Role of Histone Variant H2A.J in Fine-Tuning Chromatin Organization for the Establishment of Ionizing Radiation-Induced Senescence
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultured Lung Fibroblasts
2.2. Radiation Exposure of Cultured Lung Fibroblasts
2.3. Immunofluorescence Microscopy (IFM) Analysis
2.4. Immunhistochemistry (IHC) Analysis (SA-β-Gal)
2.5. RNA-Sequencing (RNA-Seq)
2.6. Assay for Transposase-Accessible Chromatin (ATAC-Seq)
2.7. Integrative Analysis of ATAC-Seq and mRNA-Seq Data
2.8. Enzyme-Linked ImmunoSorbent Analysis (ELISA)
2.9. Reverse-Transcriptase Quantitative Polymerase-Chain-Reaction (RT-qPCR) analysis
2.10. Radiation Schedule and Lung Tissue Sampling in Mice
2.11. IFM and IHC Analysis of Lung Tissue Sections
2.12. Digital Image Analysis of H2A.J-Immunohistochemistry
2.13. Statistical Analysis
3. Results
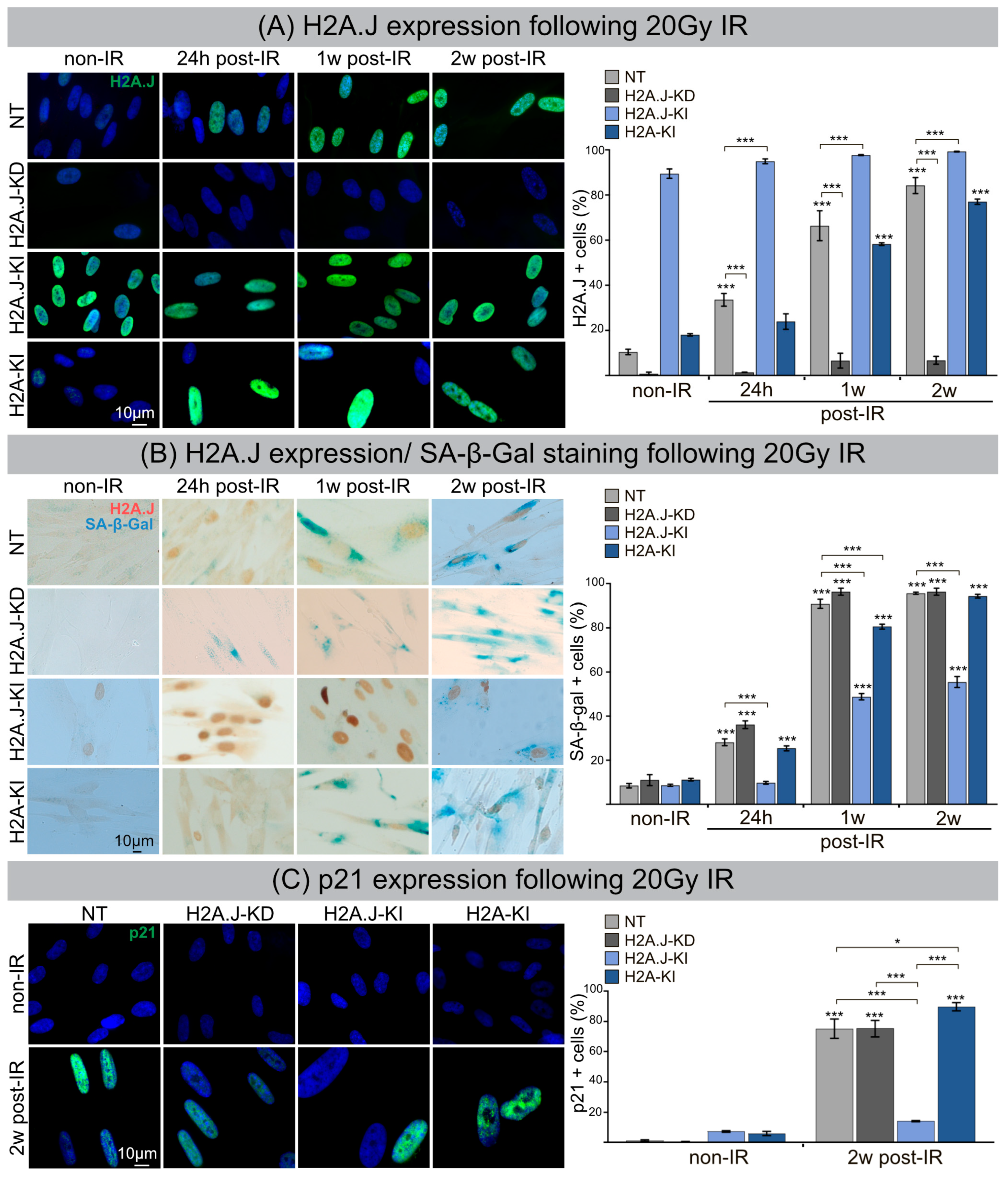
3.1. H2A.J Accumulation and Radiation-Induced Senescence
3.2. Formation of SAHF and DNA-SCARS following IR
3.3. ATAC-Seq for Chromatin Accessibility Analysis
3.4. Integrative Analysis of ATAC-Seq and RNA-Seq Datasets
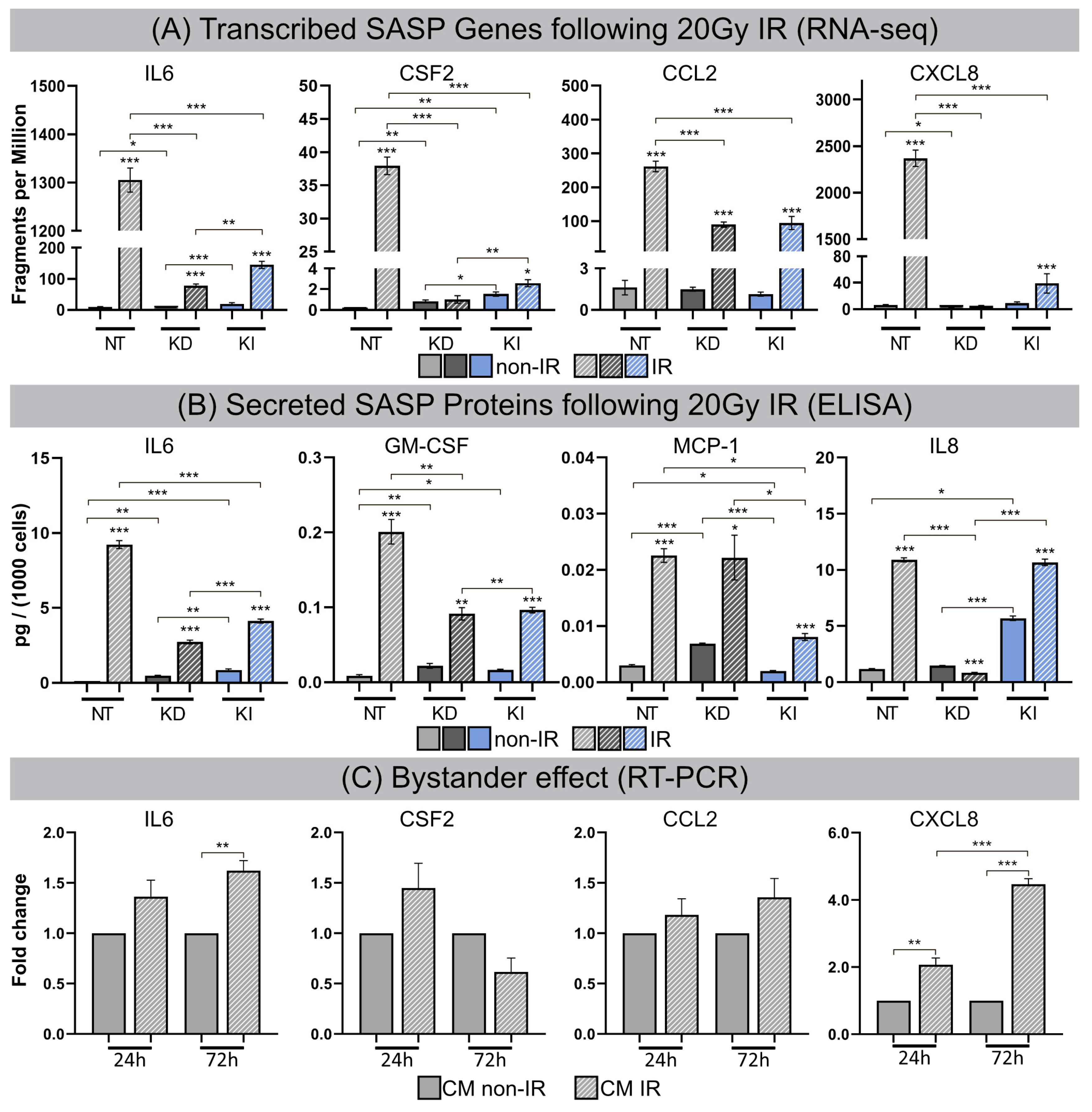
3.5. Senescence-Associated Secretory Phenotype (SASP)
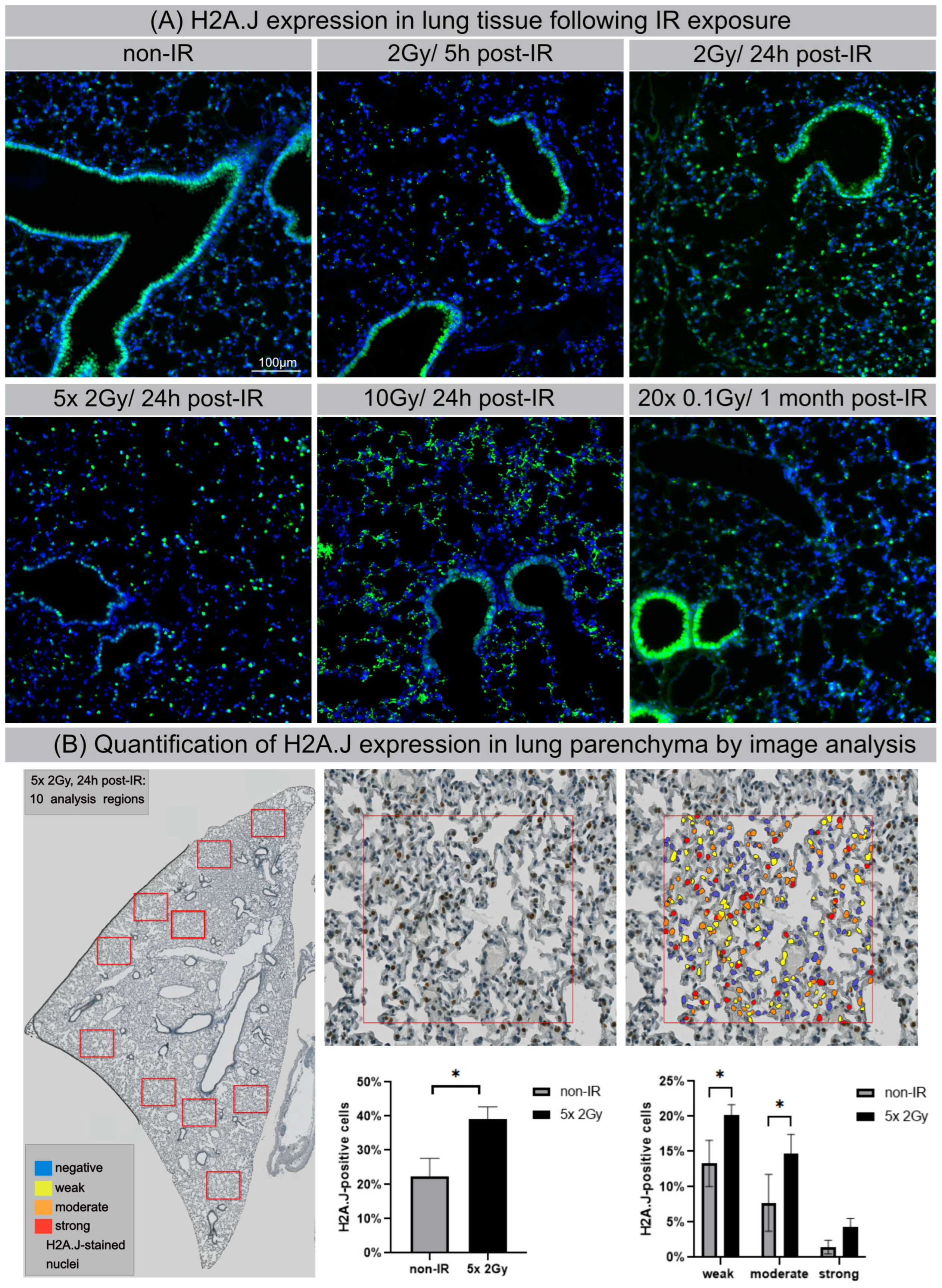
3.6. H2A.J Expression in Lung Tissue after IR Exposure

4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Di Micco, R.; Krizhanovsky, V.; Baker, D.; d’Adda di Fagagna, F. Cellular senescence in ageing: From mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell. Biol. 2021, 22, 75–95. [Google Scholar] [CrossRef] [PubMed]
- Nacarelli, T.; Liu, P.; Zhang, R. Epigenetic Basis of Cellular Senescence and Its Implications in Aging. Genes 2017, 8, 343. [Google Scholar] [CrossRef] [PubMed]
- Campisi, J.; Robert, L. Cell senescence: Role in aging and age-related diseases. Interdiscip. Top. Gerontol. 2014, 39, 45–61. [Google Scholar] [CrossRef] [PubMed]
- Price, B.D.; D’Andrea, A.D. Chromatin remodeling at DNA double-strand breaks. Cell 2013, 152, 1344–1354. [Google Scholar] [CrossRef] [PubMed]
- Panier, S.; Boulton, S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell. Biol. 2014, 15, 7–18. [Google Scholar] [CrossRef]
- Lorat, Y.; Schanz, S.; Rube, C.E. Ultrastructural Insights into the Biological Significance of Persisting DNA Damage Foci after Low Doses of Ionizing Radiation. Clin. Cancer Res. 2016, 22, 5300–5311. [Google Scholar] [CrossRef] [PubMed]
- Vancurova, M.; Hanzlikova, H.; Knoblochova, L.; Kosla, J.; Majera, D.; Mistrik, M.; Burdova, K.; Hodny, Z.; Bartek, J. PML nuclear bodies are recruited to persistent DNA damage lesions in an RNF168-53BP1 dependent manner and contribute to DNA repair. DNA Repair 2019, 78, 114–127. [Google Scholar] [CrossRef]
- Rodier, F.; Coppe, J.P.; Patil, C.K.; Hoeijmakers, W.A.; Munoz, D.P.; Raza, S.R.; Freund, A.; Campeau, E.; Davalos, A.R.; Campisi, J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 2009, 11, 973–979. [Google Scholar] [CrossRef]
- Rodier, F.; Munoz, D.P.; Teachenor, R.; Chu, V.; Le, O.; Bhaumik, D.; Coppe, J.P.; Campeau, E.; Beausejour, C.M.; Kim, S.H.; et al. DNA-SCARS: Distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J. Cell Sci. 2011, 124, 68–81. [Google Scholar] [CrossRef]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. Histone variants at a glance. J. Cell Sci. 2021, 134, jcs.244749. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Henikoff, S. Histone variants on the move: Substrates for chromatin dynamics. Nat. Rev. Mol. Cell Biol. 2017, 18, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Contrepois, K.; Coudereau, C.; Benayoun, B.A.; Schuler, N.; Roux, P.F.; Bischof, O.; Courbeyrette, R.; Carvalho, C.; Thuret, J.Y.; Ma, Z.; et al. Histone variant H2A.J accumulates in senescent cells and promotes inflammatory gene expression. Nat. Commun. 2017, 8, 14995. [Google Scholar] [CrossRef] [PubMed]
- Isermann, A.; Mann, C.; Rube, C.E. Histone Variant H2A.J Marks Persistent DNA Damage and Triggers the Secretory Phenotype in Radiation-Induced Senescence. Int. J. Mol. Sci. 2020, 21, 9130. [Google Scholar] [CrossRef]
- Rube, C.E.; Baumert, C.; Schuler, N.; Isermann, A.; Schmal, Z.; Glanemann, M.; Mann, C.; Scherthan, H. Human skin aging is associated with increased expression of the histone variant H2A.J in the epidermis. NPJ Aging Mech. Dis. 2021, 7, 7. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Res 2018, 7, 1338. [Google Scholar] [CrossRef]
- Frankish, A.; Diekhans, M.; Jungreis, I.; Lagarde, J.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Armstrong, J.; Barnes, I.; et al. Gencode 2021. Nucleic Acids Res. 2021, 49, D916–D923. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [PubMed]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed]
- Hahne, F.; Ivanek, R. Visualizing Genomic Data Using Gviz and Bioconductor. Methods Mol. Biol. 2016, 1418, 335–351. [Google Scholar] [CrossRef] [PubMed]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.; Huber, W.; Pages, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Martinez-Zamudio, R.I.; Roux, P.F.; de Freitas, J.; Robinson, L.; Dore, G.; Sun, B.; Belenki, D.; Milanovic, M.; Herbig, U.; Schmitt, C.A.; et al. AP-1 imprints a reversible transcriptional programme of senescent cells. Nat. Cell Biol. 2020, 22, 842–855. [Google Scholar] [CrossRef]
- Redon, C.E.; Schmal, Z.; Tewary, G.; Mangelinck, A.; Courbeyrette, R.; Thuret, J.Y.; Aladjem, M.I.; Bonner, W.M.; Rube, C.E.; Mann, C. Histone Variant H2A.J Is Enriched in Luminal Epithelial Gland Cells. Genes 2021, 12, 1665. [Google Scholar] [CrossRef]
- Silk, E.; Zhao, H.; Weng, H.; Ma, D. The role of extracellular histone in organ injury. Cell Death Dis. 2017, 8, e2812. [Google Scholar] [CrossRef]
- Rube, C.E.; Uthe, D.; Schmid, K.W.; Richter, K.D.; Wessel, J.; Schuck, A.; Willich, N.; Rube, C. Dose-dependent induction of transforming growth factor beta (TGF-beta) in the lung tissue of fibrosis-prone mice after thoracic irradiation. Int. J. Radiat. Oncol. Biol. Phys. 2000, 47, 1033–1042. [Google Scholar] [CrossRef]
- Rube, C.E.; Uthe, D.; Wilfert, F.; Ludwig, D.; Yang, K.; Konig, J.; Palm, J.; Schuck, A.; Willich, N.; Remberger, K.; et al. The bronchiolar epithelium as a prominent source of pro-inflammatory cytokines after lung irradiation. Int. J. Radiat. Oncol. Biol. Phys. 2005, 61, 1482–1492. [Google Scholar] [CrossRef]
- Beach, T.A.; Groves, A.M.; Johnston, C.J.; Williams, J.P.; Finkelstein, J.N. Recurrent DNA damage is associated with persistent injury in progressive radiation-induced pulmonary fibrosis. Int. J. Radiat. Biol. 2018, 94, 1104–1115. [Google Scholar] [CrossRef] [PubMed]
- Borg, M.; Jiang, D.; Berger, F. Histone variants take center stage in shaping the epigenome. Curr. Opin. Plant. Biol. 2021, 61, 101991. [Google Scholar] [CrossRef]
- Mangelinck, A.; Coudereau, C.; Courbeyrette, R.; Ourarhni, K.; Hamiche, A.; Redon, C.; Bonner, W.M.; van Dijk, E.; Derbois, C.; Olaso, R.; et al. The H2A.J histone variant contributes to Interferon-Stimulated Gene expression in senescence by its weak interaction with H1 and the derepression of repeated DNA sequences. bioRxiv 2020. [Google Scholar] [CrossRef]
- Funayama, R.; Saito, M.; Tanobe, H.; Ishikawa, F. Loss of linker histone H1 in cellular senescence. J. Cell Biol. 2006, 175, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Powell, D.R.; Curtis, D.J.; Wong, N.C. From reads to insight: A hitchhiker’s guide to ATAC-seq data analysis. Genome Biol. 2020, 21, 22. [Google Scholar] [CrossRef]
- Taghizadeh-Hesary, F.; Houshyari, M.; Farhadi, M. Mitochondrial metabolism: A predictive biomarker of radiotherapy efficacy and toxicity. J. Cancer Res. Clin. Oncol. 2023, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Poustovoitov, M.V.; Ye, X.; Santos, H.A.; Chen, W.; Daganzo, S.M.; Erzberger, J.P.; Serebriiskii, I.G.; Canutescu, A.A.; Dunbrack, R.L.; et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 2005, 8, 19–30. [Google Scholar] [CrossRef]
- Aird, K.M.; Iwasaki, O.; Kossenkov, A.V.; Tanizawa, H.; Fatkhutdinov, N.; Bitler, B.G.; Le, L.; Alicea, G.; Yang, T.L.; Johnson, F.B.; et al. HMGB2 orchestrates the chromatin landscape of senescence-associated secretory phenotype gene loci. J. Cell Biol. 2016, 215, 325–334. [Google Scholar] [CrossRef]
- Gonzalez, L.C.; Ghadaouia, S.; Martinez, A.; Rodier, F. Premature aging/senescence in cancer cells facing therapy: Good or bad? Biogerontology 2016, 17, 71–87. [Google Scholar] [CrossRef]
- Gevry, N.; Chan, H.M.; Laflamme, L.; Livingston, D.M.; Gaudreau, L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007, 21, 1869–1881. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abd Al-razaq, M.A.; Freyter, B.M.; Isermann, A.; Tewary, G.; Mangelinck, A.; Mann, C.; Rübe, C.E. Role of Histone Variant H2A.J in Fine-Tuning Chromatin Organization for the Establishment of Ionizing Radiation-Induced Senescence. Cells 2023, 12, 916. https://doi.org/10.3390/cells12060916
Abd Al-razaq MA, Freyter BM, Isermann A, Tewary G, Mangelinck A, Mann C, Rübe CE. Role of Histone Variant H2A.J in Fine-Tuning Chromatin Organization for the Establishment of Ionizing Radiation-Induced Senescence. Cells. 2023; 12(6):916. https://doi.org/10.3390/cells12060916
Chicago/Turabian StyleAbd Al-razaq, Mutaz A., Benjamin M. Freyter, Anna Isermann, Gargi Tewary, Adèle Mangelinck, Carl Mann, and Claudia E. Rübe. 2023. "Role of Histone Variant H2A.J in Fine-Tuning Chromatin Organization for the Establishment of Ionizing Radiation-Induced Senescence" Cells 12, no. 6: 916. https://doi.org/10.3390/cells12060916
APA StyleAbd Al-razaq, M. A., Freyter, B. M., Isermann, A., Tewary, G., Mangelinck, A., Mann, C., & Rübe, C. E. (2023). Role of Histone Variant H2A.J in Fine-Tuning Chromatin Organization for the Establishment of Ionizing Radiation-Induced Senescence. Cells, 12(6), 916. https://doi.org/10.3390/cells12060916





