Alternations of Lipoprotein Profiles in the Plasma as Biomarkers of Huntington’s Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement and Study Participants
2.2. Patient Recruitment and Plasma Preparation
2.3. Nuclear Magnetic Resonance Analysis (1H NMR Spectroscopic Measurements)
2.4. Statistical Analysis
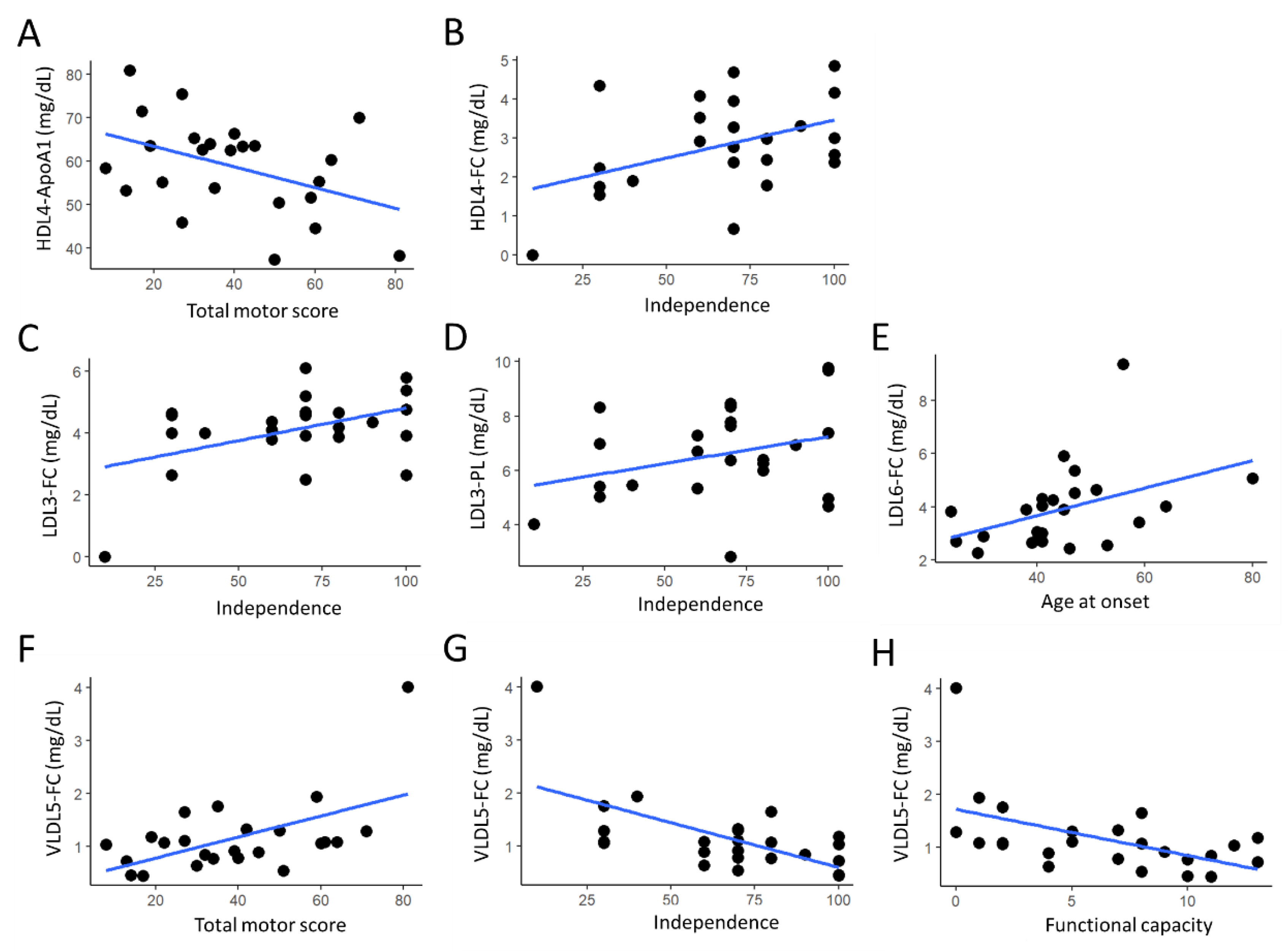
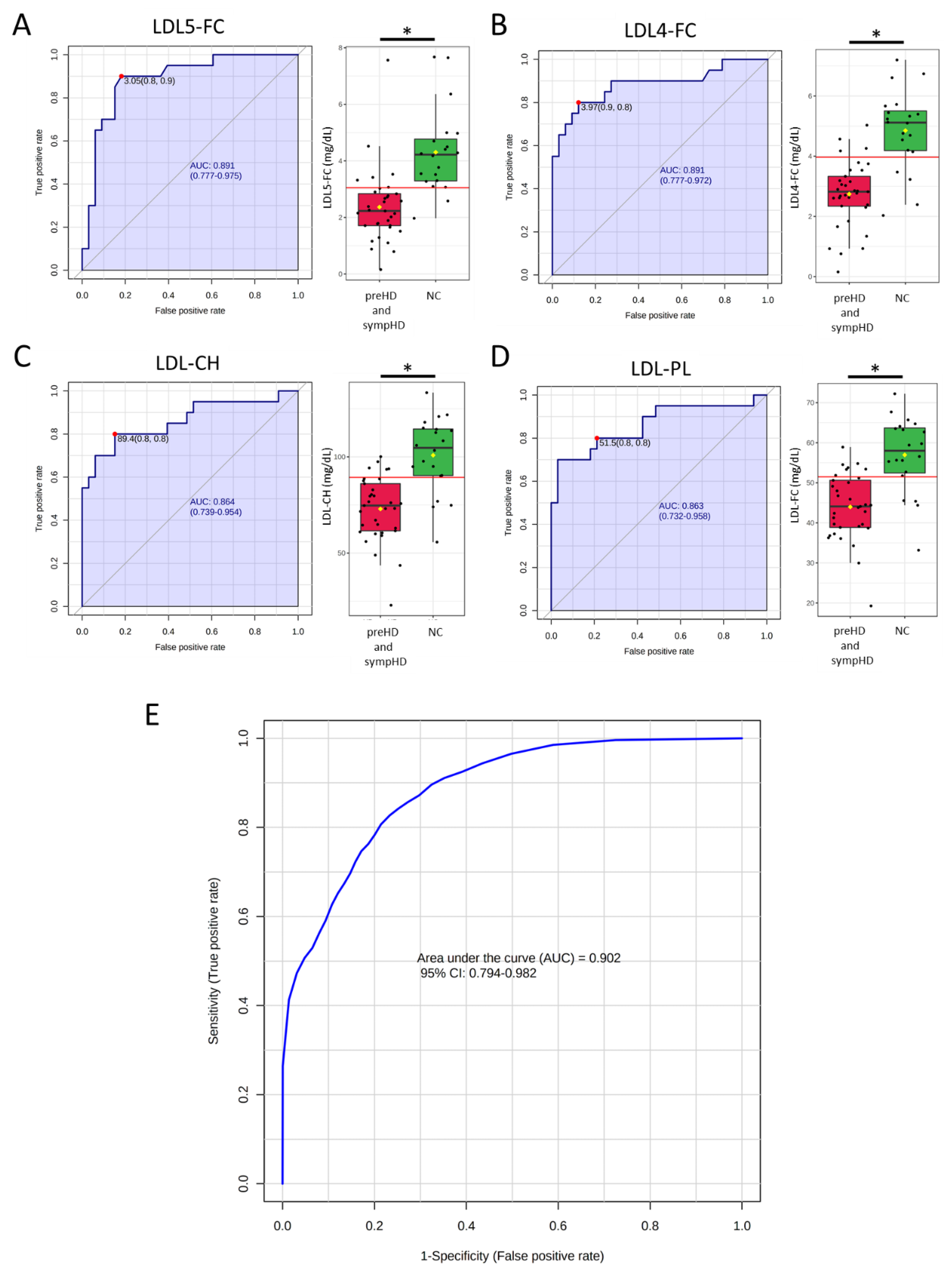
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MacDonald, M.E.; Ambrose, C.M.; Duyao, M.P.; Myers, R.H.; Lin, C.; Srinidhi, L.; Barnes, G.; Taylor, S.A.; James, M.; Groot, N.; et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993, 72, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.H. Transcriptional signatures in Huntington’s disease. Prog. Neurobiol. 2007, 83, 228–248. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.M. Mitochondrial dysfunction, metabolic deficits, and increased oxidative stress in Huntington’s disease. Chang Gung Med. J. 2011, 34, 135–152. [Google Scholar] [PubMed]
- Valera, A.G.; Diaz-Hernandez, M.; Hernandez, F.; Ortega, Z.; Lucas, J.J. The ubiquitin-proteasome system in Huntington’s disease. Neuroscientist 2005, 11, 583–594. [Google Scholar] [CrossRef] [PubMed]
- DiFiglia, M.; Sapp, E.; Chase, K.O.; Davies, S.W.; Bates, G.P.; Vonsattel, J.P.; Aronin, N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 1997, 277, 1990–1993. [Google Scholar] [CrossRef]
- Huang, Y.C.; Wu, Y.R.; Tseng, M.Y.; Chen, Y.C.; Hsieh, S.Y.; Chen, C.M. Increased prothrombin, apolipoprotein A-IV, and haptoglobin in the cerebrospinal fluid of patients with Huntington’s disease. PLoS ONE 2011, 6, e15809. [Google Scholar] [CrossRef] [PubMed]
- Mochel, F.; Charles, P.; Seguin, F.; Barritault, J.; Coussieu, C.; Perin, L.; Le Bouc, Y.; Gervais, C.; Carcelain, G.; Vassault, A.; et al. Early energy deficit in Huntington disease: Identification of a plasma biomarker traceable during disease progression. PLoS ONE 2007, 2, e647. [Google Scholar] [CrossRef]
- Underwood, B.R.; Broadhurst, D.; Dunn, W.B.; Ellis, D.I.; Michell, A.W.; Vacher, C.; Mosedale, D.E.; Kell, D.B.; Barker, R.A.; Grainger, D.J.; et al. Huntington disease patients and transgenic mice have similar pro-catabolic serum metabolite profiles. Brain 2006, 129, 877–886. [Google Scholar] [CrossRef]
- Cheng, M.L.; Chang, K.H.; Wu, Y.R.; Chen, C.M. Metabolic disturbances in plasma as biomarkers for Huntington’s disease. J. Nutr. Biochem. 2016, 31, 38–44. [Google Scholar] [CrossRef]
- McGarry, A.; Gaughan, J.; Hackmyer, C.; Lovett, J.; Khadeer, M.; Shaikh, H.; Pradhan, B.; Ferraro, T.N.; Wainer, I.W.; Moaddel, R. Cross-sectional analysis of plasma and CSF metabolomic markers in Huntington’s disease for participants of varying functional disability: A pilot study. Sci. Rep. 2020, 10, 20490. [Google Scholar] [CrossRef]
- Smith, L.C.; Pownall, H.J.; Gotto, A.M., Jr. The plasma lipoproteins: Structure and metabolism. Annu. Rev. Biochem. 1978, 47, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Camont, L.; Chapman, M.J.; Kontush, A. Biological activities of HDL subpopulations and their relevance to cardiovascular disease. Trends Mol. Med. 2011, 17, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, E.A.; Myasoedova, V.A.; Melnichenko, A.A.; Grechko, A.V.; Orekhov, A.N. Small Dense Low-Density Lipoprotein as Biomarker for Atherosclerotic Diseases. Oxid. Med. Cell. Longev. 2017, 2017, 1273042. [Google Scholar] [CrossRef]
- Steinberg, D.; Parthasarathy, S.; Carew, T.E.; Khoo, J.C.; Witztum, J.L. Beyond cholesterol. Modifications of low-density lipoprotein that increase its atherogenicity. N. Engl. J. Med. 1989, 320, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Packard, C.J.; Demant, T.; Stewart, J.P.; Bedford, D.; Caslake, M.J.; Schwertfeger, G.; Bedynek, A.; Shepherd, J.; Seidel, D. Apolipoprotein B metabolism and the distribution of VLDL and LDL subfractions. J. Lipid Res. 2000, 41, 305–318. [Google Scholar] [CrossRef]
- Krauss, R.M. Lipids and lipoproteins in patients with type 2 diabetes. Diabetes Care 2004, 27, 1496–1504. [Google Scholar] [CrossRef] [PubMed]
- Boren, J.; Chapman, M.J.; Krauss, R.M.; Packard, C.J.; Bentzon, J.F.; Binder, C.J.; Daemen, M.J.; Demer, L.L.; Hegele, R.A.; Nicholls, S.J.; et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease: Pathophysiological, genetic, and therapeutic insights: A consensus statement from the European Atherosclerosis Society Consensus Panel. Eur. Heart J. 2020, 41, 2313–2330. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Liang, Y.; Zhang, X.; Xu, J.; Lin, J.; Zhang, R.; Kang, K.; Liu, C.; Zhao, C.; Zhao, M. Low-density lipoprotein cholesterol and Alzheimer’s disease: A systematic review and meta-analysis. Front. Aging Neurosci. 2020, 12, 5. [Google Scholar] [CrossRef]
- Martinez, A.E.; Weissberger, G.; Kuklenyik, Z.; He, X.; Meuret, C.; Parekh, T.; Rees, J.C.; Parks, B.A.; Gardner, M.S.; King, S.M.; et al. The small HDL particle hypothesis of Alzheimer’s disease. Alzheimer’s Dement. 2022, 2022, 1–14. [Google Scholar] [CrossRef]
- Fang, F.; Zhan, Y.; Hammar, N.; Shen, X.; Wirdefeldt, K.; Walldius, G.; Mariosa, D. Lipids, Apolipoproteins, and the risk of Parkinson disease. Circ. Res. 2019, 125, 643–652. [Google Scholar] [CrossRef]
- Park, J.H.; Lee, C.W.; Nam, M.J.; Kim, H.; Kwon, D.Y.; Yoo, J.W.; Lee, K.N.; Han, K.; Jung, J.H.; Park, Y.G.; et al. Association of high-density lipoprotein cholesterol variability and the risk of developing Parkinson disease. Neurology 2021, 96, e1391–e1401. [Google Scholar] [CrossRef] [PubMed]
- Rozani, V.; Gurevich, T.; Giladi, N.; El-Ad, B.; Tsamir, J.; Hemo, B.; Peretz, C. Higher serum cholesterol and decreased Parkinson’s disease risk: A statin-free cohort study. Mov. Disord. 2018, 33, 1298–1305. [Google Scholar] [CrossRef] [PubMed]
- Huntington Study Group. Unified Huntington’s Disease Rating Scale: Reliability and consistency. Mov. Disord. 1996, 11, 136–142. [Google Scholar] [CrossRef]
- Oosterloo, M.; de Greef, B.T.A.; Bijlsma, E.K.; Durr, A.; Tabrizi, S.J.; Estevez-Fraga, C.; de Die-Smulders, C.E.M.; Roos, R.A.C. Disease onset in Huntington’s disease: When is the conversion? Mov. Disord. Clin. Pract. 2021, 8, 352–360. [Google Scholar] [CrossRef]
- Jimenez, B.; Holmes, E.; Heude, C.; Tolson, R.F.; Harvey, N.; Lodge, S.L.; Chetwynd, A.J.; Cannet, C.; Fang, F.; Pearce, J.T.M.; et al. Quantitative lipoprotein subclass and low molecular weight metabolite analysis in human serum and plasma by (1)H NMR spectroscopy in a multilaboratory trial. Anal. Chem. 2018, 90, 11962–11971. [Google Scholar] [CrossRef] [PubMed]
- Loo, R.L.; Lodge, S.; Kimhofer, T.; Bong, S.H.; Begum, S.; Whiley, L.; Gray, N.; Lindon, J.C.; Nitschke, P.; Lawler, N.G.; et al. Quantitative in-vitro diagnostic NMR spectroscopy for lipoprotein and metabolite measurements in plasma and serum: Recommendations for analytical artifact minimization with special reference to COVID-19/SARS-CoV-2 samples. J. Proteome Res. 2020, 19, 4428–4441. [Google Scholar] [CrossRef] [PubMed]
- Woudberg, N.J.; Pedretti, S.; Lecour, S.; Schulz, R.; Vuilleumier, N.; James, R.W.; Frias, M.A. Pharmacological intervention to modulate HDL: What do we target? Front. Pharmacol. 2017, 8, 989. [Google Scholar] [CrossRef] [PubMed]
- Barzilai, N.; Atzmon, G.; Derby, C.A.; Bauman, J.M.; Lipton, R.B. A genotype of exceptional longevity is associated with preservation of cognitive function. Neurology 2006, 67, 2170–2175. [Google Scholar] [CrossRef] [PubMed]
- van Exel, E.; de Craen, A.J.; Gussekloo, J.; Houx, P.; Bootsma-van der Wiel, A.; Macfarlane, P.W.; Blauw, G.J.; Westendorp, R.G. Association between high-density lipoprotein and cognitive impairment in the oldest old. Ann. Neurol. 2002, 51, 716–721. [Google Scholar] [CrossRef]
- Singh-Manoux, A.; Gimeno, D.; Kivimaki, M.; Brunner, E.; Marmot, M.G. Low HDL cholesterol is a risk factor for deficit and decline in memory in midlife: The Whitehall II study. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 1556–1562. [Google Scholar] [CrossRef]
- Song, F.; Poljak, A.; Crawford, J.; Kochan, N.A.; Wen, W.; Cameron, B.; Lux, O.; Brodaty, H.; Mather, K.; Smythe, G.A.; et al. Plasma apolipoprotein levels are associated with cognitive status and decline in a community cohort of older individuals. PLoS ONE 2012, 7, e34078. [Google Scholar] [CrossRef] [PubMed]
- Partonen, T.; Haukka, J.; Virtamo, J.; Taylor, P.R.; Lonnqvist, J. Association of low serum total cholesterol with major depression and suicide. Br. J. Psychiatry 1999, 175, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Almeida, O.P.; Yeap, B.B.; Hankey, G.J.; Golledge, J.; Flicker, L. HDL cholesterol and the risk of depression over 5 years. Mol. Psychiatry 2014, 19, 637–638. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Ma, X.; Xu, J.; Song, Q.; Hu, M.; Gu, X.; Zhang, Q.; Hou, L.; Chen, L.; Huang, Y.; et al. The shape effect of reconstituted high-density lipoprotein nanocarriers on brain delivery and Aβ clearance. Nano Res. 2018, 11, 5615–5628. [Google Scholar] [CrossRef]
- Zhang, X.; Yin, X.; Yu, H.; Liu, X.; Yang, F.; Yao, J.; Jin, H.; Yang, P. Quantitative proteomic analysis of serum proteins in patients with Parkinson’s disease using an isobaric tag for relative and absolute quantification labeling, two-dimensional liquid chromatography, and tandem mass spectrometry. Analyst 2012, 137, 490–495. [Google Scholar] [CrossRef]
- Yin, G.N.; Lee, H.W.; Cho, J.Y.; Suk, K. Neuronal pentraxin receptor in cerebrospinal fluid as a potential biomarker for neurodegenerative diseases. Brain Res. 2009, 1265, 158–170. [Google Scholar] [CrossRef]
- Qiang, J.K.; Wong, Y.C.; Siderowf, A.; Hurtig, H.I.; Xie, S.X.; Lee, V.M.; Trojanowski, J.Q.; Yearout, D.; Leverenz, J.B.; Montine, T.J.; et al. Plasma apolipoprotein A1 as a biomarker for Parkinson disease. Ann. Neurol. 2013, 74, 119–127. [Google Scholar] [CrossRef]
- Huang, X.; Abbott, R.D.; Petrovitch, H.; Mailman, R.B.; Ross, G.W. Low LDL cholesterol and increased risk of Parkinson’s disease: Prospective results from Honolulu-Asia Aging Study. Mov. Disord. 2008, 23, 1013–1018. [Google Scholar] [CrossRef]
- Thongtang, N.; Diffenderfer, M.R.; Ooi, E.M.M.; Barrett, P.H.R.; Turner, S.M.; Le, N.A.; Brown, W.V.; Schaefer, E.J. Metabolism and proteomics of large and small dense LDL in combined hyperlipidemia: Effects of rosuvastatin. J. Lipid Res. 2017, 58, 1315–1324. [Google Scholar] [CrossRef]
- Yu, X.; Yu, Y.; Wei, C.; Wang, L.; Jiang, J.; Zhang, R.; Dai, Q.; Kang, Y.; Chen, X. Association between small dense low-density lipoprotein cholesterol and neuroimaging markers of cerebral small vessel disease in middle-aged and elderly Chinese populations. BMC Neurol. 2021, 21, 436. [Google Scholar] [CrossRef]
- Abi-Ayad, M.; Abbou, A.; Abi-Ayad, F.Z.; Behadada, O.; Benyoucef, M. HDL-C, ApoA1 and VLDL-TG as biomarkers for the carotid plaque presence in patients with metabolic syndrome. Diabetes Metab. Syndr. 2018, 12, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Gentile, M.; Iannuzzi, A.; Giallauria, F.; D’Andrea, A.; Venturini, E.; Pacileo, M.; Covetti, G.; Panico, C.; Mattiello, A.; Vitale, G.; et al. Association between very low-density lipoprotein cholesterol (VLDL-C) and carotid intima-media thickness in postmenopausal women without overt cardiovascular disease and on LDL-C target levels. J. Clin. Med. 2020, 9, 1422. [Google Scholar] [CrossRef] [PubMed]
- Li, C.L.; Chu, C.H.; Lee, H.C.; Chou, M.C.; Liu, C.K.; Chen, C.H.; Ke, L.Y.; Chen, S.L. Immunoregulatory effects of very low density lipoprotein from healthy individuals and metabolic syndrome patients on glial cells. Immunobiology 2019, 224, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.S.; Liu, C.K.; Lee, H.C.; Chou, M.C.; Ke, L.Y.; Chen, C.H.; Chen, S.L. Electronegative very-low-density lipoprotein induces brain inflammation and cognitive dysfunction in mice. Sci. Rep. 2021, 11, 6013. [Google Scholar] [CrossRef]
- Loving, B.A.; Bruce, K.D. Lipid and Lipoprotein Metabolism in Microglia. Front. Physiol. 2020, 11, 393. [Google Scholar] [CrossRef]
- Ren, L.; Ren, X. Meta-analyses of four polymorphisms of lipoprotein lipase associated with the risk of Alzheimer’s disease. Neurosci. Lett. 2016, 619, 73–78. [Google Scholar] [CrossRef]
- Yvan-Charvet, L.; Wang, N.; Tall, A.R. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 139–143. [Google Scholar] [CrossRef]
- Zhang, M.; Zhai, X.; Li, J.; Albers, J.J.; Vuletic, S.; Ren, G. Structural basis of the lipid transfer mechanism of phospholipid transfer protein (PLTP). Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2018, 1863, 1082–1094. [Google Scholar] [CrossRef]
- Gonzalez-Guevara, E.; Cardenas, G.; Perez-Severiano, F.; Martinez-Lazcano, J.C. Dysregulated brain cholesterol metabolism is linked to neuroinflammation in Huntington’s disease. Mov. Disord. 2020, 35, 1113–1127. [Google Scholar] [CrossRef]
- Chung, S.; Timmins, J.M.; Duong, M.; Degirolamo, C.; Rong, S.; Sawyer, J.K.; Singaraja, R.R.; Hayden, M.R.; Maeda, N.; Rudel, L.L.; et al. Targeted deletion of hepatocyte ABCA1 leads to very low density lipoprotein triglyceride overproduction and low density lipoprotein hypercatabolism. J. Biol. Chem. 2010, 285, 12197–12209. [Google Scholar] [CrossRef]
- Fitz, N.F.; Carter, A.Y.; Tapias, V.; Castranio, E.L.; Kodali, R.; Lefterov, I.; Koldamova, R. ABCA1 deficiency affects basal cognitive deficits and dendritic density in mice. J. Alzheimer’s Dis. 2017, 56, 1075–1085. [Google Scholar] [CrossRef] [PubMed]


| NC | HD | |||
|---|---|---|---|---|
| (n = 20) | preHD (n = 9) | sympHD (n = 24) | All (n = 53) | |
| Age (years) | 52.70 ± 8.95 | 32.00 ± 7.93 * | 50.00 ± 10.81 | 47.96 ± 12.14 |
| Male (%) | 10 (50.00) | 1 (11.11) ** | 16 (66.66) | 427(50.94) |
| BMI | 23.42 ± 2.57 | 20.24 ± 2.41 | 21.87 ± 2.54 | 22.16 ± 2.77 |
| Pre-prandial glucose (mg/dL) | 93.30 ± 10.74 | 87.80 ± 7.93 | 98.34 ± 16.59 | 93.38 ± 12.15 |
| UHDRS | ||||
| Total motor score | 0 | 39.21 ± 19.30 | ||
| Independence scale | 100 | 66.67 ± 25.93 | ||
| Functional capacity | 13 | 6.38 ± 4.16 | ||
| Disease burden | 272.56 ± 182.39 | 457.50 ± 124.61 | ||
| Medications | ||||
| Tetrabenazine (%) | 0 | 0 | 6 (25.00) | |
| Benzodiazepines (%) | 3 (15.00) | 0 | 14 (58.33) | |
| Antipsychotics (%) | 0 | 0 | 11 (45.83) | |
| Antidepressants (%) | 0 | 0 | 8 (33.33) | |
| Ubidecarenone (%) | 0 | 0 | 10 (41.67) | |
| Metabolite Name | NC | HD | ||
|---|---|---|---|---|
| (n = 20) | preHD (n = 9) | sympHD (n = 24) | All (n = 33) | |
| ApoB (mg/dL) | 77.39 ± 13.06 | 58.96 ± 8.63 ** | 63.32 ± 11.52 ** | 62.13 ± 10.98 * |
| ApoB-PN (nmol/L) | 1407.08 ± 237.50 | 1072.05 ± 157.06 ** | 1151.33 ± 209.48 ** | 1129.71 ± 199.72 * |
| CH (mg/dL) | 178.49 ± 25.35 | 152.77 ± 19.12 ** | 155.05 ± 21.88 ** | 154.43 ± 21.19 * |
| LDL-ApoB (mg/dL) | 61.00 ± 10.23 | 48.32 ± 9.09 ** | 49.83 ± 9.54 ** | 49.42 ± 9.44 * |
| LDL-CH (mg/dL) | 100.83 ± 19.08 | 78.80 ± 16.14 ** | 71.86 ± 16.64 ** | 72.94 ± 16.60 * |
| LDL-FC (mg/dL) | 30.67 ± 5.68 | 24.15 ± 4.90 ** | 22.83 ± 5.70 ** | 23.19 ± 5.53 * |
| LDL-PL (mg/dL) | 56.96 ± 9.23 | 44.93 ± 7.92 ** | 43.64 ± 8.28 ** | 43.99 ± 8.20 * |
| LDL-PN (nmol/L) | 1163.74 ± 186.01 | 878.58 ± 165.33 ** | 906.04 ± 173.41 ** | 898.55 ± 171.68 * |
| LDL3-ApoB (mg/dL) | 9.34 ± 3.33 | 8.10 ± 2.07 | 6.54 ± 2.19 ** | 6.96 ± 2.26 * |
| LDL3-CH (mg/dL) | 16.02 ± 6.96 | 13.92 ± 4.05 | 10.82 ± 3.94 ** | 11.67 ± 4.21 * |
| LDL3-FC (mg/dL) | 5.76 ± 2.02 | 4.97 ± 1.22 | 4.11 ± 1.22 ** | 4.34 ± 1.28 * |
| LDL3-PL (mg/dL) | 9.05 ± 3.40 | 7.93 ± 1.99 | 6.42 ± 2.08 ** | 6.83 ± 2.16 * |
| LDL3-PN (nmol/L) | 169.85 ± 60.53 | 147.23 ± 37.73 | 118.89 ± 39.74 ** | 126.62 ± 41.18 * |
| LDL4-ApoB (mg/dL) | 8.50 ± 2.70 | 4.68 ± 2.26 ** | 4.32 ± 2.72 ** | 4.41 ± 2.61 * |
| LDL4-CH (mg/dL) | 14.29 ± 5.03 | 7.76 ± 3.25 ** | 6.89 ± 4.36 ** | 7.13 ± 4.11 * |
| LDL4-FC (mg/dL) | 4.85 ± 1.31 | 3.00 ± 0.72 ** | 2.64 ± 1.19 ** | 2.74 ± 1.10 * |
| LDL4-PL (mg/dL) | 8.04 ± 2.55 | 4.76 ± 1.67 ** | 4.36 ± 2.45 ** | 4.47 ± 2.27 * |
| LDL4-PN (nmol/L) | 154.61 ± 49.05 | 85.00 ± 41.15 ** | 78.47 ± 49.51 ** | 80.25 ± 47.47 * |
| LDL4-TG (mg/dL) | 1.85 ± 0.74 | 1.11 ± 0.68 | 1.29 ± 0.67 | 1.24 ± 0.67 * |
| LDL5-ApoB (mg/dL) | 9.46 ± 4.68 | 3.85 ± 1.41 ** | 5.39 ± 3.76 ** | 4.97 ± 3.36 * |
| LDL5-CH (mg/dL) | 13.88 ± 6.51 | 5.37 ± 2.16 ** | 7.33 ± 5.50 ** | 6.80 ± 4.90 * |
| LDL5-FC (mg/dL) | 4.31 ± 1.46 | 2.12 ± 0.51 ** | 2.45 ± 1.47 ** | 2.36 ± 1.29 * |
| LDL5-PL (mg/dL) | 7.73 ± 3.28 | 3.41 ± 1.08 ** | 4.42 ± 2.83 ** | 4.14 ± 2.52 * |
| LDL5-PN (nmol/L) | 172.02 ± 85.22 | 70.04 ± 25.68 ** | 97.98 ± 68.34 ** | 90.36 ± 61.08 * |
| LDL6-FC (mg/dL) | 5.25 ± 1.65 | 3.19 ± 0.91 ** | 3.90 ± 1.50 ** | 3.70 ± 1.40 * |
| HDL3-FC (mg/dL) | 2.04 ± 0.59 | 1.76 ± 0.38 | 1.54 ± 0.55 | 1.60 ± 0.52 * |
| HDL4-ApoA1 (mg/dL) | 70.93 ± 11.06 | 60.42 ± 5.64 ** | 58.92 ± 10.66 ** | 59.33 ± 9.58 * |
| HDL4-CH (mg/dL) | 18.51 ± 3.92 | 15.76 ± 2.95 | 14.90 ± 4.01 | 15.13 ± 3.77 * |
| HDL4-FC (mg/dL) | 3.87 ± 1.06 | 2.99 ± 0.60 | 2.81 ± 1.19 | 2.86 ± 1.06 * |
| VLDL5-FC (mg/dL) | 0.60 ± 0.39 | 0.90 ± 0.35 | 1.16 ± 0.71 ** | 1.09 ± 0.64 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, K.-H.; Cheng, M.-L.; Lo, C.-J.; Fan, C.-M.; Wu, Y.-R.; Chen, C.-M. Alternations of Lipoprotein Profiles in the Plasma as Biomarkers of Huntington’s Disease. Cells 2023, 12, 385. https://doi.org/10.3390/cells12030385
Chang K-H, Cheng M-L, Lo C-J, Fan C-M, Wu Y-R, Chen C-M. Alternations of Lipoprotein Profiles in the Plasma as Biomarkers of Huntington’s Disease. Cells. 2023; 12(3):385. https://doi.org/10.3390/cells12030385
Chicago/Turabian StyleChang, Kuo-Hsuan, Mei-Ling Cheng, Chi-Jen Lo, Chun-Ming Fan, Yih-Ru Wu, and Chiung-Mei Chen. 2023. "Alternations of Lipoprotein Profiles in the Plasma as Biomarkers of Huntington’s Disease" Cells 12, no. 3: 385. https://doi.org/10.3390/cells12030385
APA StyleChang, K.-H., Cheng, M.-L., Lo, C.-J., Fan, C.-M., Wu, Y.-R., & Chen, C.-M. (2023). Alternations of Lipoprotein Profiles in the Plasma as Biomarkers of Huntington’s Disease. Cells, 12(3), 385. https://doi.org/10.3390/cells12030385






