Unraveling Presenilin 2 Functions in a Knockout Zebrafish Line to Shed Light into Alzheimer’s Disease Pathogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Generation of psen2−/− Zebrafish Line
2.3. Genomic DNA Extraction from Embryos, Larvae, and Fin Clips
2.4. RNA Isolation and Quantitative Real-Time Reverse Transcription PCR
2.5. Birefringence Assay
2.6. Whole-Mount in Situ Hybridization (WMISH)
2.7. Generation of Mutant psen2 Transgenic Zebrafish Lines
2.8. Behavioral Assays
2.9. Generation of Expression Plasmids
2.10. psen2 Knockdown
2.11. PS2 Overexpression
2.12. Transient Expression of SPLICS Probe
2.13. In Vivo Mitochondrial Axonal Transport Imaging
2.14. Kymograph Construction and Analysis
2.15. Measurement of OCR
2.16. Drug Treatment
2.17. Protein Extraction and Western Blotting
2.18. Statistical Analysis
3. Results
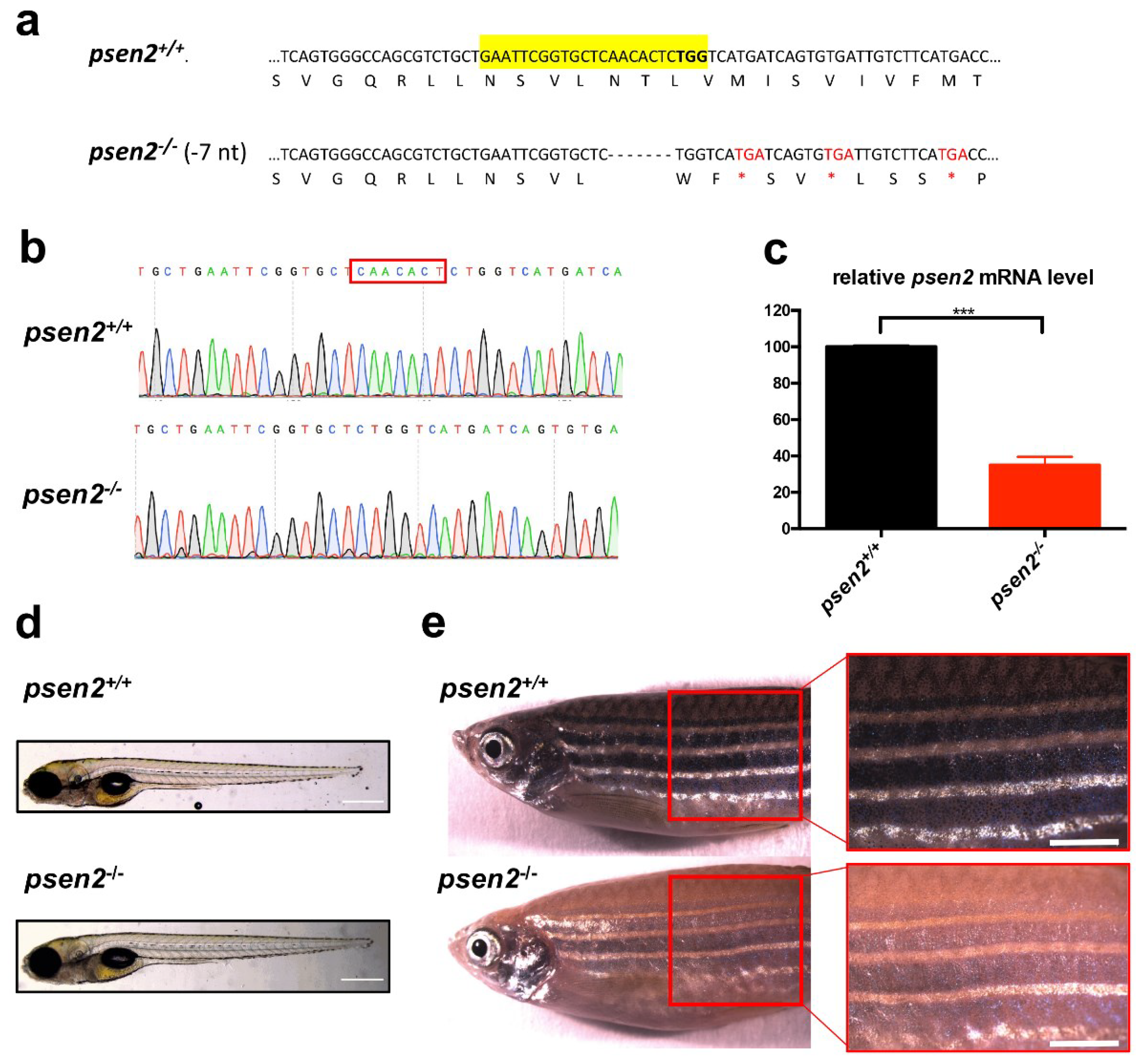
3.1. Generation of a psen2 Knockout (KO) Zebrafish Line
3.2. Psen2 Ablation Does Not Affect Notch Signaling at Early Developmental Stage
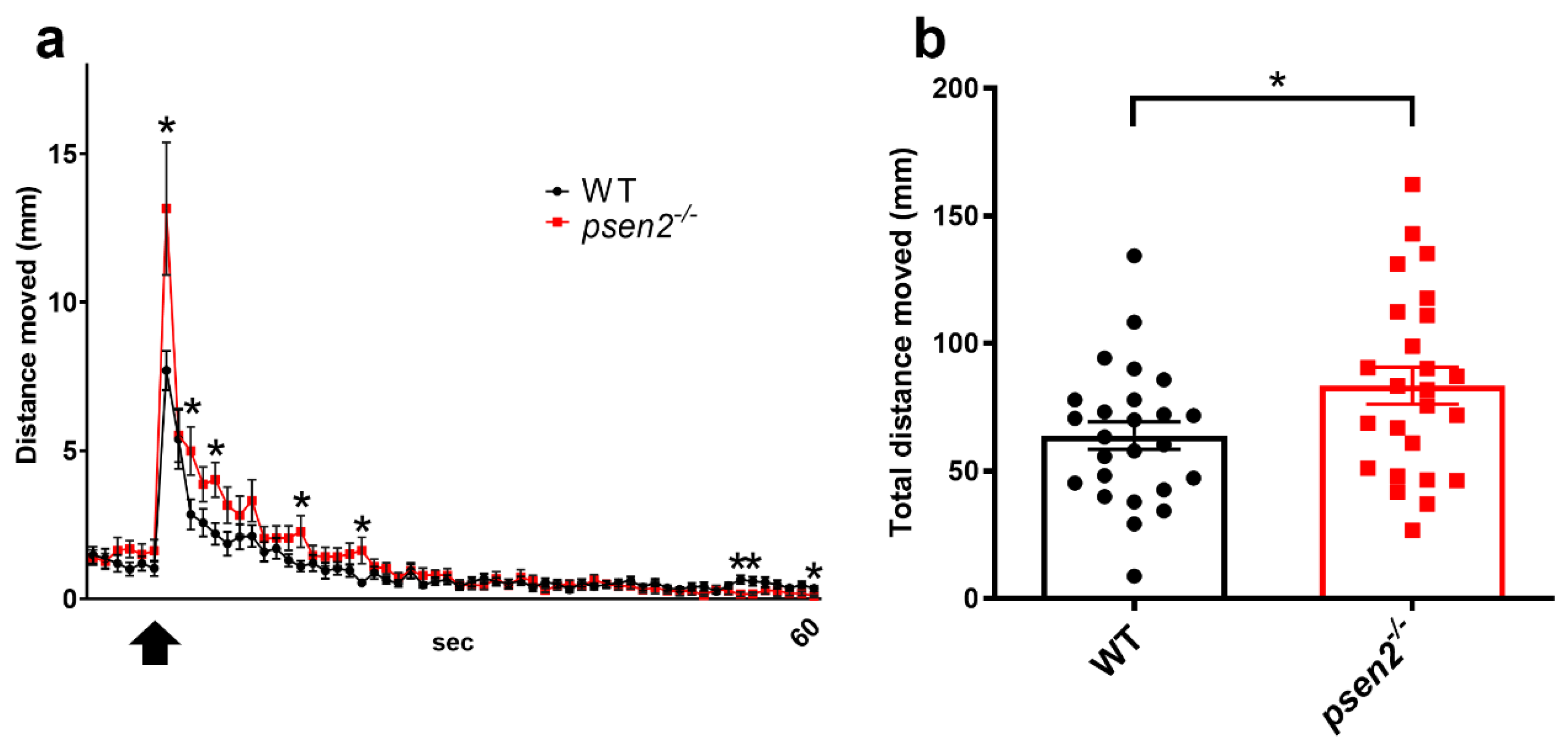
3.3. Loss of Psen2 Alters Zebrafish Locomotor Behavior
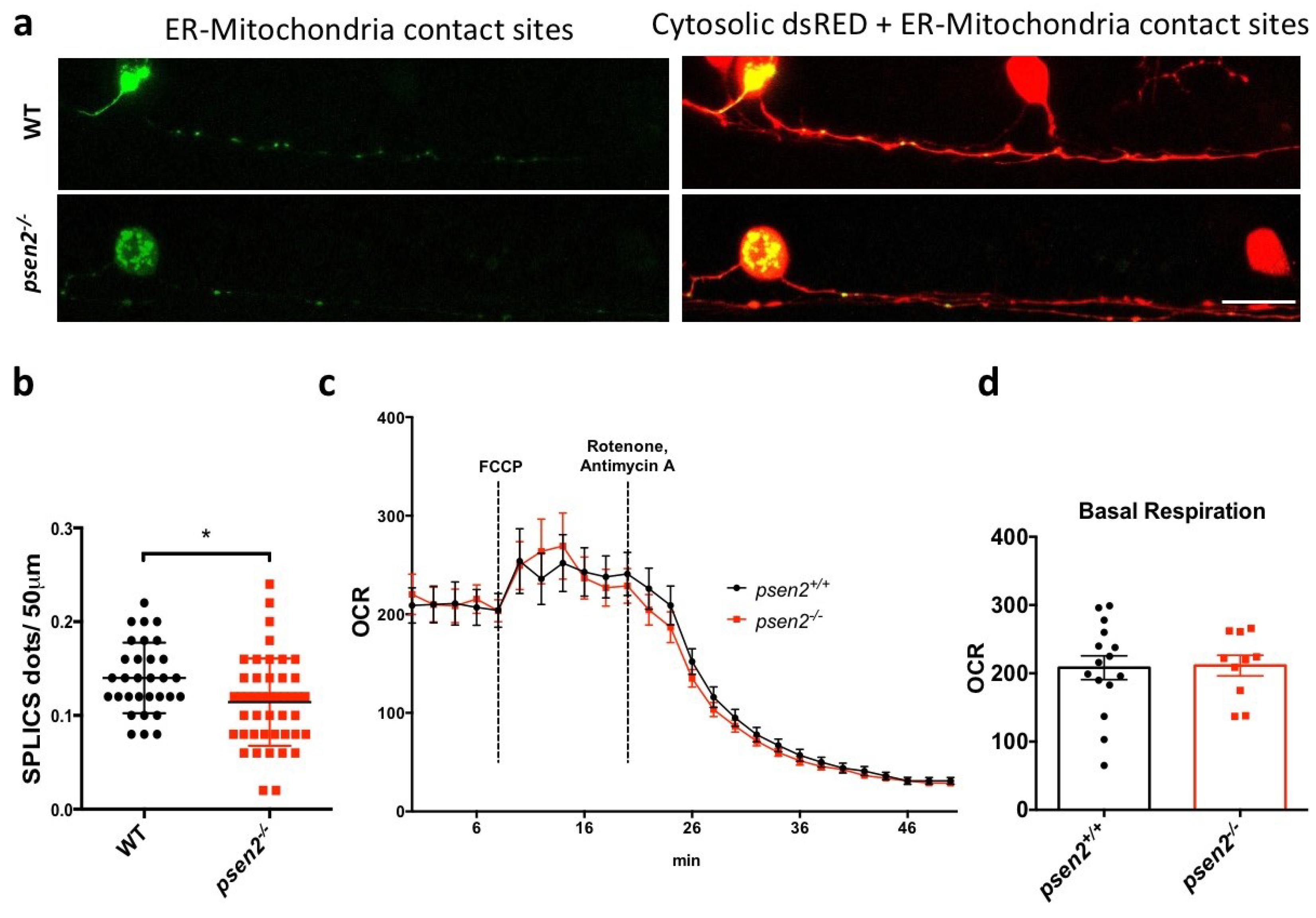
3.4. Psen2 Ablation Reduces Neuronal ER-Mitochondria Contacts But Not Mitochondrial Respiration
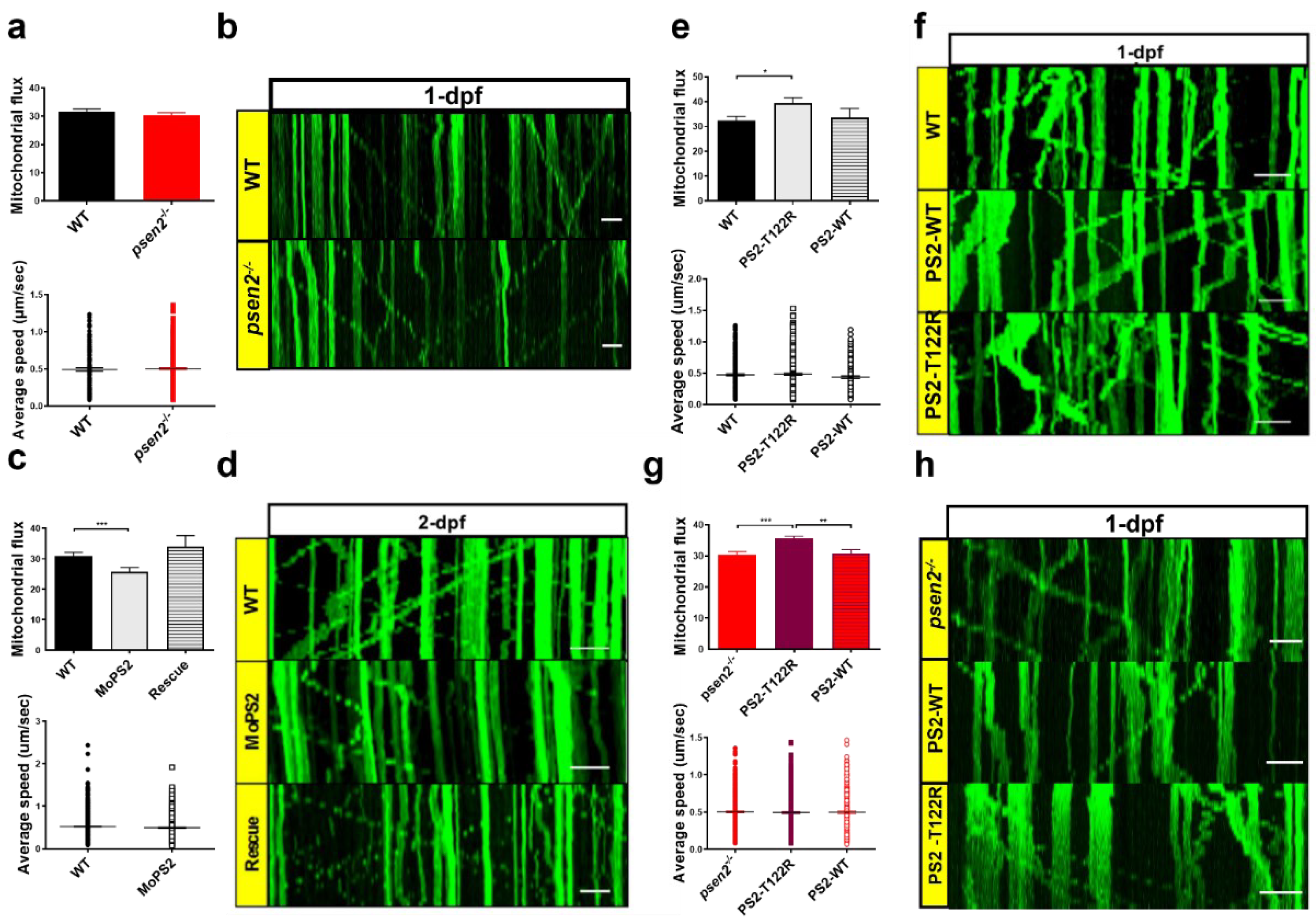
3.5. Psen2 Has a Key Role in Mitochondrial Axonal Transport
3.6. Psen2 Absence Causes a Dysregulated Autophagy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bateman, R.J.; Xiong, C.; Benzinger, T.L.S.; Fagan, A.M.; Goate, A.; Fox, N.C.; Marcus, D.S.; Cairns, N.J.; Xie, X.; Blazey, T.M.; et al. Clinical and Biomarker Changes in Dominantly Inherited Alzheimer’s Disease. N. Engl. J. Med. 2012, 367, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Mendez, M.F. Early–Onset Alzheimer Disease. Neurol. Clin 2017, 35, 263–281. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Liu, H.; Liu, X.; Zhang, W.; Zhang, S.; Jiao, B. APP, PSEN1, and PSEN2 Variants in Alzheimer’s Disease: Systematic Re–evaluation According to ACMG Guidelines. Front. Aging Neurosci. 2021, 13, 695808. [Google Scholar] [CrossRef] [PubMed]
- Sannerud, R.; Esselens, C.; Ejsmont, P.; Mattera, R.; Rochin, L.; Tharkeshwar, K.; De, G.; De, V.; Habets, R.; Baert, V.; et al. Restricted Location of PSEN2/gamma-Secretase Determines Substrate Specificity and Generates an Intracellular Abeta Pool. Cell 2016, 166, 193–208. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, M.; Cai, F.; Song, W. Biological function of Presenilin and its role in AD pathogenesis. Transl. Neurodegener. 2013, 2, 15. [Google Scholar] [CrossRef]
- Area-Gomez, E.; de Groof, A.J.C.; Boldogh, I.; Bird, T.D.; Gibson, G.E.; Koehler, C.M.; Yu, W.H.; Duff, K.E.; Yaffe, M.P.; Pon, L.A.; et al. Presenilins Are Enriched in Endoplasmic Reticulum Membranes Associated with Mitochondria. Am. J. Pathol. 2009, 175, 1810–1816. [Google Scholar] [CrossRef]
- Zampese, E.; Fasolato, C.; Kipanyula, M.J.; Bortolozzi, M.; Pozzan, T.; Pizzo, P. Presenilin 2 modulates endoplasmic reticulum (ER)-mitochondria interactions and Ca2+ cross-talk. Proc. Natl. Acad. Sci. USA 2011, 108, 2777–2782. [Google Scholar] [CrossRef]
- Filadi, R.; Greotti, E.; Turacchio, G.; Luini, A.; Pozzan, T.; Pizzo, P. Presenilin 2 Modulates Endoplasmic Reticulum-Mitochondria Coupling by Tuning the Antagonistic Effect of Mitofusin 2. Cell Rep. 2016, 15, 2226–2238. [Google Scholar] [CrossRef]
- Filadi, R.; Greotti, E.; Turacchio, G.; Luini, A.; Pozzan, T.; Pizzo, P. Mitofusin 2 ablation increases endoplasmic reticulum–mitochondria coupling. Proc. Natl. Acad. Sci. USA 2015, 112, E2174–E2181. [Google Scholar] [CrossRef]
- Zatti, G.; Ghidoni, R.; Barbiero, L.; Binetti, G.; Pozzan, T.; Fasolato, C.; Pizzo, P. The presenilin 2 M239I mutation associated with familial Alzheimer’s disease reduces Ca2+ release from intracellular stores. Neurobiol. Dis. 2004, 15, 269–278. [Google Scholar] [CrossRef]
- Giacomello, M.; Barbiero, L.; Zatti, G.; Squitti, R.; Binetti, G.; Pozzan, T.; Fasolato, C.; Ghidoni, R.; Pizzo, P. Reduction of Ca2+ stores and capacitative Ca2+ entry is associated with the familial Alzheimer’s disease presenilin-2 T122R mutation and anticipates the onset of dementia. Neurobiol. Dis. 2005, 18, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Zatti, G.; Burgo, A.; Giacomello, M.; Barbiero, L.; Ghidoni, R.; Sinigaglia, G.; Florean, C.; Bagnoli, S.; Binetti, G.; Sorbi, S.; et al. Presenilin mutations linked to familial Alzheimer’s disease reduce endoplasmic reticulum and Golgi apparatus calcium levels. Cell Calcium 2006, 39, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Brunello, L.; Zampese, E.; Florean, C.; Pozzan, T.; Pizzo, P.; Fasolato, C. Presenilin-2 dampens intracellular Ca2+ stores by increasing Ca2+ leakage and reducing Ca2+ uptake. J. Cell. Mol. Med. 2009, 13, 3358–3369. [Google Scholar] [CrossRef]
- Kipanyula, M.J.; Contreras, L.; Zampese, E.; Lazzari, C.; Wong, A.K.C.; Pizzo, P.; Fasolato, C.; Pozzan, T. Ca2+ dysregulation in neurons from transgenic mice expressing mutant presenilin 2. Aging Cell 2012, 11, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Greotti, E.; Capitanio, P.; Wong, A.; Pozzan, T.; Pizzo, P.; Pendin, D. Familial Alzheimer’s disease-linked presenilin mutants and intracellular Ca(2+) handling: A single-organelle, FRET-based analysis. Cell Calcium 2019, 79, 44–56. [Google Scholar] [CrossRef]
- Fedeli, C.; Filadi, R.; Rossi, A.; Mammucari, C.; Pizzo, P. PSEN2 (presenilin 2) mutants linked to familial Alzheimer disease impair autophagy by altering Ca2+ homeostasis. Autophagy 2019, 15, 2044–2062. [Google Scholar] [CrossRef]
- Rossi, A.; Rigotto, G.; Valente, G.; Giorgio, V.; Basso, E.; Filadi, R.; Pizzo, P. Defective Mitochondrial Pyruvate Flux Affects Cell Bioenergetics in Alzheimer’s Disease-Related Models. Cell Rep. 2020, 30, 2332–2348.e10. [Google Scholar] [CrossRef]
- Theurey, P.; Connolly, N.; Fortunati, I.; Basso, E.; Lauwen, S.; Ferrante, C.; Pinho, C.M.; Joselin, A.; Gioran, A.; Bano, D.; et al. Systems biology identifies preserved integrity but impaired metabolism of mitochondria due to a glycolytic defect in Alzheimer’s disease neurons. Aging Cell 2019, 18, e12924. [Google Scholar] [CrossRef]
- Groth, C.; Nornes, S.; McCarty, R.; Tamme, R.; Lardelli, M. Identification of a second presenilin gene in zebrafish with similarity to the human Alzheimer’s disease gene presenilin2. Dev. Genes Evol. 2002, 212, 486–490. [Google Scholar] [CrossRef]
- Leimer, U.; Lun, K.; Romig, H.; Walter, J.; Grünberg, J.; Brand, M.; Haass, C. Zebrafish (Danio rerio) presenilin promotes aberrant amyloid beta-peptide production and requires a critical aspartate residue for its function in amyloidogenesis. Biochemistry 1999, 38, 13602–13609. [Google Scholar] [CrossRef]
- Campbell, W.A.; Yang, H.; Zetterberg, H.; Baulac, S.; Sears, J.A.; Liu, T.; Wong, S.T.C.; Zhong, T.P.; Xia, W. Zebrafish lacking Alzheimer presenilin enhancer 2 (Pen-2) demonstrate excessive p53-dependent apoptosis and neuronal loss. J. Neurochem. 2006, 96, 1423–1440. [Google Scholar] [CrossRef] [PubMed]
- Sager, J.J.; Bai, Q.; Burton, E.A. Transgenic zebrafish models of neurodegenerative diseases. Anat. Embryol. 2010, 214, 285–302. [Google Scholar] [CrossRef]
- Nornes, S. Developmental control of Presenilin1 expression, endoproteolysis, and interaction in zebrafish embryos. Exp. Cell Res. 2003, 289, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Nornes, S.; Newman, M.; Verdile, G.; Wells, S.; Stoick-Cooper, C.L.; Tucker, B.; Frederich-Sleptsova, I.; Martins, R.; Lardelli, M. Interference with splicing of Presenilin transcripts has potent dominant negative effects on Presenilin activity. Hum. Mol. Genet. 2008, 17, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Newman, M.; Lardelli, M. The zebrafish orthologue of familial Alzheimer’s disease gene PRESENILIN 2 is required for normal adult melanotic skin pigmentation. PLoS ONE 2018, 13, e0206155. [Google Scholar] [CrossRef]
- Barthelson, K.; Pederson, S.M.; Newman, M.; Jiang, H.; Lardelli, M. In-Frame and Frameshift Mutations in Zebrafish Presenilin 2 Affect Different Cellular Functions in Young Adult Brains. J. Alzheimer’s Dis. Rep. 2021, 5, 395–404. [Google Scholar] [CrossRef]
- Jao, L.-E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef]
- Meeker, N.D.; Hutchinson, S.A.; Ho, L.; Trede, N.S. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Biotechniques 2007, 43, 610–614. [Google Scholar] [CrossRef]
- Thisse, C.; Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008, 3, 59–69. [Google Scholar] [CrossRef]
- Nornes, S.; Newman, M.; Wells, S.; Verdile, G.; Martins, R.N.; Lardelli, M. Independent and cooperative action of Psen2 with Psen1 in zebrafish embryos. Exp. Cell Res. 2009, 315, 2791–2801. [Google Scholar] [CrossRef]
- Vallese, F.; Catoni, C.; Cieri, D.; Barazzuol, L.; Ramirez, O.; Calore, V.; Bonora, M.; Giamogante, F.; Pinton, P.; Brini, M.; et al. An expanded palette of improved SPLICS reporters detects multiple organelle contacts in vitro and in vivo. Nat. Commun. 2020, 11, 6069. [Google Scholar] [CrossRef]
- Cribbs, J.T.; Strack, S. Functional characterization of phosphorylation sites in dynamin-related protein 1. Methods Enzymol. 2009, 457, 231–253. [Google Scholar] [PubMed]
- Moro, E.; Vettori, A.; Porazzi, P.; Schiavone, M.; Rampazzo, E.; Casari, A.; Ek, O.; Facchinello, N.; Astone, M.; Zancan, I.; et al. Generation and application of signaling pathway reporter lines in zebrafish. Mol. Genet. Genom. 2013, 288, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Babin, P.J.; Goizet, C.; Raldúa, D. Zebrafish models of human motor neuron diseases: Advantages and limitations. Prog. Neurobiol. 2014, 118, 36–58. [Google Scholar] [CrossRef] [PubMed]
- Faria, M.; Prats, E.; Novoa-Luna, K.A.; Bedrossiantz, J.; Gómez-Canela, C.; Gómez-Oliván, L.M.; Raldúa, D. Development of a vibrational startle response assay for screening environmental pollutants and drugs impairing predator avoidance. Sci. Total. Environ. 2019, 650, 87–96. [Google Scholar] [CrossRef]
- Theodoridi, A.; Dinarello, A.; Badenetti, L.; Pavlidis, M.; Valle, L.D.; Tsalafouta, A. Knockout of the hsd11b2 Gene Extends the Cortisol Stress Response in Both Zebrafish Larvae and Adults. Int. J. Mol. Sci. 2021, 22, 12525. [Google Scholar] [CrossRef]
- Takahashi, H.; Kamio, Y. Acoustic startle response and its modulation in schizophrenia and autism spectrum disorder in Asian subjects. Schizophr. Res. 2018, 198, 16–20. [Google Scholar] [CrossRef]
- Liska, G.M.; Lee, J.-Y.; Xu, K.; Sanberg, P.R.; Borlongan, C.V. Suppressed acoustic startle response in traumatic brain injury masks post-traumatic stress disorder hyper-responsivity. Neuroreport 2018, 29, 939–944. [Google Scholar] [CrossRef]
- Pang, K.C.; Sinha, S.; Avcu, P.; Roland, J.J.; Nadpara, N.; Pfister, B.; Long, M.; Santhakumar, V.; Servatius, R.J. Long-Lasting Suppression of Acoustic Startle Response after Mild Traumatic Brain Injury. J. Neurotrauma 2015, 32, 801–810. [Google Scholar] [CrossRef]
- Rossi, A.; Galla, L.; Gomiero, C.; Zentilin, L.; Giacca, M.; Giorgio, V.; Calì, T.; Pozzan, T.; Greotti, E.; Pizzo, P. Calcium Signaling and Mitochondrial Function in Presenilin 2 Knock-Out Mice: Looking for Any Loss-of-Function Phenotype Related to Alzheimer’s Disease. Cells 2021, 10, 204. [Google Scholar] [CrossRef]
- Cieri, D.; Vicario, M.; Giacomello, M.; Vallese, F.; Filadi, R.; Wagner, T.; Pozzan, T.; Pizzo, P.; Scorrano, L.; Brini, M.; et al. SPLICS: A split green fluorescent protein-based contact site sensor for narrow and wide heterotypic organelle juxtaposition. Cell Death Differ. 2018, 25, 1131–1145. [Google Scholar] [CrossRef] [PubMed]
- Calì, T.; Brini, M. Quantification of organelle contact sites by split-GFP-based contact site sensors (SPLICS) in living cells. Nat. Protoc. 2021, 16, 5287–5308. [Google Scholar] [CrossRef] [PubMed]
- Marchi, S.; Bittremieux, M.; Missiroli, S.; Morganti, C.; Patergnani, S.; Sbano, L.; Rimessi, A.; Kerkhofs, M.; Parys, J.B.; Bultynck, G.; et al. Endoplasmic Reticulum-Mitochondria Communication Through Ca2+ Signaling: The Importance of Mitochondria-Associated Membranes (MAMs). Adv. Exp. Med. Biol. 2017, 997, 49–67. [Google Scholar] [CrossRef]
- Aoyama-Ishiwatari, S.; Hirabayashi, Y. Endoplasmic Reticulum–Mitochondria Contact Sites—Emerging Intracellular Signaling Hubs. Front. Cell Dev. Biol. 2021, 9, 653828. [Google Scholar] [CrossRef]
- Yang, M.; Li, C.; Yang, S.; Xiao, Y.; Xiong, X.; Chen, W.; Zhao, H.; Zhang, Q.; Han, Y.; Sun, L. Mitochondria-Associated ER Membranes—The Origin Site of Autophagy. Front. Cell Dev. Biol. 2020, 8, 595. [Google Scholar] [CrossRef] [PubMed]
- Filadi, R.; Theurey, P.; Pizzo, P. The endoplasmic reticulum-mitochondria coupling in health and disease: Molecules, functions and significance. Cell Calcium 2017, 62, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2020, 17, 1–382. [Google Scholar] [CrossRef]
- Zhang, Z.; Gong, J.; Sviderskaya, E.V.; Wei, A.; Li, W. Mitochondrial NCKX5 regulates melanosomal biogenesis and pigment production. J. Cell Sci. 2019, 132, 232009. [Google Scholar] [CrossRef]
- Pizzo, P.; Basso, E.; Filadi, R.; Greotti, E.; Leparulo, A.; Pendin, D.; Redolfi, N.; Rossini, M.; Vajente, N.; Pozzan, T.; et al. Presenilin-2 and Calcium Handling: Molecules, Organelles, Cells and Brain Networks. Cells 2020, 9, 2166. [Google Scholar] [CrossRef]
- De Strooper, B.; Vassar, R.; Golde, T. The secretases: Enzymes with therapeutic potential in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 99–107. [Google Scholar] [CrossRef]
- Barazzuol, L.; Giamogante, F.; Calì, T. Mitochondria Associated Membranes (MAMs): Architecture and physiopathological role. Cell Calcium 2021, 94, 102343. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, W.; Li, L.; Perry, G.; Lee, H.-G.; Zhu, X. Oxidative stress and mitochondrial dysfunction in Alzheimer’s disease. Biochim. Biophys. Mol. Basis Dis. 2014, 1842, 1240–1247. [Google Scholar] [CrossRef] [PubMed]
- Di Meco, A.; Curtis, M.E.; Lauretti, E.; Praticò, D. Autophagy Dysfunction in Alzheimer’s Disease: Mechanistic Insights and New Therapeutic Opportunities. Biol. Psychiatry 2020, 87, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Filadi, R.; Pizzo, P. Defective autophagy and Alzheimer’s disease: Is calcium the key? Neural Regen. Res. 2019, 14, 2081–2082. [Google Scholar] [CrossRef]
- Sucularli, C.; Shehwana, H.; Kuscu, C.; Dungul, D.C.; Ozdag, H.; Konu, O. Functionally conserved effects of rapamycin exposure on zebrafish. Mol. Med. Rep. 2016, 13, 4421–4430. [Google Scholar] [CrossRef]
- Zhu, W.; Zhao, Z.; Cheng, B. The role of autophagy in skin pigmentation. Eur. J. Dermatol. 2020, 30, 655–662. [Google Scholar] [CrossRef]
- Hamada, H.; Watanabe, M.; Lau, H.E.; Nishida, T.; Hasegawa, T.; Parichy, D.M.; Kondo, S. Involvement of Delta/Notch signaling in zebrafish adult pigment stripe patterning. Development 2014, 141, 318–324. [Google Scholar] [CrossRef]
- Quevedo, W.C. Normal pigmentation: Histology, cellular biology, and biochemistry. Ala. J. Med Sci. 1979, 16, 305–313. [Google Scholar]
- McNiven, M.A.; Wang, M.; Porter, K.R. Microtubule polarity and the direction of pigment transport reverse simultaneously in surgically severed melanophore arms. Cell 1984, 37, 753–765. [Google Scholar] [CrossRef]
- Rawls, J.F.; Mellgren, E.M.; Johnson, S.L. How the Zebrafish Gets Its Stripes. Dev. Biol. 2001, 240, 301–314. [Google Scholar] [CrossRef]
- Daniele, T.; Hurbain, I.; Vago, R.; Casari, G.; Raposo, G.; Tacchetti, C.; Schiaffino, M.V. Mitochondria and Melanosomes Establish Physical Contacts Modulated by Mfn2 and Involved in Organelle Biogenesis. Curr. Biol. 2014, 24, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Sundvik, M.; Chen, Y.-C.; Panula, P. Presenilin1 Regulates Histamine Neuron Development and Behavior in Zebrafish, Danio rerio. J. Neurosci. 2013, 33, 1589–1597. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barazzuol, L.; Cieri, D.; Facchinello, N.; Calì, T.; Washbourne, P.; Argenton, F.; Pizzo, P. Unraveling Presenilin 2 Functions in a Knockout Zebrafish Line to Shed Light into Alzheimer’s Disease Pathogenesis. Cells 2023, 12, 376. https://doi.org/10.3390/cells12030376
Barazzuol L, Cieri D, Facchinello N, Calì T, Washbourne P, Argenton F, Pizzo P. Unraveling Presenilin 2 Functions in a Knockout Zebrafish Line to Shed Light into Alzheimer’s Disease Pathogenesis. Cells. 2023; 12(3):376. https://doi.org/10.3390/cells12030376
Chicago/Turabian StyleBarazzuol, Lucia, Domenico Cieri, Nicola Facchinello, Tito Calì, Philip Washbourne, Francesco Argenton, and Paola Pizzo. 2023. "Unraveling Presenilin 2 Functions in a Knockout Zebrafish Line to Shed Light into Alzheimer’s Disease Pathogenesis" Cells 12, no. 3: 376. https://doi.org/10.3390/cells12030376
APA StyleBarazzuol, L., Cieri, D., Facchinello, N., Calì, T., Washbourne, P., Argenton, F., & Pizzo, P. (2023). Unraveling Presenilin 2 Functions in a Knockout Zebrafish Line to Shed Light into Alzheimer’s Disease Pathogenesis. Cells, 12(3), 376. https://doi.org/10.3390/cells12030376









