.; Melnikova, I.N.; Keller, S.R.; A Speck, N. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol. Cell. Biol. 1993, 13, 3324–3339. [Google Scholar] [CrossRef] [PubMed]
- Kamachi, Y.; Ogawa, E.; Asano, M.; Ishida, S.; Murakami, Y.; Satake, M.; Ito, Y.; Shigesada, K. Purification of a mouse nuclear factor that binds to both the A and B cores of the polyomavirus enhancer. J. Virol. 1990, 64, 4808–4819. [Google Scholar] [CrossRef]
- Wang, S.W.; A Speck, N. Purification of core-binding factor, a protein that binds the conserved core site in murine leukemia virus enhancers. Mol. Cell. Biol. 1992, 12, 89–102. [Google Scholar] [CrossRef]
- Mevel, R.; Draper, J.E.; Lie-A-Ling, M.; Kouskoff, V.; Lacaud, G. RUNX transcription factors: Orchestrators of development. Development 2019, 146, dev148296. [Google Scholar] [CrossRef] [PubMed]
- Voon, D.; Hor, Y.T.; Ito, Y. The RUNX complex: Reaching beyond haematopoiesis into immunity. Immunology 2015, 146, 523–536. [Google Scholar] [CrossRef]
- De Bruijn, M.; Dzierzak, E. Runx transcription factors in the development and function of the definitive hematopoietic system. Blood 2017, 129, 2061–2069. [Google Scholar] [CrossRef]
- Komori, T. Regulation of skeletal development by the Runx family of transcription factors. J. Cell. Biochem. 2005, 95, 445–453. [Google Scholar] [CrossRef]
- Komori, T. Regulation of Proliferation, Differentiation and Functions of Osteoblasts by Runx2. Int. J. Mol. Sci. 2019, 20, 1694. [Google Scholar] [CrossRef]
- Inoue, K.-I.; Ozaki, S.; Shiga, T.; Ito, K.; Masuda, T.; Okado, N.; Iseda, T.; Kawaguchi, S.; Ogawa, M.; Bae, S.-C.; et al. Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nat. Neurosci. 2002, 5, 946–954. [Google Scholar] [CrossRef]
- Levanon, D.; Bettoun, D.; Harris-Cerruti, C.; Woolf, E.; Negreanu, V.; Eilam, R.; Bernstein, Y.; Goldenberg, D.; Xiao, C.; Fliegauf, M.; et al. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 2002, 21, 3454–3463. [Google Scholar] [CrossRef]
- Boto, P.; Csuth, T.I.; Szatmári, I. RUNX3-Mediated Immune Cell Development and Maturation. Crit. Rev. Immunol. 2018, 38, 63–78. [Google Scholar] [CrossRef]
- Malik, N.; Yan, H.; Moshkovich, N.; Palangat, M.; Yang, H.; Sanchez, V.; Cai, Z.; Peat, T.J.; Jiang, S.; Liu, C.; et al. The transcription factor CBFB suppresses breast cancer through orchestrating translation and transcription. Nat. Commun. 2019, 10, 2071. [Google Scholar] [CrossRef]
- Chimge, N.-O.; Frenkel, B. The RUNX family in breast cancer: Relationships with estrogen signaling. Oncogene 2013, 32, 2121–2130. [Google Scholar] [CrossRef]
- Blyth, K.; Cameron, E.R.; Neil, J.C. The runx genes: Gain or loss of function in cancer. Nat. Rev. Cancer 2005, 5, 376–387. [Google Scholar] [CrossRef]
- Kagoshima, H.; Shigesada, K.; Satake, M.; Ito, Y.; Miyoshi, H.; Ohki, M.; Pepling, M.; Gergen, P. The runt domain identifies a new family of heterometric transcriptional regulators. Trends Genet. 1993, 9, 338–341. [Google Scholar] [CrossRef]
- Bravo, J.; Li, Z.; A Speck, N.; Warren, A.J. The leukemia-associated AML1 (Runx1)--CBFβ complex functions as a DNA-induced molecular clamp. Nat. Struct. Biol. 2001, 8, 371–378. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Hajra, A.; Collins, F.S. Structure of the leukemia-associated human CBFB gene. Genomics 1995, 26, 571–579. [Google Scholar] [CrossRef]
- Tahirov, T.H.; Inoue-Bungo, T.; Morii, H.; Fujikawa, A.; Sasaki, M.; Kimura, K.; Shiina, M.; Sato, K.; Kumasaka, T.; Yamamoto, M.; et al. Structural Analyses of DNA Recognition by the AML1/Runx-1 Runt Domain and Its Allosteric Control by CBFβ. Cell 2001, 104, 755–767. [Google Scholar] [CrossRef]
- Wang, Q.; Stacy, T.; Miller, J.D.; Lewis, A.F.; Gu, T.-L.; Huang, X.; Bushweller, J.H.; Bories, J.-C.; Alt, F.W.; Ryan, G.; et al. The CBFβ Subunit Is Essential for CBFα2 (AML1) Function In Vivo. Cell 1996, 87, 697–708. [Google Scholar] [CrossRef]
- Ogawa, E.; Inuzuka, M.; Maruyama, M.; Satake, M.; Fujimoto, M.N.; Ito, Y.; Shigesada, K. Molecular Cloning and Characterization of PEBP2β, the Heterodimeric Partner of a Novel Drosophila runt-Related DNA Binding Protein PEBP2α. Virology 1993, 194, 314–331. [Google Scholar] [CrossRef]
- Bushweller, J.H.; Huang, X.; Peng, J.W.; Speck, N.A. Solution structure of core binding factor β and map of the CBFα binding site. Nat. Struct. Biol. 1999, 6, 624–627. [Google Scholar] [CrossRef]
- Yan, J.; Liu, Y.; Lukasik, S.M.; A Speck, N.; Bushweller, J.H. CBFβ allosterically regulates the Runx1 Runt domain via a dynamic conformational equilibrium. Nat. Struct. Mol. Biol. 2004, 11, 901–906. [Google Scholar] [CrossRef]
- Gu, T.-L.; Goetz, T.L.; Graves, B.J.; Speck, N.A. Auto-Inhibition and Partner Proteins, Core-Binding Factor β (CBFβ) and Ets-1, Modulate DNA Binding by CBFα2 (AML1). Mol. Cell. Biol. 2000, 20, 91–103. [Google Scholar] [CrossRef]
- Huang, G.; Shigesada, K.; Ito, K.; Wee, H.; Yokomizo, T.; Ito, Y. Dimerization with PEBP2β protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 2001, 20, 723–733. [Google Scholar] [CrossRef]
- Otto, F.; Lübbert, M.; Stock, M. Upstream and downstream targets of RUNX proteins. J. Cell. Biochem. 2003, 89, 9–18. [Google Scholar] [CrossRef]
- The UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bye-A-Jee, H.; et al. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Kurokawa, M.; Imai, Y.; Izutsu, K.; Asai, T.; Ichikawa, M.; Yamamoto, G.; Nitta, E.; Yamagata, T.; Sasaki, K.; et al. AML1 Is Functionally Regulated through p300-mediated Acetylation on Specific Lysine Residues. J. Biol. Chem. 2004, 279, 15630–15638. [Google Scholar] [CrossRef]
- Jin, Y.-H.; Jeon, E.-J.; Li, Q.-L.; Lee, Y.H.; Choi, J.-K.; Kim, W.-J.; Lee, K.-Y.; Bae, S.-C. Transforming Growth Factor-β Stimulates p300-dependent RUNX3 Acetylation, Which Inhibits Ubiquitination-mediated Degradation. J. Biol. Chem. 2004, 279, 29409–29417. [Google Scholar] [CrossRef]
- Westendorf, J.J.; Hiebert, S.W. Mammalian runt-domain proteins and their roles in hematopoiesis, osteogenesis, and leukemia. J. Cell. Biochem. 1999, 75, 51–58. [Google Scholar] [CrossRef]
- Chuang, L.S.H.; Ito, K.; Ito, Y. RUNX family: Regulation and diversification of roles through interacting proteins. Int. J. Cancer 2013, 132, 1260–1271. [Google Scholar] [CrossRef]
- Aikawa, Y.; Nguyen, L.A.; Isono, K.; Takakura, N.; Tagata, Y.; Schmitz, M.L.; Koseki, H.; Kitabayashi, I. Roles of HIPK1 and HIPK2 in AML1- and p300-dependent transcription, hematopoiesis and blood vessel formation. EMBO J. 2006, 25, 3955–3965. [Google Scholar] [CrossRef]
- Tanaka, T.; Kurokawa, M.; Ueki, K.; Tanaka, K.; Imai, Y.; Mitani, K.; Okazaki, K.; Sagata, N.; Yazaki, Y.; Shibata, Y.; et al. The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol. Cell. Biol. 1996, 16, 3967–3979. [Google Scholar] [CrossRef]
- Aho, T.L.; Sandholm, J.; Peltola, K.J.; Ito, Y.; Koskinen, P.J. Pim-1 kinase phosphorylates RUNX family transcription factors and enhances their activity. BMC Cell Biol. 2006, 7, 21. [Google Scholar] [CrossRef]
- Kim, H.-R.; Oh, B.-C.; Choi, J.-K.; Bae, S.-C. Pim-1 kinase phosphorylates and stabilizes RUNX3 and alters its subcellular localization. J. Cell. Biochem. 2008, 105, 1048–1058. [Google Scholar] [CrossRef]
- Wee, H.-J.; Voon, D.C.-C.; Bae, S.-C.; Ito, Y. PEBP2-β/CBF-β–dependent phosphorylation of RUNX1 and p300 by HIPK2: Implications for leukemogenesis. Blood 2008, 112, 3777–3787. [Google Scholar] [CrossRef]
- Jeon, E.-J.; Lee, K.-Y.; Choi, N.-S.; Lee, M.-H.; Kim, H.-N.; Jin, Y.-H.; Ryoo, H.-M.; Choi, J.-Y.; Yoshida, M.; Nishino, N.; et al. Bone Morphogenetic Protein-2 Stimulates Runx2 Acetylation. J. Biol. Chem. 2006, 281, 16502–16511. [Google Scholar] [CrossRef]
- Zhao, X.; Jankovic, V.; Gural, A.; Huang, G.; Pardanani, A.; Menendez, S.; Zhang, J.; Dunne, R.; Xiao, A.; Erdjument-Bromage, H.; et al. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008, 22, 640–653. [Google Scholar] [CrossRef]
- Chi, X.-Z.; Kim, J.; Lee, Y.-H.; Lee, J.-W.; Lee, K.-S.; Wee, H.; Kim, W.-J.; Park, W.-Y.; Oh, B.-C.; Stein, G.S.; et al. Runt-Related Transcription Factor RUNX3 Is a Target of MDM2-Mediated Ubiquitination. Cancer Res 2009, 69, 8111–8119. [Google Scholar] [CrossRef]
- Huang, G.; Zhao, X.; Wang, L.; Elf, S.; Xu, H.; Zhao, X.; Sashida, G.; Zhang, Y.; Liu, Y.; Lee, J.; et al. The ability of MLL to bind RUNX1 and methylate H3K4 at PU.1 regulatory regions is impaired by MDS/AML-associated RUNX1/AML1 mutations. Blood 2011, 118, 6544–6552. [Google Scholar] [CrossRef]
- Zhao, M.; Qiao, M.; Oyajobi, B.O.; Mundy, G.R.; Chen, D. E3 Ubiquitin Ligase Smurf1 Mediates Core-binding Factor α1/Runx2 Degradation and Plays A Specific Role in Osteoblast Differentiation. J. Biol. Chem. 2003, 278, 27939–27944. [Google Scholar] [CrossRef]
- Pelletier, N.; Champagne, N.; Stifani, S.; Yang, X.-J. MOZ and MORF histone acetyltransferases interact with the Runt-domain transcription factor Runx2. Oncogene 2002, 21, 2729–2740. [Google Scholar] [CrossRef]
- Kitabayashi, I.; Aikawa, Y.; Nguyen, L.A.; Yokoyama, A.; Ohki, M. Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ-CBP fusion protein. EMBO J. 2001, 20, 7184–7196. [Google Scholar] [CrossRef]
- Bakshi, R.; Hassan, M.Q.; Pratap, J.; Lian, J.B.; Montecino, M.A.; van Wijnen, A.J.; Stein, J.L.; Imbalzano, A.N.; Stein, G.S. The human SWI/SNF complex associates with RUNX1 to control transcription of hematopoietic target genes. J. Cell. Physiol. 2010, 225, 569–576. [Google Scholar] [CrossRef]
- Villagra, A.; Cruzat, F.; Carvallo, L.; Paredes, R.; Olate, J.; van Wijnen, A.J.; Stein, G.S.; Lian, J.B.; Stein, J.L.; Imbalzano, A.N.; et al. Chromatin Remodeling and Transcriptional Activity of the Bone-specific Osteocalcin Gene Require CCAAT/Enhancer-binding Protein β-dependent Recruitment of SWI/SNF Activity. J. Biol. Chem. 2006, 281, 22695–22706. [Google Scholar] [CrossRef]
- Martinez, M.; Hinojosa, M.; Trombly, D.; Morin, V.; Stein, J.; Stein, G.; Javed, A.; Gutierrez, S.E. Transcriptional Auto-Regulation of RUNX1 P1 Promoter. PLoS ONE 2016, 11, e0149119. [Google Scholar] [CrossRef] [PubMed]
- Ghozi, M.C.; Bernstein, Y.; Negreanu, V.; Levanon, D.; Groner, Y. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc. Natl. Acad. Sci. USA 1996, 93, 1935–1940. [Google Scholar] [CrossRef] [PubMed]
- Spender, L.C.; Whiteman, H.J.; Karstegl, C.E.; Farrell, P.J. Transcriptional cross-regulation of RUNX1 by RUNX3 in human B cells. Oncogene 2005, 24, 1873–1881. [Google Scholar] [CrossRef] [PubMed]
- Okuda, T.; van Deursen, J.; Hiebert, S.W.; Grosveld, G.; Downing, J.R. AML1, the Target of Multiple Chromosomal Translocations in Human Leukemia, Is Essential for Normal Fetal Liver Hematopoiesis. Cell 1996, 84, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Stacy, T.; Binder, M.; Marin-Padilla, M.; Sharpe, A.H.; Speck, N.A. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA 1996, 93, 3444–3449. [Google Scholar] [CrossRef]
- Komori, T.; Yagi, H.; Nomura, S.; Yamaguchi, A.; Sasaki, K.; Deguchi, K.; Shimizu, Y.; Bronson, R.T.; Gao, Y.H.; Inada, M.; et al. Targeted Disruption of Cbfa1 Results in a Complete Lack of Bone Formation owing to Maturational Arrest of Osteoblasts. Cell 1997, 89, 755–764. [Google Scholar] [CrossRef]
- Otto, F.; Thornell, A.P.; Crompton, T.; Denzel, A.; Gilmour, K.C.; Rosewell, I.R.; Stamp, G.W.; Beddington, R.S.; Mundlos, S.; Olsen, B.R.; et al. Cbfa1, a Candidate Gene for Cleidocranial Dysplasia Syndrome, Is Essential for Osteoblast Differentiation and Bone Development. Cell 1997, 89, 765–771. [Google Scholar] [CrossRef]
- Kundu, M.; Javed, A.; Jeon, J.-P.; Horner, A.; Shum, L.; Eckhaus, M.; Muenke, M.; Lian, J.B.; Yang, Y.; Nuckolls, G.H.; et al. Cbfβ interacts with Runx2 and has a critical role in bone development. Nat. Genet. 2002, 32, 639–644. [Google Scholar] [CrossRef]
- Yoshida, C.A.; Furuichi, T.; Fujita, T.; Fukuyama, R.; Kanatani, N.; Kobayashi, S.; Satake, M.; Takada, K.; Komori, T. Core-binding factor β interacts with Runx2 and is required for skeletal development. Nat. Genet. 2002, 32, 633–638. [Google Scholar] [CrossRef]
- Bauer, O.; Sharir, A.; Kimura, A.; Hantisteanu, S.; Takeda, S.; Groner, Y. Loss of Osteoblast Runx3 Produces Severe Congenital Osteopenia. Mol. Cell. Biol. 2015, 35, 1097–1109. [Google Scholar] [CrossRef]
- Woolf, E.; Xiao, C.; Fainaru, O.; Lotem, J.; Rosen, D.; Negreanu, V.; Bernstein, Y.; Goldenberg, D.; Brenner, O.; Berke, G.; et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl. Acad. Sci. USA 2003, 100, 7731–7736. [Google Scholar] [CrossRef]
- Brenner, O.; Levanon, D.; Negreanu, V.; Golubkov, O.; Fainaru, O.; Woolf, E.; Groner, Y. Loss of Runx3 function in leukocytes is associated with spontaneously developed colitis and gastric mucosal hyperplasia. Proc. Natl. Acad. Sci. USA 2004, 101, 16016–16021. [Google Scholar] [CrossRef] [PubMed]
- Fainaru, O.; Woolf, E.; Lotem, J.; Yarmus, M.; Brenner, O.; Goldenberg, D.; Negreanu, V.; Bernstein, Y.; Levanon, D.; Jung, S.; et al. Runx3 regulates mouse TGF-β-mediated dendritic cell function and its absence results in airway inflammation. EMBO J. 2004, 23, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Taniuchi, I.; Osato, M.; Egawa, T.; Sunshine, M.J.; Bae, S.C.; Komori, T.; Ito, Y.; Littman, D.R. Differential Requirements for Runx Proteins in CD4 Repression and Epigenetic Silencing during T Lymphocyte Development. Cell 2002, 111, 621–633. [Google Scholar] [CrossRef]
- Rooney, N.; Riggio, A.I.; Mendoza-Villanueva, D.; Shore, P.; Cameron, E.R.; Blyth, K. Runx Genes in Breast Cancer and the Mammary Lineage. In Advances in Experimental Medicine and Biology; Springer: Singapore, 2017; Volume 962, pp. 353–368. [Google Scholar] [CrossRef]
- Rojas, A.; Otálora-Otálora, B.A.; Henríquez, B.; López-Kleine, L. RUNX family: Oncogenes or tumor suppressors (Review). Oncol. Rep. 2019, 42, 3–19. [Google Scholar] [CrossRef]
- Ito, Y.; Bae, S.-C.; Chuang, L.S.H. The RUNX family: Developmental regulators in cancer. Nat. Rev. Cancer 2015, 15, 81–95. [Google Scholar] [CrossRef]
- Chuang, L.S.H.; Ito, K.; Ito, Y. Roles of RUNX in Solid Tumors. In Advances in Experimental Medicine and Biology; Springer New York LLC: New York, NY, USA, 2017; Volume 962, pp. 299–320. [Google Scholar] [CrossRef]
- Sweeney, K.; Cameron, E.R.; Blyth, K. Complex Interplay between the RUNX Transcription Factors and Wnt/β-Catenin Pathway in Cancer: A Tango in the Night. Mol. Cells 2020, 43, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Chimge, N.-O.; Ahmed-Alnassar, S.; Frenkel, B. Relationship between RUNX1 and AXIN1 in ER-negative versus ER-positive Breast Cancer. Cell Cycle 2017, 16, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Riggio, A.I.; Blyth, K. The enigmatic role of RUNX1 in female-related cancers—current knowledge & future perspectives. FEBS J. 2017, 284, 2345–2362. [Google Scholar] [CrossRef]
- Ramaswamy, S.; Ross, K.N.; Lander, E.S.; Golub, T.R. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 2002, 33, 49–54. [Google Scholar] [CrossRef]
- Ellis, M.J.; Ding, L.; Shen, D.; Luo, J.; Suman, V.J.; Wallis, J.W.; Van Tine, B.A.; Hoog, J.; Goiffon, R.J.; Goldstein, T.C.; et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 2012, 486, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Banerji, S.; Cibulskis, K.; Rangel-Escareno, C.; Brown, K.K.; Carter, S.L.; Frederick, A.M.; Lawrence, M.S.; Sivachenko, A.Y.; Sougnez, C.; Zou, L.; et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 2012, 486, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Kadota, M.; Yang, H.H.; Gomez, B.; Sato, M.; Clifford, R.J.; Meerzaman, D.; Dunn, B.K.; Wakefield, L.; Lee, M.P. Delineating Genetic Alterations for Tumor Progression in the MCF10A Series of Breast Cancer Cell Lines. PLoS ONE 2010, 5, e9201. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Messier, T.L.; Tye, C.E.; Dobson, J.R.; Fritz, A.J.; Sikora, K.R.; Browne, G.; Stein, J.L.; Lian, J.B.; Stein, G.S. Runx1 stabilizes the mammary epithelial cell phenotype and prevents epithelial to mesenchymal transition. Oncotarget 2017, 8, 17610–17627. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Brugge, J.S.; Janes, K.A. Intersection of FOXO- and RUNX1-mediated gene expression programs in single breast epithelial cells during morphogenesis and tumor progression. Proc. Natl. Acad. Sci. USA 2011, 108, E803–E812. [Google Scholar] [CrossRef]
- Chimge, N.-O.; Little, G.H.; Baniwal, S.K.; Adisetiyo, H.; Xie, Y.; Zhang, T.; O’Laughlin, A.; Liu, Z.Y.; Ulrich, P.; Martin, A.; et al. RUNX1 prevents oestrogen-mediated AXIN1 suppression and β-catenin activation in ER-positive breast cancer. Nat. Commun. 2016, 7, 10751. [Google Scholar] [CrossRef]
- Kulkarni, M.; Tan, T.Z.; Sulaiman, N.B.S.; Lamar, J.M.; Bansal, P.; Cui, J.; Qiao, Y.; Ito, Y. RUNX1 and RUNX3 protect against YAP-mediated EMT, stem-ness and shorter survival outcomes in breast cancer. Oncotarget 2018, 9, 14175–14192. [Google Scholar] [CrossRef]
- Berx, G.; Cleton-Jansen, A.M.; Nollet, F.; de Leeuw, W.J.; van de Vijver, M.; Cornelisse, C.; van Roy, F. E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. EMBO J. 1995, 14, 6107–6115. [Google Scholar] [CrossRef]
- Birchmeier, W.; Behrens, J. Cadherin expression in carcinomas: Role in the formation of cell junctions and the prevention of invasiveness. Biochim. Biophys. Acta 1994, 1198, 11–26. [Google Scholar] [CrossRef]
- Padmanaban, V.; Krol, I.; Suhail, Y.; Szczerba, B.M.; Aceto, N.; Bader, J.S.; Ewald, A.J. E-cadherin is required for metastasis in multiple models of breast cancer. Nature 2019, 573, 439–444. [Google Scholar] [CrossRef]
- Liu, Y.-N.; Lee, W.-W.; Wang, C.-Y.; Chao, T.-H.; Chen, Y.; Chen, J.H. Regulatory mechanisms controlling human E-cadherin gene expression. Oncogene 2005, 24, 8277–8290. [Google Scholar] [CrossRef] [PubMed]
- Karn, T.; Pusztai, L.; Holtrich, U.; Iwamoto, T.; Shiang, C.Y.; Schmidt, M.; Müller, V.; Solbach, C.; Gaetje, R.; Hanker, L.; et al. Homogeneous Datasets of Triple Negative Breast Cancers Enable the Identification of Novel Prognostic and Predictive Signatures. PLoS ONE 2011, 6, e28403. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [PubMed]
- Rody, A.; Karn, T.; Liedtke, C.; Pusztai, L.; Ruckhaeberle, E.; Hanker, L.; Gaetje, R.; Solbach, C.; Ahr, A.; Metzler, D.; et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011, 13, R97. [Google Scholar] [CrossRef]
- Scheitz, C.J.F.; Lee, T.S.; McDermitt, D.J.; Tumbar, T. Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J. 2012, 31, 4124–4139. [Google Scholar] [CrossRef]
- Ferrari, N.; Mohammed, Z.M.A.; Nixon, C.; Mason, S.M.; Mallon, E.; McMillan, N.C.; Morris, J.S.; Cameron, E.R.; Edwards, J.; Blyth, K. Expression of RUNX1 Correlates with Poor Patient Prognosis in Triple Negative Breast Cancer. PLoS ONE 2014, 9, e100759. [Google Scholar] [CrossRef]
- Browne, G.; Dragon, J.A.; Hong, D.; Messier, T.L.; Gordon, J.A.R.; Farina, N.H.; Boyd, J.R.; VanOudenhove, J.J.; Perez, A.W.; Zaidi, S.K.; et al. MicroRNA-378-mediated suppression of Runx1 alleviates the aggressive phenotype of triple-negative MDA-MB-231 human breast cancer cells. Tumor Biol. 2016, 37, 8825–8839. [Google Scholar] [CrossRef]
- Browne, G.; Taipaleenmäki, H.; Bishop, N.M.; Madasu, S.C.; Shaw, L.M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. Runx1 is associated with breast cancer progression in MMTV-PyMT transgenic mice and its depletion in vitro inhibits migration and invasion. J. Cell. Physiol. 2015, 230, 2522–2532. [Google Scholar] [CrossRef]
- Hnisz, D.; Abraham, B.J.; Lee, T.I.; Lau, A.; Saint-André, V.; Sigova, A.A.; Hoke, H.A.; Young, R.A. Super-Enhancers in the Control of Cell Identity and Disease. Cell 2013, 155, 934–947. [Google Scholar] [CrossRef]
- Recouvreux, M.S.; Grasso, E.N.; Echeverria, P.C.; Rocha-Viegas, L.; Castilla, L.H.; Schere-Levy, C.; Tocci, J.M.; Kordon, E.C.; Rubinstein, N. RUNX1 and FOXP3 interplay regulates expression of breast cancer related genes. Oncotarget 2016, 7, 6552–6565. [Google Scholar] [CrossRef]
- Daino, K.; Nishimura, M.; Imaoka, T.; Takabatake, M.; Morioka, T.; Nishimura, Y.; Shimada, Y.; Kakinuma, S. Epigenetic dysregulation of key developmental genes in radiation-induced rat mammary carcinomas. Int. J. Cancer 2018, 143, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Janes, K.A. RUNX1 and its understudied role in breast cancer. Cell Cycle 2011, 10, 3461–3465. [Google Scholar] [CrossRef] [PubMed]
- Chimge, N.-O.; Baniwal, S.K.; Luo, J.; Coetzee, S.; Khalid, O.; Berman, B.P.; Tripathy, D.; Ellis, M.J.; Frenkel, B. Opposing Effects of Runx2 and Estradiol on Breast Cancer Cell Proliferation: In Vitro Identification of Reciprocally Regulated Gene Signature Related to Clinical Letrozole Responsiveness. Clin. Cancer Res. 2012, 18, 901–911. [Google Scholar] [CrossRef]
- Khalid, O.; Baniwal, S.K.; Purcell, D.J.; Leclerc, N.; Gabet, Y.; Stallcup, M.R.; Coetzee, G.A.; Frenkel, B. Modulation of Runx2 Activity by Estrogen Receptor-α: Implications for Osteoporosis and Breast Cancer. Endocrinology 2008, 149, 5984–5995. [Google Scholar] [CrossRef]
- Pratap, J.; Lian, J.B.; Javed, A.; Barnes, G.L.; Van Wijnen, A.J.; Stein, J.L.; Stein, G.S. Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metastasis Rev. 2006, 25, 589–600. [Google Scholar] [CrossRef]
- Ferrari, N.; McDonald, L.; Morris, J.S.; Cameron, E.R.; Blyth, K. RUNX2 in mammary gland development and breast cancer. J. Cell. Physiol. 2013, 228, 1137–1142. [Google Scholar] [CrossRef]
- Wysokinski, D.; Blasiak, J.; Pawlowska, E. Role of RUNX2 in Breast Carcinogenesis. Int. J. Mol. Sci. 2015, 16, 20969–20993. [Google Scholar] [CrossRef] [PubMed]
- Javed, A.; Barnes, G.L.; Pratap, J.; Antkowiak, T.; Gerstenfeld, L.C.; van Wijnen, A.J.; Stein, J.L.; Lian, J.B.; Stein, G.S. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc. Natl. Acad. Sci. USA 2005, 102, 1454–1459. [Google Scholar] [CrossRef]
- Pratap, J.; Wixted, J.J.; Gaur, T.; Zaidi, S.K.; Dobson, J.; Gokul, K.D.; Hussain, S.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; et al. Runx2 Transcriptional Activation of Indian Hedgehog and a Downstream Bone Metastatic Pathway in Breast Cancer Cells. Cancer Res 2008, 68, 7795–7802. [Google Scholar] [CrossRef]
- Pratap, J.; Lian, J.B.; Stein, G.S. Metastatic bone disease: Role of transcription factors and future targets. Bone 2011, 48, 30–36. [Google Scholar] [CrossRef]
- Leong, D.T.; Lim, J.; Goh, X.; Pratap, J.; Pereira, B.P.; Kwok, H.S.; Nathan, S.S.; Dobson, J.R.; Lian, J.B.; Ito, Y.; et al. Cancer-related ectopic expression of the bone-related transcription factor RUNX2 in non-osseous metastatic tumor cells is linked to cell proliferation and motility. Breast Cancer Res. 2010, 12, R89. [Google Scholar] [CrossRef] [PubMed]
- Pande, S.; Browne, G.; Padmanabhan, S.; Zaidi, S.K.; Lian, J.B.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S. Oncogenic cooperation between PI3K/Akt signaling and transcription factor Runx2 promotes the invasive properties of metastatic breast cancer cells. J. Cell. Physiol. 2013, 228, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Barnes, G.L.; Hebert, K.E.; Kamal, M.; Javed, A.; Einhorn, T.A.; Lian, J.B.; Stein, G.S.; Gerstenfeld, L.C. Fidelity of Runx2 Activity in Breast Cancer Cells Is Required for the Generation of Metastases-Associated Osteolytic Disease. Cancer Res. 2004, 64, 4506–4513. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Villanueva, D.; Zeef, L.; Shore, P. Metastatic breast cancer cells inhibit osteoblast differentiation through the Runx2/CBFβ-dependent expression of the Wnt antagonist, sclerostin. Breast Cancer Res. 2011, 13, R106. [Google Scholar] [CrossRef]
- Rutkovskiy, A.; Stensløkken, K.-O.; Vaage, I.J. Osteoblast Differentiation at a Glance. Med. Sci. Monit. Basic Res. 2016, 22, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Pratap, J.; Javed, A.; Languino, L.R.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. The Runx2 Osteogenic Transcription Factor Regulates Matrix Metalloproteinase 9 in Bone Metastatic Cancer Cells and Controls Cell Invasion. Mol. Cell. Biol. 2005, 25, 8581–8591. [Google Scholar] [CrossRef]
- Baniwal, S.K.; Khalid, O.; Gabet, Y.; Shah, R.R.; Purcell, D.J.; Mav, D.; E Kohn-Gabet, A.; Shi, Y.; A Coetzee, G.; Frenkel, B. Runx2 transcriptome of prostate cancer cells: Insights into invasiveness and bone metastasis. Mol. Cancer 2010, 9, 258. [Google Scholar] [CrossRef]
- Akech, J.; Wixted, J.J.; Bedard, K.; Van Der Deen, M.; Hussain, S.; Guise, T.A.; Van Wijnen, A.J.; Stein, J.L.; Languino, L.R.; Altieri, D.C.; et al. Runx2 association with progression of prostate cancer in patients: Mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene 2010, 29, 811–821. [Google Scholar] [CrossRef]
- Bonewald, L.F.; Johnson, M.L. Osteocytes, mechanosensing and Wnt signaling. Bone 2008, 42, 606–615. [Google Scholar] [CrossRef]
- Kang, Y.; Siegel, P.M.; Shu, W.; Drobnjak, M.; Kakonen, S.M.; Cordón-Cardo, C.; Guise, T.A.; Massagué, J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003, 3, 537–549. [Google Scholar] [CrossRef]
- Onodera, Y.; Miki, Y.; Suzuki, T.; Takagi, K.; Akahira, J.-I.; Sakyu, T.; Watanabe, M.; Inoue, S.; Ishida, T.; Ohuchi, N.; et al. Runx2 in human breast carcinoma: Its potential roles in cancer progression. Cancer Sci. 2010, 101, 2670–2675. [Google Scholar] [CrossRef] [PubMed]
- Lau, Q.C.; Raja, E.; Salto-Tellez, M.; Liu, Q.; Ito, K.; Inoue, M.; Putti, T.C.; Loh, M.; Ko, T.K.; Huang, C.; et al. RUNX3 Is Frequently Inactivated by Dual Mechanisms of Protein Mislocalization and Promoter Hypermethylation in Breast Cancer. Cancer Res 2006, 66, 6512–6520. [Google Scholar] [CrossRef] [PubMed]
- McDonald, L.; Ferrari, N.; Terry, A.; Bell, M.; Mohammed, Z.M.; Orange, C.; Jenkins, A.; Muller, W.J.; Gusterson, B.A.; Neil, J.C.; et al. RUNX2 correlates with subtype-specific breast cancer in a human tissue microarray, and ectopic expression of Runx2 perturbs differentiation in the mouse mammary gland. Dis. Model. Mech. 2014, 7, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Chimge, N.-O.; Baniwal, S.K.; Little, G.H.; Chen, Y.-B.; Kahn, M.; Tripathy, D.; Borok, Z.; Frenkel, B. Regulation of breast cancer metastasis by Runx2 and estrogen signaling: The role of SNAI2. Breast Cancer Res. 2011, 13, R127. [Google Scholar] [CrossRef]
- Brusgard, J.L.; Choe, M.; Chumsri, S.; Renoud, K.; MacKerell, A.D., Jr.; Sudol, M.; Passaniti, A. RUNX2 and TAZ-dependent signaling pathways regulate soluble E-Cadherin levels and tumorsphere formation in breast cancer cells. Oncotarget 2015, 6, 28132–28150. [Google Scholar] [CrossRef]
- Kim, W.-J.; Kim, E.-J.; Jeong, P.; Quan, C.; Kim, J.; Li, Q.-L.; Yang, J.-O.; Ito, Y.; Bae, S.-C. RUNX3 Inactivation by Point Mutations and Aberrant DNA Methylation in Bladder Tumors. Cancer Res 2005, 65, 9347–9354. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.L.; Ito, K.; Sakakura, C.; Fukamachi, H.; Inoue, K.-I.; Chi, X.-Z.; Lee, K.-Y.; Nomura, S.; Lee, C.-W.; Han, S.-B.; et al. Causal Relationship between the Loss of RUNX3 Expression and Gastric Cancer. Cell 2002, 109, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-S.; Lee, Y.-S.; Lee, J.-M.; Ito, K.; Cinghu, S.; Kim, J.-H.; Jang, J.-W.; Li, Y.-H.; Goh, Y.-M.; Chi, X.-Z.; et al. Runx3 is required for the differentiation of lung epithelial cells and suppression of lung cancer. Oncogene 2010, 29, 3349–3361. [Google Scholar] [CrossRef]
- Weisenberger, D.J.; Siegmund, K.D.; Campan, M.; Young, J.; Long, T.I.; Faasse, M.A.; Kang, G.H.; Widschwendter, M.; Weener, D.; Buchanan, D.; et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006, 38, 787–793. [Google Scholar] [CrossRef]
- Nomoto, S.; Haruki, N.; Tatematsu, Y.; Konishi, H.; Mitsudomi, T.; Takahashi, T.; Takahashi, T. Frequent allelic imbalance suggests involvement of a tumor suppressor gene at 1p36 in the pathogenesis of human lung cancers. Genes, Chromosom. Cancer 2000, 28, 342–346. [Google Scholar] [CrossRef]
- Schwab, M.; Praml, C.; Amler, L.C. Genomic instability in Ip and human malignancies. Genes, Chromosom. Cancer 1996, 16, 211–229. [Google Scholar] [CrossRef]
- Ezaki, T.; Yanagisawa, A.; Ohta, K.; Aiso, S.; Watanabe, M.; Hibi, T.; Kato, Y.; Nakajima, T.E.; Ariyama, T.; Inazawa, J.; et al. Deletion mapping on chromosome 1p in well-differentiated gastric cancer. Br. J. Cancer 1996, 73, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, H.; Matsuyama, H.; Fukunaga, K.; Yoshihiro, S.; Wada, T.; Naito, K. Allelic imbalance at 1p36 may predict prognosis of chemoradiation therapy for bladder preservation in patients with invasive bladder cancer. Br. J. Cancer 2004, 91, 1025–1031. [Google Scholar] [CrossRef]
- Subramaniam, M.M.; Chan, J.Y.; Soong, R.; Ito, K.; Ito, Y.; Yeoh, K.G.; Salto-Tellez, M.; Putti, T.C. RUNX3 inactivation by frequent promoter hypermethylation and protein mislocalization constitute an early event in breast cancer progression. Breast Cancer Res. Treat. 2008, 113, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Tong, D.; Lou, G.; Zhang, Y.; Geng, J. Expression of RUNX3 Gene, Methylation Status and Clinicopathological Significance in Breast Cancer and Breast Cancer Cell Lines. Pathobiology 2008, 75, 244–251. [Google Scholar] [CrossRef]
- Huang, B.; Qu, Z.; Ong, C.W.; Tsang, Y.-H.N.; Xiao, G.; Shapiro, D.; Salto-Tellez, M.; Ito, K.; Ito, Y.; Chen, L.-F. RUNX3 acts as a tumor suppressor in breast cancer by targeting estrogen receptor α. Oncogene 2012, 31, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chen, C.; Ma, D.; Li, Y.; Yin, Q.; Li, Q.; Xiang, C. Inhibition of PIM1 attenuates the stem cell–like traits of breast cancer cells by promoting RUNX3 nuclear retention. J. Cell. Mol. Med. 2020, 24, 6308–6323. [Google Scholar] [CrossRef]
- Fu, J.; Sun, H.; Xu, F.; Chen, R.; Wang, X.; Ding, Q.; Xia, T. RUNX regulated immune-associated genes predicts prognosis in breast cancer. Front. Genet. 2022, 13, 2204. [Google Scholar] [CrossRef]
- Levanon, D.; Bernstein, Y.; Negreanu, V.; Bone, K.R.; Pozner, A.; Eilam, R.; Lotem, J.; Brenner, O.; Groner, Y. Absence of Runx3 expression in normal gastrointestinal epithelium calls into question its tumour suppressor function. EMBO Mol. Med. 2011, 3, 593–604. [Google Scholar] [CrossRef]
- Finak, G.; Bertos, N.; Pepin, F.; Sadekova, S.; Souleimanova, M.; Zhao, H.; Chen, H.; Omeroglu, G.; Meterissian, S.; Omeroglu, A.; et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat. Med. 2008, 14, 518–527. [Google Scholar] [CrossRef]
- Fritz, A.J.; Hong, D.; Boyd, J.; Kost, J.; Finstaad, K.H.; Fitzgerald, M.P.; Hanna, S.; Abuarqoub, A.H.; Malik, M.; Bushweller, J.; et al. RUNX1 and RUNX2 transcription factors function in opposing roles to regulate breast cancer stem cells. J. Cell. Physiol. 2020, 235, 7261–7272. [Google Scholar] [CrossRef] [PubMed]
- van Bragt, M.P.; Hu, X.; Xie, Y.; Li, Z. RUNX1, a transcription factor mutated in breast cancer, controls the fate of ER-positive mammary luminal cells. Elife 2014, 3, e03881. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, N.; Riggio, A.I.; Mason, S.; McDonald, L.; King, A.; Higgins, T.; Rosewell, I.; Neil, J.C.; Smalley, M.J.; Sansom, O.J.; et al. Runx2 contributes to the regenerative potential of the mammary epithelium. Sci. Rep. 2015, 5, 15658. [Google Scholar] [CrossRef]
- Sokol, E.S.; Sanduja, S.; Jin, D.X.; Miller, D.H.; Mathis, R.A.; Gupta, P.B. Perturbation-Expression Analysis Identifies RUNX1 as a Regulator of Human Mammary Stem Cell Differentiation. PLOS Comput. Biol. 2015, 11, e1004161. [Google Scholar] [CrossRef]
- Hong, D.; Fritz, A.J.; Finstad, K.H.; Fitzgerald, M.P.; Weinheimer, A.; Viens, A.L.; Ramsey, J.; Stein, J.L.; Lian, J.B.; Stein, G.S. Suppression of Breast Cancer Stem Cells and Tumor Growth by the RUNX1 Transcription Factor. Mol. Cancer Res. 2018, 16, 1952–1964. [Google Scholar] [CrossRef]
- Matsuo, J.; Mon, N.N.; Douchi, D.; Yamamura, A.; Kulkarni, M.; Heng, D.L.; Chen, S.; Nuttonmanit, N.; Li, Y.; Yang, H.; et al. A Runx1-enhancer Element eR1 Identified Lineage Restricted Mammary Luminal Stem Cells. STEM CELLS 2022, 40, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Kouros-Mehr, H.; Werb, Z. Candidate regulators of mammary branching morphogenesis identified by genome-wide transcript analysis. Dev. Dyn. 2006, 235, 3404–3412. [Google Scholar] [CrossRef] [PubMed]
- Owens, T.W.; Rogers, R.L.; Best, S.A.; Ledger, A.; Mooney, A.-M.; Ferguson, A.; Shore, P.; Swarbrick, A.; Ormandy, C.J.; Simpson, P.T.; et al. Runx2 Is a Novel Regulator of Mammary Epithelial Cell Fate in Development and Breast Cancer. Cancer Res 2014, 74, 5277–5286. [Google Scholar] [CrossRef] [PubMed]
- Blyth, K.; Vaillant, F.; Jenkins, A.; McDonald, L.; Pringle, M.A.; Huser, C.; Stein, T.; Neil, J.; Cameron, E.R. Runx2 in normal tissues and cancer cells: A developing story. Blood Cells, Mol. Dis. 2010, 45, 117–123. [Google Scholar] [CrossRef]
- Lipovka, Y.; Konhilas, J.P. The complex nature of oestrogen signalling in breast cancer: Enemy or ally? Biosci. Rep. 2016, 36, e00352. [Google Scholar] [CrossRef]
- Kleuser, B.; Malek, D.; Gust, R.; Pertz, H.H.; Potteck, H. 17-β-Estradiol Inhibits Transforming Growth Factor-β Signaling and Function in Breast Cancer Cells via Activation of Extracellular Signal-Regulated Kinase through the G Protein-Coupled Receptor 30. Mol. Pharmacol. 2008, 74, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Stender, J.D.; Kim, K.; Charn, T.H.; Komm, B.; Chang, K.C.N.; Kraus, W.L.; Benner, C.; Glass, C.K.; Katzenellenbogen, B.S. Genome-Wide Analysis of Estrogen Receptor α DNA Binding and Tethering Mechanisms Identifies Runx1 as a Novel Tethering Factor in Receptor-Mediated Transcriptional Activation. Mol. Cell. Biol. 2010, 30, 3943–3955. [Google Scholar] [CrossRef] [PubMed]
- Lambertini, E.; Penolazzi, L.; Tavanti, E.; Schincaglia, G.P.; Zennaro, M.; Gambari, R.; Piva, R. Human estrogen receptor α gene is a target of Runx2 transcription factor in osteoblasts. Exp. Cell Res. 2007, 313, 1548–1560. [Google Scholar] [CrossRef] [PubMed]
- Kammerer, M.; Gutzwiller, S.; Stauffer, D.; Delhon, I.; Seltenmeyer, Y.; Fournier, B. Estrogen Receptor α (ERα) and Estrogen Related Receptor α (ERRα) are both transcriptional regulators of the Runx2-I isoform. Mol. Cell. Endocrinol. 2013, 369, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Sakakura, C.; Hagiwara, A.; Miyagawa, K.; Nakashima, S.; Yoshikawa, T.; Kin, S.; Nakase, Y.; Ito, K.; Yamagishi, H.; Yazumi, S.; et al. Frequent downregulation of the runt domain transcription factorsRUNX1,RUNX3 and their cofactorCBFB in gastric cancer. Int. J. Cancer 2005, 113, 221–228. [Google Scholar] [CrossRef]
- Carlton, A.L.; Illendula, A.; Gao, Y.; Llaneza, D.C.; Boulton, A.; Shah, A.; Rajewski, R.A.; Landen, C.N.; Wotton, D.; Bushweller, J.H. Small molecule inhibition of the CBFβ/RUNX interaction decreases ovarian cancer growth and migration through alterations in genes related to epithelial-to-mesenchymal transition. Gynecol. Oncol. 2018, 149, 350–360. [Google Scholar] [CrossRef]
- Chen, X.; Chen, Z.; Yu, S.; Nie, F.; Yan, S.; Ma, P.; Chen, Q.; Wei, C.; Fu, H.; Xu, T.; et al. Long Noncoding RNA LINC01234 Functions as a Competing Endogenous RNA to Regulate CBFB Expression by Sponging miR-204-5p in Gastric Cancer. Clin. Cancer Res. 2018, 24, 2002–2014. [Google Scholar] [CrossRef]
- Miyagawa, K.; Sakakura, C.; Nakashima, S.; Yoshikawa, T.; Kin, S.; Nakase, Y.; Ito, K.; Yamagishi, H.; Ida, H.; Yazumi, S.; et al. Down-Regulation of RUNX1, RUNX3 and CBFβ in Hepatocellular Carcinomas in an Early Stage of Hepatocarcinogenesis. Anticancer. Res. 2006, 26, 3633–3643. [Google Scholar]
- Andersen, C.L.; Christensen, L.L.; Thorsen, K.; Schepeler, T.; Sørensen, F.B.; Verspaget, H.W.; Simon, R.; Kruhøffer, M.; Aaltonen, L.; Laurberg, S.; et al. Dysregulation of the transcription factors SOX4, CBFB and SMARCC1 correlates with outcome of colorectal cancer. Br. J. Cancer 2009, 100, 511–523. [Google Scholar] [CrossRef]
- Davis, J.N.; Rogers, D.; Adams, L.; Yong, T.; Jung, J.S.; Cheng, B.; Fennell, K.; Borazanci, E.; Moustafa, Y.W.; Sun, A.; et al. Association of core-binding factor β with the malignant phenotype of prostate and ovarian cancer cells. J. Cell. Physiol. 2010, 225, 875–887. [Google Scholar] [CrossRef]
- Ojesina, A.I.; Lichtenstein, L.; Freeman, S.S.; Pedamallu, C.S.; Imaz-Rosshandler, I.; Pugh, T.J.; Cherniack, A.D.; Ambrogio, L.; Cibulskis, K.; Bertelsen, B.; et al. Landscape of genomic alterations in cervical carcinomas. Nature 2014, 506, 371–375. [Google Scholar] [CrossRef]
- Wang, C.; Shi, Z.; Zhang, Y.; Li, M.; Zhu, J.; Huang, Z.; Zhang, J.; Chen, J. CBFβ promotes colorectal cancer progression through transcriptionally activating OPN, FAM129A, and UPP1 in a RUNX2-dependent manner. Cell Death Differ. 2021, 28, 3176–3192. [Google Scholar] [CrossRef] [PubMed]
- Griffith, O.L.; Spies, N.C.; Anurag, M.; Griffith, M.; Luo, J.; Tu, D.; Yeo, B.; Kunisaki, J.; A Miller, C.; Krysiak, K.; et al. The prognostic effects of somatic mutations in ER-positive breast cancer. Nat. Commun. 2018, 9, 3476. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, C.; Bachelot, T.; Filleron, T.; Pedrero, M.; Campone, M.; Soria, J.-C.; Massard, C.; Lévy, C.; Arnedos, M.; Lacroix-Triki, M.; et al. Mutational Profile of Metastatic Breast Cancers: A Retrospective Analysis. PLOS Med. 2016, 13, e1002201. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Jiang, B.; Guo, J.; Shao, H.; Del Priore, I.S.; Chang, Q.; Kudo, R.; Li, Z.; Razavi, P.; Liu, B.; et al. INK4 Tumor Suppressor Proteins Mediate Resistance to CDK4/6 Kinase Inhibitors. Cancer Discov. 2021, 12, 356–371. [Google Scholar] [CrossRef] [PubMed]
- Stella, S.; Vitale, S.R.; Massimino, M.; Motta, G.; Longhitano, C.; Lanzafame, K.; Martorana, F.; Fazzari, C.; Vecchio, G.M.; Tirrò, E.; et al. Molecular Analysis of Luminal Androgen Receptor Reveals Activated Pathways and Potential Therapeutic Targets in Breast Cancer. Cancer Genom. Proteom. 2022, 19, 464–476. [Google Scholar] [CrossRef]
- Rajendran, B.K.; Deng, C.-X. Characterization of potential driver mutations involved in human breast cancer by computational approaches. Oncotarget 2017, 8, 50252–50272. [Google Scholar] [CrossRef]
- Pegg, H.J.; Harrison, H.; Rogerson, C.; Shore, P. The RUNX Transcriptional Coregulator, CBFβ, Suppresses Migration of ER+ Breast Cancer Cells by Repressing ERα-Mediated Expression of the Migratory Factor TFF1. Mol. Cancer Res. 2019, 17, 1015–1023. [Google Scholar] [CrossRef]
- Malik, N.; Yan, H.; Yang, H.H.; Ayaz, G.; DuBois, W.; Tseng, Y.-C.; Kim, Y.-I.; Jiang, S.; Liu, C.; Lee, M.; et al. CBFB cooperates with p53 to maintain TAp73 expression and suppress breast cancer. PLOS Genet. 2021, 17, e1009553. [Google Scholar] [CrossRef]
- Hsu, C.-H.; Ma, H.-P.; Ong, J.R.; Hsieh, M.-S.; Yadav, V.K.; Yeh, C.-T.; Chao, T.-Y.; Lee, W.-H.; Huang, W.-C.; Kuo, K.-T.; et al. Cancer-Associated Exosomal CBFB Facilitates the Aggressive Phenotype, Evasion of Oxidative Stress, and Preferential Predisposition to Bone Prometastatic Factor of Breast Cancer Progression. Dis. Markers 2022, 2022, 8446629. [Google Scholar] [CrossRef]
- Mendoza-Villanueva, D.; Deng, W.; Lopez-Camacho, C.; Shore, P. The Runx transcriptional co-activator, CBFβ, is essential for invasion of breast cancer cells. Mol. Cancer 2010, 9, 171. [Google Scholar] [CrossRef]
- Ran, R.; Harrison, H.; Ariffin, N.S.; Ayub, R.; Pegg, H.J.; Deng, W.; Mastro, A.; Ottewell, P.D.; Mason, S.M.; Blyth, K.; et al. A role for CBFβ in maintaining the metastatic phenotype of breast cancer cells. Oncogene 2020, 39, 2624–2637. [Google Scholar] [CrossRef] [PubMed]
- Pulido, C.; Vendrell, I.; Ferreira, A.R.; Casimiro, S.; Mansinho, A.; Alho, I.; Costa, L. Bone metastasis risk factors in breast cancer. Ecancermedicalscience 2017, 11, 715. [Google Scholar] [CrossRef] [PubMed]
- Illendula, A.; Gilmour, J.; Grembecka, J.; Tirumala, V.S.S.; Boulton, A.; Kuntimaddi, A.; Schmidt, C.; Wang, L.; Pulikkan, J.A.; Zong, H.; et al. Small Molecule Inhibitor of CBFβ-RUNX Binding for RUNX Transcription Factor Driven Cancers. Ebiomedicine 2016, 8, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Gernapudi, R.; Choi, E.Y.; Lapidus, R.G.; Passaniti, A. Characterization of CADD522, a small molecule that inhibits RUNX2-DNA binding and exhibits antitumor activity. Oncotarget 2017, 8, 70916–70940. [Google Scholar] [CrossRef]
- Oo, Z.M.; Illendula, A.; Grembecka, J.; Schmidt, C.; Zhou, Y.; Esain, V.; Kwan, W.; Frost, I.; North, T.E.; Rajewski, R.A.; et al. A tool compound targeting the core binding factor Runt domain to disrupt binding to CBFβ in leukemic cells. Leuk. Lymphoma 2018, 59, 2188–2200. [Google Scholar] [CrossRef]
- Halperin, C.; Hey, J.; Weichenhan, D.; Stein, Y.; Mayer, S.; Lutsik, P.; Plass, C.; Scherz-Shouval, R. Global DNA Methylation Analysis of Cancer-Associated Fibroblasts Reveals Extensive Epigenetic Rewiring Linked with RUNX1 Upregulation in Breast Cancer Stroma. Cancer Res 2022, 82, 4139–4152. [Google Scholar] [CrossRef]
- Gao, L.; Zhou, F. Comprehensive Analysis of RUNX and TGF-β Mediated Regulation of Immune Cell Infiltration in Breast Cancer. Front. Cell Dev. Biol. 2021, 9, 730380. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, Y.; Shen, P.; Gong, L. Prognostic value and immune characteristics of RUNX gene family in human cancers: A pan-cancer analysis. Aging 2022, 14, 4014–4035. [Google Scholar] [CrossRef]
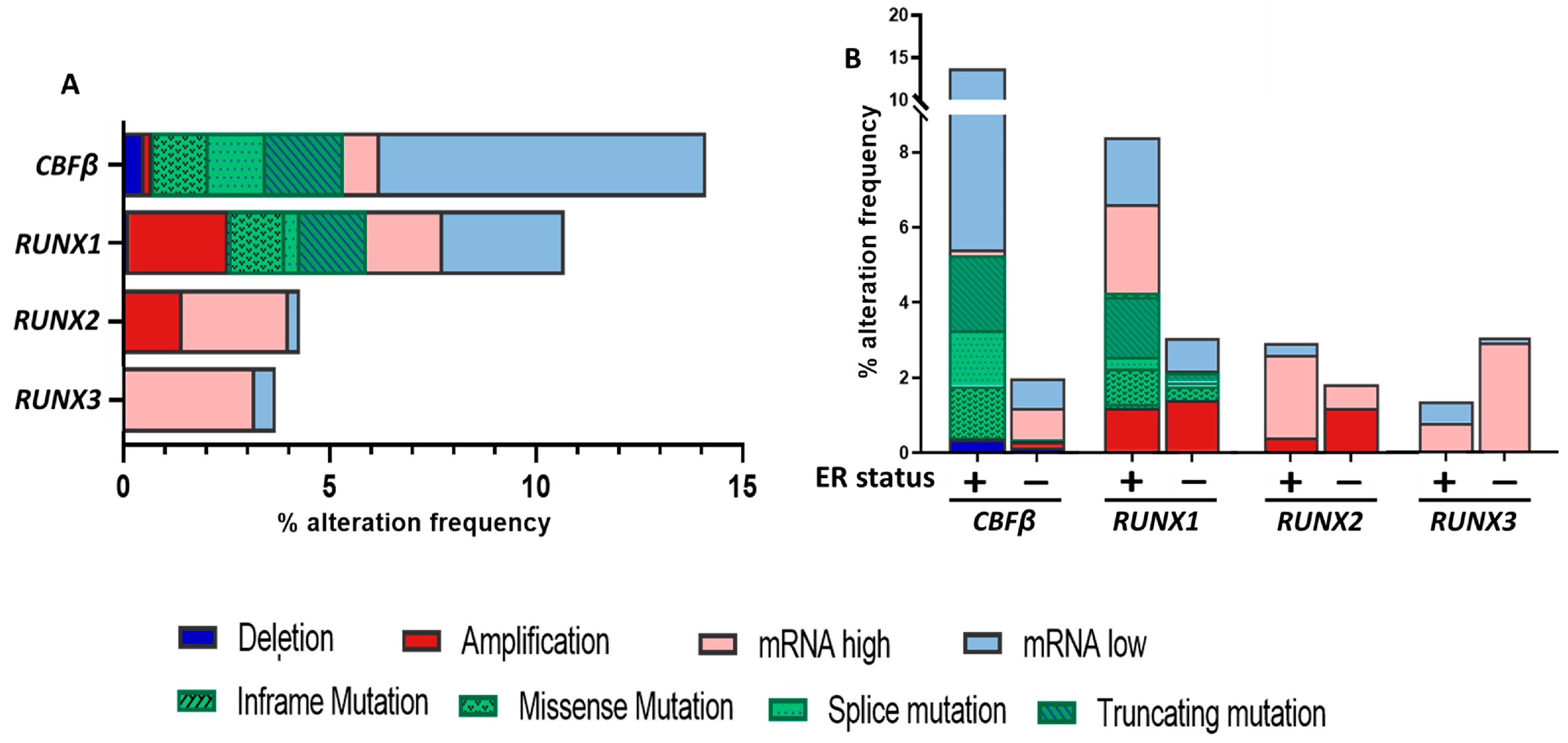
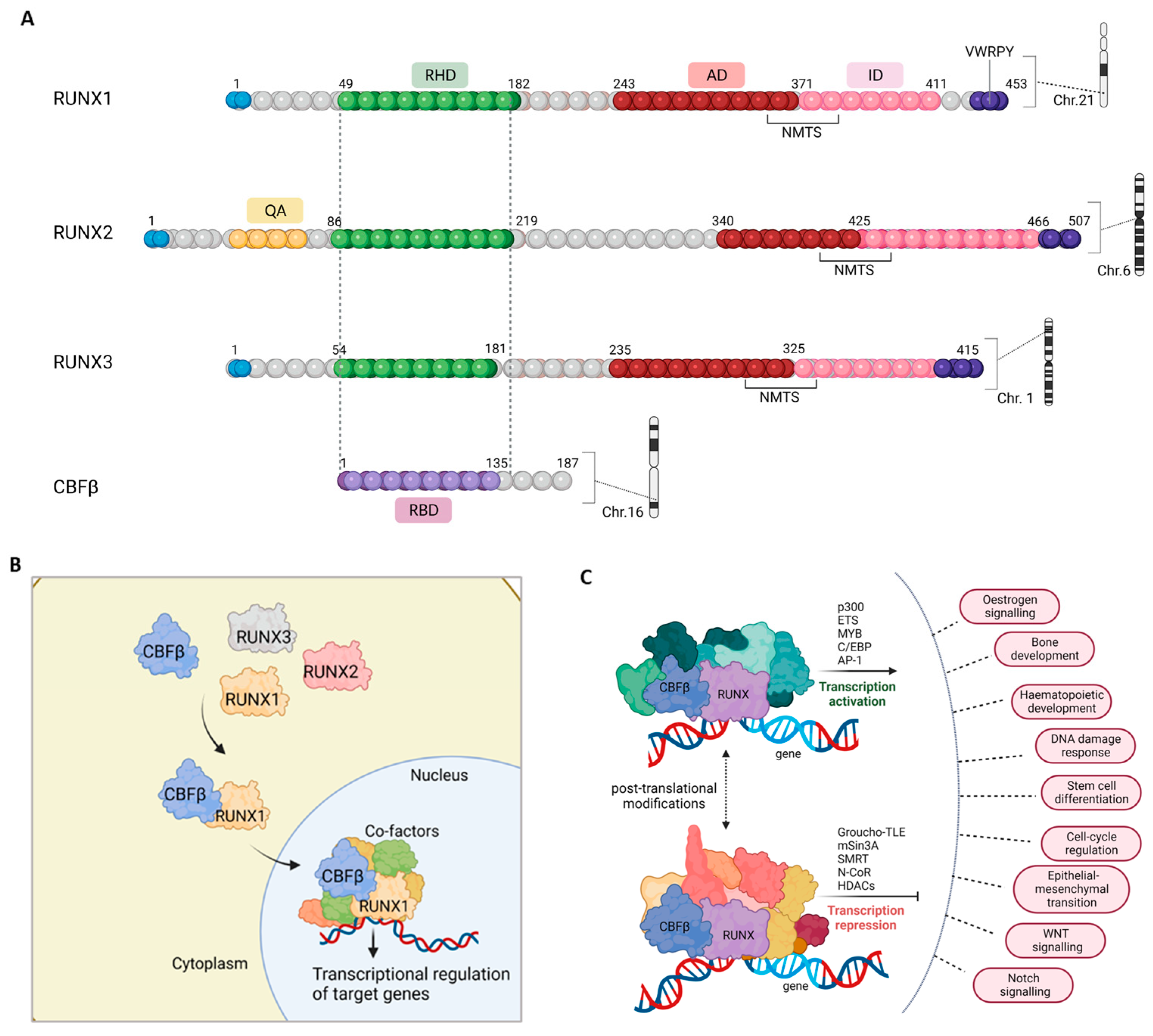

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, A.S.; Campbell, K.J.; Cameron, E.R.; Blyth, K. The RUNX/CBFβ Complex in Breast Cancer: A Conundrum of Context. Cells 2023, 12, 641. https://doi.org/10.3390/cells12040641
Khan AS, Campbell KJ, Cameron ER, Blyth K. The RUNX/CBFβ Complex in Breast Cancer: A Conundrum of Context. Cells. 2023; 12(4):641. https://doi.org/10.3390/cells12040641
Chicago/Turabian StyleKhan, Adiba S., Kirsteen J. Campbell, Ewan R. Cameron, and Karen Blyth. 2023. "The RUNX/CBFβ Complex in Breast Cancer: A Conundrum of Context" Cells 12, no. 4: 641. https://doi.org/10.3390/cells12040641
APA StyleKhan, A. S., Campbell, K. J., Cameron, E. R., & Blyth, K. (2023). The RUNX/CBFβ Complex in Breast Cancer: A Conundrum of Context. Cells, 12(4), 641. https://doi.org/10.3390/cells12040641






