Targeting Transcription Factors ATF5, CEBPB and CEBPD with Cell-Penetrating Peptides to Treat Brain and Other Cancers
Abstract
1. Introduction
2. A Brief Introduction to ATF5, CEBPB and CEBPD
3. ATF5: Recognition as a Target for Brain and Other Cancers
3.1. The Role of ATF5 in Growth and Differentiation of Neuroprogenitor Cells Suggests a Potential Role in Brain Cancers
3.2. ATF5 as an Anti-Apoptotic Factor
3.3. Additional Evidence That Identifies ATF5 as a Potential Target for Treatment of Brain Cancers
3.3.1. ATF5 Expression in Brain Tumors
3.3.2. ATF5 Expression and Patient Survival
3.3.3. Evidence from Knockdown Studies Verifies ATF5 as a Potential Target in Brain Cancers
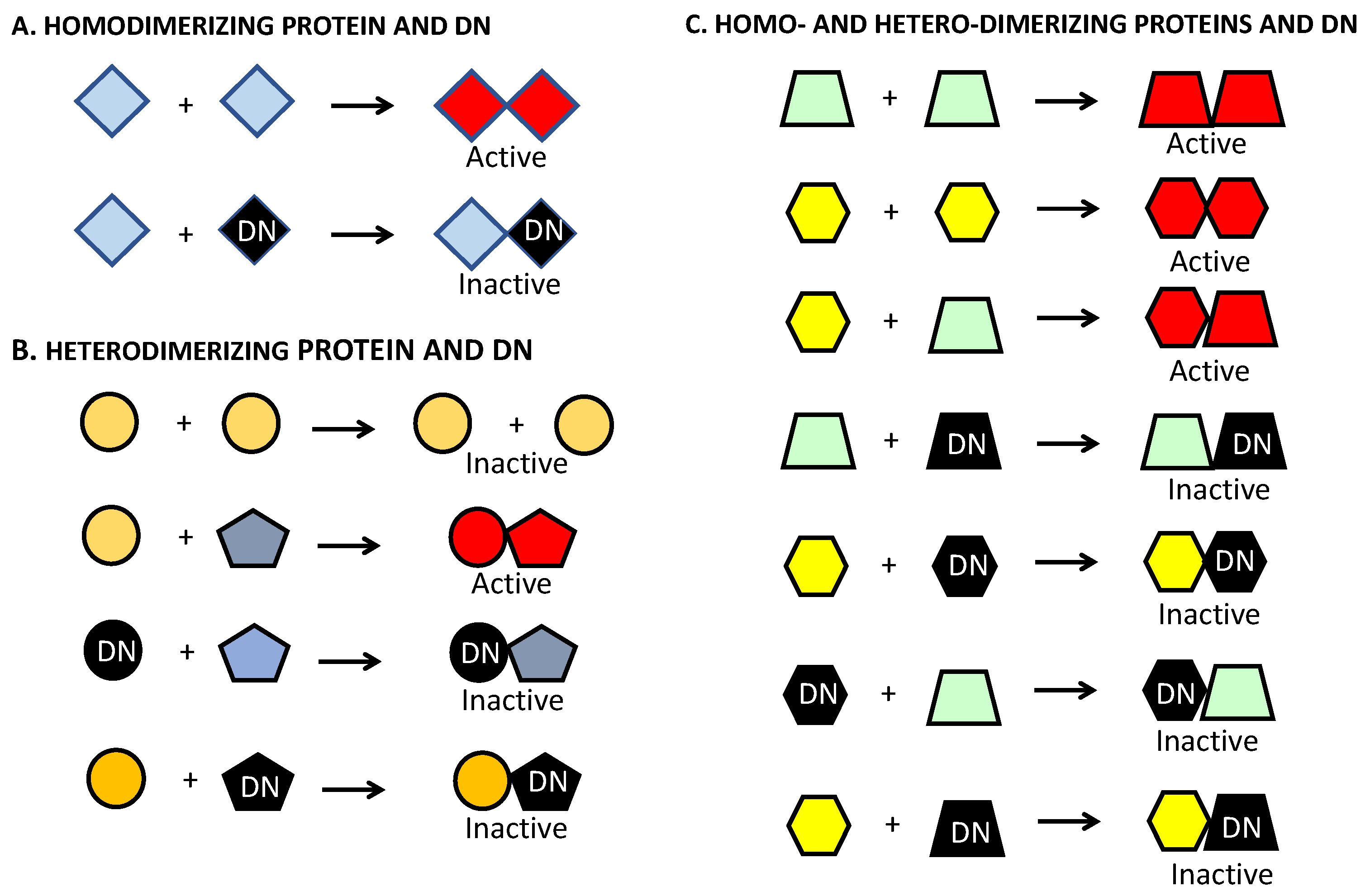
4. Dominant-Negative Constructs as a Strategy to Target ATF5 and Other Basic Leucine Zipper Proteins in Brain and Other Cancers
4.1. Development of Dominant-Negative ATF5 Constructs
4.2. DN-ATF5 Constructs and Effects on GBM Cells In Vitro and In Vivo
5. Drugging ATF5 Signaling in Brain and Other Tumors with Cell-Penetrating DN-ATF5 (CP-DN-ATF5): Design, Efficacy and Safety of CP-DN-ATF5 as a Potential Drug
6. DN–ATF5 Targets bZIP Transfection Factors CEBPB and CEBPD, but Not ATF5
7. CEBPB and CEBPD as Additional Targets for Treatment of Gliomas and Other Cancers
7.1. CEBPB/CEBPD Expression and Patient Prognosis
7.2. CEBPB and CEBPD and Transition to the Mesenchymal Phenotype in GBM
7.3. CEBPB/CEBPD and Glioma Cell Survival, Growth and Invasive Behavior
7.4. CEBPB/CEBPD and Glioma Stem Cell Renewal and Growth
7.5. CEBPB/CEBPD and Temozolomide Resistance in Gliomas
7.6. Regulation of CEBPB/CEBPD in Gliomas by Oncogenic Drivers
8. ATF5, CEBPB and CEBPD as Targets for Treatment of Non-Brain Cancers
9. Targeting ATF5, CEBPB and CEBPD with NEXT Generation Cell-Penetrating Leucine Zipper peptides: Bpep and Dpep
9.1. Potential Advantages of Directly Targeting ATF5, CEBPB and CEBPD Simultaneously
9.2. Dpep and Bpep as New Cell-Penetrating Peptides to Simultaneously Target ATF5, CEBPB and CEBPD and as Potential Therapeutic Treatments for Brain and Other Cancers
10. ST101
11. Mechanisms of Action of CP-DN-ATF5, Bpep, Dpep and ST101 on Brain and Other Tumor Cells
11.1. The Peptides Promote Tumor Cell Apoptosis
11.2. Dysregulation of Cell Pro- and Anti-Apoptotic Proteins BCL2, MCL1, Survivin and BMF
11.3. Consideration of Proliferation and Cell Cycle
11.4. Proximal Transcriptional Actions
11.5. Potential Interference with Additional Transcription Factors
12. Combination Therapies Employing CP-DN-ATF5, Bpep, Dpep and ST101
12.1. BH3-Mimetics
12.2. TRAIL
12.3. Radiotherapy
12.4. Temozolomide (TMZ)
12.5. Paclitaxel
12.6. Chloroquine
12.7. Doxorubicin
13. Clinical Trial with ST101 for Recurrent GBM
14. Conclusions and Perspectives—What Is Next?
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reinke, A.W.; Baek, J.; Ashenberg, O.; Keating, A.E. Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science 2013, 340, 730–734. [Google Scholar] [CrossRef]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science 1989, 243, 1681–1688. [Google Scholar] [CrossRef]
- Vinson, C.R.; Garcia, K.C. Molecular model for DNA recognition by the family of basic-helix-loop-helix-zipper proteins. New Biol. 1992, 4, 396–403. [Google Scholar]
- Lekstrom-Himes, J.; Xanthopoulos, K.G. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 1998, 273, 28545–28548. [Google Scholar] [CrossRef]
- Agre, P.; Johnson, P.F.; McKnight, S.L. Cognate DNA binding specificity retained after leucine zipper exchange between GCN4 and C/EBP. Science 1989, 246, 922–926. [Google Scholar] [CrossRef]
- Vinson, C.R.; Hai, T.; Boyd, S.M. Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: Prediction and rational design. Genes Dev. 1993, 7, 1047–1058. [Google Scholar] [CrossRef]
- Newman, J.R.; Keating, A.E. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 2003, 300, 2097–2101. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Sears, T.K.; Angelastro, J.M. The transcription factor ATF5: Role in cellular differentiation, stress responses, and cancer. Oncotarget 2017, 8, 84595–84609. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.; Haynes, C.M. Mitochondrial dysfunction in cancer: Potential roles of ATF5 and the mitochondrial UPR. Semin. Cancer Biol. 2017, 47, 43–49. [Google Scholar] [CrossRef]
- Paerhati, P.; Liu, J.; Jin, Z.; Jakos, T.; Zhu, S.; Qian, L.; Zhu, J.; Yuan, Y. Advancements in Activating Transcription Factor 5 Function in Regulating Cell Stress and Survival. Int. J. Mol. Sci. 2022, 23, 7129. [Google Scholar] [CrossRef]
- Sebastian, T.; Johnson, P.F. Stop and go: Anti-proliferative and mitogenic functions of the transcription factor C/EBPbeta. Cell Cycle. 2006, 5, 953–957. [Google Scholar] [CrossRef]
- Smink, J.J.; Leutz, A. Instruction of mesenchymal cell fate by the transcription factor C/EBPbeta. Gene 2012, 497, 10–17. [Google Scholar] [CrossRef]
- van der Krieken, S.E.; Popeijus, H.E.; Mensink, R.P.; Plat, J. CCAAT/enhancer binding protein beta in relation to ER stress, inflammation, and metabolic disturbances. Biomed. Res. Int. 2015, 2015, 324815. [Google Scholar] [CrossRef] [PubMed]
- Pulido-Salgado, M.; Vidal-Taboada, J.M.; Saura, J. C/EBPbeta and C/EBPdelta transcription factors: Basic biology and roles in the CNS. Prog. Neurobiol. 2015, 132, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Miller, M. Interactions of CCAAT/enhancer-binding protein beta with transcriptional coregulators. Postepy Biochem. 2016, 62, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Spike, A.J.; Rosen, J.M. C/EBPss Isoform Specific Gene Regulation: It’s a Lot more Complicated than you Think! J. Mammary Gland. Biol. Neoplasia 2020, 25, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, K.; Sterneck, E. The many faces of C/EBPdelta and their relevance for inflammation and cancer. Int. J. Biol. Sci. 2013, 9, 917–933. [Google Scholar] [CrossRef] [PubMed]
- Angelastro, J.M.; Klimaschewski, L.; Tang, S.; Vitolo, O.V.; Weissman, T.A.; Donlin, L.T.; Shelanski, M.L.; Greene, L.A. Identification of diverse nerve growth factor-regulated genes by serial analysis of gene expression (SAGE) profiling. Proc. Natl. Acad. Sci. USA 2000, 97, 10424–10429. [Google Scholar] [CrossRef]
- Greene, L.A.; Aletta, J.M.; Rukenstein, A.; Green, S.H. PC12 pheochromocytoma cells: Culture, nerve growth factor treatment, and experimental exploitation. Methods Enzymol. 1987, 147, 207–216. [Google Scholar]
- Angelastro, J.M.; Ignatova, T.N.; Kukekov, V.G.; Steindler, D.A.; Stengren, G.B.; Mendelsohn, C.; Greene, L.A. Regulated expression of ATF5 is required for the progression of neural progenitor cells to neurons. J. Neurosci. 2003, 23, 4590–4600. [Google Scholar] [CrossRef]
- Angelastro, J.M.; Mason, J.L.; Ignatova, T.N.; Kukekov, V.G.; Stengren, G.B.; Goldman, J.E.; Greene, L.A. Downregulation of activating transcription factor 5 is required for differentiation of neural progenitor cells into astrocytes. J. Neurosci. 2005, 25, 3889–3899. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.L.; Angelastro, J.M.; Ignatova, T.N.; Kukekov, V.G.; Lin, G.; Greene, L.A.; Goldman, J.E. ATF5 regulates the proliferation and differentiation of oligodendrocytes. Mol. Cell. Neurosci. 2005, 29, 372–380. [Google Scholar] [CrossRef]
- Umemura, M.; Ogura, T.; Matsuzaki, A.; Nakano, H.; Takao, K.; Miyakawa, T.; Takahashi, Y. Comprehensive Behavioral Analysis of Activating Transcription Factor 5-Deficient Mice. Front. Behav. Neurosci. 2017, 11, 125. [Google Scholar] [CrossRef]
- Umemura, M.; Kaneko, Y.; Tanabe, R.; Takahashi, Y. ATF5 deficiency causes abnormal cortical development. Sci. Rep. 2021, 11, 7295. [Google Scholar] [CrossRef]
- Devireddy, L.R.; Teodoro, J.G.; Richard, F.A.; Green, M.R. Induction of apoptosis by a secreted lipocalin that is transcriptionally regulated by IL-3 deprivation. Science 2001, 293, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Persengiev, S.P.; Devireddy, L.R.; Green, M.R. Inhibition of apoptosis by ATFx: A novel role for a member of the ATF/CREB family of mammalian bZIP transcription factors. Genes Dev. 2002, 16, 1806–1814. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Z.; Li, L.; Zhu, L.J.; Smith, T.W.; Demers, A.; Ross, A.H.; Moser, R.P.; Green, M.R. A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nat. Med. 2010, 16, 671–677. [Google Scholar] [CrossRef]
- Angelastro, J.M.; Canoll, P.D.; Kuo, J.; Weicker, M.; Costa, A.; Bruce, J.N.; Greene, L.A. Selective destruction of glioblastoma cells by interference with the activity or expression of ATF5. Oncogene 2006, 25, 907–916. [Google Scholar] [CrossRef]
- Huang, R.; Qian, D.; Hu, M.; Zhang, X.; Song, J.; Li, L.; Chen, H.; Wang, B. Association between human cytomegalovirus infection and histone acetylation level in various histological types of glioma. Oncol. Lett. 2015, 10, 2812–2820. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Wang, B.; Qian, D.; Wang, M.; Huang, R.; Wei, L.; Li, L.; Zhang, L.; Liu, D.X. Human cytomegalovirus immediate-early protein promotes survival of glioma cells through interacting and acetylating ATF5. Oncotarget 2017, 8, 32157–32170. [Google Scholar] [CrossRef] [PubMed]
- Feldheim, J.; Kessler, A.F.; Schmitt, D.; Wilczek, L.; Linsenmann, T.; Dahlmann, M.; Monoranu, C.M.; Ernestus, R.I.; Hagemann, C.; Lohr, M. Expression of activating transcription factor 5 (ATF5) is increased in astrocytomas of different WHO grades and correlates with survival of glioblastoma patients. OncoTargets Ther. 2018, 11, 8673–8684. [Google Scholar] [CrossRef] [PubMed]
- York, D.; Sproul, C.D.; Chikere, N.; Dickinson, P.J.; Angelastro, J.M. Expression and targeting of transcription factor ATF5 in dog gliomas. Vet. Comp. Oncol. 2018, 16, 102–107. [Google Scholar] [CrossRef]
- Wang, T.; Qian, D.; Hu, M.; Li, L.; Zhang, L.; Chen, H.; Yang, R.; Wang, B. Human cytomegalovirus inhibits apoptosis by regulating the activating transcription factor 5 signaling pathway in human malignant glioma cells. Oncol. Lett. 2014, 8, 1051–1057. [Google Scholar] [CrossRef]
- Huang, J.L.; Jiang, G.; Song, Q.X.; Gu, X.; Hu, M.; Wang, X.L.; Song, H.H.; Chen, L.P.; Lin, Y.Y.; Jiang, D.; et al. Lipoprotein-biomimetic nanostructure enables efficient targeting delivery of siRNA to Ras-activated glioblastoma cells via macropinocytosis. Nat. Commun. 2017, 8, 15144. [Google Scholar] [CrossRef]
- Zhou, D.; Palam, L.R.; Jiang, L.; Narasimhan, J.; Staschke, K.A.; Wek, R.C. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J. Biol. Chem. 2008, 283, 7064–7073. [Google Scholar] [CrossRef]
- Costa-Mattioli, M.; Walter, P. The integrated stress response: From mechanism to disease. Science 2020, 368, eaat5314. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.M.; Wang, J.; Qian, D.M.; Song, J.Y.; Chen, H.; Zhu, X.L.; Zhou, R.; Zhao, Y.D.; Zhou, X.Z.; Li, L.; et al. DNA methylation level of promoter region of activating transcription factor 5 in glioma. J. Zhejiang Univ. Sci. B 2015, 16, 757–762. [Google Scholar] [CrossRef]
- Wang, M.; Hu, M.; Li, Z.; Qian, D.; Wang, B.; Liu, D.X. miR-141-3p functions as a tumor suppressor modulating activating transcription factor 5 in glioma. Biochem. Biophys. Res. Commun. 2017, 490, 1260–1267. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Li, H.; Xie, H.; Fan, M.; Wang, J.; Zhang, N.; Ma, J.; Che, S. ELF1 Transcription Factor Enhances the Progression of Glioma via ATF5 promoter. ACS Chem. Neurosci. 2021, 12, 1252–1261. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y. Role of ATF5 During Cerebellar Development. Ph.D. Thesis, Columbia University, Ann Arbor, MI, USA, 2011. [Google Scholar]
- Monaco, S.E.; Angelastro, J.M.; Szabolcs, M.; Greene, L.A. The transcription factor ATF5 is widely expressed in carcinomas, and interference with its function selectively kills neoplastic, but not nontransformed, breast cell lines. Int. J. Cancer 2007, 120, 1883–1890. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Nutt, C.L.; Betensky, R.A.; Stemmer-Rachamimov, A.O.; Denko, N.C.; Ligon, K.L.; Rowitch, D.H.; Louis, D.N. Histology-based expression profiling yields novel prognostic markers in human glioblastoma. J. Neuropathol. Exp. Neurol. 2005, 64, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hu, M.; Xing, F.; Wang, M.; Wang, B.; Qian, D. Human cytomegalovirus infection promotes the stemness of U251 glioma cells. J. Med. Virol. 2017, 89, 878–886. [Google Scholar] [CrossRef] [PubMed]
- Herskowitz, I. Functional inactivation of genes by dominant negative mutations. Nature 1987, 329, 219–222. [Google Scholar] [CrossRef]
- Nguyen, B.N.; Elmore, L.W.; Holt, S.E. Mechanism of dominant-negative telomerase function. Cell Cycle 2009, 8, 3227–3233. [Google Scholar] [CrossRef]
- Dejager, S.; Mietus-Snyder, M.; Friera, A.; Pitas, R.E. Dominant negative mutations of the scavenger receptor. Native receptor inactivation by expression of truncated variants. J. Clin. Investig. 1993, 92, 894–902. [Google Scholar] [CrossRef]
- Krylov, D.; Olive, M.; Vinson, C. Extending dimerization interfaces: The bZIP basic region can form a coiled coil. EMBO J. 1995, 14, 5329–5337. [Google Scholar] [CrossRef]
- Olive, M.; Williams, S.C.; Dezan, C.; Johnson, P.F.; Vinson, C. Design of a C/EBP-specific, dominant-negative bZIP protein with both inhibitory and gain-of-function properties. J. Biol. Chem. 1996, 271, 2040–2047. [Google Scholar] [CrossRef]
- Olive, M.; Krylov, D.; Echlin, D.R.; Gardner, K.; Taparowsky, E.; Vinson, C. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J. Biol. Chem. 1997, 272, 18586–18594. [Google Scholar] [CrossRef]
- Arias, A.; Lame, M.W.; Santarelli, L.; Hen, R.; Greene, L.A.; Angelastro, J.M. Regulated ATF5 loss-of-function in adult mice blocks formation and causes regression/eradication of gliomas. Oncogene 2012, 31, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Assanah, M.; Lochhead, R.; Ogden, A.; Bruce, J.; Goldman, J.; Canoll, P. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J. Neurosci. 2006, 26, 6781–6790. [Google Scholar] [CrossRef] [PubMed]
- Hesselager, G.; Uhrbom, L.; Westermark, B.; Nister, M. Complementary effects of platelet-derived growth factor autocrine stimulation and p53 or Ink4a-Arf deletion in a mouse glioma model. Cancer Res. 2003, 63, 4305–4309. [Google Scholar] [PubMed]
- Vale, N.; Duarte, D.; Silva, S.; Correia, A.S.; Costa, B.; Gouveia, M.J.; Ferreira, A. Cell-penetrating peptides in oncologic pharmacotherapy: A review. Pharmacol. Res. 2020, 162, 105231. [Google Scholar] [CrossRef] [PubMed]
- Dupont, E.; Prochiantz, A.; Joliot, A. Penetratin Story: An Overview. Methods Mol. Biol. 2015, 1324, 29–37. [Google Scholar]
- Cates, C.C.; Arias, A.D.; Nakayama Wong, L.S.; Lame, M.W.; Sidorov, M.; Cayanan, G.; Rowland, D.J.; Fung, J.; Karpel-Massler, G.; Siegelin, M.D.; et al. Regression/eradication of gliomas in mice by a systemically-deliverable ATF5 dominant-negative peptide. Oncotarget 2016, 7, 12718–12730. [Google Scholar] [CrossRef]
- Ciaccio, N.A.; Reynolds, T.S.; Middaugh, C.R.; Laurence, J.S. Influence of the valine zipper region on the structure and aggregation of the basic leucine zipper (bZIP) domain of activating transcription factor 5 (ATF5). Mol. Pharm. 2012, 9, 3190–3199. [Google Scholar] [CrossRef]
- Karpel-Massler, G.; Horst, B.A.; Shu, C.; Chau, L.; Tsujiuchi, T.; Bruce, J.N.; Canoll, P.; Greene, L.A.; Angelastro, J.M.; Siegelin, M.D. A Synthetic Cell-Penetrating Dominant-Negative ATF5 Peptide Exerts Anticancer Activity against a Broad Spectrum of Treatment-Resistant Cancers. Clin. Cancer Res. 2016, 22, 4698–4711. [Google Scholar] [CrossRef] [PubMed]
- Peters, C.S.; Liang, X.; Li, S.; Kannan, S.; Peng, Y.; Taub, R.; Diamond, R.H. ATF-7, a novel bZIP protein, interacts with the PRL-1 protein-tyrosine phosphatase. J. Biol. Chem. 2001, 276, 13718–13726. [Google Scholar] [CrossRef]
- Ciaccio, N.A.; Moreno, M.L.; Bauer, R.L.; Laurence, J.S. High-yield expression in E. coli and refolding of the bZIP domain of activating transcription factor 5. Protein. Expr. Purif. 2008, 62, 235–243. [Google Scholar] [CrossRef]
- Sun, X.; Jefferson, P.; Zhou, Q.; Angelastro, J.M.; Greene, L.A. Dominant-Negative ATF5 Compromises Cancer Cell Survival by Targeting CEBPB and CEBPD. Mol. Cancer Res. 2020, 18, 216–228. [Google Scholar] [CrossRef]
- Nakajima, T.; Kinoshita, S.; Sasagawa, T.; Sasaki, K.; Naruto, M.; Kishimoto, T.; Akira, S. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc. Natl. Acad. Sci. USA 1993, 90, 2207–2211. [Google Scholar] [CrossRef]
- Homma, J.; Yamanaka, R.; Yajima, N.; Tsuchiya, N.; Genkai, N.; Sano, M.; Tanaka, R. Increased expression of CCAAT/enhancer binding protein beta correlates with prognosis in glioma patients. Oncol. Rep. 2006, 15, 595–601. [Google Scholar]
- Carro, M.S.; Lim, W.K.; Alvarez, M.J.; Bollo, R.J.; Zhao, X.; Snyder, E.Y.; Sulman, E.P.; Anne, S.L.; Doetsch, F.; Colman, H.; et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature 2010, 463, 318–325. [Google Scholar] [CrossRef]
- Cooper, L.A.; Gutman, D.A.; Chisolm, C.; Appin, C.; Kong, J.; Rong, Y.; Kurc, T.; Van Meir, E.G.; Saltz, J.H.; Moreno, C.S.; et al. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am. J. Pathol. 2012, 180, 2108–2119. [Google Scholar] [CrossRef]
- Chu, T.; Rice, E.J.; Booth, G.T.; Salamanca, H.H.; Wang, Z.; Core, L.J.; Longo, S.L.; Corona, R.J.; Chin, L.S.; Lis, J.T.; et al. Chromatin run-on and sequencing maps the transcriptional regulatory landscape of glioblastoma multiforme. Nat. Genet. 2018, 50, 1553–1564. [Google Scholar] [CrossRef]
- Kudo, T.; Prentzell, M.T.; Mohapatra, S.R.; Sahm, F.; Zhao, Z.; Grummt, I.; Wick, W.; Opitz, C.A.; Platten, M.; Green, E.W. Constitutive Expression of the Immunosuppressive Tryptophan Dioxygenase TDO2 in Glioblastoma Is Driven by the Transcription Factor C/EBPbeta. Front. Immunol. 2020, 11, 657. [Google Scholar] [CrossRef]
- Lei, K.; Xia, Y.; Wang, X.C.; Ahn, E.H.; Jin, L.; Ye, K. C/EBPbeta mediates NQO1 and GSTP1 anti-oxidative reductases expression in glioblastoma, promoting brain tumor proliferation. Redox Biol. 2020, 34, 101578. [Google Scholar] [CrossRef]
- Wang, D.; Ruan, X.; Liu, X.; Xue, Y.; Shao, L.; Yang, C.; Zhu, L.; Yang, Y.; Li, Z.; Yu, B.; et al. SUMOylation of PUM2 promotes the vasculogenic mimicry of glioma cells via regulating CEBPD. Clin. Transl. Med. 2020, 10, e168. [Google Scholar] [CrossRef]
- Wang, S.M.; Lin, W.C.; Lin, H.Y.; Chen, Y.L.; Ko, C.Y.; Wang, J.M. CCAAT/Enhancer-binding protein delta mediates glioma stem-like cell enrichment and ATP-binding cassette transporter ABCA1 activation for temozolomide resistance in glioblastoma. Cell Death Discov. 2021, 7, 8. [Google Scholar] [CrossRef]
- Halliday, J.; Helmy, K.; Pattwell, S.S.; Pitter, K.L.; LaPlant, Q.; Ozawa, T.; Holland, E.C. In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proc. Natl. Acad. Sci. USA 2014, 111, 5248–5253. [Google Scholar] [CrossRef]
- Minata, M.; Audia, A.; Shi, J.; Lu, S.; Bernstock, J.; Pavlyukov, M.S.; Das, A.; Kim, S.H.; Shin, Y.J.; Lee, Y.; et al. Phenotypic Plasticity of Invasive Edge Glioma Stem-like Cells in Response to Ionizing Radiation. Cell. Rep. 2019, 26, 1893–1905.e7. [Google Scholar] [CrossRef]
- Yin, J.; Oh, Y.T.; Kim, J.Y.; Kim, S.S.; Choi, E.; Kim, T.H.; Hong, J.H.; Chang, N.; Cho, H.J.; Sa, J.K.; et al. Transglutaminase 2 Inhibition Reverses Mesenchymal Transdifferentiation of Glioma Stem Cells by Regulating C/EBPbeta Signaling. Cancer Res. 2017, 77, 4973–4984. [Google Scholar] [CrossRef]
- Aguilar-Morante, D.; Cortes-Canteli, M.; Sanz-Sancristobal, M.; Santos, A.; Perez-Castillo, A. Decreased CCAAT/enhancer binding protein beta expression inhibits the growth of glioblastoma cells. Neuroscience 2011, 176, 110–119. [Google Scholar] [CrossRef]
- Aguilar-Morante, D.; Morales-Garcia, J.A.; Santos, A.; Perez-Castillo, A. CCAAT/enhancer binding protein beta induces motility and invasion of glioblastoma cells through transcriptional regulation of the calcium binding protein S100A4. Oncotarget 2015, 6, 4369–4384. [Google Scholar] [CrossRef]
- Di Pascale, F.; Nama, S.; Muhuri, M.; Quah, S.; Ismail, H.M.; Chan, X.H.D.; Sundaram, G.M.; Ramalingam, R.; Burke, B.; Sampath, P. C/EBPbeta mediates RNA polymerase III-driven transcription of oncomiR-138 in malignant gliomas. Nucleic Acids Res. 2018, 46, 336–349. [Google Scholar] [CrossRef]
- Wang, S.M.; Lin, H.Y.; Chen, Y.L.; Hsu, T.I.; Chuang, J.Y.; Kao, T.J.; Ko, C.Y. CCAAT/enhancer-binding protein delta regulates the stemness of glioma stem-like cells through activating PDGFA expression upon inflammatory stimulation. J. Neuroinflamm. 2019, 16, 146. [Google Scholar] [CrossRef]
- Lin, H.Y.; Lim, S.W.; Hsu, T.I.; Yang, W.B.; Huang, C.C.; Tsai, Y.T.; Chang, W.C.; Ko, C.Y. CCAAT/Enhancer-Binding Protein Delta Regulates Glioblastoma Survival through Catalase-Mediated Hydrogen Peroxide Clearance. Oxid. Med. Cell. Longev. 2022, 2022, 4081380. [Google Scholar] [CrossRef]
- Biserova, K.; Jakovlevs, A.; Uljanovs, R.; Strumfa, I. Cancer Stem Cells: Significance in Origin, Pathogenesis and Treatment of Glioblastoma. Cells 2021, 10, 621. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, B.; Feng, L.; Sun, B.; He, S.; Yang, Y.; Wu, G.; E, G.; Liu, C.; Gao, Y.; et al. Targeting JUN, CEBPB, and HDAC3: A Novel Strategy to Overcome Drug Resistance in Hypoxic Glioblastoma. Front. Oncol. 2019, 9, 33. [Google Scholar] [CrossRef]
- Gao, Z.; Xu, J.; Fan, Y.; Qi, Y.; Wang, S.; Zhao, S.; Guo, X.; Xue, H.; Deng, L.; Zhao, R.; et al. PDIA3P1 promotes Temozolomide resistance in glioblastoma by inhibiting C/EBPbeta degradation to facilitate proneural-to-mesenchymal transition. J. Exp. Clin. Cancer Res. 2022, 41, 223. [Google Scholar] [CrossRef]
- Selagea, L.; Mishra, A.; Anand, M.; Ross, J.; Tucker-Burden, C.; Kong, J.; Brat, D.J. EGFR and C/EBP-beta oncogenic signaling is bidirectional in human glioma and varies with the C/EBP-beta isoform. FASEB J. 2016, 30, 4098–4108. [Google Scholar] [CrossRef]
- Kong, X.; Meng, W.; Zhou, Z.; Li, Y.; Zhou, B.; Wang, R.; Zhan, L. Overexpression of activating transcription factor 5 in human rectal cancer. Exp. Ther. Med. 2011, 2, 827–831. [Google Scholar] [CrossRef]
- Wang, D.; Yang, L.; Yu, W.; Wu, Q.; Lian, J.; Li, F.; Liu, S.; Li, A.; He, Z.; Liu, J.; et al. Colorectal cancer cell-derived CCL20 recruits regulatory T cells to promote chemoresistance via FOXO1/CEBPB/NF-kappaB signaling. J. Immunother. Cancer 2019, 7, 215. [Google Scholar] [CrossRef]
- Shao, K.; Pu, W.; Zhang, J.; Guo, S.; Qian, F.; Glurich, I.; Jin, Q.; Ma, Y.; Ju, S.; Zhang, Z.; et al. DNA hypermethylation contributes to colorectal cancer metastasis by regulating the binding of CEBPB and TFCP2 to the CPEB1 promoter. Clin. Epigenetics 2021, 13, 89. [Google Scholar] [CrossRef]
- Tang, X.; Liang, Y.; Sun, G.; He, Q.; Qu, H.; Gao, P. UBQLN4 is activated by C/EBPbeta and exerts oncogenic effects on colorectal cancer via the Wnt/beta-catenin signaling pathway. Cell Death Discov. 2021, 7, 398. [Google Scholar] [CrossRef]
- Zhou, Z.; Shu, Y.; Bao, H.; Han, S.; Liu, Z.; Zhao, N.; Yuan, W.; Jian, C.; Shu, X. Stress-induced epinephrine promotes epithelial-to-mesenchymal transition and stemness of CRC through the CEBPB/TRIM2/P53 axis. J. Transl. Med. 2022, 20, 262. [Google Scholar] [CrossRef]
- Wang, Z.; Pang, J.; Wang, L.; Dong, Q.; Jin, D. CEBPB regulates the bile acid receptor FXR to accelerate colon cancer progression by modulating aerobic glycolysis. J. Clin. Lab. Anal. 2022, 36, e24703. [Google Scholar] [CrossRef]
- Salaroglio, I.C.; Belisario, D.C.; Akman, M.; La Vecchia, S.; Godel, M.; Anobile, D.P.; Ortone, G.; Digiovanni, S.; Fontana, S.; Costamagna, C.; et al. Mitochondrial ROS drive resistance to chemotherapy and immune-killing in hypoxic non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2022, 41, 243. [Google Scholar] [CrossRef]
- Okazaki, K.; Anzawa, H.; Liu, Z.; Ota, N.; Kitamura, H.; Onodera, Y.; Alam, M.M.; Matsumaru, D.; Suzuki, T.; Katsuoka, F.; et al. Enhancer remodeling promotes tumor-initiating activity in NRF2-activated non-small cell lung cancers. Nat. Commun. 2020, 11, 5911. [Google Scholar] [CrossRef]
- Okazaki, K.; Anzawa, H.; Katsuoka, F.; Kinoshita, K.; Sekine, H.; Motohashi, H. CEBPB is required for NRF2-mediated drug resistance in NRF2-activated non-small cell lung cancer cells. J. Biochem. 2022, 171, 567–578. [Google Scholar] [CrossRef]
- Liu, M.; Li, R.; Wang, M.; Liu, T.; Zhou, Q.; Zhang, D.; Wang, J.; Shen, M.; Ren, X.; Sun, Q. PGAM1 regulation of ASS1 contributes to the progression of breast cancer through the cAMP/AMPK/CEBPB pathway. Mol. Oncol. 2022, 16, 2843–2860. [Google Scholar] [CrossRef]
- Sterken, B.A.; Ackermann, T.; Muller, C.; Zuidhof, H.R.; Kortman, G.; Hernandez-Segura, A.; Broekhuis, M.; Spierings, D.; Guryev, V.; Calkhoven, C.F. C/EBPbeta isoform-specific regulation of migration and invasion in triple-negative breast cancer cells. NPJ Breast Cancer 2022, 8, 11. [Google Scholar] [CrossRef]
- Liu, X.Z.; Rulina, A.; Choi, M.H.; Pedersen, L.; Lepland, J.; Takle, S.T.; Madeleine, N.; Peters, S.D.; Wogsland, C.E.; Grondal, S.M.; et al. C/EBPB-dependent adaptation to palmitic acid promotes tumor formation in hormone receptor negative breast cancer. Nat. Commun. 2022, 13, 69. [Google Scholar] [CrossRef]
- Wang, S.; Xia, D.; Wang, X.; Cao, H.; Wu, C.; Sun, Z.; Zhang, D.; Liu, H. C/EBPbeta regulates the JAK/STAT signaling pathway in triple-negative breast cancer. FEBS Open Bio 2021, 11, 1250–1258. [Google Scholar] [CrossRef]
- Lee, L.L.; Kim, S.J.; Hahn, Y.I.; Jang, J.H.; Saeidi, S.; Surh, Y.J. Stabilization of C/EBPbeta through direct interaction with STAT3 in H-Ras transformed human mammary epithelial cells. Biochem. Biophys. Res. Commun. 2021, 546, 130–137. [Google Scholar] [CrossRef]
- Wu, H.; Gu, J.; Zhou, D.; Cheng, W.; Wang, Y.; Wang, Q.; Wang, X. LINC00160 mediated paclitaxel-And doxorubicin-resistance in breast cancer cells by regulating TFF3 via transcription factor C/EBPbeta. J. Cell. Mol. Med. 2020, 24, 8589–8602. [Google Scholar] [CrossRef]
- Zhang, R.; Li, X.; Liu, Z.; Wang, Y.; Zhang, H.; Xu, H. EZH2 inhibitors-mediated epigenetic reactivation of FOSB inhibits triple-negative breast cancer progress. Cancer Cell. Int. 2020, 20, 175. [Google Scholar] [CrossRef]
- Wang, F.; Gao, Y.; Tang, L.; Ning, K.; Geng, N.; Zhang, H.; Li, Y.; Li, Y.; Liu, F.; Li, F. A novel PAK4-CEBPB-CLDN4 axis involving in breast cancer cell migration and invasion. Biochem. Biophys. Res. Commun. 2019, 511, 404–408. [Google Scholar] [CrossRef]
- Balamurugan, K.; Mendoza-Villanueva, D.; Sharan, S.; Summers, G.H.; Dobrolecki, L.E.; Lewis, M.T.; Sterneck, E. C/EBPdelta links IL-6 and HIF-1 signaling to promote breast cancer stem cell-associated phenotypes. Oncogene 2019, 38, 3765–3780. [Google Scholar] [CrossRef]
- Cao, J.; Wang, M.; Wang, T. CCAAT enhancer binding protein beta has a crucial role in regulating breast cancer cell growth via activating the TGF-beta-Smad3 signaling pathway. Exp. Ther. Med. 2017, 14, 1554–1560. [Google Scholar] [CrossRef]
- Ben-Shmuel, S.; Rashed, R.; Rostoker, R.; Isakov, E.; Shen-Orr, Z.; LeRoith, D. Activating Transcription Factor-5 Knockdown Reduces Aggressiveness of Mammary Tumor Cells and Attenuates Mammary Tumor Growth. Front. Endocrinol. (Lausanne) 2017, 8, 173. [Google Scholar] [CrossRef]
- Kurzejamska, E.; Johansson, J.; Jirstrom, K.; Prakash, V.; Ananthaseshan, S.; Boon, L.; Fuxe, J.; Religa, P. C/EBPbeta expression is an independent predictor of overall survival in breast cancer patients by MHCII/CD4-dependent mechanism of metastasis formation. Oncogenesis 2014, 3, e125. [Google Scholar] [CrossRef]
- Park, B.H.; Kook, S.; Lee, S.; Jeong, J.H.; Brufsky, A.; Lee, B.C. An isoform of C/EBPbeta, LIP, regulates expression of the chemokine receptor CXCR4 and modulates breast cancer cell migration. J. Biol. Chem. 2013, 288, 28656–28667. [Google Scholar] [CrossRef]
- Chen, A.; Qian, D.; Wang, B.; Hu, M.; Lu, J.; Qi, Y.; Liu, D.X. ATF5 is overexpressed in epithelial ovarian carcinomas and interference with its function increases apoptosis through the downregulation of Bcl-2 in SKOV-3 cells. Int. J. Gynecol. Pathol. 2012, 31, 532–537. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, X.X.; Li, M.C.; Cao, C.H.; Wan, D.Y.; Xi, B.X.; Tan, J.H.; Wang, J.; Yang, Z.Y.; Feng, X.X.; et al. C/EBPbeta enhances platinum resistance of ovarian cancer cells by reprogramming H3K79 methylation. Nat. Commun. 2018, 9, 1739. [Google Scholar] [CrossRef]
- Tan, J.; Zheng, X.; Li, M.; Ye, F.; Song, C.; Xu, C.; Zhang, X.; Li, W.; Wang, Y.; Zeng, S.; et al. C/EBPbeta promotes poly(ADP-ribose) polymerase inhibitor resistance by enhancing homologous recombination repair in high-grade serous ovarian cancer. Oncogene 2021, 40, 3845–3858. [Google Scholar] [CrossRef]
- Yang, S.; Wang, J.; Cheng, R.; Pang, B.; Sun, P. LINC00035 Transcriptional Regulation of SLC16A3 via CEBPB Affects Glycolysis and Cell Apoptosis in Ovarian Cancer. Evid. Based Complement. Alternat. Med. 2021, 2021, 5802082. [Google Scholar] [CrossRef] [PubMed]
- Hour, T.C.; Lai, Y.L.; Kuan, C.I.; Chou, C.K.; Wang, J.M.; Tu, H.Y.; Hu, H.T.; Lin, C.S.; Wu, W.J.; Pu, Y.S.; et al. Transcriptional up-regulation of SOD1 by CEBPD: A potential target for cisplatin resistant human urothelial carcinoma cells. Biochem. Pharmacol. 2010, 80, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Gong, Y.; Li, H.; Jiao, L.; Xin, D.; Gong, Y.; He, Z.; Zhou, L.; Jin, Y.; Wang, X.; et al. C/EBPbeta promotes the viability of human bladder cancer cell by contributing to the transcription of bladder cancer specific lncRNA UCA1. Biochem. Biophys. Res. Commun. 2018, 506, 674–679. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Tian, H.; Zhi, X.; Xiao, Z.; Chen, T.; Yuan, H.; Chen, Q.; Chen, M.; Yang, J.; Zhou, Q.; et al. Activating transcription factor 5 (ATF5) promotes tumorigenic capability and activates the Wnt/b-catenin pathway in bladder cancer. Cancer Cell Int. 2021, 21, 660. [Google Scholar] [CrossRef]
- Regalo, G.; Canedo, P.; Suriano, G.; Resende, C.; Campos, M.L.; Oliveira, M.J.; Figueiredo, C.; Rodrigues-Pereira, P.; Blin, N.; Seruca, R.; et al. C/EBPbeta is over-expressed in gastric carcinogenesis and is associated with COX-2 expression. J. Pathol. 2006, 210, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Yang, Z.; Lu, X.; Yousuf, S.; Zhao, M.; Li, W.; Miao, J.; Wang, X.; Yu, H.; Zhu, X.; et al. Anoikis resistant gastric cancer cells promote angiogenesis and peritoneal metastasis through C/EBPbeta-mediated PDGFB autocrine and paracrine signaling. Oncogene 2021, 40, 5764–5779. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Minton, A.Z.; Agrawal, V. C/EBPbeta regulates metastatic gene expression and confers TNF-alpha resistance to prostate cancer cells. Prostate 2009, 69, 1435–1447. [Google Scholar] [CrossRef]
- Barakat, D.J.; Mendonca, J.; Barberi, T.; Zhang, J.; Kachhap, S.K.; Paz-Priel, I.; Friedman, A.D. C/EBPbeta regulates sensitivity to bortezomib in prostate cancer cells by inducing REDD1 and autophagosome-lysosome fusion. Cancer Lett. 2016, 375, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Barakat, D.J.; Zhang, J.; Barberi, T.; Denmeade, S.R.; Friedman, A.D.; Paz-Priel, I. CCAAT/Enhancer binding protein beta controls androgen-deprivation-induced senescence in prostate cancer cells. Oncogene 2015, 34, 5912–5922. [Google Scholar] [CrossRef]
- Li, D.; Liu, J.; Huang, S.; Bi, X.; Wang, B.; Chen, Q.; Chen, H.; Pu, X. CCAAT enhancer binding protein beta promotes tumor growth and inhibits apoptosis in prostate cancer by methylating estrogen receptor beta. Neoplasma 2018, 65, 34–41. [Google Scholar] [CrossRef]
- Adamo, H.; Hammarsten, P.; Hagglof, C.; Dahl Scherdin, T.; Egevad, L.; Stattin, P.; Halin Bergstrom, S.; Bergh, A. Prostate cancer induces C/EBPbeta expression in surrounding epithelial cells which relates to tumor aggressiveness and patient outcome. Prostate 2019, 79, 435–445. [Google Scholar] [CrossRef]
- Wang, W.J.; Lai, H.Y.; Zhang, F.; Shen, W.J.; Chu, P.Y.; Liang, H.Y.; Liu, Y.B.; Wang, J.M. MCL1 participates in leptin-promoted mitochondrial fusion and contributes to drug resistance in gallbladder cancer. JCI Insight 2021, 6, 15. [Google Scholar] [CrossRef]
- Hu, M.; Wang, B.; Qian, D.; Li, L.; Zhang, L.; Song, X.; Liu, D.X. Interference with ATF5 function enhances the sensitivity of human pancreatic cancer cells to paclitaxel-induced apoptosis. Anticancer Res. 2012, 32, 4385–4394. [Google Scholar]
- Huang, C.S.; Chu, J.; Zhu, X.X.; Li, J.H.; Huang, X.T.; Cai, J.P.; Zhao, W.; Yin, X.Y. The C/EBPbeta-LINC01133 axis promotes cell proliferation in pancreatic ductal adenocarcinoma through upregulation of CCNG1. Cancer Lett. 2018, 421, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Sterneck, E.; Zhu, S.; Ramirez, A.; Jorcano, J.L.; Smart, R.C. Conditional ablation of C/EBP beta demonstrates its keratinocyte-specific requirement for cell survival and mouse skin tumorigenesis. Oncogene 2006, 25, 1272–1276. [Google Scholar] [CrossRef] [PubMed]
- Messenger, Z.J.; Hall, J.R.; Jima, D.D.; House, J.S.; Tam, H.W.; Tokarz, D.A.; Smart, R.C. C/EBPbeta deletion in oncogenic Ras skin tumors is a synthetic lethal event. Cell Death Dis. 2018, 9, 1054. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, F.; Xu, Q.; Yang, B.; Li, X.; Jiang, S.; Hu, L.; Zhang, X.; Zhu, L.; Li, Q.; et al. Inhibiting Importin 4-mediated nuclear import of CEBPD enhances chemosensitivity by repression of PRKDC-driven DNA damage repair in cervical cancer. Oncogene 2020, 39, 5633–5648. [Google Scholar] [CrossRef] [PubMed]
- Shuai, Y.; Fan, E.; Zhong, Q.; Feng, G.; Chen, Q.; Gou, X.; Zhang, G. ATF5 involved in radioresistance in nasopharyngeal carcinoma by promoting epithelial-to-mesenchymal phenotype transition. Eur. Arch. Otorhinolaryngol. 2020, 277, 2869–2879. [Google Scholar] [CrossRef]
- Du, Q.; Tan, Z.; Shi, F.; Tang, M.; Xie, L.; Zhao, L.; Li, Y.; Hu, J.; Zhou, M.; Bode, A.; et al. PGC1alpha/CEBPB/CPT1A axis promotes radiation resistance of nasopharyngeal carcinoma through activating fatty acid oxidation. Cancer Sci. 2019, 110, 2050–2062. [Google Scholar] [CrossRef]
- Liu, H.; Huang, Q.; Lv, Y.; Dong, Y.; Song, D. CEBPB knockdown sensitizes nasopharyngeal carcinoma cells to cisplatin by promoting the expression of serine protease inhibitor Kazal-type 5. Anticancer Drugs 2022, 33, e327–e335. [Google Scholar] [CrossRef]
- Xu, C.; Shen, Y.; Shi, Y.; Zhang, M.; Zhou, L. Eukaryotic translation initiation factor 3 subunit B promotes head and neck cancer via CEBPB translation. Cancer Cell Int. 2022, 22, 161. [Google Scholar] [CrossRef]
- Rong, L.; Chen, B.; Liu, K.; Liu, B.; He, X.; Liu, J.; Li, J.; He, M.; Zhu, L.; Liu, K.; et al. CircZDBF2 up-regulates RNF145 by ceRNA model and recruits CEBPB to accelerate oral squamous cell carcinoma progression via NFkappaB signaling pathway. J. Transl. Med. 2022, 20, 148. [Google Scholar] [CrossRef]
- Li, H.; Yang, F.; Chai, L.; Zhang, L.; Li, S.; Xu, Z.; Kong, L. CCAAT/Enhancer Binding Protein beta-Mediated MMP3 Upregulation Promotes Esophageal Squamous Cell Cancer Invasion In Vitro and Is Associated with Metastasis in Human Patients. Genet. Test. Mol. Biomarkers 2019, 23, 304–309. [Google Scholar] [CrossRef]
- Cao, L.J.; Zhang, Y.J.; Dong, S.Q.; Li, X.Z.; Tong, X.T.; Chen, D.; Wu, Z.Y.; Zheng, X.H.; Xue, W.Q.; Jia, W.H.; et al. ATAD2 interacts with C/EBPbeta to promote esophageal squamous cell carcinoma metastasis via TGF-beta1/Smad3 signaling. J. Exp. Clin. Cancer Res. 2021, 40, 109. [Google Scholar] [CrossRef] [PubMed]
- Qin, M.; Han, F.; Wu, J.; Gao, F.X.; Li, Y.; Yan, D.X.; He, X.M.; Long, Y.; Tang, X.P.; Ren, D.L.; et al. KDM6B promotes ESCC cell proliferation and metastasis by facilitating C/EBPbeta transcription. BMC Cancer 2021, 21, 559. [Google Scholar]
- He, F.; Xiao, H.; Cai, Y.; Zhang, N. ATF5 and HIF1alpha cooperatively activate HIF1 signaling pathway in esophageal cancer. Cell. Commun. Signal. 2021, 19, 53. [Google Scholar] [CrossRef] [PubMed]
- Swoboda, A.; Soukup, R.; Eckel, O.; Kinslechner, K.; Wingelhofer, B.; Schorghofer, D.; Sternberg, C.; Pham, H.T.T.; Vallianou, M.; Horvath, J.; et al. STAT3 promotes melanoma metastasis by CEBP-induced repression of the MITF pathway. Oncogene 2021, 40, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Ambrosini, G.; Do, C.; Tycko, B.; Realubit, R.B.; Karan, C.; Musi, E.; Carvajal, R.D.; Chua, V.; Aplin, A.E.; Schwartz, G.K. Inhibition of NF-kappaB-Dependent Signaling Enhances Sensitivity and Overcomes Resistance to BET Inhibition in Uveal Melanoma. Cancer Res. 2019, 79, 2415–2425. [Google Scholar] [CrossRef] [PubMed]
- Nachiyappan, A.; Soon, J.L.J.; Lim, H.J.; Lee, V.K.; Taneja, R. EHMT1 promotes tumor progression and maintains stemness by regulating ALDH1A1 expression in alveolar rhabdomyosarcoma. J. Pathol. 2022, 256, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.; Zhang, J.; Tan, Z.; Shen, L.F.; Zhou, R.R.; Zhang, Y.Y. Maf1 suppression of ATF5-dependent mitochondrial unfolded protein response contributes to rapamycin-induced radio-sensitivity in lung cancer cell line A549. Aging (Albany NY) 2021, 13, 7300–7313. [Google Scholar] [CrossRef] [PubMed]
- Rajbhandari, P.; Lopez, G.; Capdevila, C.; Salvatori, B.; Yu, J.; Rodriguez-Barrueco, R.; Martinez, D.; Yarmarkovich, M.; Weichert-Leahey, N.; Abraham, B.J.; et al. Cross-Cohort Analysis Identifies a TEAD4-MYCN Positive Feedback Loop as the Core Regulatory Element of High-Risk Neuroblastoma. Cancer Discov. 2018, 8, 582–599. [Google Scholar] [CrossRef]
- Hua, Z.Y.; Hansen, J.N.; He, M.; Dai, S.K.; Choi, Y.; Fulton, M.D.; Lloyd, S.M.; Szemes, M.; Sen, J.; Ding, H.F.; et al. PRMT1 promotes neuroblastoma cell survival through ATF5. Oncogenesis 2020, 9, 50. [Google Scholar] [CrossRef]
- Gardiner, J.D.; Abegglen, L.M.; Huang, X.; Carter, B.E.; Schackmann, E.A.; Stucki, M.; Paxton, C.N.; Lor Randall, R.; Amatruda, J.F.; Putnam, A.R.; et al. C/EBPbeta-1 promotes transformation and chemoresistance in Ewing sarcoma cells. Oncotarget 2017, 8, 26013–26026. [Google Scholar] [CrossRef]
- Wang, Y.H.; Wu, W.J.; Wang, W.J.; Huang, H.Y.; Li, W.M.; Yeh, B.W.; Wu, T.F.; Shiue, Y.L.; Sheu, J.J.; Wang, J.M.; et al. CEBPD amplification and overexpression in urothelial carcinoma: A driver of tumor metastasis indicating adverse prognosis. Oncotarget 2015, 6, 31069–31084. [Google Scholar] [CrossRef]
- Wang, W.J.; Li, C.F.; Chu, Y.Y.; Wang, Y.H.; Hour, T.C.; Yen, C.J.; Chang, W.C.; Wang, J.M. Inhibition of the EGFR/STAT3/CEBPD Axis Reverses Cisplatin Cross-resistance with Paclitaxel in the Urothelial Carcinoma of the Urinary Bladder. Clin. Cancer Res. 2017, 23, 503–513. [Google Scholar] [CrossRef]
- Chan, T.C.; Hsing, C.H.; Shiue, Y.L.; Huang, S.K.; Hsieh, K.L.; Kuo, Y.H.; Li, C.F. Angiogenesis Driven by the CEBPD-hsa-miR-429-VEGFA Signaling Axis Promotes Urothelial Carcinoma Progression. Cells 2022, 11, 638. [Google Scholar] [CrossRef]
- Yusenko, M.V.; Trentmann, A.; Casolari, D.A.; Abdel Ghani, L.; Lenz, M.; Horn, M.; Dorner, W.; Klempnauer, S.; Mootz, H.D.; Arteaga, M.F.; et al. C/EBPbeta is a MYB- and p300-cooperating pro-leukemogenic factor and promising drug target in acute myeloid leukemia. Oncogene 2021, 40, 4746–4758. [Google Scholar] [CrossRef]
- Klempnauer, K.H. C/EBPbeta sustains the oncogenic program of AML cells by cooperating with MYB and co-activator p300 in a transcriptional module. Exp. Hematol. 2022, 108, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Abdel Ghani, L.; Yusenko, M.V.; Frank, D.; Moorthy, R.; Widen, J.C.; Dorner, W.; Khandanpour, C.; Harki, D.A.; Klempnauer, K.H. A synthetic covalent ligand of the C/EBPbeta transactivation domain inhibits acute myeloid leukemia cells. Cancer Lett. 2022, 530, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, J.; Gagne, V.; Labuda, M.; Beaubois, C.; Sinnett, D.; Laverdiere, C.; Moghrabi, A.; Sallan, S.E.; Silverman, L.B.; Neuberg, D.; et al. ATF5 polymorphisms influence ATF function and response to treatment in children with childhood acute lymphoblastic leukemia. Blood 2011, 118, 5883–5890. [Google Scholar] [CrossRef]
- Youssef, Y.H.; Makkeyah, S.M.; Soliman, A.F.; Meky, N.H. Influence of genetic variants in asparaginase pathway on the susceptibility to asparaginase-related toxicity and patients’ outcome in childhood acute lymphoblastic leukemia. Cancer Chemother. Pharmacol. 2021, 88, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Kurata, M.; Onishi, I.; Takahara, T.; Yamazaki, Y.; Ishibashi, S.; Goitsuka, R.; Kitamura, D.; Takita, J.; Hayashi, Y.; Largaesapda, D.A.; et al. C/EBPbeta induces B-cell acute lymphoblastic leukemia and cooperates with BLNK mutations. Cancer Sci. 2021, 112, 4920–4930. [Google Scholar] [CrossRef]
- Duprez, E. A new role for C/EBPbeta in acute promyelocytic leukemia. Cell Cycle 2004, 3, 389–390. [Google Scholar] [CrossRef]
- Mittal, A.K.; Hegde, G.V.; Aoun, P.; Bociek, R.G.; Dave, B.J.; Joshi, A.D.; Sanger, W.G.; Weisenburger, D.D.; Joshi, S.S. Molecular basis of aggressive disease in chronic lymphocytic leukemia patients with 11q deletion and trisomy 12 chromosomal abnormalities. Int. J. Mol. Med. 2007, 20, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Pal, R.; Janz, M.; Galson, D.L.; Gries, M.; Li, S.; Johrens, K.; Anagnostopoulos, I.; Dorken, B.; Mapara, M.Y.; Borghesi, L.; et al. C/EBPbeta regulates transcription factors critical for proliferation and survival of multiple myeloma cells. Blood 2009, 114, 3890–3898. [Google Scholar] [CrossRef] [PubMed]
- Piva, R.; Pellegrino, E.; Mattioli, M.; Agnelli, L.; Lombardi, L.; Boccalatte, F.; Costa, G.; Ruggeri, B.A.; Cheng, M.; Chiarle, R.; et al. Functional validation of the anaplastic lymphoma kinase signature identifies CEBPB and BCL2A1 as critical target genes. J. Clin. Investig. 2006, 116, 3171–3182. [Google Scholar] [CrossRef] [PubMed]
- Bisikirska, B.; Bansal, M.; Shen, Y.; Teruya-Feldstein, J.; Chaganti, R.; Califano, A. Elucidation and Pharmacological Targeting of Novel Molecular Drivers of Follicular Lymphoma Progression. Cancer Res. 2016, 76, 664–674. [Google Scholar] [CrossRef]
- White, J.H.; McIllhinney, R.A.; Wise, A.; Ciruela, F.; Chan, W.Y.; Emson, P.C.; Billinton, A.; Marshall, F.H. The GABAB receptor interacts directly with the related transcription factors CREB2 and ATFx. Proc. Natl. Acad. Sci. USA 2000, 97, 13967–13972. [Google Scholar] [CrossRef]
- Jiang, X.; Su, L.; Zhang, Q.; He, C.; Zhang, Z.; Yi, P.; Liu, J. GABAB receptor complex as a potential target for tumor therapy. J. Histochem. Cytochem. 2012, 60, 269–279. [Google Scholar] [CrossRef]
- Shu, Q.; Liu, J.; Liu, X.; Zhao, S.; Li, H.; Tan, Y.; Xu, J. GABAB R/GSK-3beta/NF-kappaB signaling pathway regulates the proliferation of colorectal cancer cells. Cancer Med. 2016, 5, 1259–1267. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.A.; Kandpal, G.; Ma, L.; Austin, C.P. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: Regulation and loss of interaction with mutation. Hum. Mol. Genet. 2003, 12, 1591–1608. [Google Scholar] [CrossRef]
- Kakiuchi, C.; Ishiwata, M.; Nanko, S.; Kunugi, H.; Minabe, Y.; Nakamura, K.; Mori, N.; Fujii, K.; Yamada, K.; Yoshikawa, T.; et al. Association analysis of ATF4 and ATF5, genes for interacting-proteins of DISC1, in bipolar disorder. Neurosci. Lett. 2007, 417, 316–321. [Google Scholar] [CrossRef]
- Gao, X.; Mi, Y.; Guo, N.; Hu, Z.; Hu, F.; Liu, D.; Gao, L.; Gou, X.; Jin, W. Disrupted in schizophrenia 1 (DISC1) inhibits glioblastoma development by regulating mitochondria dynamics. Oncotarget 2016, 7, 85963–85974. [Google Scholar] [CrossRef] [PubMed]
- Madarampalli, B.; Yuan, Y.; Liu, D.; Lengel, K.; Xu, Y.; Li, G.; Yang, J.; Liu, X.; Lu, Z.; Liu, D.X. ATF5 Connects the Pericentriolar Materials to the Proximal End of the Mother Centriole. Cell 2015, 162, 580–592. [Google Scholar] [CrossRef]
- Fiorese, C.J.; Schulz, A.M.; Lin, Y.F.; Rosin, N.; Pellegrino, M.W.; Haynes, C.M. The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr. Biol. 2016, 26, 2037–2043. [Google Scholar] [CrossRef] [PubMed]
- Siegelin, M.D.; Dohi, T.; Raskett, C.M.; Orlowski, G.M.; Powers, C.M.; Gilbert, C.A.; Ross, A.H.; Plescia, J.; Altieri, D.C. Exploiting the mitochondrial unfolded protein response for cancer therapy in mice and human cells. J. Clin. Investig. 2011, 121, 1349–1360. [Google Scholar] [CrossRef] [PubMed]
- Keerthiga, R.; Pei, D.S.; Fu, A. Mitochondrial dysfunction, UPR(mt) signaling, and targeted therapy in metastasis tumor. Cell Biosci. 2021, 11, 186. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Y.D.; Zhang, Y.Y.; Qian, S.W.; Zhang, Z.C.; Li, S.F.; Guo, L.; Liu, Y.; Wen, B.; Lei, Q.Y.; et al. p300-dependent acetylation of activating transcription factor 5 enhances C/EBPbeta transactivation of C/EBPalpha during 3T3-L1 differentiation. Mol. Cell. Biol. 2014, 34, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Khramushin, A.; Ben-Aharon, Z.; Tsaban, T.; Varga, J.K.; Avraham, O.; Schueler-Furman, O. Matching protein surface structural patches for high-resolution blind peptide docking. Proc. Natl. Acad. Sci. USA 2022, 119, e2121153119. [Google Scholar] [CrossRef]
- Zhou, Q.; Sun, X.; Pasquier, N.; Jefferson, P.; Nguyen, T.T.T.; Siegelin, M.D.; Angelastro, J.M.; Greene, L.A. Cell-Penetrating CEBPB and CEBPD Leucine Zipper Decoys as Broadly Acting Anti-Cancer Agents. Cancers 2021, 13, 2504. [Google Scholar] [CrossRef] [PubMed]
- Darvishi, E.; Ghamsari, L.; Leong, S.F.; Ramirez, R.; Koester, M.; Gallagher, E.; Yu, M.; Mason, J.M.; Merutka, G.; Kappel, B.J.; et al. Anticancer Activity of ST101, A Novel Antagonist of CCAAT/Enhancer Binding Protein beta. Mol. Cancer Ther. 2022, 21, 1632–1644. [Google Scholar] [CrossRef]
- Dluzen, D.; Li, G.; Tacelosky, D.; Moreau, M.; Liu, D.X. BCL-2 is a downstream target of ATF5 that mediates the prosurvival function of ATF5 in a cell type-dependent manner. J. Biol. Chem. 2011, 286, 7705–7713. [Google Scholar] [CrossRef]
- Wheatley, S.P.; Altieri, D.C. Survivin at a glance. J. Cell. Sci. 2019, 132, jcs223826. [Google Scholar] [CrossRef] [PubMed]
- Brun, S.N.; Markant, S.L.; Esparza, L.A.; Garcia, G.; Terry, D.; Huang, J.M.; Pavlyukov, M.S.; Li, X.N.; Grant, G.A.; Crawford, J.R.; et al. Survivin as a therapeutic target in Sonic hedgehog-driven medulloblastoma. Oncogene 2015, 34, 3770–3779. [Google Scholar] [CrossRef]
- Frazzi, R. BIRC3 and BIRC5: Multi-faceted inhibitors in cancer. Cell Biosci. 2021, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Yang, P.; Wang, K.; Liu, Y.; Liu, X.; Shan, X.; Huang, R.; Zhang, K.; Wang, J. Survivin is a prognostic indicator in glioblastoma and may be a target of microRNA-218. Oncol. Lett. 2019, 18, 359–367. [Google Scholar] [CrossRef]
- Sun, X.; Angelastro, J.M.; Merino, D.; Zhou, Q.; Siegelin, M.D.; Greene, L.A. Dominant-negative ATF5 rapidly depletes survivin in tumor cells. Cell Death Dis. 2019, 10, 709. [Google Scholar] [CrossRef] [PubMed]
- Putavet, D.A.; de Keizer, P.L.J. Residual Disease in Glioma Recurrence: A Dangerous Liaison with Senescence. Cancers 2021, 13, 1560. [Google Scholar] [CrossRef]
- Bousset, L.; Gil, J. Targeting senescence as an anticancer therapy. Mol. Oncol. 2022, 16, 3855–3880. [Google Scholar] [CrossRef] [PubMed]
- Salotti, J.; Sakchaisri, K.; Tourtellotte, W.G.; Johnson, P.F. An Arf-Egr-C/EBPbeta pathway linked to ras-induced senescence and cancer. Mol. Cell. Biol. 2015, 35, 866–883. [Google Scholar] [CrossRef]
- Podust, L.M.; Krezel, A.M.; Kim, Y. Crystal structure of the CCAAT box/enhancer-binding protein beta activating transcription factor-4 basic leucine zipper heterodimer in the absence of DNA. J. Biol. Chem. 2001, 276, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, H.; Maeda, S.; Hayashi, M.; Takeda, S.; Akira, S.; Komiya, S.; Nakamura, T.; Akiyama, H.; Imamura, T. CCAAT/enhancer-binding protein beta promotes osteoblast differentiation by enhancing Runx2 activity with ATF4. Mol. Biol. Cell. 2008, 19, 5373–5386. [Google Scholar] [CrossRef]
- Mann, I.K.; Chatterjee, R.; Zhao, J.; He, X.; Weirauch, M.T.; Hughes, T.R.; Vinson, C. CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome. Res. 2013, 23, 988–997. [Google Scholar] [CrossRef]
- Ebert, S.M.; Bullard, S.A.; Basisty, N.; Marcotte, G.R.; Skopec, Z.P.; Dierdorff, J.M.; Al-Zougbi, A.; Tomcheck, K.C.; DeLau, A.D.; Rathmacher, J.A.; et al. Activating transcription factor 4 (ATF4) promotes skeletal muscle atrophy by forming a heterodimer with the transcriptional regulator C/EBPbeta. J. Biol. Chem. 2020, 295, 2787–2803. [Google Scholar] [CrossRef]
- Wortel, I.M.N.; van der Meer, L.T.; Kilberg, M.S.; van Leeuwen, F.N. Surviving Stress: Modulation of ATF4-Mediated Stress Responses in Normal and Malignant Cells. Trends. Endocrinol. Metab. 2017, 28, 794–806. [Google Scholar] [CrossRef]
- Moeckel, S.; LaFrance, K.; Wetsch, J.; Seliger, C.; Riemenschneider, M.J.; Proescholdt, M.; Hau, P.; Vollmann-Zwerenz, A. ATF4 contributes to autophagy and survival in sunitinib treated brain tumor initiating cells (BTICs). Oncotarget 2019, 10, 368–382. [Google Scholar] [CrossRef]
- Lorenz, N.I.; Sittig, A.C.M.; Urban, H.; Luger, A.L.; Engel, A.L.; Munch, C.; Steinbach, J.P.; Ronellenfitsch, M.W. Activating transcription factor 4 mediates adaptation of human glioblastoma cells to hypoxia and temozolomide. Sci. Rep. 2021, 11, 14161. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, Z.; Liu, C.; Wang, B.; Liu, P.; Fang, S.; Yang, F.; You, Y.; Li, X. ATF4-dependent fructolysis fuels growth of glioblastoma multiforme. Nat. Commun. 2022, 13, 6108. [Google Scholar] [CrossRef]
- Kaspar, S.; Oertlin, C.; Szczepanowska, K.; Kukat, A.; Senft, K.; Lucas, C.; Brodesser, S.; Hatzoglou, M.; Larsson, O.; Topisirovic, I.; et al. Adaptation to mitochondrial stress requires CHOP-directed tuning of ISR. Sci. Adv. 2021, 7, eabf0971. [Google Scholar] [CrossRef] [PubMed]
- Parkin, S.E.; Baer, M.; Copeland, T.D.; Schwartz, R.C.; Johnson, P.F. Regulation of CCAAT/enhancer-binding protein (C/EBP) activator proteins by heterodimerization with C/EBPgamma (Ig/EBP). J. Biol. Chem. 2002, 277, 23563–23572. [Google Scholar] [CrossRef]
- Huggins, C.J.; Malik, R.; Lee, S.; Salotti, J.; Thomas, S.; Martin, N.; Quinones, O.A.; Alvord, W.G.; Olanich, M.E.; Keller, J.R.; et al. C/EBPgamma suppresses senescence and inflammatory gene expression by heterodimerizing with C/EBPbeta. Mol. Cell. Biol. 2013, 33, 3242–3258. [Google Scholar] [CrossRef]
- Renfro, Z.; White, B.E.; Stephens, K.E. CCAAT enhancer binding protein gamma (C/EBP-gamma): An understudied transcription factor. Adv. Biol. Regul. 2022, 84, 100861. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.M.; Cory, S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 2007, 26, 1324–1337. [Google Scholar] [CrossRef] [PubMed]
- Karpel-Massler, G.; Ishida, C.T.; Zhang, Y.; Halatsch, M.E.; Westhoff, M.A.; Siegelin, M.D. Targeting intrinsic apoptosis and other forms of cell death by BH3-mimetics in glioblastoma. Expert. Opin. Drug Discov. 2017, 12, 1031–1040. [Google Scholar] [CrossRef]
- Kaloni, D.; Diepstraten, S.T.; Strasser, A.; Kelly, G.L. BCL-2 protein family: Attractive targets for cancer therapy. Apoptosis 2022, 1–19. [Google Scholar] [CrossRef]
- Diepstraten, S.T.; Anderson, M.A.; Czabotar, P.E.; Lessene, G.; Strasser, A.; Kelly, G.L. The manipulation of apoptosis for cancer therapy using BH3-mimetic drugs. Nat. Rev. Cancer 2022, 22, 45–64. [Google Scholar] [CrossRef] [PubMed]
- Koessinger, A.L.; Cloix, C.; Koessinger, D.; Heiland, D.H.; Bock, F.J.; Strathdee, K.; Kinch, K.; Martinez-Escardo, L.; Paul, N.R.; Nixon, C.; et al. Increased apoptotic sensitivity of glioblastoma enables therapeutic targeting by BH3-mimetics. Cell Death Differ. 2022, 29, 2089–2104. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Li, X.; Morsch, M.; Ismail, M.; Liu, Y.; Rehman, F.U.; Zhang, D.; Wang, Y.; Zheng, M.; Chung, R.; et al. Brain-Targeted Codelivery of Bcl-2/Bcl-xl and Mcl-1 Inhibitors by Biomimetic Nanoparticles for Orthotopic Glioblastoma Therapy. ACS Nano 2022, 16, 6293–6308. [Google Scholar] [CrossRef]
- Wendt, M.D. Discovery of ABT-263, a Bcl-family protein inhibitor: Observations on targeting a large protein-protein interaction. Expert. Opin. Drug Discov. 2008, 3, 1123–1143. [Google Scholar] [CrossRef]
- Wong, S.H.M.; Kong, W.Y.; Fang, C.M.; Loh, H.S.; Chuah, L.H.; Abdullah, S.; Ngai, S.C. The TRAIL to cancer therapy: Hindrances and potential solutions. Crit. Rev. Oncol. Hematol. 2019, 143, 81–94. [Google Scholar] [CrossRef]
- Deng, L.; Zhai, X.; Liang, P.; Cui, H. Overcoming TRAIL Resistance for Glioblastoma Treatment. Biomolecules 2021, 11, 572. [Google Scholar] [CrossRef]
- Ishihara, S.; Yasuda, M.; Ishizu, A.; Ishikawa, M.; Shirato, H.; Haga, H. Activating transcription factor 5 enhances radioresistance and malignancy in cancer cells. Oncotarget 2015, 6, 4602–4614. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Aykin-Burns, N.; Krager, K.J.; Shah, S.K.; Melnyk, S.B.; Hauer-Jensen, M.; Pawar, S.A. Loss of C/EBPdelta enhances IR-induced cell death by promoting oxidative stress and mitochondrial dysfunction. Free Radic. Biol. Med. 2016, 99, 296–307. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Dmello, C.; Chen, L.; Arrieta, V.A.; Gonzalez-Buendia, E.; Kane, J.R.; Magnusson, L.P.; Baran, A.; James, C.D.; Horbinski, C.; et al. Ultrasound-mediated Delivery of Paclitaxel for Glioma: A Comparative Study of Distribution, Toxicity, and Efficacy of Albumin-bound Versus Cremophor Formulations. Clin. Cancer Res. 2020, 26, 477–486. [Google Scholar] [CrossRef]
- Ullah, I.; Chung, K.; Bae, S.; Li, Y.; Kim, C.; Choi, B.; Nam, H.Y.; Kim, S.H.; Yun, C.O.; Lee, K.Y.; et al. Nose-to-Brain Delivery of Cancer-Targeting Paclitaxel-Loaded Nanoparticles Potentiates Antitumor Effects in Malignant Glioblastoma. Mol. Pharm. 2020, 17, 1193–1204. [Google Scholar] [CrossRef]
- Weyerhauser, P.; Kantelhardt, S.R.; Kim, E.L. Re-purposing Chloroquine for Glioblastoma: Potential Merits and Confounding Variables. Front. Oncol. 2018, 8, 335. [Google Scholar] [CrossRef]
- Caron, N.J.; Quenneville, S.P.; Tremblay, J.P. Endosome disruption enhances the functional nuclear delivery of Tat-fusion proteins. Biochem. Biophys. Res. Commun. 2004, 319, 12–20. [Google Scholar] [CrossRef] [PubMed]
- van der Zanden, S.Y.; Qiao, X.; Neefjes, J. New insights into the activities and toxicities of the old anticancer drug doxorubicin. FEBS J. 2020, 288, 6095–6111. [Google Scholar] [CrossRef] [PubMed]
- Aragon-Sanabria, V.; Aditya, A.; Zhang, L.; Chen, F.; Yoo, B.; Cao, T.; Madajewski, B.; Lee, R.; Turker, M.Z.; Ma, K.; et al. Ultrasmall Nanoparticle Delivery of Doxorubicin Improves Therapeutic Index for High-Grade Glioma. Clin. Cancer Res. 2022, 28, 2938–2952. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, F.; Gondi, V.; Butowski, N.; Falchook, G.; Williams, A.; Peters, K.; Evans, J.; Lakhani, N.; McKean, M.; Symeonides, S.; et al. CTNI-49. Early signal of activity from a phase 2 study of ST101, a first-in-class peptide antagonist of CCAAT/enhancer-binding protein β (C/EBPβ), in recurrent glioblastoma (GBM). Neuro-Oncol. 2022, 24 (Suppl. S7), vii83. [Google Scholar] [CrossRef]
- Dauparas, J.; Anishchenko, I.; Bennett, N.; Bai, H.; Ragotte, R.J.; Milles, L.F.; Wicky, B.I.M.; Courbet, A.; de Haas, R.J.; Bethel, N.; et al. Robust deep learning-based protein sequence design using ProteinMPNN. Science 2022, 378, 49–56. [Google Scholar] [CrossRef]
- Chang, L.; Mondal, A.; Perez, A. Towards rational computational peptide design. Front. Bioinform. 2022, 2, 1046493. [Google Scholar] [CrossRef]
- Bottens, R.A.; Yamada, T. Cell-Penetrating Peptides (CPPs) as Therapeutic and Diagnostic Agents for Cancer. Cancers 2022, 14, 5546. [Google Scholar] [CrossRef]
- Shoari, A.; Tooyserkani, R.; Tahmasebi, M.; Lowik, D. Delivery of Various Cargos into Cancer Cells and Tissues via Cell-Penetrating Peptides: A Review of the Last Decade. Pharmaceutics 2021, 13, 1391. [Google Scholar] [CrossRef] [PubMed]
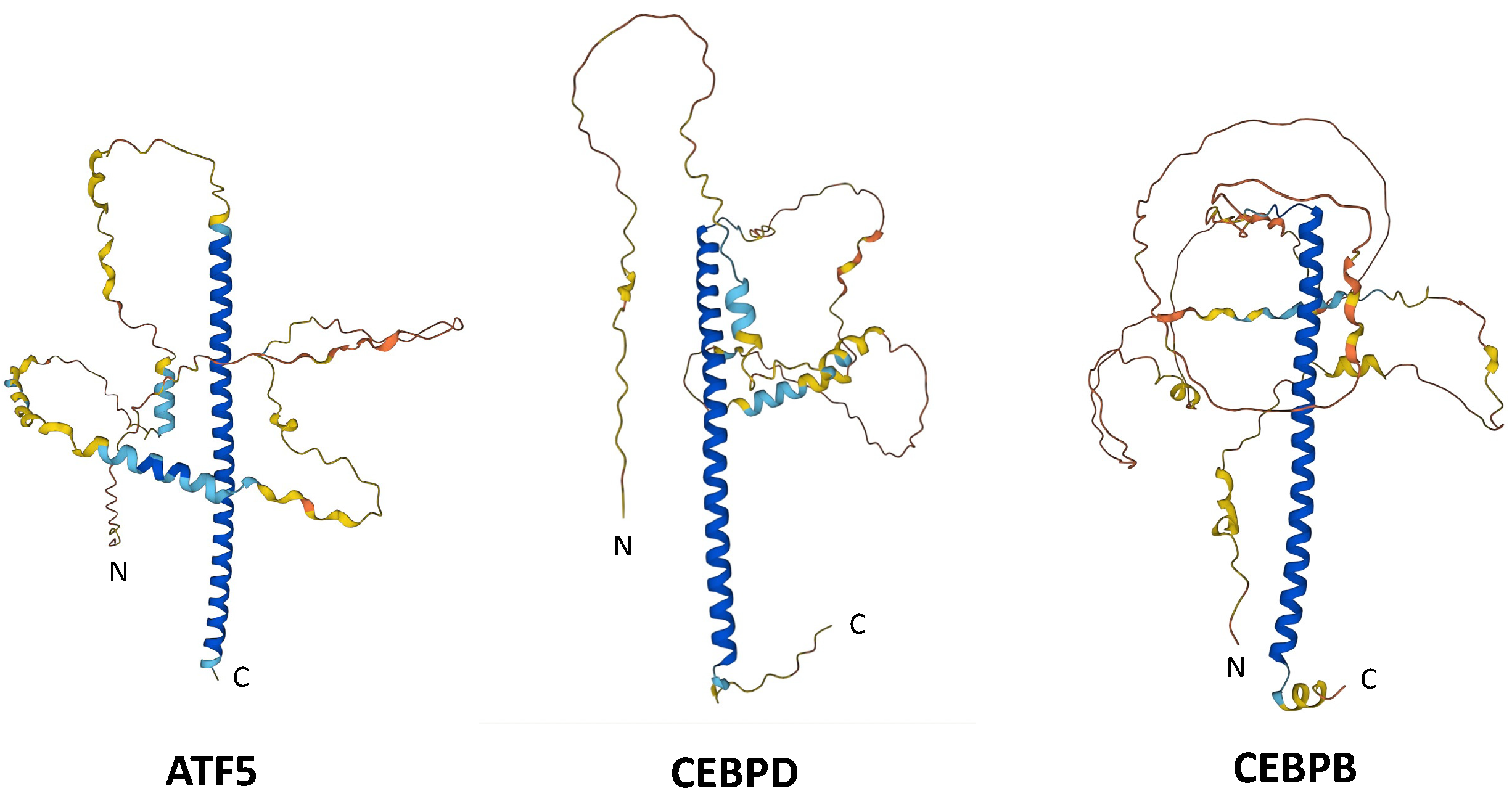

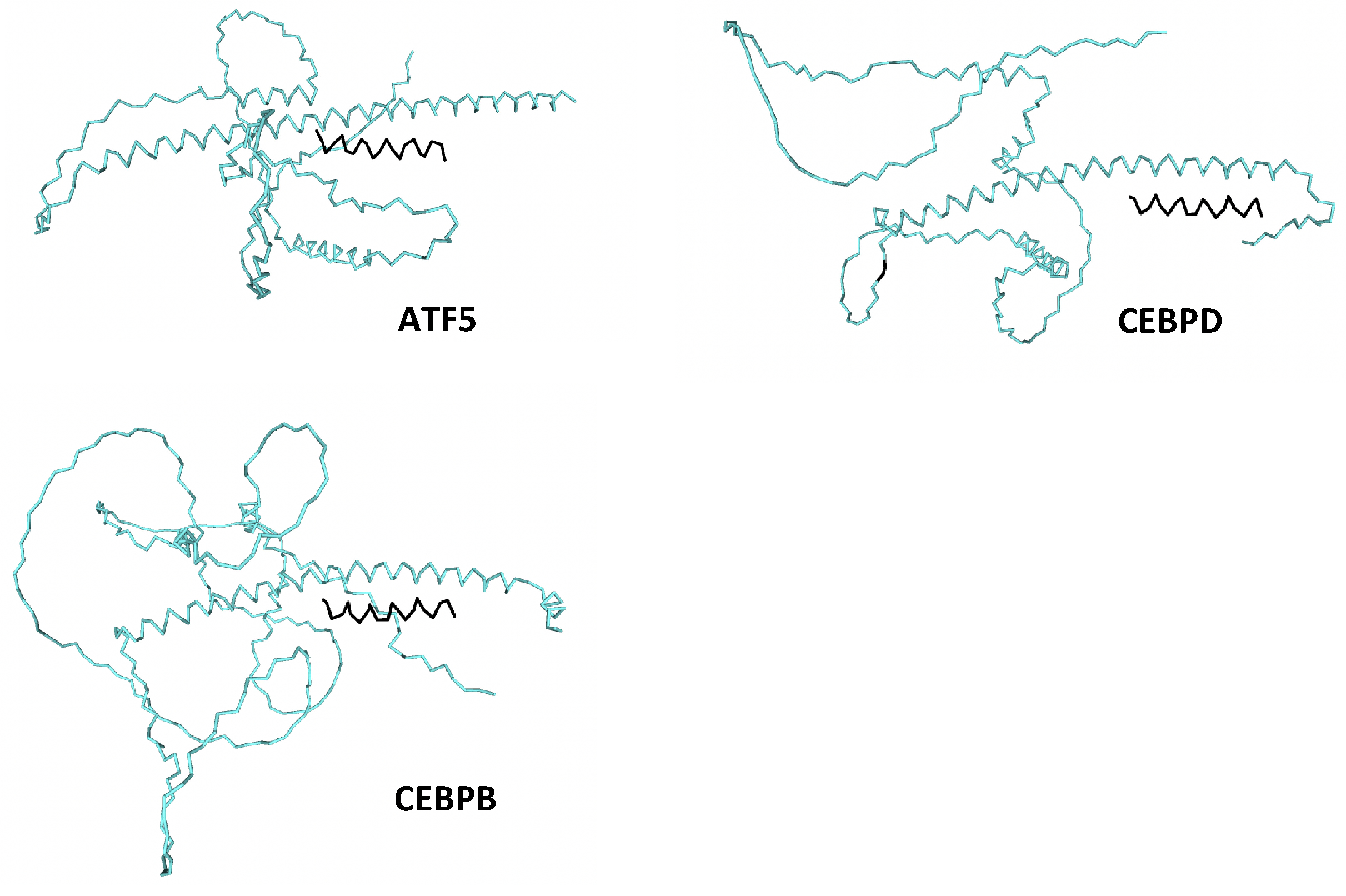
| Peptide | ATF5 | CEBPB | CEBPD |
|---|---|---|---|
| CP-DN-ATF5 | − | + | + |
| Dpep | + | + | + |
| Bpep | + | + | + |
| ST101 | − | + | +? |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greene, L.A.; Zhou, Q.; Siegelin, M.D.; Angelastro, J.M. Targeting Transcription Factors ATF5, CEBPB and CEBPD with Cell-Penetrating Peptides to Treat Brain and Other Cancers. Cells 2023, 12, 581. https://doi.org/10.3390/cells12040581
Greene LA, Zhou Q, Siegelin MD, Angelastro JM. Targeting Transcription Factors ATF5, CEBPB and CEBPD with Cell-Penetrating Peptides to Treat Brain and Other Cancers. Cells. 2023; 12(4):581. https://doi.org/10.3390/cells12040581
Chicago/Turabian StyleGreene, Lloyd A., Qing Zhou, Markus D. Siegelin, and James M. Angelastro. 2023. "Targeting Transcription Factors ATF5, CEBPB and CEBPD with Cell-Penetrating Peptides to Treat Brain and Other Cancers" Cells 12, no. 4: 581. https://doi.org/10.3390/cells12040581
APA StyleGreene, L. A., Zhou, Q., Siegelin, M. D., & Angelastro, J. M. (2023). Targeting Transcription Factors ATF5, CEBPB and CEBPD with Cell-Penetrating Peptides to Treat Brain and Other Cancers. Cells, 12(4), 581. https://doi.org/10.3390/cells12040581







