Targeting the ‘Undruggable’ Driver Protein, KRAS, in Epithelial Cancers: Current Perspective
Abstract
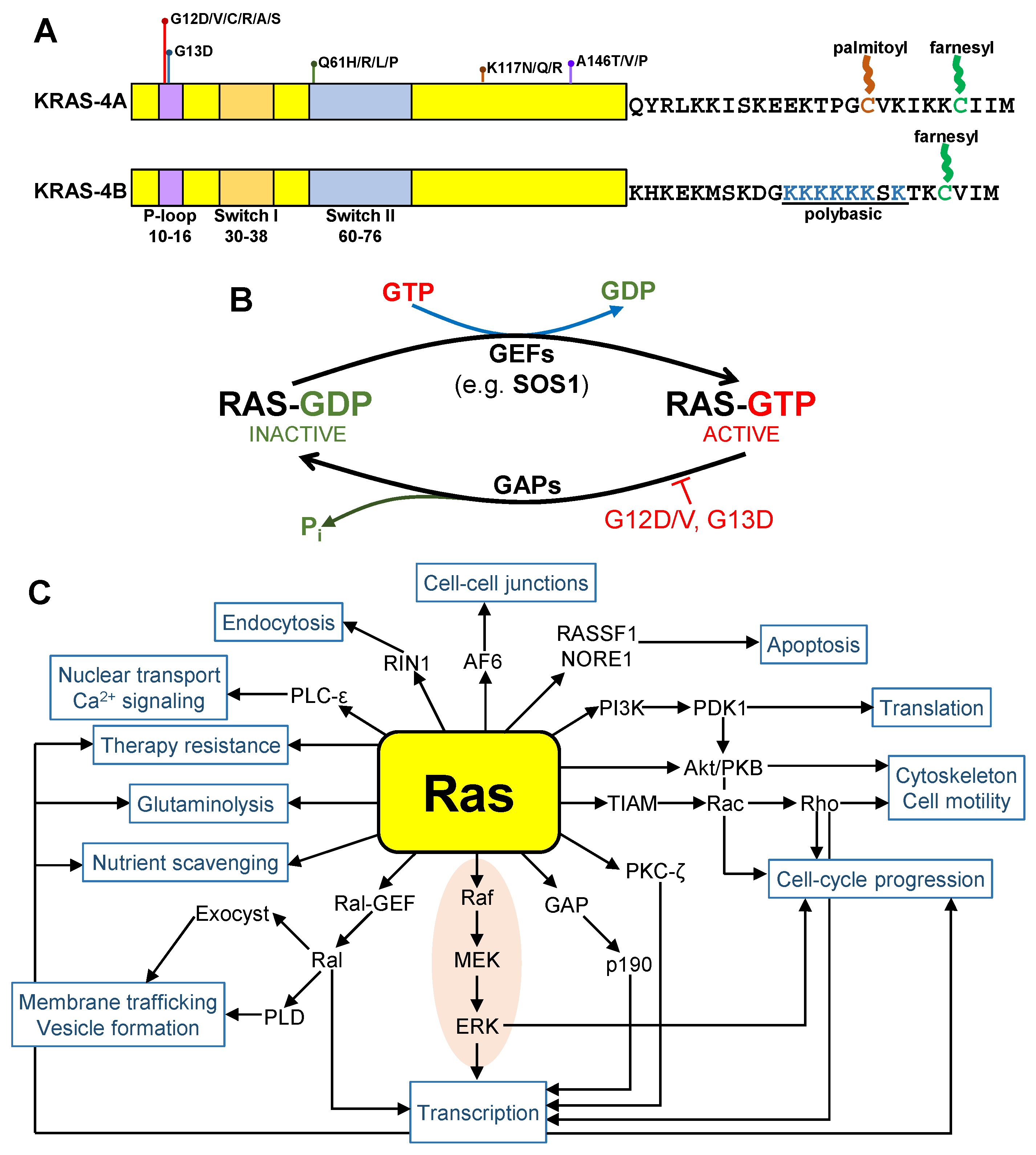
1. Introduction
2. Synthetic Drugs
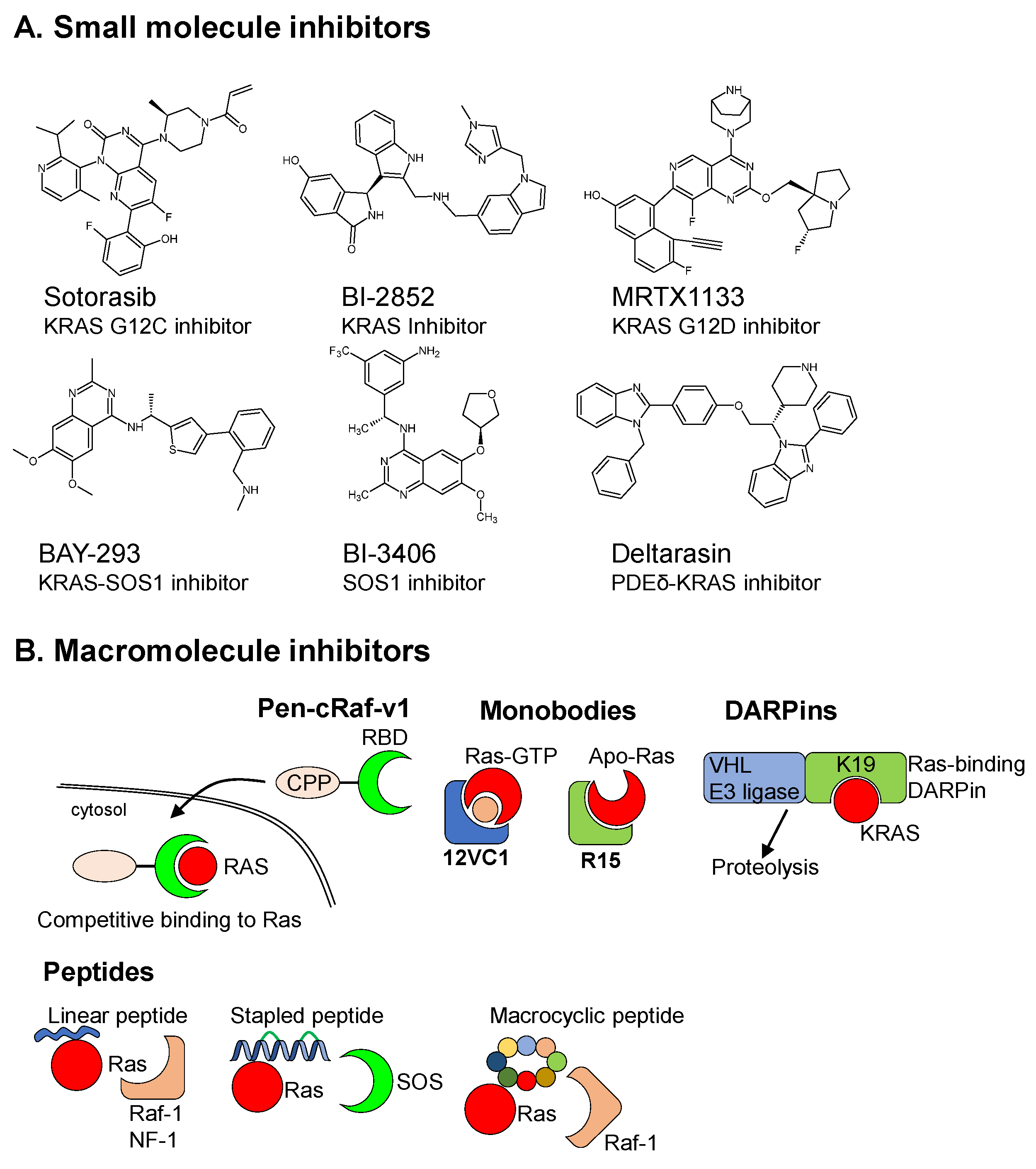
2.1. Small Molecule Inhibitors (SMIs)
2.2. Macromolecular Drug/Protein Scaffold
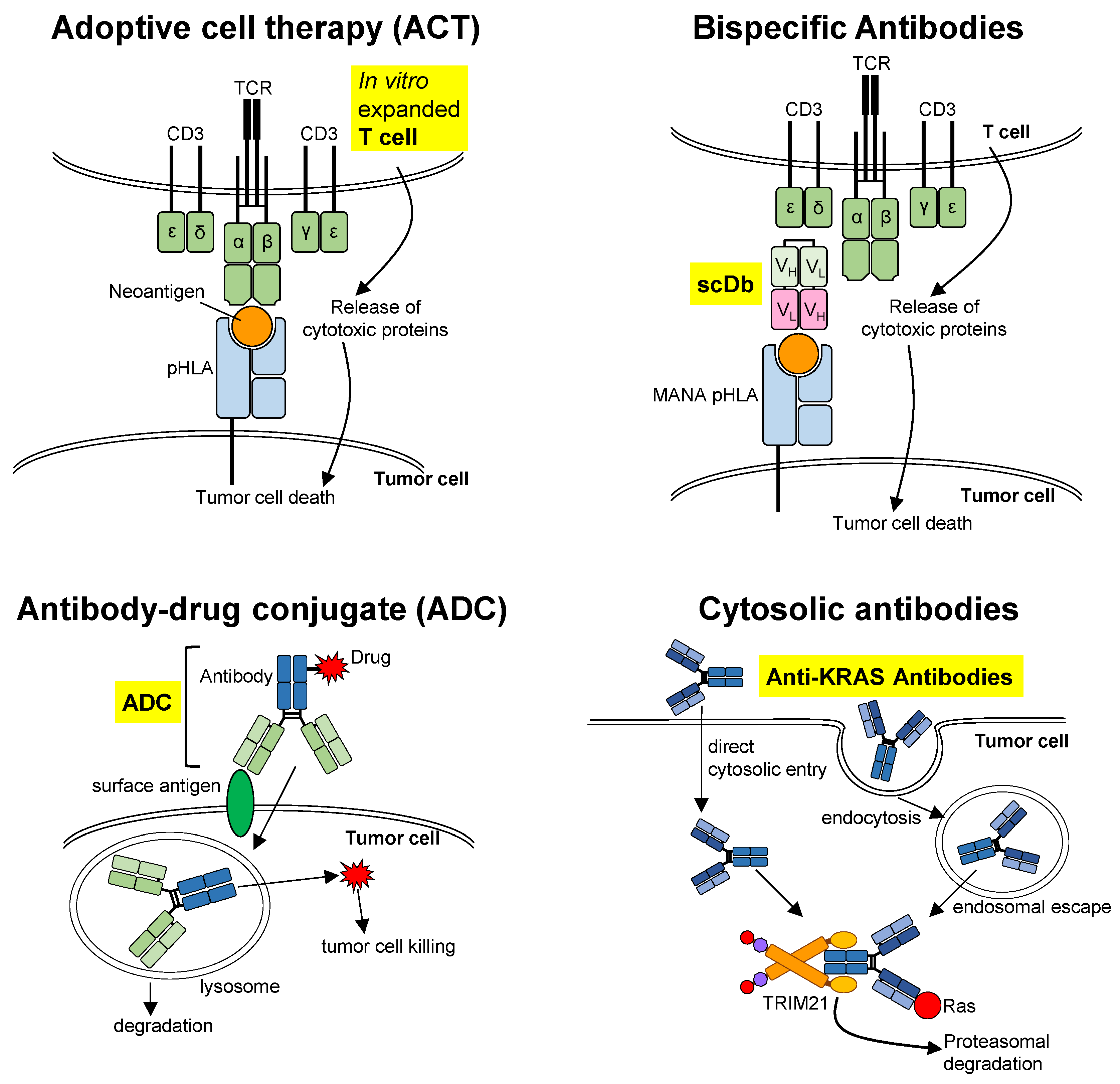
3. Biologics
3.1. Adoptive Cell Therapy (ACT)
3.2. Bispecific Antibodies
3.3. Antibody–Drug Conjugate (ADC)
3.4. Cytosolic Antibodies
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- SEER. Cancer Stat Facts: Colorectal Cancer. Available online: https://seer.cancer.gov/statfacts/html/colorect.html (accessed on 10 September 2022).
- Gill, S.; Loprinzi, C.L.; Sargent, D.J.; Thome, S.D.; Alberts, S.R.; Haller, D.G.; Benedetti, J.; Francini, G.; Shepherd, L.E.; Francois Seitz, J.; et al. Pooled analysis of fluorouracil-based adjuvant therapy for stage ii and iii colon cancer: Who benefits and by how much? J. Clin. Oncol. 2004, 22, 1797–1806. [Google Scholar] [CrossRef] [PubMed]
- Goyle, S.; Maraveyas, A. Chemotherapy for colorectal cancer. Dig. Surg. 2005, 22, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Bleiberg, H. Adjuvant treatment of colon cancer. Curr. Opin. Oncol. 2005, 17, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aranda Aguilar, E.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. Esmo consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef]
- Benson, A.B.; Venook, A.P.; Al-Hawary, M.M.; Arain, M.A.; Chen, Y.J.; Ciombor, K.K.; Cohen, S.; Cooper, H.S.; Deming, D.; Farkas, L.; et al. Colon cancer, version 2.2021, nccn clinical practice guidelines in oncology. J. Natl. Compr. Canc. Netw 2021, 19, 329–359. [Google Scholar] [CrossRef]
- Misale, S.; Yaeger, R.; Hobor, S.; Scala, E.; Janakiraman, M.; Liska, D.; Valtorta, E.; Schiavo, R.; Buscarino, M.; Siravegna, G.; et al. Emergence of kras mutations and acquired resistance to anti-egfr therapy in colorectal cancer. Nature 2012, 486, 532–536. [Google Scholar] [CrossRef]
- Diaz, L.A., Jr.; Williams, R.T.; Wu, J.; Kinde, I.; Hecht, J.R.; Berlin, J.; Allen, B.; Bozic, I.; Reiter, J.G.; Nowak, M.A.; et al. The molecular evolution of acquired resistance to targeted egfr blockade in colorectal cancers. Nature 2012, 486, 537–540. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. Pd-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- Prior, I.A.; Hood, F.E.; Hartley, J.L. The frequency of ras mutations in cancer. Cancer Res. 2020, 80, 2969–2974. [Google Scholar] [CrossRef]
- Tsai, F.D.; Lopes, M.S.; Zhou, M.; Court, H.; Ponce, O.; Fiordalisi, J.J.; Gierut, J.J.; Cox, A.D.; Haigis, K.M.; Philips, M.R. K-ras4a splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc. Natl. Acad. Sci. USA 2015, 112, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.; Burkhart, D.L.; Haigis, K.M. Classification of kras-activating mutations and the implications for therapeutic intervention. Cancer Discov. 2022, 12, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Scheffzek, K.; Ahmadian, M.R.; Kabsch, W.; Wiesmuller, L.; Lautwein, A.; Schmitz, F.; Wittinghofer, A. The ras-rasgap complex: Structural basis for gtpase activation and its loss in oncogenic ras mutants. Science 1997, 277, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Scheffzek, K.; Ahmadian, M.R.; Wittinghofer, A. Gtpase-activating proteins: Helping hands to complement an active site. Trends Biochem Sci. 1998, 23, 257–262. [Google Scholar] [CrossRef]
- Wang, W.; Fang, G.; Rudolph, J. Ras inhibition via direct ras binding--is there a path forward? Bioorg. Med Chem. Lett. 2012, 22, 5766–5776. [Google Scholar] [CrossRef]
- Cox, A.D.; Fesik, S.W.; Kimmelman, A.C.; Luo, J.; Der, C.J. Drugging the undruggable ras: Mission possible? Nat. Rev. Drug Discov. 2014, 13, 828–851. [Google Scholar] [CrossRef]
- Stephen, A.G.; Esposito, D.; Bagni, R.K.; McCormick, F. Dragging ras back in the ring. Cancer Cell 2014, 25, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.Y.; Richardson, B.C. The mapk signalling pathways and colorectal cancer. Lancet Oncol. 2005, 6, 322–327. [Google Scholar] [CrossRef]
- Drosten, M.; Barbacid, M. Targeting the mapk pathway in kras-driven tumors. Cancer Cell 2020, 37, 543–550. [Google Scholar] [CrossRef]
- Lam, K.K.; Tang, C.L.; Tan, E.; Wong, S.H.; Cheah, P.Y. Kras mutation-independent downregulation of mapk/pi3k signaling in colorectal cancer. Mol. Oncol. 2022, 16, 1171–1183. [Google Scholar] [CrossRef]
- Weinberg, R.A. The Biology of Cancer; Garland Science: New York, NY, USA, 2007. [Google Scholar]
- Mukhopadhyay, S.; Vander Heiden, M.G.; McCormick, F. The metabolic landscape of ras-driven cancers from biology to therapy. Nat Cancer 2021, 2, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Ostrem, J.M.; Peters, U.; Sos, M.L.; Wells, J.A.; Shokat, K.M. K-ras(g12c) inhibitors allosterically control gtp affinity and effector interactions. Nature 2013, 503, 548–551. [Google Scholar] [CrossRef] [PubMed]
- Koga, T.; Suda, K.; Fujino, T.; Ohara, S.; Hamada, A.; Nishino, M.; Chiba, M.; Shimoji, M.; Takemoto, T.; Arita, T.; et al. Kras secondary mutations that confer acquired resistance to kras g12c inhibitors, sotorasib and adagrasib, and overcoming strategies: Insights from in vitro experiments. J. Thorac. Oncol. 2021, 16, 1321–1332. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Lin, J.J.; Li, C.; Ryan, M.B.; Zhang, J.; Kiedrowski, L.A.; Michel, A.G.; Syed, M.U.; Fella, K.A.; Sakhi, M.; et al. Clinical acquired resistance to kras(g12c) inhibition through a novel kras switch-ii pocket mutation and polyclonal alterations converging on ras-mapk reactivation. Cancer Discov. 2021, 11, 1913–1922. [Google Scholar] [CrossRef]
- Awad, M.M.; Liu, S.; Rybkin, I.; Arbour, K.C.; Dilly, J.; Zhu, V.W.; Johnson, M.L.; Heist, R.S.; Patil, T.; Riely, G.J.; et al. Acquired resistance to kras(g12c) inhibition in cancer. N. Engl. J. Med. 2021, 384, 2382–2393. [Google Scholar] [CrossRef]
- Moore, A.R.; Malek, S. The promise and peril of kras g12c inhibitors. Cancer Cell 2021, 39, 1059–1061. [Google Scholar] [CrossRef]
- Punekar, S.R.; Velcheti, V.; Neel, B.G.; Wong, K.K. The current state of the art and future trends in ras-targeted cancer therapies. Nat. Rev. Clin. Oncol. 2022, 19, 637–655. [Google Scholar] [CrossRef]
- Herdeis, L.; Gerlach, D.; McConnell, D.B.; Kessler, D. Stopping the beating heart of cancer: Kras reviewed. Curr. Opin. Struct. Biol. 2021, 71, 136–147. [Google Scholar] [CrossRef]
- Kessler, D.; Gmachl, M.; Mantoulidis, A.; Martin, L.J.; Zoephel, A.; Mayer, M.; Gollner, A.; Covini, D.; Fischer, S.; Gerstberger, T.; et al. Drugging an undruggable pocket on kras. Proc. Natl. Acad. Sci. USA 2019, 116, 15823–15829. [Google Scholar] [CrossRef]
- Tran, T.H.; Alexander, P.; Dharmaiah, S.; Agamasu, C.; Nissley, D.V.; McCormick, F.; Esposito, D.; Simanshu, D.K.; Stephen, A.G.; Balius, T.E. The small molecule bi-2852 induces a nonfunctional dimer of kras. Proc. Natl. Acad. Sci. USA 2020, 117, 3363–3364. [Google Scholar] [CrossRef]
- Wang, C.X.; Wang, T.T.; Zhang, K.D.; Li, M.Y.; Shen, Q.C.; Lu, S.Y.; Zhang, J. Pan-kras inhibitors suppress proliferation through feedback regulation in pancreatic ductal adenocarcinoma. Acta Pharmacol. Sin. 2022, 43, 2696–2708. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Allen, S.; Blake, J.F.; Bowcut, V.; Briere, D.M.; Calinisan, A.; Dahlke, J.R.; Fell, J.B.; Fischer, J.P.; Gunn, R.J.; et al. Identification of mrtx1133, a noncovalent, potent, and selective kras(g12d) inhibitor. J. Med. Chem. 2022, 65, 3123–3133. [Google Scholar] [CrossRef]
- Hallin, J.; Bowcut, V.; Calinisan, A.; Briere, D.M.; Hargis, L.; Engstrom, L.D.; Laguer, J.; Medwid, J.; Vanderpool, D.; Lifset, E.; et al. Anti-tumor efficacy of a potent and selective non-covalent kras(g12d) inhibitor. Nat. Med. 2022, 28, 2171–2182. [Google Scholar] [CrossRef] [PubMed]
- Hillig, R.C.; Sautier, B.; Schroeder, J.; Moosmayer, D.; Hilpmann, A.; Stegmann, C.M.; Werbeck, N.D.; Briem, H.; Boemer, U.; Weiske, J.; et al. Discovery of potent sos1 inhibitors that block ras activation via disruption of the ras-sos1 interaction. Proc. Natl. Acad. Sci. USA 2019, 116, 2551–2560. [Google Scholar] [CrossRef] [PubMed]
- Plangger, A.; Rath, B.; Hochmair, M.; Funovics, M.; Hamilton, G. Cytotoxicity of combinations of the pan-kras inhibitor bay-293 against primary non-small lung cancer cells. Transl. Oncol. 2021, 14, 101230. [Google Scholar] [CrossRef] [PubMed]
- Plangger, A.; Rath, B.; Stickler, S.; Hochmair, M.; Lang, C.; Weigl, L.; Funovics, M.; Hamilton, G. Cytotoxicity of combinations of the pan-kras sos1 inhibitor bay-293 against pancreatic cancer cell lines. Discov. Oncol. 2022, 13, 84. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, C.; Su, R.; Yin, Z.; Huang, G.; Yang, J.; Li, Z.; Zhang, K.; Fei, J. Targeting sos1 overcomes imatinib resistance with bcr-abl independence through uptake transporter slc22a4 in cml. Mol. Ther. Oncolytics 2021, 23, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.H.; Gmachl, M.; Ramharter, J.; Savarese, F.; Gerlach, D.; Marszalek, J.R.; Sanderson, M.P.; Kessler, D.; Trapani, F.; Arnhof, H.; et al. Bi-3406, a potent and selective sos1-kras interaction inhibitor, is effective in kras-driven cancers through combined mek inhibition. Cancer Discov. 2021, 11, 142–157. [Google Scholar] [CrossRef]
- Zhao, Y.; Xue, J.Y.; Lito, P. Suppressing nucleotide exchange to inhibit kras-mutant tumors. Cancer Discov. 2021, 11, 17–19. [Google Scholar] [CrossRef]
- Martin, E.W.; Mittag, T. Dwelling at membranes promotes decisive signaling. Science 2019, 363, 1036–1037. [Google Scholar] [CrossRef]
- Chandra, A.; Grecco, H.E.; Pisupati, V.; Perera, D.; Cassidy, L.; Skoulidis, F.; Ismail, S.A.; Hedberg, C.; Hanzal-Bayer, M.; Venkitaraman, A.R.; et al. The gdi-like solubilizing factor pdedelta sustains the spatial organization and signalling of ras family proteins. Nat. Cell Biol. 2011, 14, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Schmick, M.; Vartak, N.; Papke, B.; Kovacevic, M.; Truxius, D.C.; Rossmannek, L.; Bastiaens, P.I.H. Kras localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell 2014, 157, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, G.; Papke, B.; Ismail, S.; Vartak, N.; Chandra, A.; Hoffmann, M.; Hahn, S.A.; Triola, G.; Wittinghofer, A.; Bastiaens, P.I.; et al. Small molecule inhibition of the kras-pdedelta interaction impairs oncogenic kras signalling. Nature 2013, 497, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Papke, B.; Murarka, S.; Vogel, H.A.; Martin-Gago, P.; Kovacevic, M.; Truxius, D.C.; Fansa, E.K.; Ismail, S.; Zimmermann, G.; Heinelt, K.; et al. Identification of pyrazolopyridazinones as pdedelta inhibitors. Nat. Commun. 2016, 7, 11360. [Google Scholar] [CrossRef] [PubMed]
- Martin-Gago, P.; Fansa, E.K.; Klein, C.H.; Murarka, S.; Janning, P.; Schurmann, M.; Metz, M.; Ismail, S.; Schultz-Fademrecht, C.; Baumann, M.; et al. A pde6delta-kras inhibitor chemotype with up to seven h-bonds and picomolar affinity that prevents efficient inhibitor release by arl2. Angew. Chem. Int. Ed. Engl. 2017, 56, 2423–2428. [Google Scholar] [CrossRef] [PubMed]
- Leung, E.L.H.; Luo, L.X.; Liu, Z.Q.; Wong, V.K.W.; Lu, L.L.; Xie, Y.; Zhang, N.; Qu, Y.Q.; Fan, X.X.; Li, Y.; et al. Inhibition of kras-dependent lung cancer cell growth by deltarasin: Blockage of autophagy increases its cytotoxicity. Cell Death Dis. 2018, 9, 216. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.H.; Truxius, D.C.; Vogel, H.A.; Harizanova, J.; Murarka, S.; Martin-Gago, P.; Bastiaens, P.I.H. Pdedelta inhibition impedes the proliferation and survival of human colorectal cancer cell lines harboring oncogenic kras. Int. J. Cancer 2019, 144, 767–776. [Google Scholar] [CrossRef]
- Zhang, H.; Hosier, S.; Terew, J.M.; Zhang, K.; Cote, R.H.; Baehr, W. Assay and functional properties of prbp(pdedelta), a prenyl-binding protein interacting with multiple partners. Methods Enzymol. 2005, 403, 42–56. [Google Scholar]
- Ling, X.; Wu, W.; Aljahdali, I.A.M.; Liao, J.; Santha, S.; Fountzilas, C.; Boland, P.M.; Li, F. Fl118, acting as a ‘molecular glue degrader’, binds to dephosphorylates and degrades the oncoprotein ddx5 (p68) to control c-myc, survivin and mutant kras against colorectal and pancreatic cancer with high efficacy. Clin. Transl. Med. 2022, 12, e881. [Google Scholar] [CrossRef]
- Tolani, B.; Celli, A.; Yao, Y.; Tan, Y.Z.; Fetter, R.; Liem, C.R.; de Smith, A.J.; Vasanthakumar, T.; Bisignano, P.; Cotton, A.D.; et al. Ras-mutant cancers are sensitive to small molecule inhibition of v-type atpases in mice. Nat. Biotechnol 2022, 40, 1834–1844. [Google Scholar] [CrossRef]
- Chatani, P.D.; Yang, J.C. Mutated ras: Targeting the “untargetable” with t cells. Clin. Cancer Res. 2020, 26, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Damnernsawad, A.; Kong, G.; Wen, Z.; Liu, Y.; Rajagopalan, A.; You, X.; Wang, J.; Zhou, Y.; Ranheim, E.A.; Luo, H.R.; et al. Kras is required for adult hematopoiesis. Stem Cells 2016, 34, 1859–1871. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.H.; Gerlach, D.; Misale, S.; Petronczki, M.; Kraut, N. Expanding the reach of precision oncology by drugging all kras mutants. Cancer Discov. 2022, 12, 924–937. [Google Scholar] [CrossRef]
- Lin, A.; Giuliano, C.J.; Palladino, A.; John, K.M.; Abramowicz, C.; Yuan, M.L.; Sausville, E.L.; Lukow, D.A.; Liu, L.; Chait, A.R.; et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med. 2019, 11, eaaw8412. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.K.; Heishima, K.; Sugito, N.; Sugawara, R.; Ueda, H.; Yukihiro, A.; Honda, R. Specific inhibition of oncogenic ras using cell-permeable ras-binding domains. Cell Chem. Biol. 2021, 28, 1581–1589.e6. [Google Scholar] [CrossRef] [PubMed]
- Teng, K.W.; Tsai, S.T.; Hattori, T.; Fedele, C.; Koide, A.; Yang, C.; Hou, X.; Zhang, Y.; Neel, B.G.; O’Bryan, J.P.; et al. Selective and noncovalent targeting of ras mutants for inhibition and degradation. Nat. Commun. 2021, 12, 2656. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Koide, A.; Zuberi, M.; Ketavarapu, G.; Denbaum, E.; Teng, K.W.; Rhett, J.M.; Spencer-Smith, R.; Hobbs, G.A.; Camp, E.R.; et al. Identification of the nucleotide-free state as a therapeutic vulnerability for inhibition of selected oncogenic ras mutants. Cell Rep. 2022, 38, 110322. [Google Scholar] [CrossRef]
- Pei, D.; Chen, K.; Liao, H. Targeting ras with macromolecules. Cold Spring Harb. Perspect. Med. 2018, 8, a031476. [Google Scholar] [CrossRef]
- Clark, G.J.; Drugan, J.K.; Terrell, R.S.; Bradham, C.; Der, C.J.; Bell, R.M.; Campbell, S. Peptides containing a consensus ras binding sequence from raf-1 and thegtpase activating protein nf1 inhibit ras function. Proc. Natl. Acad. Sci. USA 1996, 93, 1577–1581. [Google Scholar] [CrossRef]
- Barnard, D.; Sun, H.; Baker, L.; Marshall, M.S. In vitro inhibition of ras-raf association by short peptides. Biochem. Biophys. Res. Commun. 1998, 247, 176–180. [Google Scholar] [CrossRef]
- Xu, C.W.; Luo, Z. Inactivation of ras function by allele-specific peptide aptamers. Oncogene 2002, 21, 5753–5757. [Google Scholar] [CrossRef] [PubMed]
- Gareiss, P.C.; Schneekloth, A.R.; Salcius, M.J.; Seo, S.Y.; Crews, C.M. Identification and characterization of a peptidic ligand for ras. Chembiochem 2010, 11, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Patgiri, A.; Yadav, K.K.; Arora, P.S.; Bar-Sagi, D. An orthosteric inhibitor of the ras-sos interaction. Nat. Chem. Biol. 2011, 7, 585–587. [Google Scholar] [CrossRef] [PubMed]
- Leshchiner, E.S.; Parkhitko, A.; Bird, G.H.; Luccarelli, J.; Bellairs, J.A.; Escudero, S.; Opoku-Nsiah, K.; Godes, M.; Perrimon, N.; Walensky, L.D. Direct inhibition of oncogenic kras by hydrocarbon-stapled sos1 helices. Proc. Natl. Acad. Sci. USA 2015, 112, 1761–1766. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Upadhyaya, P.; Villalona-Calero, M.A.; Briesewitz, R.; Pei, D. Inhibition of ras-effector interaction by cyclic peptides. Medchemcomm 2013, 4, 378–382. [Google Scholar] [CrossRef]
- Upadhyaya, P.; Qian, Z.; Habir, N.A.; Pei, D. Direct ras inhibitors identified from a structurally rigidified bicyclic peptide library. Tetrahedron 2014, 70, 7714–7720. [Google Scholar] [CrossRef]
- Upadhyaya, P.; Qian, Z.; Selner, N.G.; Clippinger, S.R.; Wu, Z.; Briesewitz, R.; Pei, D. Inhibition of ras signaling by blocking ras-effector interactions with cyclic peptides. Angew. Chem. Int. Ed. Engl. 2015, 54, 7602–7606. [Google Scholar] [CrossRef]
- Trinh, T.B.; Upadhyaya, P.; Qian, Z.; Pei, D. Discovery of a direct ras inhibitor by screening a combinatorial library of cell-permeable bicyclic peptides. ACS Comb. Sci. 2016, 18, 75–85. [Google Scholar] [CrossRef]
- Sakamoto, K.; Kamada, Y.; Sameshima, T.; Yaguchi, M.; Niida, A.; Sasaki, S.; Miwa, M.; Ohkubo, S.; Sakamoto, J.I.; Kamaura, M.; et al. K-ras(g12d)-selective inhibitory peptides generated by random peptide t7 phage display technology. Biochem. Biophys. Res. Commun. 2017, 484, 605–611. [Google Scholar] [CrossRef]
- Stumpp, M.T.; Binz, H.K.; Amstutz, P. Darpins: A new generation of protein therapeutics. Drug Discov. Today 2008, 13, 695–701. [Google Scholar] [CrossRef]
- Bery, N.; Legg, S.; Debreczeni, J.; Breed, J.; Embrey, K.; Stubbs, C.; Kolasinska-Zwierz, P.; Barrett, N.; Marwood, R.; Watson, J.; et al. Kras-specific inhibition using a darpin binding to a site in the allosteric lobe. Nat. Commun. 2019, 10, 2607. [Google Scholar] [CrossRef] [PubMed]
- Bery, N.; Miller, A.; Rabbitts, T. A potent kras macromolecule degrader specifically targeting tumours with mutant kras. Nat. Commun. 2020, 11, 3233. [Google Scholar] [CrossRef] [PubMed]
- Stumpp, M.T.; Dawson, K.M.; Binz, H.K. Beyond antibodies: The darpin((r)) drug platform. BioDrugs 2020, 34, 423–433. [Google Scholar] [CrossRef]
- Leko, V.; Rosenberg, S.A. Identifying and targeting human tumor antigens for t cell-based immunotherapy of solid tumors. Cancer Cell 2020, 38, 454–472. [Google Scholar] [CrossRef] [PubMed]
- Chandran, S.S.; Klebanoff, C.A. T cell receptor-based cancer immunotherapy: Emerging efficacy and pathways of resistance. Immunol. Rev. 2019, 290, 127–147. [Google Scholar] [CrossRef] [PubMed]
- Leidner, R.; Sanjuan Silva, N.; Huang, H.; Sprott, D.; Zheng, C.; Shih, Y.P.; Leung, A.; Payne, R.; Sutcliffe, K.; Cramer, J.; et al. Neoantigen t-cell receptor gene therapy in pancreatic cancer. N. Engl. J. Med. 2022, 386, 2112–2119. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.P.; Vale, N.R.; Zacharakis, N.; Krishna, S.; Yu, Z.; Gasmi, B.; Gartner, J.J.; Sindiri, S.; Malekzadeh, P.; Deniger, D.C.; et al. Adoptive cellular therapy with autologous tumor-infiltrating lymphocytes and t-cell receptor-engineered t cells targeting common p53 neoantigens in human solid tumors. Cancer Immunol. Res. 2022, 10, 932–946. [Google Scholar] [CrossRef]
- Wherry, E.J.; Kurachi, M. Molecular and cellular insights into t cell exhaustion. Nat. Rev. Immunol. 2015, 15, 486–499. [Google Scholar] [CrossRef]
- Cafri, G.; Gartner, J.J.; Zaks, T.; Hopson, K.; Levin, N.; Paria, B.C.; Parkhurst, M.R.; Yossef, R.; Lowery, F.J.; Jafferji, M.S.; et al. Mrna vaccine-induced neoantigen-specific t cell immunity in patients with gastrointestinal cancer. J. Clin. Investig. 2020, 130, 5976–5988. [Google Scholar] [CrossRef]
- Weidanz, J. Targeting cancer with bispecific antibodies. Science 2021, 371, 996–997. [Google Scholar] [CrossRef]
- Hsiue, E.H.; Wright, K.M.; Douglass, J.; Hwang, M.S.; Mog, B.J.; Pearlman, A.H.; Paul, S.; DiNapoli, S.R.; Konig, M.F.; Wang, Q.; et al. Targeting a neoantigen derived from a common tp53 mutation. Science 2021, 371, eabc8697. [Google Scholar] [CrossRef] [PubMed]
- Douglass, J.; Hsiue, E.H.; Mog, B.J.; Hwang, M.S.; DiNapoli, S.R.; Pearlman, A.H.; Miller, M.S.; Wright, K.M.; Azurmendi, P.A.; Wang, Q.; et al. Bispecific antibodies targeting mutant ras neoantigens. Sci. Immunol. 2021, 6, eabd5515. [Google Scholar] [CrossRef] [PubMed]
- Seliger, B.; Ferrone, S. Hla class i antigen processing machinery defects in cancer cells-frequency, functional significance, and clinical relevance with special emphasis on their role in t cell-based immunotherapy of malignant disease. Methods Mol. Biol. 2020, 2055, 325–350. [Google Scholar] [PubMed]
- Autio, K.A.; Boni, V.; Humphrey, R.W.; Naing, A. Probody therapeutics: An emerging class of therapies designed to enhance on-target effects with reduced off-tumor toxicity for use in immuno-oncology. Clin. Cancer Res. 2020, 26, 984–989. [Google Scholar] [CrossRef]
- Boustany, L.M.; LaPorte, S.L.; Wong, L.; White, C.; Vinod, V.; Shen, J.; Yu, W.; Koditek, D.; Winter, M.B.; Moore, S.J.; et al. A probody t cell-engaging bispecific antibody targeting egfr and cd3 inhibits colon cancer growth with limited toxicity. Cancer Res. 2022, 82, 4288–4298. [Google Scholar] [CrossRef] [PubMed]
- Hattori, T.; Maso, L.; Araki, K.Y.; Koide, A.; Hayman, J.; Akkapeddi, P.; Bang, I.; Neel, B.G.; Koide, S. Creating mhc-restricted neoantigens with covalent inhibitors that can be targeted by immune therapy. Cancer Discov. 2023, 13, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.D.; Ting, J.P.; Der, C.J. Convergence of targeted and immune therapies for the treatment of oncogene-driven cancers. Cancer Discov. 2023, 13, 19–22. [Google Scholar] [CrossRef]
- McDaid, W.J.; Greene, M.K.; Johnston, M.C.; Pollheimer, E.; Smyth, P.; McLaughlin, K.; Van Schaeybroeck, S.; Straubinger, R.M.; Longley, D.B.; Scott, C.J. Repurposing of cetuximab in antibody-directed chemotherapy-loaded nanoparticles in egfr therapy-resistant pancreatic tumours. Nanoscale 2019, 11, 20261–20273. [Google Scholar] [CrossRef]
- Greene, M.K.; Chen, T.; Robinson, E.; Straubinger, N.L.; Minx, C.; Chan, D.K.W.; Wang, J.; Burrows, J.F.; Van Schaeybroeck, S.; Baker, J.R.; et al. Controlled coupling of an ultrapotent auristatin warhead to cetuximab yields a next-generation antibody-drug conjugate for egfr-targeted therapy of kras mutant pancreatic cancer. Br. J. Cancer 2020, 123, 1502–1512. [Google Scholar] [CrossRef]
- Sorbara, M.; Cordelier, P.; Bery, N. Antibody-based approaches to target pancreatic tumours. Antibodies 2022, 11, 47. [Google Scholar] [CrossRef]
- Ha, S.Y.Y.; Anami, Y.; Yamazaki, C.M.; Xiong, W.; Haase, C.M.; Olson, S.D.; Lee, J.; Ueno, N.T.; Zhang, N.; An, Z.; et al. An enzymatically cleavable tripeptide linker for maximizing the therapeutic index of antibody-drug conjugates. Mol. Cancer Ther. 2022, 21, 1449–1461. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.F.; Xiong, H.L.; Cao, J.L.; Wang, S.J.; Guo, X.R.; Lin, B.Y.; Zhang, Y.; Zhao, J.H.; Wang, Y.B.; Zhang, T.Y.; et al. A cell-penetrating whole molecule antibody targeting intracellular hbx suppresses hepatitis b virus via trim21-dependent pathway. Theranostics 2018, 8, 549–562. [Google Scholar] [CrossRef]
- Ruiz-Arguelles, A.; Alarcon-Segovia, D. Penetration of autoantibodies into living cells, 2000. Isr. Med. Assoc. J. 2001, 3, 121–126. [Google Scholar] [PubMed]
- Scaletti, F.; Hardie, J.; Lee, Y.W.; Luther, D.C.; Ray, M.; Rotello, V.M. Protein delivery into cells using inorganic nanoparticle-protein supramolecular assemblies. Chem. Soc. Rev. 2018, 47, 3421–3432. [Google Scholar] [CrossRef] [PubMed]
- Gordon, R.E.; Nemeth, J.F.; Singh, S.; Lingham, R.B.; Grewal, I.S. Harnessing sle autoantibodies for intracellular delivery of biologic therapeutics. Trends Biotechnol. 2021, 39, 298–310. [Google Scholar] [CrossRef]
- Pei, D.; Buyanova, M. Overcoming endosomal entrapment in drug delivery. Bioconjug Chem. 2019, 30, 273–283. [Google Scholar] [CrossRef]
- Choi, D.K.; Bae, J.; Shin, S.M.; Shin, J.Y.; Kim, S.; Kim, Y.S. A general strategy for generating intact, full-length igg antibodies that penetrate into the cytosol of living cells. MAbs 2014, 6, 1402–1414. [Google Scholar] [CrossRef]
- Shin, S.M.; Choi, D.K.; Jung, K.; Bae, J.; Kim, J.S.; Park, S.W.; Song, K.H.; Kim, Y.S. Antibody targeting intracellular oncogenic ras mutants exerts anti-tumour effects after systemic administration. Nat. Commun. 2017, 8, 15090. [Google Scholar] [CrossRef]
- Shin, S.M.; Kim, J.S.; Park, S.W.; Jun, S.Y.; Kweon, H.J.; Choi, D.K.; Lee, D.; Cho, Y.B.; Kim, Y.S. Direct targeting of oncogenic ras mutants with a tumor-specific cytosol-penetrating antibody inhibits ras mutant-driven tumor growth. Sci. Adv. 2020, 6, eaay2174. [Google Scholar] [CrossRef]
- Fujioka, A.; Terai, K.; Itoh, R.E.; Aoki, K.; Nakamura, T.; Kuroda, S.; Nishida, E.; Matsuda, M. Dynamics of the ras/erk mapk cascade as monitored by fluorescent probes. J. Biol. Chem. 2006, 281, 8917–8926. [Google Scholar] [CrossRef]
- Foss, S.; Bottermann, M.; Jonsson, A.; Sandlie, I.; James, L.C.; Andersen, J.T. Trim21-from intracellular immunity to therapy. Front. Immunol. 2019, 10, 2049. [Google Scholar] [CrossRef]
- Gaston, J.; Maestrali, N.; Lalle, G.; Gagnaire, M.; Masiero, A.; Dumas, B.; Dabdoubi, T.; Radosevic, K.; Berne, P.F. Intracellular delivery of therapeutic antibodies into specific cells using antibody-peptide fusions. Sci. Rep. 2019, 9, 18688. [Google Scholar] [CrossRef]
- Avci, F.G.; Akbulut, B.S.; Ozkirimli, E. Membrane active peptides and their biophysical characterization. Biomolecules 2018, 8, 77. [Google Scholar] [CrossRef]
- Zhou, M.; Zou, X.; Cheng, K.; Zhong, S.; Su, Y.; Wu, T.; Tao, Y.; Cong, L.; Yan, B.; Jiang, Y. The role of cell-penetrating peptides in potential anti-cancer therapy. Clin. Transl. Med. 2022, 12, e822. [Google Scholar] [CrossRef]
- Niu, Y.; Yu, M.; Zhang, J.; Yang, Y.; Xu, C.; Yeh, M.; Taran, E.; Hou, J.J.C.; Gray, P.P.; Yu, C. Synthesis of silica nanoparticles with controllable surface roughness for therapeutic protein delivery. J. Mater. Chem. B 2015, 3, 8477–8485. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, M.; Song, H.; Yu, C. Silica-based nanoparticles for biomedical applications: From nanocarriers to biomodulators. Acc. Chem. Res. 2020, 53, 1545–1556. [Google Scholar] [CrossRef]
- Mout, R.; Ray, M.; Tay, T.; Sasaki, K.; Yesilbag Tonga, G.; Rotello, V.M. General strategy for direct cytosolic protein delivery via protein-nanoparticle co-engineering. ACS Nano 2017, 11, 6416–6421. [Google Scholar] [CrossRef]
- Dolgin, E. Better lipids to power next generation of mrna vaccines. Science 2022, 376, 680–681. [Google Scholar] [CrossRef]
- Chan, A.; Wang, H.H.; Haley, R.M.; Song, C.; Gonzalez-Martinez, D.; Bugaj, L.; Mitchell, M.J.; Tsourkas, A. Cytosolic delivery of small protein scaffolds enables efficient inhibition of ras and myc. Mol. Pharm. 2022, 19, 1104–1116. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.H.; Tsourkas, A. Cytosolic delivery of inhibitory antibodies with cationic lipids. Proc. Natl. Acad. Sci. USA 2019, 116, 22132–22139. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mrna delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef] [PubMed]
- Bedard, P.L.; Tabernero, J.; Janku, F.; Wainberg, Z.A.; Paz-Ares, L.; Vansteenkiste, J.; Van Cutsem, E.; Perez-Garcia, J.; Stathis, A.; Britten, C.D.; et al. A phase ib dose-escalation study of the oral pan-pi3k inhibitor buparlisib (bkm120) in combination with the oral mek1/2 inhibitor trametinib (gsk1120212) in patients with selected advanced solid tumors. Clin. Cancer Res. 2015, 21, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Tolcher, A.W.; Patnaik, A.; Papadopoulos, K.P.; Rasco, D.W.; Becerra, C.R.; Allred, A.J.; Orford, K.; Aktan, G.; Ferron-Brady, G.; Ibrahim, N.; et al. Phase i study of the mek inhibitor trametinib in combination with the akt inhibitor afuresertib in patients with solid tumors and multiple myeloma. Cancer Chemother. Pharmacol. 2015, 75, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Tolcher, A.W.; Bendell, J.C.; Papadopoulos, K.P.; Burris, H.A., 3rd; Patnaik, A.; Jones, S.F.; Rasco, D.; Cox, D.S.; Durante, M.; Bellew, K.M.; et al. A phase ib trial of the oral mek inhibitor trametinib (gsk1120212) in combination with everolimus in patients with advanced solid tumors. Ann. Oncol. 2015, 26, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, G.I.; LoRusso, P.; Kwak, E.; Pandya, S.; Rudin, C.M.; Kurkjian, C.; Cleary, J.M.; Pilat, M.J.; Jones, S.; de Crespigny, A.; et al. Phase ib study of the mek inhibitor cobimetinib (gdc-0973) in combination with the pi3k inhibitor pictilisib (gdc-0941) in patients with advanced solid tumors. Investig. New Drugs 2020, 38, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.R.; Rosenberg, S.C.; McCormick, F.; Malek, S. Ras-targeted therapies: Is the undruggable drugged? Nat. Rev. Drug Discov. 2020, 19, 533–552. [Google Scholar] [CrossRef]
- Marin, I.; Boix, O.; Garcia-Garijo, A.; Sirois, I.; Caballe, A.; Zarzuela, E.; Ruano, I.; Stephan-Otto Attolini, C.; Prats, N.; Lopez-Dominguez, J.A.; et al. Cellular senescence is immunogenic and promotes anti-tumor immunity. Cancer Discov. 2022, 13, 410–431. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lam, K.K.; Wong, S.H.; Cheah, P.Y. Targeting the ‘Undruggable’ Driver Protein, KRAS, in Epithelial Cancers: Current Perspective. Cells 2023, 12, 631. https://doi.org/10.3390/cells12040631
Lam KK, Wong SH, Cheah PY. Targeting the ‘Undruggable’ Driver Protein, KRAS, in Epithelial Cancers: Current Perspective. Cells. 2023; 12(4):631. https://doi.org/10.3390/cells12040631
Chicago/Turabian StyleLam, Kuen Kuen, Siew Heng Wong, and Peh Yean Cheah. 2023. "Targeting the ‘Undruggable’ Driver Protein, KRAS, in Epithelial Cancers: Current Perspective" Cells 12, no. 4: 631. https://doi.org/10.3390/cells12040631
APA StyleLam, K. K., Wong, S. H., & Cheah, P. Y. (2023). Targeting the ‘Undruggable’ Driver Protein, KRAS, in Epithelial Cancers: Current Perspective. Cells, 12(4), 631. https://doi.org/10.3390/cells12040631





