Merkel Cell Polyomavirus Large T Antigen Induces Cellular Senescence for Host Growth Arrest and Viral Genome Persistence through Its Unique Domain
Abstract
1. Introduction
2. Results
2.1. MCPyV LT Mediates Cellular Senescence in Human Fibroblast Cells through Its Unique Domain
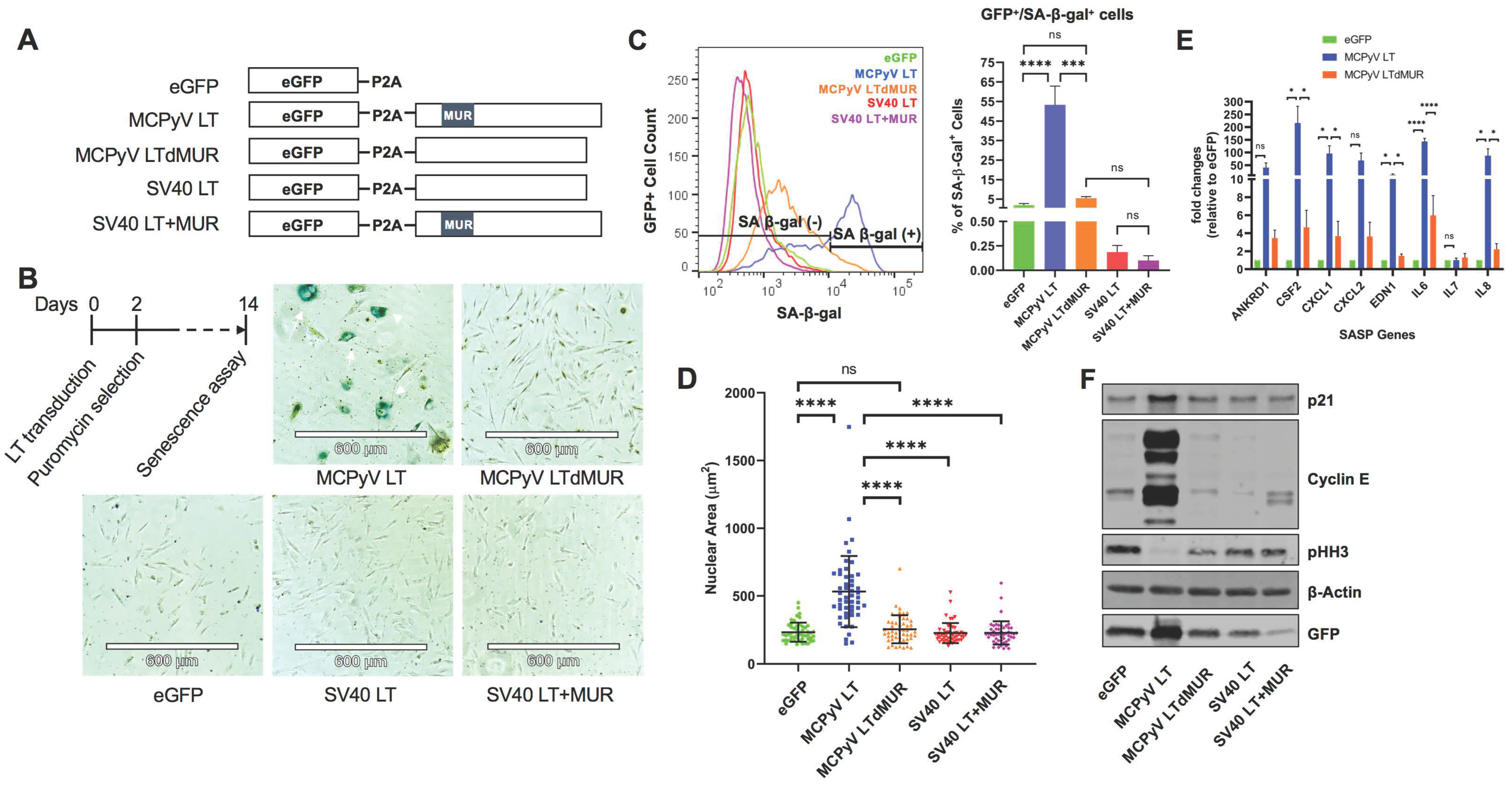
2.2. MCPyV LT-Mediated Senescence Is DDR-Independent

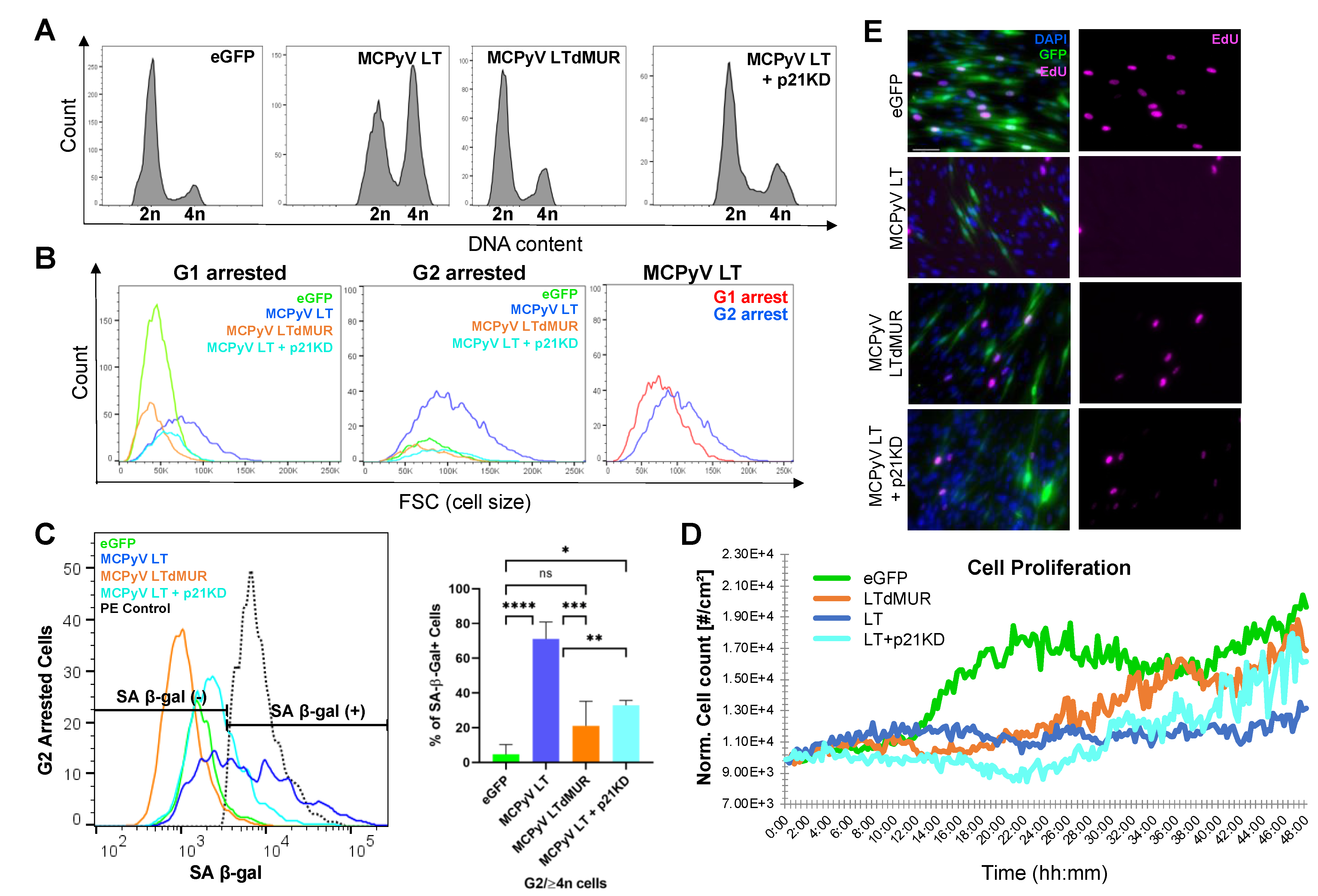
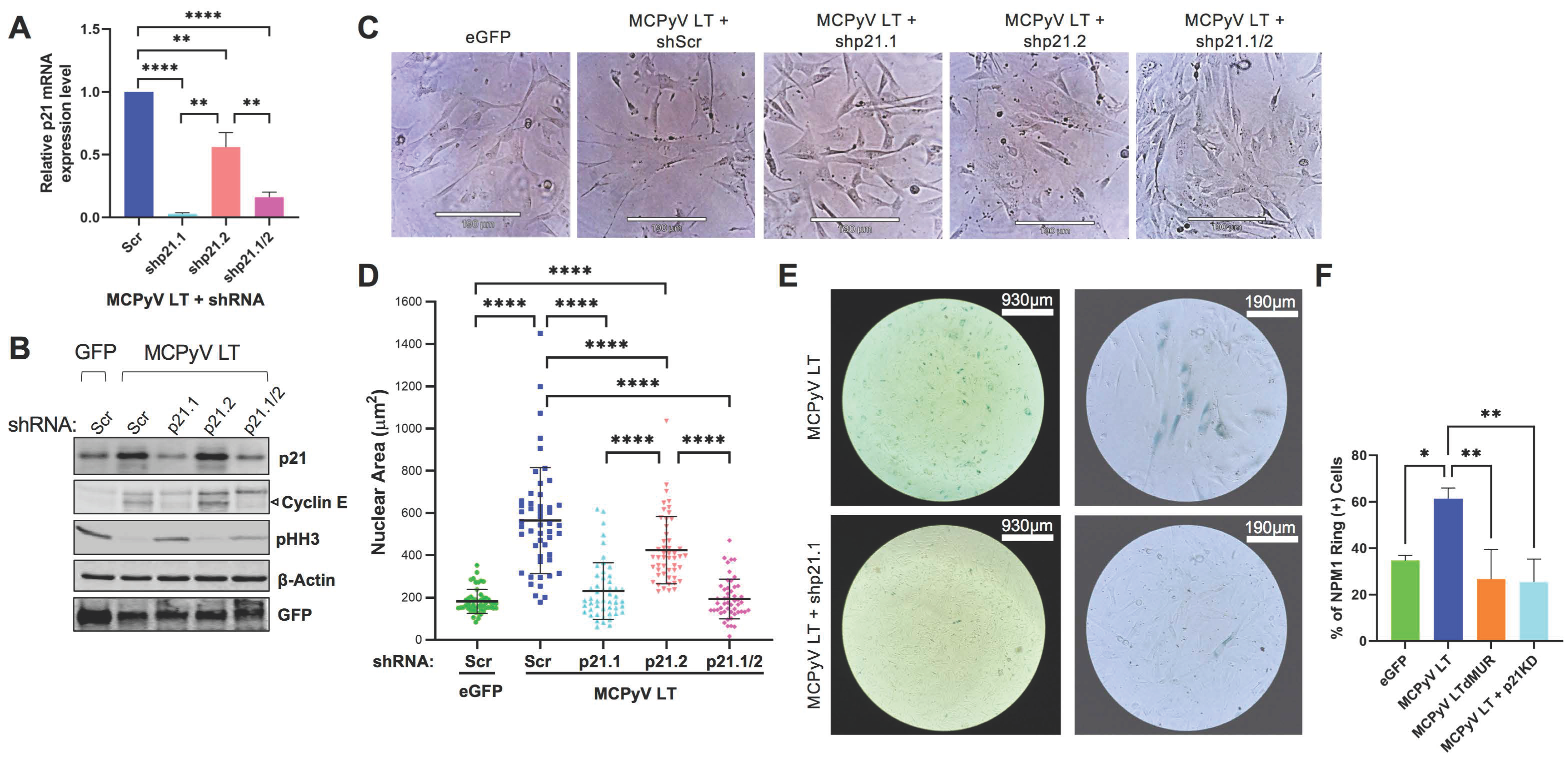
2.3. MCPyV LT-Induced Senescence Is p21-Dependent
2.4. Senescent MCPyV LT-Expressing Cells Are Arrested in G2 Phase
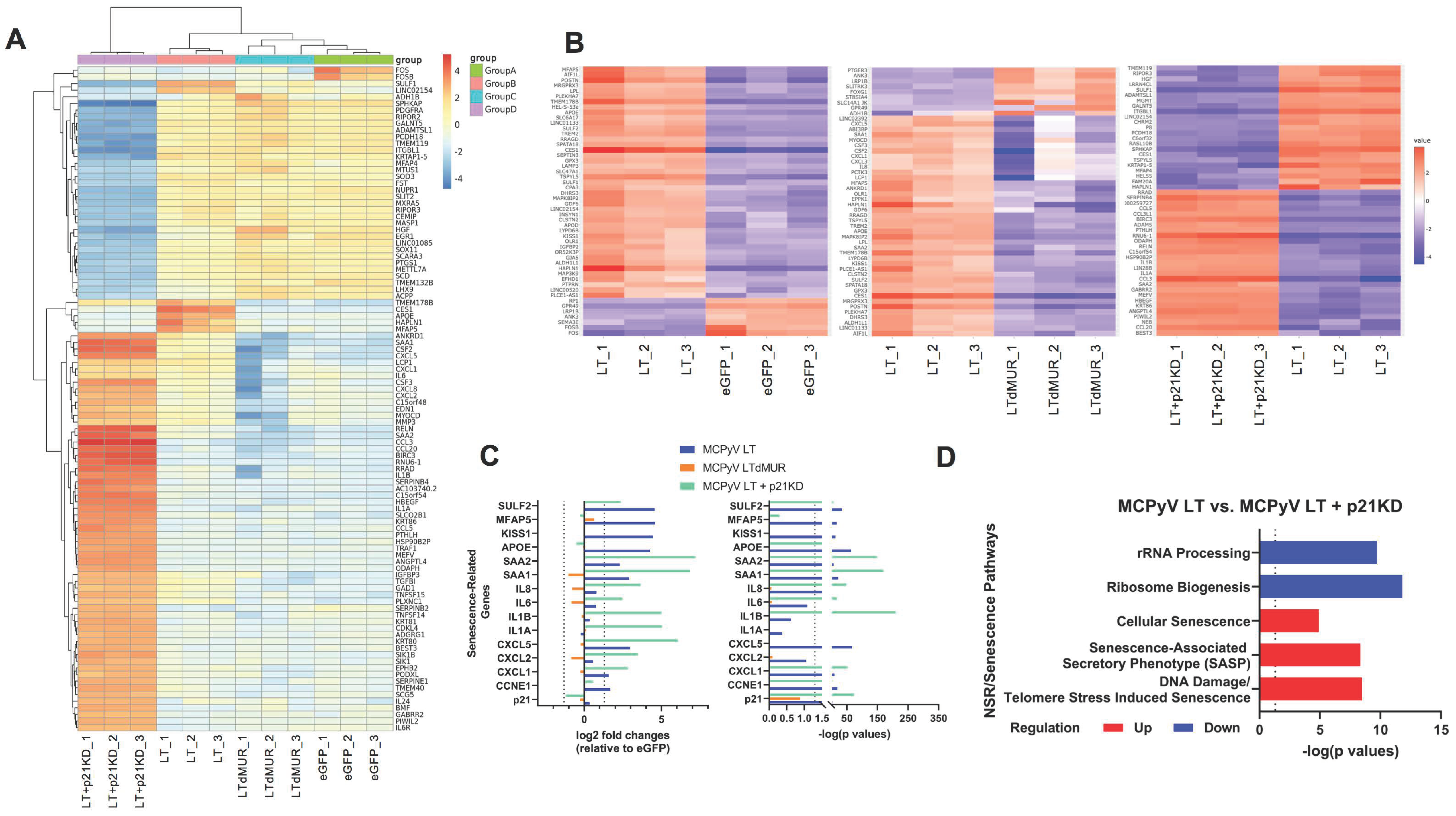
2.5. MCPyV LT-Induced Senescence Alters Host Gene Expression Profile
2.6. MCPyV LT-Mediated Senescence Maintains MCPyV Genome
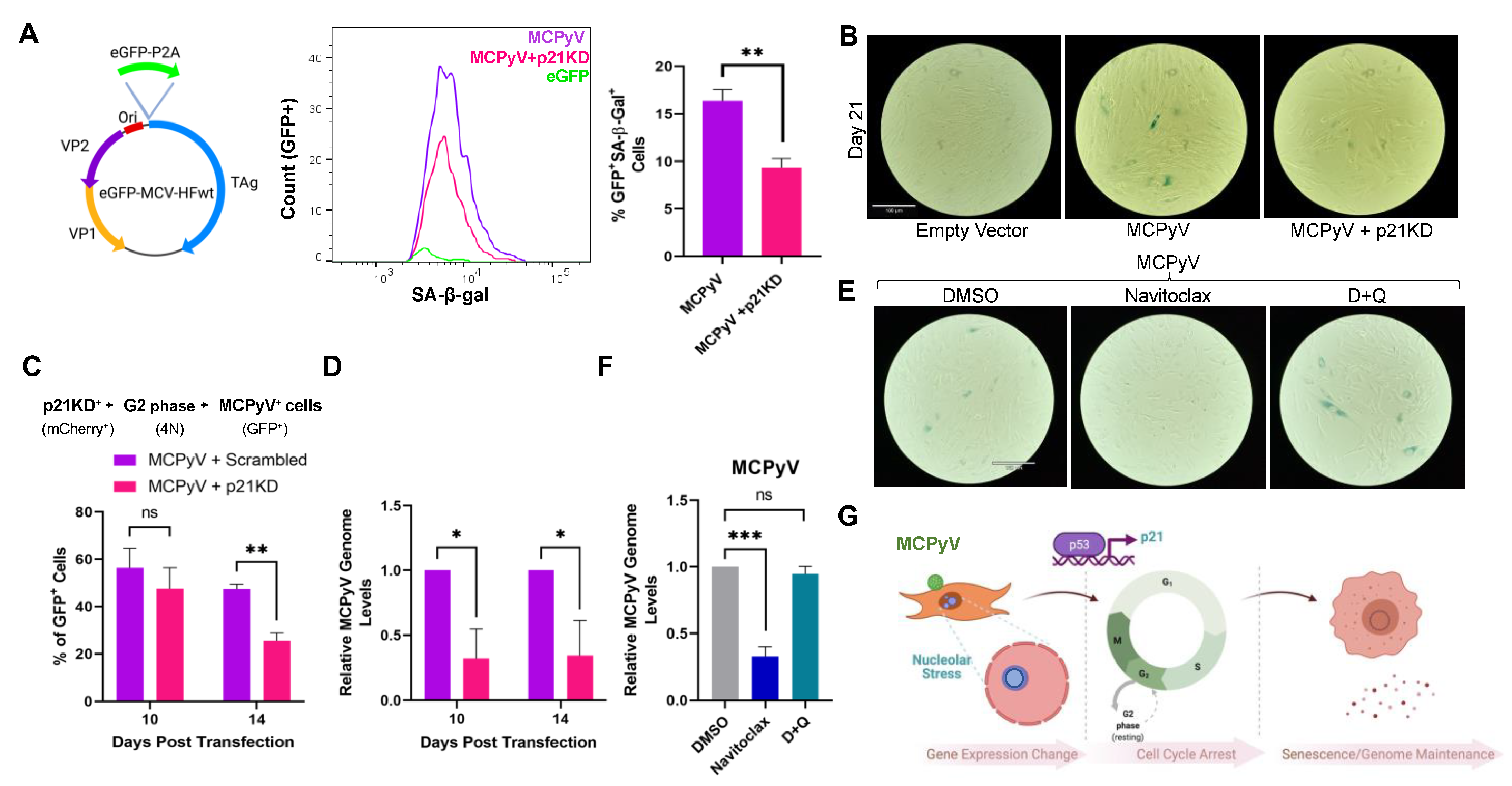
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Plasmids
4.2. Lentiviral Transduction and p21 Knockdown
4.3. Quantitative Immunoblotting and Antibodies
4.4. Immunofluorescence Assay
4.5. Senescence Assay
4.6. SASP Analysis by RT-qPCR
4.7. Cell Cycle Analysis
4.8. EdU Incorporation Assay
4.9. Cell Proliferation Assay
4.10. RNA Sequencing Analysis
4.11. Flow Cytometric Analysis of MCPyV Genome
4.12. Senolytic Treatment
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Di Micco, R.; Krizhanovsky, V.; Baker, D.; d’Adda di Fagagna, F. Cellular senescence in ageing: From mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 2021, 22, 75–95. [Google Scholar] [CrossRef] [PubMed]
- Baz-Martínez, M.; Da Silva-Álvarez, S.; Rodríguez, E.; Guerra, J.; El Motiam, A.; Vidal, A.; García-Caballero, T.; González-Barcia, M.; Sánchez, L.; Muñoz-Fontela, C.; et al. Cell senescence is an antiviral defense mechanism. Sci. Rep. 2016, 6, 37007. [Google Scholar] [CrossRef] [PubMed]
- van Deursen, J.M. The role of senescent cells in ageing. Nature 2014, 509, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Herranz, N.; Gil, J. Mechanisms and functions of cellular senescence. J. Clin. Invest. 2018, 128, 1238–1246. [Google Scholar] [CrossRef] [PubMed]
- Kelley, W.J.; Zemans, R.L.; Goldstein, D.R. Cellular senescence: Friend or foe to respiratory viral infections? Eur. Respir. J. 2020, 56, 2002708. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Seong, R.K.; Shin, O.S. Enhanced Viral Replication by Cellular Replicative Senescence. Immune Netw. 2016, 16, 286–295. [Google Scholar] [CrossRef]
- Seoane, R.; Vidal, S.; Bouzaher, Y.H.; El Motiam, A.; Rivas, C. The Interaction of Viruses with the Cellular Senescence Response. Biology 2020, 9, 455. [Google Scholar] [CrossRef] [PubMed]
- Chuprin, A.; Gal, H.; Biron-Shental, T.; Biran, A.; Amiel, A.; Rozenblatt, S.; Krizhanovsky, V. Cell fusion induced by ERVWE1 or measles virus causes cellular senescence. Genes Dev. 2013, 27, 2356–2366. [Google Scholar] [CrossRef]
- Hsieh, T.H.; Tsai, T.T.; Chen, C.L.; Shen, T.J.; Jhan, M.K.; Tseng, P.C.; Lin, C.F. Senescence in Monocytes Facilitates Dengue Virus Infection by Increasing Infectivity. Front. Cell. Infect. Microbiol. 2020, 10, 375. [Google Scholar] [CrossRef]
- Camell, C.D.; Yousefzadeh, M.J.; Zhu, Y.; Prata, L.G.P.L.; Huggins, M.A.; Pierson, M.; Zhang, L.; O’Kelly, R.D.; Pirtskhalava, T.; Xun, P.; et al. Senolytics reduce coronavirus-related mortality in old mice. Science 2021, 373, eabe4832. [Google Scholar] [CrossRef]
- Chang, Y.; Moore, P.S. Merkel cell carcinoma: A virus-induced human cancer. Annu Rev. Pathol. 2012, 7, 123–144. [Google Scholar] [CrossRef] [PubMed]
- Kwun, H.J.; Guastafierro, A.; Shuda, M.; Meinke, G.; Bohm, A.; Moore, P.S.; Chang, Y. The minimum replication origin of merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J. Virol. 2009, 83, 12118–12128. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, X.; Diaz, J.; Tsang, S.H.; Buck, C.B.; You, J. Merkel cell polyomavirus large T antigen disrupts host genomic integrity and inhibits cellular proliferation. J. Virol. 2013, 87, 9173–9188. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Rozenblatt-Rosen, O.; Paulson, K.G.; Nghiem, P.; DeCaprio, J.A. Merkel cell polyomavirus large T antigen has growth-promoting and inhibitory activities. J. Virol. 2013, 87, 6118–6126. [Google Scholar] [CrossRef] [PubMed]
- Nwogu, N.; Ortiz, L.E.; Kwun, H.J. Merkel Cell Polyomavirus Large T Antigen Unique Domain Regulates Its Own Protein Stability and Cell Growth. Viruses 2020, 12, 1043. [Google Scholar] [CrossRef]
- Houben, R.; Angermeyer, S.; Haferkamp, S.; Aue, A.; Goebeler, M.; Schrama, D.; Hesbacher, S. Characterization of functional domains in the Merkel cell polyoma virus Large T antigen. Int. J. Cancer 2015, 136, E290–E300. [Google Scholar] [CrossRef]
- Kwun, H.J.; Chang, Y.; Moore, P.S. Protein-mediated viral latency is a novel mechanism for Merkel cell polyomavirus persistence. Proc. Natl. Acad. Sci. USA 2017, 114, E4040–E4047. [Google Scholar] [CrossRef]
- Ortiz, L.E.; Pham, A.M.; Kwun, H.J. Identification of the Merkel Cell Polyomavirus Large Tumor Antigen Ubiquitin Conjugation Residue. Int. J. Mol. Sci. 2021, 22, 7169. [Google Scholar] [CrossRef]
- Kwun, H.J.; Shuda, M.; Feng, H.; Camacho, C.J.; Moore, P.S.; Chang, Y. Merkel cell polyomavirus small T antigen controls viral replication and oncoprotein expression by targeting the cellular ubiquitin ligase SCFFbw7. Cell Host Microbe 2013, 14, 125–135. [Google Scholar] [CrossRef]
- Siebels, S.; Czech-Sioli, M.; Spohn, M.; Schmidt, C.; Theiss, J.; Indenbirken, D.; Günther, T.; Grundhoff, A.; Fischer, N. Merkel Cell Polyomavirus DNA Replication Induces Senescence in Human Dermal Fibroblasts in a Kap1/Trim28-Dependent Manner. mBio 2020, 11, e00142-20. [Google Scholar] [CrossRef]
- Moran, J.M.T.; Biecek, P.; Donizy, P.; Wu, C.L.; Kopczynski, J.; Pieniazek, M.; Ryś, J.; Hoang, M.P. Large nuclear size correlated with better overall survival, Merkel cell polyomavirus positivity, and terminal deoxynucleotidyl transferase expression in Merkel cell carcinoma. J. Am. Acad. Dermatol. 2021, 84, 550–552. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, S.; DuPree, E.L.; Almasan, A. A dual role of cyclin E in cell proliferation and apoptosis may provide a target for cancer therapy. Curr. Cancer Drug Targets 2004, 4, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Koff, A.; Cross, F.; Fisher, A.; Schumacher, J.; Leguellec, K.; Philippe, M.; Roberts, J.M. Human cyclin E, a new cyclin that interacts with two members of the CDC2 gene family. Cell 1991, 66, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Dulić, V.; Lees, E.; Reed, S.I. Association of human cyclin E with a periodic G1-S phase protein kinase. Science 1992, 257, 1958–1961. [Google Scholar] [CrossRef] [PubMed]
- Dulić, V.; Drullinger, L.F.; Lees, E.; Reed, S.I.; Stein, G.H. Altered regulation of G1 cyclins in senescent human diploid fibroblasts: Accumulation of inactive cyclin E-Cdk2 and cyclin D1-Cdk2 complexes. Proc. Natl. Acad. Sci. USA 1993, 90, 11034–11038. [Google Scholar] [CrossRef]
- Van Hooser, A.; Goodrich, D.W.; Allis, C.D.; Brinkley, B.R.; Mancini, M.A. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. J. Cell Sci. 1998, 111, 3497–3506. [Google Scholar] [CrossRef]
- Ren, J.; Tang, C.Z.; Li, X.D.; Niu, Z.B.; Zhang, B.Y.; Zhang, T.; Gao, M.J.; Ran, X.Z.; Su, Y.P.; Wang, F.C. Identification of G2/M phase transition by sequential nuclear and cytoplasmic changes and molecular markers in mice intestinal epithelial cells. Cell Cycle 2018, 17, 780–791. [Google Scholar] [CrossRef]
- Hendzel, M.J.; Wei, Y.; Mancini, M.A.; Van Hooser, A.; Ranalli, T.; Brinkley, B.R.; Bazett-Jones, D.P.; Allis, C.D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 1997, 106, 348–360. [Google Scholar] [CrossRef]
- Guo, C.Y.; Mizzen, C.; Wang, Y.; Larner, J.M. Histone H1 and H3 dephosphorylation are differentially regulated by radiation-induced signal transduction pathways. Cancer Res. 2000, 60, 5667–5672. [Google Scholar]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef]
- Rubbi, C.P.; Milner, J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003, 22, 6068–6077. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Yang, J.; Yi, J. Nucleolar Stress: Hallmarks, sensing mechanism and diseases. Cell Stress 2018, 2, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Wang, M.; Zhao, Y.; Sun, X.; Yang, Y.; Li, X.; Zhou, A.; Chu, H.; Zhou, H.; Xu, J.; et al. A redox mechanism underlying nucleolar stress sensing by nucleophosmin. Nat. Commun. 2016, 7, 13599. [Google Scholar] [CrossRef] [PubMed]
- Potapova, T.A.; Gerton, J.L. Ribosomal DNA and the nucleolus in the context of genome organization. Chromosome Res. 2019, 27, 109–127. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, P.; Real-Calderón, P.; Flor-Parra, I.; Salas-Pino, S.; Daga, R.R. Acute Heat Stress Leads to Reversible Aggregation of Nuclear Proteins into Nucleolar Rings in Fission Yeast. Cell Rep. 2020, 33, 108377. [Google Scholar] [CrossRef] [PubMed]
- Hein, J.; Boichuk, S.; Wu, J.; Cheng, Y.; Freire, R.; Jat, P.S.; Roberts, T.M.; Gjoerup, O.V. Simian virus 40 large T antigen disrupts genome integrity and activates a DNA damage response via Bub1 binding. J. Virol. 2009, 83, 117–127. [Google Scholar] [CrossRef]
- Mölder, A.; Sebesta, M.; Gustafsson, M.; Gisselson, L.; Wingren, A.G.; Alm, K. Non-invasive, label-free cell counting and quantitative analysis of adherent cells using digital holography. J. Microsc. 2008, 232, 240–247. [Google Scholar] [CrossRef]
- Iarovaia, O.V.; Minina, E.P.; Sheval, E.V.; Onichtchouk, D.; Dokudovskaya, S.; Razin, S.V.; Vassetzky, Y.S. Nucleolus: A Central Hub for Nuclear Functions. Trends Cell. Biol. 2019, 29, 647–659. [Google Scholar] [CrossRef]
- Paluvai, H.; Di Giorgio, E.; Brancolini, C. The Histone Code of Senescence. Cells 2020, 9, 466. [Google Scholar] [CrossRef]
- Chau, B.N.; Diaz, R.L.; Saunders, M.A.; Cheng, C.; Chang, A.N.; Warrener, P.; Bradshaw, J.; Linsley, P.S.; Cleary, M.A. Identification of SULF2 as a novel transcriptional target of p53 by use of integrated genomic analyses. Cancer Res. 2009, 69, 1368–1374. [Google Scholar] [CrossRef]
- Voutetakis, K.; Chatziioannou, A.; Gonos, E.S.; Trougakos, I.P. Comparative Meta-Analysis of Transcriptomics Data during Cellular Senescence and In Vivo Tissue Ageing. Oxid. Med. Cell Longev. 2015, 2015, 732914. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, I.; Desroziers, E. Development and Aging of the Kisspeptin-GPR54 System in the Mammalian Brain: What are the Impacts on Female Reproductive Function? Front. Endocrinol. 2013, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, C.G.; Hamby, M.E.; McReynolds, M.L.; Ray, W.J. The Role of APOE4 in Disrupting the Homeostatic Functions of Astrocytes and Microglia in Aging and Alzheimer’s Disease. Front. Aging Neurosci. 2019, 11, 14. [Google Scholar] [CrossRef]
- Nishimura, K.; Taguchi, K.; Kishi, S.; Brooks, C.R.; Ochi, A.; Kadoya, H.; Ikeda, Y.; Miyoshi, M.; Tamaki, M.; Abe, H.; et al. Dual disruption of eNOS and ApoE gene accelerates kidney fibrosis and senescence after injury. Biochem. Biophys. Res. Commun. 2021, 556, 142–148. [Google Scholar] [CrossRef]
- Lee, S.; Yu, Y.; Trimpert, J.; Benthani, F.; Mairhofer, M.; Richter-Pechanska, P.; Wyler, E.; Belenki, D.; Kaltenbrunner, S.; Pammer, M.; et al. Virus-induced senescence is a driver and therapeutic target in COVID-19. Nature 2021, 599, 283–289. [Google Scholar] [CrossRef]
- Zhu, Y.; Tchkonia, T.; Fuhrmann-Stroissnigg, H.; Dai, H.M.; Ling, Y.Y.; Stout, M.B.; Pirtskhalava, T.; Giorgadze, N.; Johnson, K.O.; Giles, C.B.; et al. Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell 2016, 15, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wang, Z.; Ghosh, S.; Zhou, Z. Defective ATM-Kap-1-mediated chromatin remodeling impairs DNA repair and accelerates senescence in progeria mouse model. Aging Cell 2013, 12, 316–318. [Google Scholar] [CrossRef]
- Leon, K.E.; Buj, R.; Lesko, E.; Dahl, E.S.; Chen, C.W.; Tangudu, N.K.; Imamura-Kawasawa, Y.; Kossenkov, A.V.; Hobbs, R.P.; Aird, K.M. DOT1L modulates the senescence-associated secretory phenotype through epigenetic regulation of IL1A. J. Cell. Biol. 2021, 220, e202008101. [Google Scholar] [CrossRef]
- Aird, K.M.; Worth, A.J.; Snyder, N.W.; Lee, J.V.; Sivanand, S.; Liu, Q.; Blair, I.A.; Wellen, K.E.; Zhang, R. ATM couples replication stress and metabolic reprogramming during cellular senescence. Cell. Rep. 2015, 11, 893–901. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Ren, J.; Chen, Q.; Chen, Z.J. cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. USA 2017, 114, E4612–E4620. [Google Scholar] [CrossRef]
- de Magalhães, J.P.; Chainiaux, F.; Remacle, J.; Toussaint, O. Stress-induced premature senescence in BJ and hTERT-BJ1 human foreskin fibroblasts. FEBS Lett. 2002, 523, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Gorbunova, V.; Seluanov, A.; Pereira-Smith, O.M. Expression of human telomerase (hTERT) does not prevent stress-induced senescence in normal human fibroblasts but protects the cells from stress-induced apoptosis and necrosis. J. Biol. Chem. 2002, 277, 38540–38549. [Google Scholar] [CrossRef] [PubMed]
- Speck, S.H.; Ganem, D. Viral latency and its regulation: Lessons from the gamma-herpesviruses. Cell. Host Microbe 2010, 8, 100–115. [Google Scholar] [CrossRef] [PubMed]
- Harms, P.W.; Harms, K.L.; Moore, P.S.; DeCaprio, J.A.; Nghiem, P.; Wong, M.K.K.; Brownell, I.; International Workshop on Merkel Cell Carcinoma Research (IWMCC) Working Group. The biology and treatment of Merkel cell carcinoma: Current understanding and research priorities. Nat. Rev. Clin. Oncol. 2018, 15, 763–776. [Google Scholar] [CrossRef]
- Gelgor, A.; Gam Ze Letova, C.; Yegorov, Y.; Kalt, I.; Sarid, R. Nucleolar stress enhances lytic reactivation of the Kaposi’s sarcoma-associated herpesvirus. Oncotarget 2018, 9, 13822–13833. [Google Scholar] [CrossRef]
- Nwogu, N.; Ortiz, L.E.; Kwun, H.J. Surface charge of Merkel cell polyomavirus small T antigen determines cell transformation through allosteric FBW7 WD40 domain targeting. Oncogenesis 2020, 9, 53. [Google Scholar] [CrossRef]
- Alvarez Orellana, J.; Kwun, H.J.; Artusi, S.; Chang, Y.; Moore, P.S. Sirolimus and other mTOR inhibitors directly activate latent pathogenic human polyomavirus replication. J. Infect. Dis. 2020, 224, 1160–1169. [Google Scholar] [CrossRef]
- Moore, P.S.; Chang, Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer 2010, 10, 878–889. [Google Scholar] [CrossRef]
- Moens, U.; Macdonald, A. Effect of the Large and Small T-Antigens of Human Polyomaviruses on Signaling Pathways. Int. J. Mol. Sci. 2019, 20, 3914. [Google Scholar] [CrossRef]
- Lill, C.; Schneider, S.; Item, C.B.; Loewe, R.; Houben, R.; Halbauer, D.; Heiduschka, G.; Brunner, M.; Thurnher, D. P53 mutation is a rare event in Merkel cell carcinoma of the head and neck. Eur. Arch. Otorhinolaryngol. 2011, 268, 1639–1646. [Google Scholar] [CrossRef]
- Houben, R.; Dreher, C.; Angermeyer, S.; Borst, A.; Utikal, J.; Haferkamp, S.; Peitsch, W.K.; Schrama, D.; Hesbacher, S. Mechanisms of p53 restriction in Merkel cell carcinoma cells are independent of the Merkel cell polyoma virus T antigens. J. Invest. Dermatol. 2013, 133, 2453–2460. [Google Scholar] [CrossRef] [PubMed]
- Bonifacio, L.N.; Jarstfer, M.B. MiRNA profile associated with replicative senescence, extended cell culture, and ectopic telomerase expression in human foreskin fibroblasts. PLoS ONE 2010, 5, e12519. [Google Scholar] [CrossRef] [PubMed]
- Yosef, R.; Pilpel, N.; Papismadov, N.; Gal, H.; Ovadya, Y.; Vadai, E.; Miller, S.; Porat, Z.; Ben-Dor, S.; Krizhanovsky, V. p21 maintains senescent cell viability under persistent DNA damage response by restraining JNK and caspase signaling. EMBO J. 2017, 36, 2280–2295. [Google Scholar] [CrossRef] [PubMed]
- Stein, G.H.; Dulić, V. Origins of G1 arrest in senescent human fibroblasts. Bioessays 1995, 17, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Prata, L.G.P.L.; Ovsyannikova, I.G.; Tchkonia, T.; Kirkland, J.L. Senescent cell clearance by the immune system: Emerging therapeutic opportunities. Semin. Immunol. 2018, 40, 101275. [Google Scholar] [CrossRef] [PubMed]
- Verhaegen, M.E.; Mangelberger, D.; Weick, J.W.; Vozheiko, T.D.; Harms, P.W.; Nash, K.T.; Quintana, E.; Baciu, P.; Johnson, T.M.; Bichakjian, C.K.; et al. Merkel cell carcinoma dependence on bcl-2 family members for survival. J. Invest. Dermatol. 2014, 134, 2241–2250. [Google Scholar] [CrossRef] [PubMed]
- Chteinberg, E.; Wetzels, S.; Gerritsen, W.; Temmerman, L.; van den Oord, J.; Biessen, E.; Kurz, A.K.; Winnepenninckx, V.; Zenke, M.; Speel, E.J.; et al. Navitoclax combined with Alpelisib effectively inhibits Merkel cell carcinoma cell growth. Ther. Adv. Med. Oncol. 2020, 12, 1758835920975621. [Google Scholar] [CrossRef]
- Noren Hooten, N.; Evans, M.K. Techniques to Induce and Quantify Cellular Senescence. J. Vis. Exp. 2017, 123, e55533. [Google Scholar] [CrossRef]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef]
- Feng, H.; Kwun, H.J.; Liu, X.; Gjoerup, O.; Stolz, D.B.; Chang, Y.; Moore, P.S. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS ONE 2011, 6, e22468. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pham, A.M.; Ortiz, L.E.; Lukacher, A.E.; Kwun, H.J. Merkel Cell Polyomavirus Large T Antigen Induces Cellular Senescence for Host Growth Arrest and Viral Genome Persistence through Its Unique Domain. Cells 2023, 12, 380. https://doi.org/10.3390/cells12030380
Pham AM, Ortiz LE, Lukacher AE, Kwun HJ. Merkel Cell Polyomavirus Large T Antigen Induces Cellular Senescence for Host Growth Arrest and Viral Genome Persistence through Its Unique Domain. Cells. 2023; 12(3):380. https://doi.org/10.3390/cells12030380
Chicago/Turabian StylePham, Alexander M., Luz E. Ortiz, Aron E. Lukacher, and Hyun Jin Kwun. 2023. "Merkel Cell Polyomavirus Large T Antigen Induces Cellular Senescence for Host Growth Arrest and Viral Genome Persistence through Its Unique Domain" Cells 12, no. 3: 380. https://doi.org/10.3390/cells12030380
APA StylePham, A. M., Ortiz, L. E., Lukacher, A. E., & Kwun, H. J. (2023). Merkel Cell Polyomavirus Large T Antigen Induces Cellular Senescence for Host Growth Arrest and Viral Genome Persistence through Its Unique Domain. Cells, 12(3), 380. https://doi.org/10.3390/cells12030380





