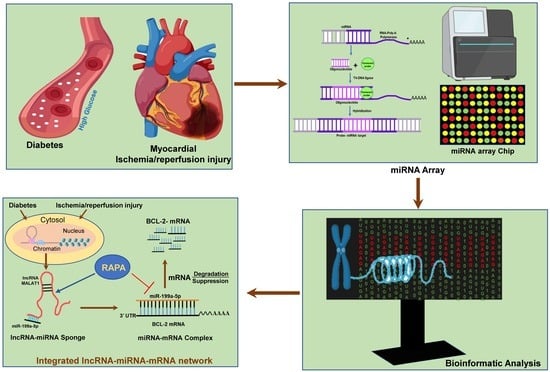
Integrated Analysis of lncRNA–miRNA–mRNA Regulatory Network in Rapamycin-Induced Cardioprotection against Ischemia/Reperfusion Injury in Diabetic Rabbits
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Care and Ethic Statement
2.2. Rabbit Model of Type I Diabetes
2.3. Conscious Ischemia/Reperfusion Injury
2.4. RNA Isolation and miRNA Array
2.5. miRNA Normalization and Differential Expression Analysis
2.6. Heat Map and Volcano Plot
2.7. miRNA–mRNA Target Prediction
2.8. LncRNA–miRNA Target Prediction
2.9. GO and KEGG miRNA Enrichment Analysis of DE miRNAs
2.10. Reverse Transcription and Real-Time PCR
2.11. Protein Expression–Western Blot
2.12. Data Availability
2.13. Statistical Analysi
3. Results
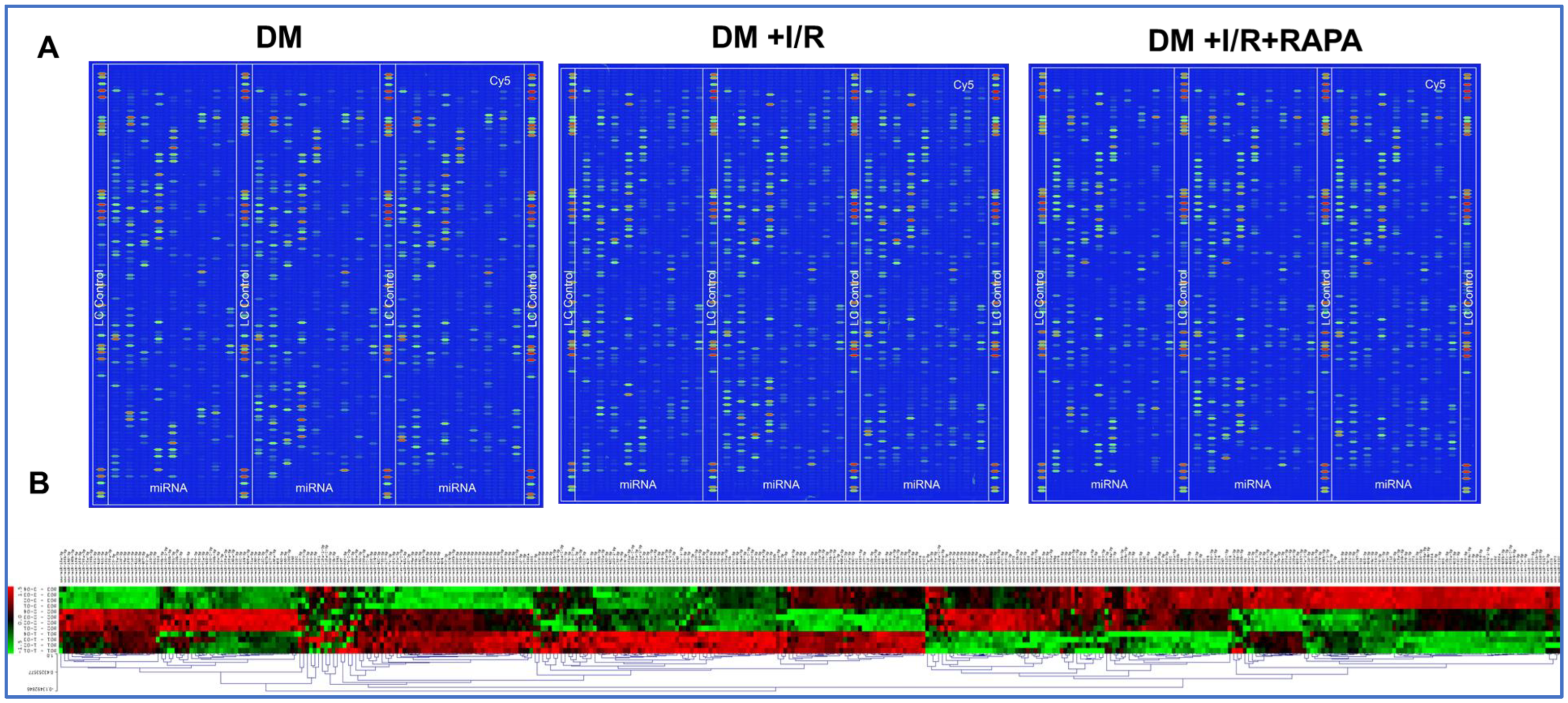
3.1. miRNA Array Analysis
3.2. Heat Map and Volcano Plot
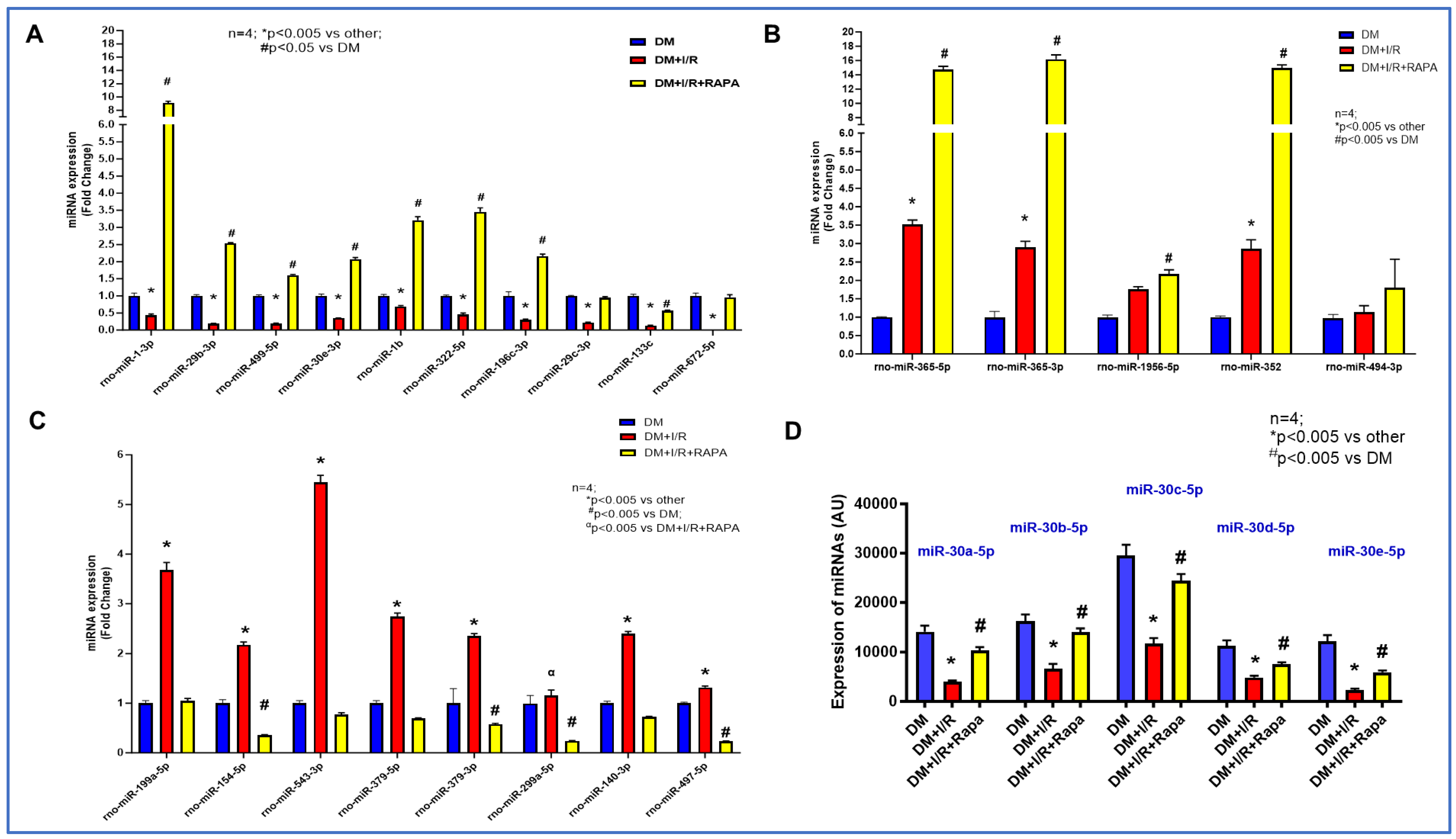
3.3. Rapamycin Alters the Expression of miRNAs
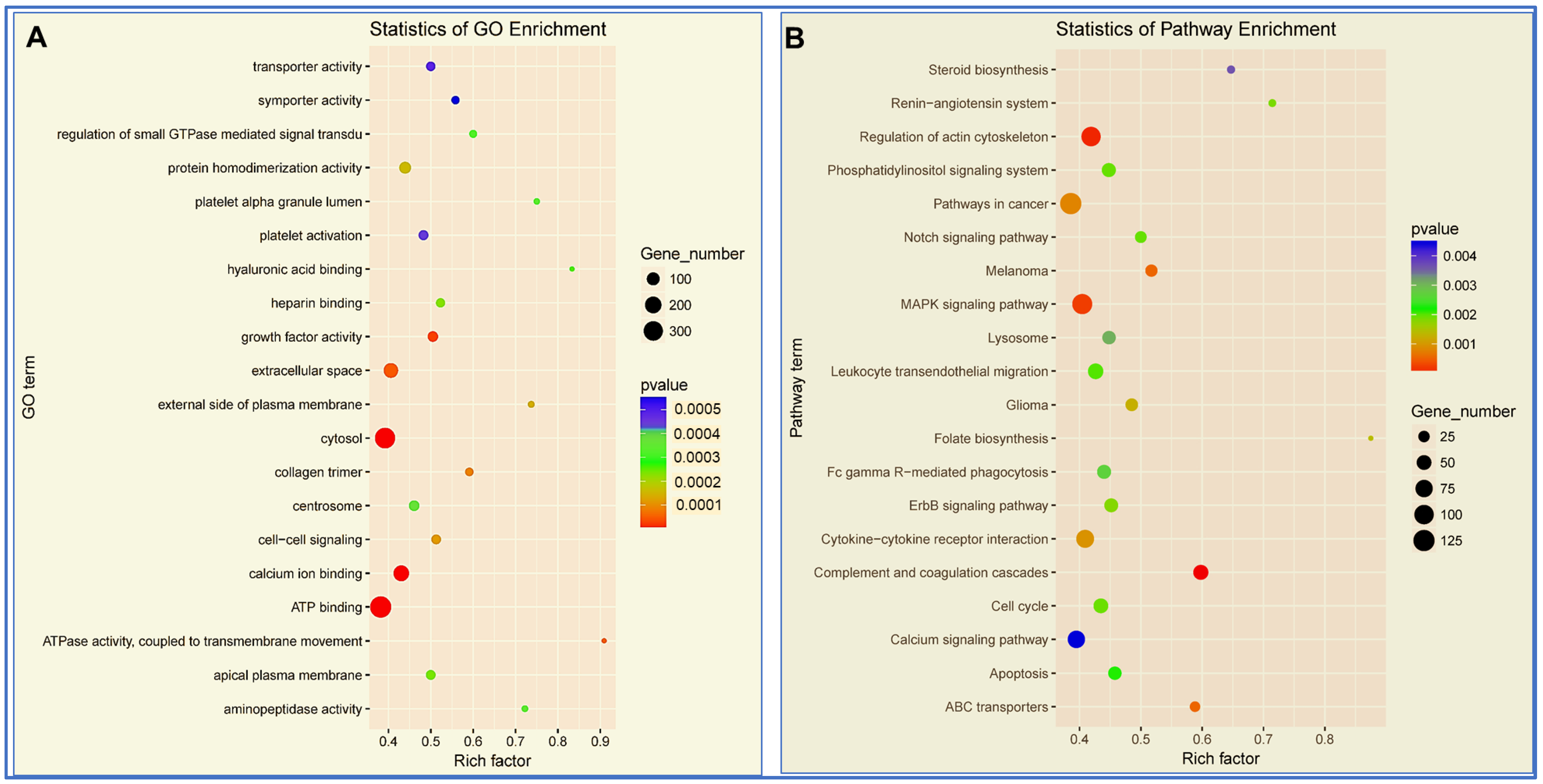
3.4. Enrichment Analysis of miRNAs
3.5. Target Prediction of miRNAs
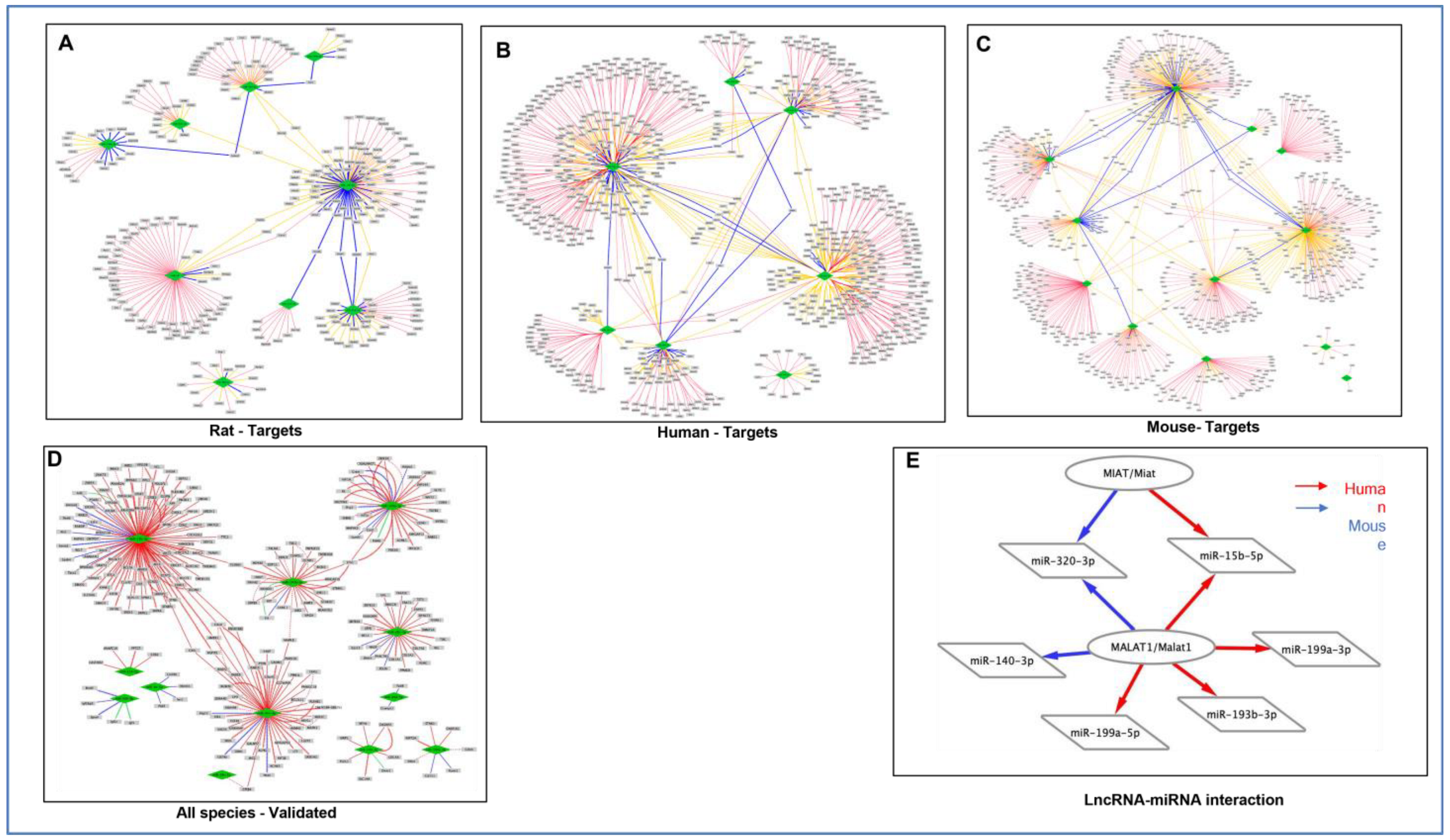
3.6. Analysis of lncRNA–miRNA Network
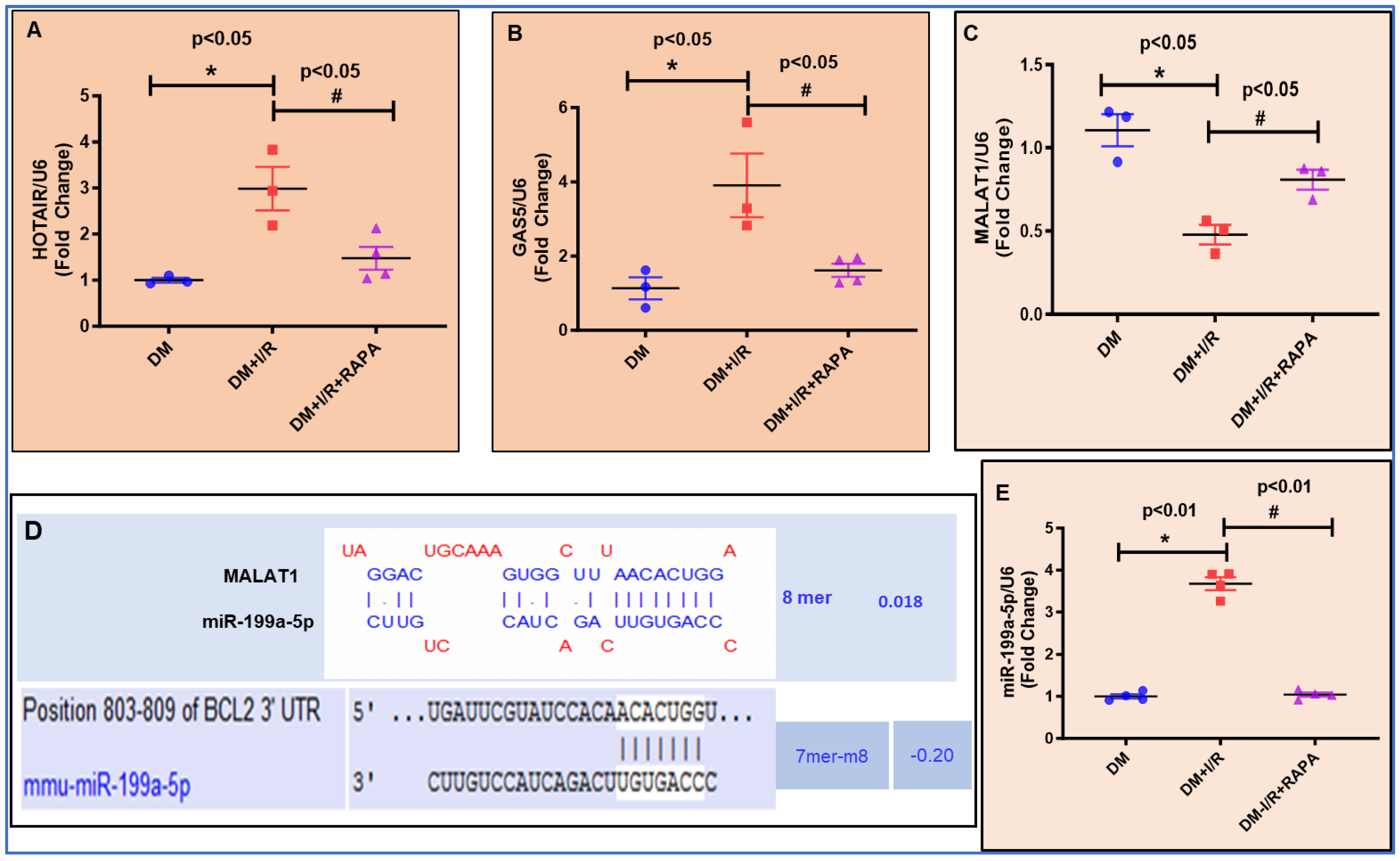
3.7. Expression of lncRNAs and miRNA by RT-QPCR
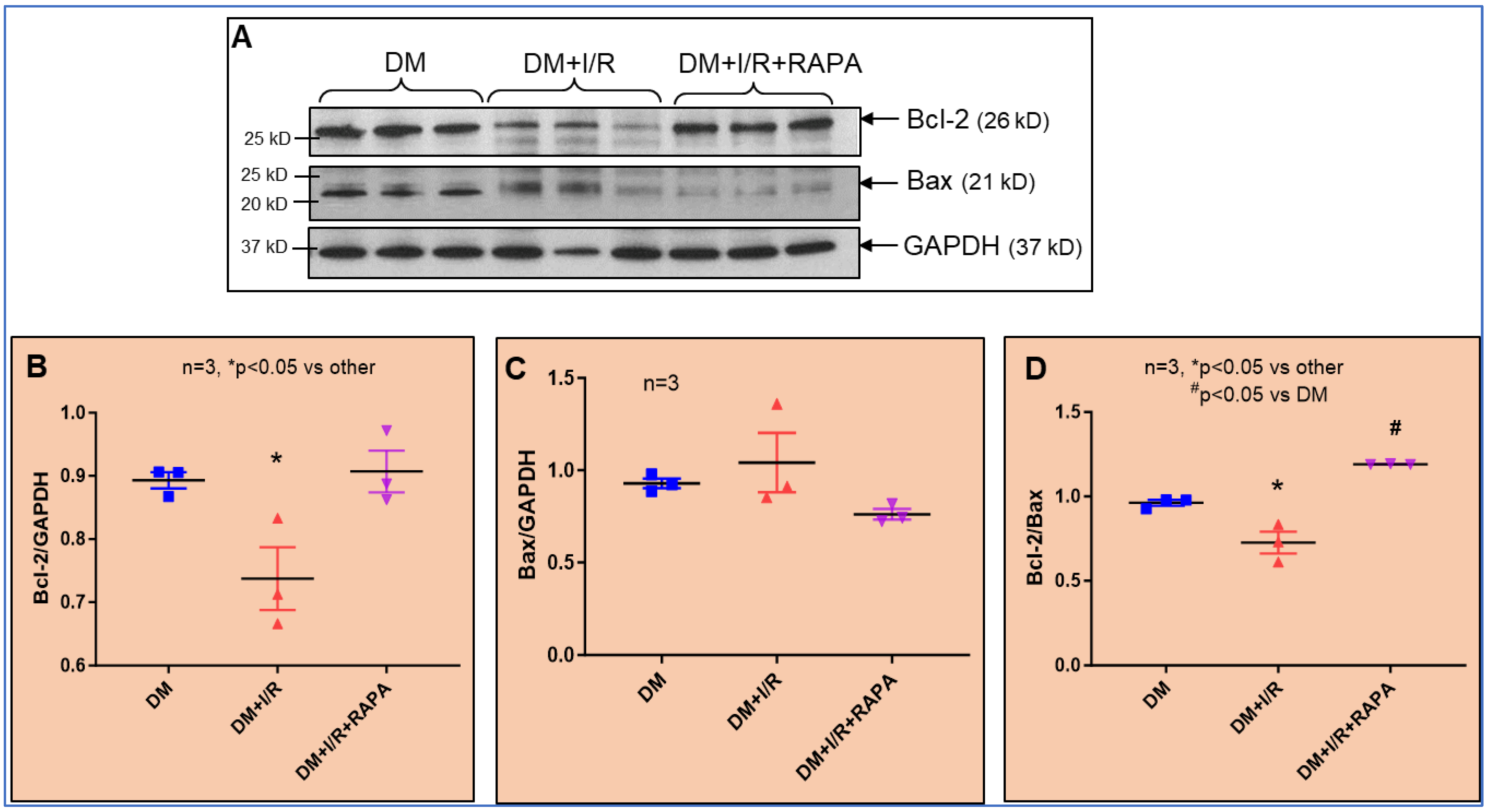
3.8. Protein Expression of Bcl2
4. Discussion
5. Limitation of the Study
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ohkuma, T.; Komorita, Y.; Peters, S.A.E.; Woodward, M. Diabetes as a risk factor for heart failure in women and men: A systematic review and meta-analysis of 47 cohorts including 12 million individuals. Diabetologia 2019, 62, 1550–1560. [Google Scholar] [CrossRef] [PubMed]
- McAllister, D.A.; Read, S.H.; Kerssens, J.; Livingstone, S.; McGurnaghan, S.; Jhund, P.; Petrie, J.; Sattar, N.; Fischbacher, C.; Kristensen, S.L.; et al. Incidence of Hospitalization for Heart Failure and Case-Fatality Among 3.25 Million People With and Without Diabetes Mellitus. Circulation 2018, 138, 2774–2786. [Google Scholar] [CrossRef] [PubMed]
- Margolis, J.R.; Kannel, W.S.; Feinleib, M.; Dawber, T.R.; McNamara, P.M. Clinical features of unrecognized myocardial infarction—silent and symptomatic. Eighteen year follow-up: The Framingham study. Am. J. Cardiol. 1973, 32, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Rawshani, A.; Rawshani, A.; Franzen, S.; Sattar, N.; Eliasson, B.; Svensson, A.M.; Zethelius, B.; Miftaraj, M.; McGuire, D.K.; Rosengren, A.; et al. Risk Factors, Mortality, and Cardiovascular Outcomes in Patients with Type 2 Diabetes. N. Engl. J. Med. 2018, 379, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Rawshani, A.; Rawshani, A.; Franzen, S.; Eliasson, B.; Svensson, A.M.; Miftaraj, M.; McGuire, D.K.; Sattar, N.; Rosengren, A.; Gudbjornsdottir, S. Mortality and Cardiovascular Disease in Type 1 and Type 2 Diabetes. N. Engl. J. Med. 2017, 376, 1407–1418. [Google Scholar] [CrossRef] [PubMed]
- Severino, P.; D’Amato, A.; Netti, L.; Pucci, M.; Infusino, F.; Maestrini, V.; Mancone, M.; Fedele, F. Myocardial Ischemia and Diabetes Mellitus: Role of Oxidative Stress in the Connection between Cardiac Metabolism and Coronary Blood Flow. J. Diabetes Res. 2019, 2019, 9489826. [Google Scholar] [CrossRef] [PubMed]
- Severino, P.; D’Amato, A.; Netti, L.; Pucci, M.; De Marchis, M.; Palmirotta, R.; Volterrani, M.; Mancone, M.; Fedele, F. Diabetes Mellitus and Ischemic Heart Disease: The Role of Ion Channels. Int. J. Mol. Sci. 2018, 19, 802. [Google Scholar] [CrossRef]
- Di Carli, M.F.; Janisse, J.; Grunberger, G.; Ager, J. Role of chronic hyperglycemia in the pathogenesis of coronary microvascular dysfunction in diabetes. J. Am. Coll. Cardiol. 2003, 41, 1387–1393. [Google Scholar] [CrossRef]
- Yahagi, K.; Kolodgie, F.D.; Lutter, C.; Mori, H.; Romero, M.E.; Finn, A.V.; Virmani, R. Pathology of Human Coronary and Carotid Artery Atherosclerosis and Vascular Calcification in Diabetes Mellitus. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 191–204. [Google Scholar] [CrossRef]
- Niakan, E.; Harati, Y.; Rolak, L.A.; Comstock, J.P.; Rokey, R. Silent myocardial infarction and diabetic cardiovascular autonomic neuropathy. Arch. Intern. Med. 1986, 146, 2229–2230. [Google Scholar] [CrossRef]
- Divers, J.; Mayer-Davis, E.J.; Lawrence, J.M.; Isom, S.; Dabelea, D.; Dolan, L.; Imperatore, G.; Marcovina, S.; Pettitt, D.J.; Pihoker, C.; et al. Trends in Incidence of Type 1 and Type 2 Diabetes Among Youths—Selected Counties and Indian Reservations, United States, 2002–2015. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Collaborators, G.B.D.D. Global, regional, and national burden of diabetes from 1990 to 2021, with projections of prevalence to 2050: A systematic analysis for the Global Burden of Disease Study 2021. Lancet 2023, 402, 203–234. [Google Scholar] [CrossRef]
- Menke, A.; Rust, K.F.; Fradkin, J.; Cheng, Y.J.; Cowie, C.C. Associations between trends in race/ethnicity, aging, and body mass index with diabetes prevalence in the United States: A series of cross-sectional studies. Ann. Intern. Med. 2014, 161, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Sheng, H.; Tan, Y.; Zhang, Q. Prevalence of diabetes in the USA from the perspective of demographic characteristics, physical indicators and living habits based on NHANES 2009–2018. Front. Endocrinol. 2023, 14, 1088882. [Google Scholar] [CrossRef]
- Cosentino, F.; Grant, P.J.; Aboyans, V.; Bailey, C.J.; Ceriello, A.; Delgado, V.; Federici, M.; Filippatos, G.; Grobbee, D.E.; Hansen, T.B.; et al. 2019 ESC Guidelines on diabetes, pre-diabetes, and cardiovascular diseases developed in collaboration with the EASD. Eur. Heart J. 2020, 41, 255–323. [Google Scholar] [CrossRef]
- Chen, H.Y.; Saczynski, J.S.; McManus, D.D.; Lessard, D.; Yarzebski, J.; Lapane, K.L.; Gore, J.M.; Goldberg, R.J. The impact of cardiac and noncardiac comorbidities on the short-term outcomes of patients hospitalized with acute myocardial infarction: A population-based perspective. Clin. Epidemiol. 2013, 5, 439–448. [Google Scholar] [CrossRef]
- Avogaro, A.; Bonora, E.; Consoli, A.; Del Prato, S.; Genovese, S.; Giorgino, F. Glucose-lowering therapy and cardiovascular outcomes in patients with type 2 diabetes mellitus and acute coronary syndrome. Diab Vasc. Dis. Res. 2019, 16, 399–414. [Google Scholar] [CrossRef]
- Ke, C.; Lipscombe, L.L.; Weisman, A.; Zhou, L.; Austin, P.C.; Shah, B.R.; Booth, G.L. Trends in the Association Between Diabetes and Cardiovascular Events, 1994–2019. JAMA 2022, 328, 1866–1869. [Google Scholar] [CrossRef]
- Orchard, T.J. Cardiovascular disease in type 1 diabetes: A continuing challenge. Lancet Diabetes Endocrinol. 2021, 9, 548–549. [Google Scholar] [CrossRef]
- Bloomgarden, Z.T. Cardiovascular disease in diabetes. Diabetes Care 2010, 33, e49–e54. [Google Scholar] [CrossRef][Green Version]
- Eliasson, L.; Esguerra, J.L.S. MicroRNA Networks in Pancreatic Islet Cells: Normal Function and Type 2 Diabetes. Diabetes 2020, 69, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.; Ronn, T. Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab. 2019, 29, 1028–1044. [Google Scholar] [CrossRef] [PubMed]
- Volkov, P.; Bacos, K.; Ofori, J.K.; Esguerra, J.L.; Eliasson, L.; Ronn, T.; Ling, C. Whole-Genome Bisulfite Sequencing of Human Pancreatic Islets Reveals Novel Differentially Methylated Regions in Type 2 Diabetes Pathogenesis. Diabetes 2017, 66, 1074–1085. [Google Scholar] [CrossRef] [PubMed]
- Hall, E.; Dekker Nitert, M.; Volkov, P.; Malmgren, S.; Mulder, H.; Bacos, K.; Ling, C. The effects of high glucose exposure on global gene expression and DNA methylation in human pancreatic islets. Mol. Cell Endocrinol. 2018, 472, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Hathaway, Q.A.; Pinti, M.V.; Durr, A.J.; Waris, S.; Shepherd, D.L.; Hollander, J.M. Regulating microRNA expression: At the heart of diabetes mellitus and the mitochondrion. Am. J. Physiol. Heart Circ. Physiol. 2018, 314, H293–H310. [Google Scholar] [CrossRef] [PubMed]
- Ordovas, J.M.; Smith, C.E. Epigenetics and cardiovascular disease. Nat. Rev. Cardiol. 2010, 7, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Dimmeler, S. Long noncoding RNAs in cardiovascular diseases. Circ. Res. 2015, 116, 737–750. [Google Scholar] [CrossRef]
- Colpaert, R.M.W.; Calore, M. Epigenetics and microRNAs in cardiovascular diseases. Genomics 2021, 113, 540–551. [Google Scholar] [CrossRef]
- Thum, T.; Mayr, M. Review focus on the role of microRNA in cardiovascular biology and disease. Cardiovasc. Res. 2012, 93, 543–544. [Google Scholar] [CrossRef]
- Anderson, K.M.; Anderson, D.M. LncRNAs at the heart of development and disease. Mamm. Genome 2022, 33, 354–365. [Google Scholar] [CrossRef]
- Das, A.; Samidurai, A.; Salloum, F.N. Deciphering Non-coding RNAs in Cardiovascular Health and Disease. Front. Cardiovasc. Med. 2018, 5, 73. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.M.; Giacca, M. Small non-coding RNA therapeutics for cardiovascular disease. Eur. Heart J. 2022, 43, 4548–4561. [Google Scholar] [CrossRef] [PubMed]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. microRNAs: Tiny regulators with great potential. Cell 2001, 107, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Lee, C.T.; Risom, T.; Strauss, W.M. Evolutionary conservation of microRNA regulatory circuits: An examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol. 2007, 26, 209–218. [Google Scholar] [CrossRef]
- Niwa, R.; Slack, F.J. The evolution of animal microRNA function. Curr. Opin. Genet. Dev. 2007, 17, 145–150. [Google Scholar] [CrossRef]
- Mao, X.; Li, L.; Cao, Y. Evolutionary comparisons of miRNA regulation system in six model organisms. Genetica 2014, 142, 109–118. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Elzo, M.A.; Gan, M.; Tang, T.; Shao, J.; Lai, T.; Ma, Y.; Jia, X.; Lai, S. Multi-Omics Analysis of Key microRNA-mRNA Metabolic Regulatory Networks in Skeletal Muscle of Obese Rabbits. Int. J. Mol. Sci. 2021, 22, 4204. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Brennecke, J.; Stark, A.; Russell, R.B.; Cohen, S.M. Principles of microRNA-target recognition. PLoS Biol. 2005, 3, e85. [Google Scholar] [CrossRef] [PubMed]
- Kehl, T.; Backes, C.; Kern, F.; Fehlmann, T.; Ludwig, N.; Meese, E.; Lenhof, H.P.; Keller, A. About miRNAs, miRNA seeds, target genes and target pathways. Oncotarget 2017, 8, 107167–107175. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Li, Y.; Zhao, Z.; Lu, J.; Chen, H.; Ding, N.; Wang, G.; Xu, J.; Li, X. Identifying and functionally characterizing tissue-specific and ubiquitously expressed human lncRNAs. Oncotarget 2016, 7, 7120–7133. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Thomson, J.M.; Wong, H.Y.; Hammond, S.M.; Hogan, B.L. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev. Biol. 2007, 310, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Liu, J.; Li, X.; Liu, X. LncRNA MIR17HG inhibits non-small cell lung cancer by upregulating miR-142-3p to downregulate Bach-1. BMC Pulm. Med. 2020, 20, 78. [Google Scholar] [CrossRef] [PubMed]
- Hube, F.; Francastel, C. Coding and Non-coding RNAs, the Frontier Has Never Been So Blurred. Front. Genet. 2018, 9, 140. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, C. Coding or Noncoding, the Converging Concepts of RNAs. Front. Genet. 2019, 10, 496. [Google Scholar] [CrossRef]
- Kageyama, Y.; Kondo, T.; Hashimoto, Y. Coding vs non-coding: Translatability of short ORFs found in putative non-coding transcripts. Biochimie 2011, 93, 1981–1986. [Google Scholar] [CrossRef]
- Leung, A.; Natarajan, R. Long Noncoding RNAs in Diabetes and Diabetic Complications. Antioxid. Redox Signal 2018, 29, 1064–1073. [Google Scholar] [CrossRef]
- Taylor, H.J.; Hung, Y.H.; Narisu, N.; Erdos, M.R.; Kanke, M.; Yan, T.; Grenko, C.M.; Swift, A.J.; Bonnycastle, L.L.; Sethupathy, P.; et al. Human pancreatic islet microRNAs implicated in diabetes and related traits by large-scale genetic analysis. Proc. Natl. Acad. Sci. USA 2023, 120, e2206797120. [Google Scholar] [CrossRef] [PubMed]
- Samidurai, A.; Ockaili, R.; Cain, C.; Roh, S.K.; Filippone, S.M.; Kraskauskas, D.; Kukreja, R.C.; Das, A. Differential Regulation of mTOR Complexes with miR-302a Attenuates Myocardial Reperfusion Injury in Diabetes. iScience 2020, 23, 101863. [Google Scholar] [CrossRef] [PubMed]
- Villa Vigil, M.A.; Alvarez Arenal, A.; Rodriguez Gonzalez, M.A. [Selection of materials and technic for single restorations in relation to different dental parameters]. Rev. Esp. Estomatol. 1987, 35, 329–336. Available online: https://www.ncbi.nlm.nih.gov/pubmed/3483496 (accessed on 12 September 2023). [PubMed]
- Xiao, H.; Zhang, M.; Wu, H.; Wu, J.; Hu, X.; Pei, X.; Li, D.; Zhao, L.; Hua, Q.; Meng, B.; et al. CIRKIL Exacerbates Cardiac Ischemia/Reperfusion Injury by Interacting With Ku70. Circ. Res. 2022, 130, e3–e17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, X.; Cai, B.; Li, Y.; Jiang, Y.; Fu, X.; Zhao, Y.; Gao, H.; Yang, Y.; Yang, J.; et al. The long noncoding RNA lncCIRBIL disrupts the nuclear translocation of Bclaf1 alleviating cardiac ischemia-reperfusion injury. Nat. Commun. 2021, 12, 522. [Google Scholar] [CrossRef] [PubMed]
- Latronico, M.V.; Catalucci, D.; Condorelli, G. Emerging role of microRNAs in cardiovascular biology. Circ. Res. 2007, 101, 1225–1236. [Google Scholar] [CrossRef] [PubMed]
- Samidurai, A.; Kukreja, R.C.; Das, A. Emerging Role of mTOR Signaling-Related miRNAs in Cardiovascular Diseases. Oxid. Med. Cell Longev. 2018, 2018, 6141902. [Google Scholar] [CrossRef]
- Das, S.; Shah, R.; Dimmeler, S.; Freedman, J.E.; Holley, C.; Lee, J.M.; Moore, K.; Musunuru, K.; Wang, D.Z.; Xiao, J.; et al. Noncoding RNAs in Cardiovascular Disease: Current Knowledge, Tools and Technologies for Investigation, and Future Directions: A Scientific Statement From the American Heart Association. Circ. Genom. Precis. Med. 2020, 13, e000062. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Hatzigeorgiou, A.G. Analyzing MiRNA-LncRNA Interactions. Methods Mol. Biol. 2016, 1402, 271–286. [Google Scholar] [CrossRef]
- Huang, Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J. Cell Mol. Med. 2018, 22, 5768–5775. [Google Scholar] [CrossRef]
- Wang, S.; Cheng, Z.; Chen, X.; Lu, G.; Zhu, X.; Xu, G. Long Noncoding RNA SNHG4 Attenuates the Injury of Myocardial Infarction via Regulating miR-148b-3p/DUSP1 Axis. Cardiovasc. Ther. 2022, 2022, 1652315. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-coding RNAs as regulators in epigenetics (Review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Deng, J.; Zhang, B.; He, X.; Meng, Z.; Li, G.; Ye, H.; Zheng, S.; Wei, L.; Deng, X.; et al. LncRNA HOTAIR epigenetically suppresses miR-122 expression in hepatocellular carcinoma via DNA methylation. EBioMedicine 2018, 36, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Durrant, D.; Koka, S.; Salloum, F.N.; Xi, L.; Kukreja, R.C. Mammalian target of rapamycin (mTOR) inhibition with rapamycin improves cardiac function in type 2 diabetic mice: Potential role of attenuated oxidative stress and altered contractile protein expression. J. Biol. Chem. 2014, 289, 4145–4160. [Google Scholar] [CrossRef] [PubMed]
- Suhara, T.; Baba, Y.; Shimada, B.K.; Higa, J.K.; Matsui, T. The mTOR Signaling Pathway in Myocardial Dysfunction in Type 2 Diabetes Mellitus. Curr. Diab Rep. 2017, 17, 38. [Google Scholar] [CrossRef]
- Kanamori, H.; Takemura, G.; Goto, K.; Tsujimoto, A.; Mikami, A.; Ogino, A.; Watanabe, T.; Morishita, K.; Okada, H.; Kawasaki, M.; et al. Autophagic adaptations in diabetic cardiomyopathy differ between type 1 and type 2 diabetes. Autophagy 2015, 11, 1146–1160. [Google Scholar] [CrossRef]
- Khan, S.; Salloum, F.; Das, A.; Xi, L.; Vetrovec, G.W.; Kukreja, R.C. Rapamycin confers preconditioning-like protection against ischemia-reperfusion injury in isolated mouse heart and cardiomyocytes. J. Mol. Cell Cardiol. 2006, 41, 256–264. [Google Scholar] [CrossRef]
- Das, A.; Salloum, F.N.; Durrant, D.; Ockaili, R.; Kukreja, R.C. Rapamycin protects against myocardial ischemia-reperfusion injury through JAK2-STAT3 signaling pathway. J. Mol. Cell Cardiol. 2012, 53, 858–869. [Google Scholar] [CrossRef]
- Samidurai, A.; Salloum, F.N.; Durrant, D.; Chernova, O.B.; Kukreja, R.C.; Das, A. Chronic treatment with novel nanoformulated micelles of rapamycin, Rapatar, protects diabetic heart against ischaemia/reperfusion injury. Br. J. Pharmacol. 2017, 174, 4771–4784. [Google Scholar] [CrossRef]
- Samidurai, A.; Saravanan, M.; Ockaili, R.; Kraskauskas, D.; Lau, S.Y.V.; Kodali, V.; Ramasamy, S.; Bhoopathi, K.; Nair, M.; Roh, S.K.; et al. Single-Dose Treatment with Rapamycin Preserves Post-Ischemic Cardiac Function through Attenuation of Fibrosis and Inflammation in Diabetic Rabbit. Int. J. Mol. Sci. 2023, 24, 8998. [Google Scholar] [CrossRef]
- Das, A.; Salloum, F.N.; Filippone, S.M.; Durrant, D.E.; Rokosh, G.; Bolli, R.; Kukreja, R.C. Inhibition of mammalian target of rapamycin protects against reperfusion injury in diabetic heart through STAT3 signaling. Basic. Res. Cardiol. 2015, 110, 31. [Google Scholar] [CrossRef] [PubMed]
- Haniff, H.S.; Liu, X.; Tong, Y.; Meyer, S.M.; Knerr, L.; Lemurell, M.; Abegg, D.; Aikawa, H.; Adibekian, A.; Disney, M.D. A structure-specific small molecule inhibits a miRNA-200 family member precursor and reverses a type 2 diabetes phenotype. Cell Chem. Biol. 2022, 29, 300–311.e310. [Google Scholar] [CrossRef] [PubMed]
- Ning, S.; Zhang, S.; Guo, Z. MicroRNA-494 regulates high glucose-induced cardiomyocyte apoptosis and autophagy by PI3K/AKT/mTOR signalling pathway. ESC Heart Fail. 2023, 10, 1401–1411. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.C.; Xu, H.M.; Guo, G.L.; Zhang, T.; Shi, J.W.; Chang, C. Long Non-Coding RNA H19 Protects H9c2 Cells against Hypoxia-Induced Injury by Targeting MicroRNA-139. Cell Physiol. Biochem. 2017, 44, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Samidurai, A.; Ockaili, R.; Cain, C.; Roh, S.K.; Filippone, S.M.; Kraskauskas, D.; Kukreja, R.C.; Das, A. Preclinical model of type 1 diabetes and myocardial ischemia/reperfusion injury in conscious rabbits-demonstration of cardioprotection with rapamycin. STAR Protoc. 2021, 2, 100772. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.P.; Tang, X.L.; Guo, Y.; Steenbergen, C.; Lefer, D.J.; Kukreja, R.C.; Kong, M.; Li, Q.; Bhushan, S.; Zhu, X.; et al. The NHLBI-sponsored Consortium for preclinicAl assESsment of cARdioprotective therapies (CAESAR): A new paradigm for rigorous, accurate, and reproducible evaluation of putative infarct-sparing interventions in mice, rabbits, and pigs. Circ. Res. 2015, 116, 572–586. [Google Scholar] [CrossRef] [PubMed]
- Torrado, J.; Cain, C.; Mauro, A.G.; Romeo, F.; Ockaili, R.; Chau, V.Q.; Nestler, J.A.; Devarakonda, T.; Ghosh, S.; Das, A.; et al. Sacubitril/Valsartan Averts Adverse Post-Infarction Ventricular Remodeling and Preserves Systolic Function in Rabbits. J. Am. Coll. Cardiol. 2018, 72, 2342–2356. [Google Scholar] [CrossRef]
- Zhou, X.; Zhu, Q.; Eicken, C.; Sheng, N.; Zhang, X.; Yang, L.; Gao, X. MicroRNA profiling using microParaflo microfluidic array technology. Methods Mol. Biol. 2012, 822, 153–182. [Google Scholar] [CrossRef]
- Bolstad, B.M.; Irizarry, R.A.; Astrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef]
- Rice, J.D.; Taylor, J.M. Locally Weighted Score Estimation for Quantile Classification in Binary Regression Models. Stat. Biosci. 2016, 8, 333–350. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Muller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Sumida, Y.; Fujibayashi, A.; Fukaguchi, K.; Sanzen, N.; Nishiuchi, R.; Sekiguchi, K. The tetraspanin CD151 regulates cell morphology and intracellular signaling on laminin-511. FEBS J. 2008, 275, 3335–3351. [Google Scholar] [CrossRef] [PubMed]
- Sadej, R.; Grudowska, A.; Turczyk, L.; Kordek, R.; Romanska, H.M. CD151 in cancer progression and metastasis: A complex scenario. Lab. Invest. 2014, 94, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Sompel, K.; Elango, A.; Smith, A.J.; Tennis, M.A. Cancer chemoprevention through Frizzled receptors and EMT. Discov. Oncol. 2021, 12, 32. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Chen, Y.; Shou, T.; Hu, J.; Qing, C. miRNA-99b-5p targets FZD8 to inhibit non-small cell lung cancer proliferation, migration and invasion. Onco Targets Ther. 2019, 12, 2615–2621. [Google Scholar] [CrossRef]
- Gonzalez-Mariscal, I.; Martin-Montalvo, A.; Vazquez-Fonseca, L.; Pomares-Viciana, T.; Sanchez-Cuesta, A.; Fernandez-Ayala, D.J.; Navas, P.; Santos-Ocana, C. The mitochondrial phosphatase PPTC7 orchestrates mitochondrial metabolism regulating coenzyme Q(10) biosynthesis. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 1235–1248. [Google Scholar] [CrossRef]
- Mao, M.; Biery, M.C.; Kobayashi, S.V.; Ward, T.; Schimmack, G.; Burchard, J.; Schelter, J.M.; Dai, H.; He, Y.D.; Linsley, P.S. T lymphocyte activation gene identification by coregulated expression on DNA microarrays. Genomics 2004, 83, 989–999. [Google Scholar] [CrossRef]
- Nielsen, J.; Christiansen, J.; Lykke-Andersen, J.; Johnsen, A.H.; Wewer, U.M.; Nielsen, F.C. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol. Cell Biol. 1999, 19, 1262–1270. [Google Scholar] [CrossRef]
- Li, Y.; Hiroi, Y.; Liao, J.K. Notch signaling as an important mediator of cardiac repair and regeneration after myocardial infarction. Trends Cardiovasc. Med. 2010, 20, 228–231. [Google Scholar] [CrossRef]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermuller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef] [PubMed]
- Brockdorff, N.; Ashworth, A.; Kay, G.F.; McCabe, V.M.; Norris, D.P.; Cooper, P.J.; Swift, S.; Rastan, S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 1992, 71, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Hung, T.; Chang, H.Y. Long noncoding RNA in genome regulation: Prospects and mechanisms. RNA Biol. 2010, 7, 582–585. [Google Scholar] [CrossRef]
- Mattick, J.S. Non-coding RNAs: The architects of eukaryotic complexity. EMBO Rep. 2001, 2, 986–991. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Schmitz, K.M.; Mayer, C.; Postepska, A.; Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes. Dev. 2010, 24, 2264–2269. [Google Scholar] [CrossRef]
- Li, Y.; Syed, J.; Sugiyama, H. RNA-DNA Triplex Formation by Long Noncoding RNAs. Cell Chem. Biol. 2016, 23, 1325–1333. [Google Scholar] [CrossRef]
- Xiang, J.F.; Yin, Q.F.; Chen, T.; Zhang, Y.; Zhang, X.O.; Wu, Z.; Zhang, S.; Wang, H.B.; Ge, J.; Lu, X.; et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014, 24, 513–531. [Google Scholar] [CrossRef]
- Bell, J.C.; Jukam, D.; Teran, N.A.; Risca, V.I.; Smith, O.K.; Johnson, W.L.; Skotheim, J.M.; Greenleaf, W.J.; Straight, A.F. Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. eLife 2018, 7, e27024. [Google Scholar] [CrossRef]
- Jakubik, D.; Fitas, A.; Eyileten, C.; Jarosz-Popek, J.; Nowak, A.; Czajka, P.; Wicik, Z.; Sourij, H.; Siller-Matula, J.M.; De Rosa, S.; et al. MicroRNAs and long non-coding RNAs in the pathophysiological processes of diabetic cardiomyopathy: Emerging biomarkers and potential therapeutics. Cardiovasc. Diabetol. 2021, 20, 55. [Google Scholar] [CrossRef]
- Santulli, G.; Totary-Jain, H. Tailoring mTOR-based therapy: Molecular evidence and clinical challenges. Pharmacogenomics 2013, 14, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Taheri, M.; Eghtedarian, R.; Ghafouri-Fard, S.; Omrani, M.D. Non-coding RNAs and type 2 diabetes mellitus. Arch. Physiol. Biochem. 2023, 129, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Kokkinopoulou, I.; Maratou, E.; Mitrou, P.; Boutati, E.; Sideris, D.C.; Fragoulis, E.G.; Christodoulou, M.I. Decreased expression of microRNAs targeting type-2 diabetes susceptibility genes in peripheral blood of patients and predisposed individuals. Endocrine 2019, 66, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Karakikes, I.; Chaanine, A.H.; Kang, S.; Mukete, B.N.; Jeong, D.; Zhang, S.; Hajjar, R.J.; Lebeche, D. Therapeutic cardiac-targeted delivery of miR-1 reverses pressure overload-induced cardiac hypertrophy and attenuates pathological remodeling. J. Am. Heart Assoc. 2013, 2, e000078. [Google Scholar] [CrossRef]
- Xue, Y.; Fan, X.; Yang, R.; Jiao, Y.; Li, Y. miR-29b-3p inhibits post-infarct cardiac fibrosis by targeting FOS. Biosci. Rep. 2020, 40, BSR20201227. [Google Scholar] [CrossRef]
- Schellinger, I.N.; Wagenhauser, M.; Chodisetti, G.; Mattern, K.; Dannert, A.; Petzold, A.; Jakubizka-Smorag, J.; Emrich, F.; Haunschild, J.; Schuster, A.; et al. MicroRNA miR-29b regulates diabetic aortic remodeling and stiffening. Mol. Ther. Nucleic Acids 2021, 24, 188–199. [Google Scholar] [CrossRef]
- Cai, Y.; Li, Y. Upregulation of miR-29b-3p protects cardiomyocytes from hypoxia-induced apoptosis by targeting TRAF5. Cell Mol. Biol. Lett. 2019, 24, 27. [Google Scholar] [CrossRef]
- Dalgaard, L.T.; Sorensen, A.E.; Hardikar, A.A.; Joglekar, M.V. The microRNA-29 family: Role in metabolism and metabolic disease. Am. J. Physiol. Cell Physiol. 2022, 323, C367–C377. [Google Scholar] [CrossRef]
- van Rooij, E.; Sutherland, L.B.; Thatcher, J.E.; DiMaio, J.M.; Naseem, R.H.; Marshall, W.S.; Hill, J.A.; Olson, E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13027–13032. [Google Scholar] [CrossRef]
- Caravia, X.M.; Fanjul, V.; Oliver, E.; Roiz-Valle, D.; Moran-Alvarez, A.; Desdin-Mico, G.; Mittelbrunn, M.; Cabo, R.; Vega, J.A.; Rodriguez, F.; et al. The microRNA-29/PGC1alpha regulatory axis is critical for metabolic control of cardiac function. PLoS Biol. 2018, 16, e2006247. [Google Scholar] [CrossRef] [PubMed]
- Jensen, D.M.; Han, P.; Mangala, L.S.; Lopez-Berestein, G.; Sood, A.K.; Liu, J.; Kriegel, A.J.; Usa, K.; Widlansky, M.E.; Liang, M. Broad-acting therapeutic effects of miR-29b-chitosan on hypertension and diabetic complications. Mol. Ther. 2022, 30, 3462–3476. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lu, J.; Bao, X.; Wang, X.; Wu, J.; Li, X.; Hong, W. MiR-499-5p protects cardiomyocytes against ischaemic injury via anti-apoptosis by targeting PDCD4. Oncotarget 2016, 7, 35607–35617. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Han, Y.; Niu, L.; Li, J.; Chen, Y. MiR-499 inhibited hypoxia/reoxygenation induced cardiomyocytes injury by targeting SOX6. Biotechnol. Lett. 2019, 41, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jia, Z.; Zhang, C.; Sun, M.; Wang, W.; Chen, P.; Ma, K.; Zhang, Y.; Li, X.; Zhou, C. miR-499 protects cardiomyocytes from H 2O 2-induced apoptosis via its effects on Pdcd4 and Pacs2. RNA Biol. 2014, 11, 339–350. [Google Scholar] [CrossRef]
- Wang, J.X.; Jiao, J.Q.; Li, Q.; Long, B.; Wang, K.; Liu, J.P.; Li, Y.R.; Li, P.F. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat. Med. 2011, 17, 71–78. [Google Scholar] [CrossRef]
- Zhu, J.; Yao, K.; Wang, Q.; Guo, J.; Shi, H.; Ma, L.; Liu, H.; Gao, W.; Zou, Y.; Ge, J. Ischemic Postconditioning-Regulated miR-499 Protects the Rat Heart Against Ischemia/Reperfusion Injury by Inhibiting Apoptosis through PDCD4. Cell Physiol. Biochem. 2016, 39, 2364–2380. [Google Scholar] [CrossRef]
- Yildirim, S.S.; Akman, D.; Catalucci, D.; Turan, B. Relationship between downregulation of miRNAs and increase of oxidative stress in the development of diabetic cardiac dysfunction: Junctin as a target protein of miR-1. Cell Biochem. Biophys. 2013, 67, 1397–1408. [Google Scholar] [CrossRef]
- Lv, H.; Tian, M.; Hu, P.; Wang, B.; Yang, L. Overexpression of miR-365a-3p relieves sepsis-induced acute myocardial injury by targeting MyD88/NF-kappaB pathway. Can. J. Physiol. Pharmacol. 2021, 99, 1007–1015. [Google Scholar] [CrossRef]
- Boada, C.; Zinger, A.; Tsao, C.; Zhao, P.; Martinez, J.O.; Hartman, K.; Naoi, T.; Sukhoveshin, R.; Sushnitha, M.; Molinaro, R.; et al. Rapamycin-Loaded Biomimetic Nanoparticles Reverse Vascular Inflammation. Circ. Res. 2020, 126, 25–37. [Google Scholar] [CrossRef]
- Ji, J.J.; Chen, S.Y.; Yang, Z.W.; Zhang, R.; Qian, L.L.; Jiang, Y.; Guo, J.Q.; Wu, Y.; Fan, Q.L.; Yao, Y.Y.; et al. Delivery of Mir-196c-3p with NIR-II light-triggered gel attenuates cardiomyocyte ferroptosis in cardiac ischemia-reperfusion injury. Nanomedicine 2023, 47, 102618. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Kong, L.; Li, J. MicroRNA-494-3p protects rat cardiomyocytes against septic shock via PTEN. Exp. Ther. Med. 2019, 17, 1706–1716. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, Y.; Yuan, J.; Gao, W.; Zhong, X.; Yao, K.; Lin, L.; Ge, J. Dendritic cell-derived exosomal miR-494-3p promotes angiogenesis following myocardial infarction. Int. J. Mol. Med. 2021, 47, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, N.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Isomi, M.; Nakashima, H.; Akiyama, M.; Wada, R.; Inagawa, K.; et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014, 33, 1565–1581. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.T.; Wu, X.D.; Lu, Y.X.; Sun, Y.H.; Zhu, H.H.; Liang, J.B.; He, W.K.; Zeng, Z.Y.; Li, L. Potential Involvement of MiR-30e-3p in Myocardial Injury Induced by Coronary Microembolization via Autophagy Activation. Cell Physiol. Biochem. 2017, 44, 1995–2004. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chang, H.; Zhang, H.; Zhang, L. MiR-30e Attenuates Isoproterenol-induced Cardiac Fibrosis Through Suppressing Snai1/TGF-beta Signaling. J. Cardiovasc. Pharmacol. 2017, 70, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Dai, R.; Ren, Y.; Lv, X.; Chang, C.; He, S.; Li, Q.; Yang, X.; Ren, L.; Wei, R.; Su, Q. MicroRNA-30e-3p reduces coronary microembolism-induced cardiomyocyte pyroptosis and inflammation by sequestering HDAC2 from the SMAD7 promoter. Am. J. Physiol. Cell Physiol. 2023, 324, C222–C235. [Google Scholar] [CrossRef]
- Chen, Z.; Su, X.; Shen, Y.; Jin, Y.; Luo, T.; Kim, I.M.; Weintraub, N.L.; Tang, Y. MiR322 mediates cardioprotection against ischemia/reperfusion injury via FBXW7/notch pathway. J. Mol. Cell Cardiol. 2019, 133, 67–74. [Google Scholar] [CrossRef]
- Bernardo, B.C.; Nguyen, S.S.; Gao, X.M.; Tham, Y.K.; Ooi, J.Y.; Patterson, N.L.; Kiriazis, H.; Su, Y.; Thomas, C.J.; Lin, R.C.; et al. Inhibition of miR-154 Protects Against Cardiac Dysfunction and Fibrosis in a Mouse Model of Pressure Overload. Sci. Rep. 2016, 6, 22442. [Google Scholar] [CrossRef]
- Xia, K.; Zhang, Y.; Sun, D. miR-217 and miR-543 downregulation mitigates inflammatory response and myocardial injury in children with viral myocarditis by regulating the SIRT1/AMPK/NF-kappaB signaling pathway. Int. J. Mol. Med. 2020, 45, 634–646. [Google Scholar] [CrossRef]
- Joshi, S.R.; Dhagia, V.; Gairhe, S.; Edwards, J.G.; McMurtry, I.F.; Gupte, S.A. MicroRNA-140 is elevated and mitofusin-1 is downregulated in the right ventricle of the Sugen5416/hypoxia/normoxia model of pulmonary arterial hypertension. Am. J. Physiol. Heart Circ. Physiol. 2016, 311, H689–H698. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.R.; Kunze, A.C.; Stenzig, J.; Eschenhagen, T.; Hirt, M.N. Blockade of miR-140-3p prevents functional deterioration in afterload-enhanced engineered heart tissue. Sci. Rep. 2019, 9, 11494. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhong, Z.; Wang, X.; Wang, Y.; Liu, Y.; Chang, Z. microRNA-497 inhibition mitigates myocardial infarction via enhancing wingless/integrated signal pathway in bone marrow mesenchymal stem cells. J. Cell Biochem. 2019, 120, 13403–13412. [Google Scholar] [CrossRef] [PubMed]
- Daneshgar, N.; Rabinovitch, P.S.; Dai, D.F. TOR Signaling Pathway in Cardiac Aging and Heart Failure. Biomolecules 2021, 11, 168. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Chen, Z.; Santulli, G.; Gu, L.; Yang, Z.G.; Yuan, Z.Q.; Zhao, Y.T.; Xin, H.B.; Deng, K.Y.; Wang, S.Q.; et al. Functional role of Calstabin2 in age-related cardiac alterations. Sci. Rep. 2014, 4, 7425. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Chen, W.; Yan, M.; Liu, J.; Luo, H.; Wang, C.; Yang, P. Rapamycin regulates the balance between cardiomyocyte apoptosis and autophagy in chronic heart failure by inhibiting mTOR signaling. Int. J. Mol. Med. 2020, 45, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.M.; O’Leary, M.N.; Zambataro, C.A.; Academia, E.C.; Presley, M.P.; Garrett, B.J.; Zykovich, A.; Mooney, S.D.; Strong, R.; Rosen, C.J.; et al. Late-life rapamycin treatment reverses age-related heart dysfunction. Aging Cell 2013, 12, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.F.; Karunadharma, P.P.; Chiao, Y.A.; Basisty, N.; Crispin, D.; Hsieh, E.J.; Chen, T.; Gu, H.; Djukovic, D.; Raftery, D.; et al. Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell 2014, 13, 529–539. [Google Scholar] [CrossRef]
- Li, Z.; Song, Y.; Liu, L.; Hou, N.; An, X.; Zhan, D.; Li, Y.; Zhou, L.; Li, P.; Yu, L.; et al. miR-199a impairs autophagy and induces cardiac hypertrophy through mTOR activation. Cell Death Differ. 2017, 24, 1205–1213. [Google Scholar] [CrossRef]
- Zhou, Y.; Jia, W.K.; Jian, Z.; Zhao, L.; Liu, C.C.; Wang, Y.; Xiao, Y.B. Downregulation of microRNA-199a-5p protects cardiomyocytes in cyanotic congenital heart disease by attenuating endoplasmic reticulum stress. Mol. Med. Rep. 2017, 16, 2992–3000. [Google Scholar] [CrossRef]
- Yan, M.; Yang, S.; Meng, F.; Zhao, Z.; Tian, Z.; Yang, P. MicroRNA 199a-5p induces apoptosis by targeting JunB. Sci. Rep. 2018, 8, 6699. [Google Scholar] [CrossRef]
- Sun, Y.; Ma, L. New Insights into Long Non-Coding RNA MALAT1 in Cancer and Metastasis. Cancers 2019, 11, 216. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wu, Z.; Wu, T.; Huang, Y.; Cheng, Z.; Li, X.; Sun, T.; Xie, X.; Zhou, Y.; Du, Z. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016, 7, e2123. [Google Scholar] [CrossRef] [PubMed]
- Kwok, Z.H.; Roche, V.; Chew, X.H.; Fadieieva, A.; Tay, Y. A non-canonical tumor suppressive role for the long non-coding RNA MALAT1 in colon and breast cancers. Int. J. Cancer 2018, 143, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sun, Y.; Zhong, L.; Xiao, Z.; Yang, M.; Chen, M.; Wang, C.; Xie, X.; Chen, X. The suppression of ox-LDL-induced inflammatory cytokine release and apoptosis of HCAECs by long non-coding RNA-MALAT1 via regulating microRNA-155/SOCS1 pathway. Nutr. Metab. Cardiovasc. Dis. 2018, 28, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Michalik, K.M.; You, X.; Manavski, Y.; Doddaballapur, A.; Zornig, M.; Braun, T.; John, D.; Ponomareva, Y.; Chen, W.; Uchida, S.; et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 2014, 114, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wu, X.; Han, Y.; Tian, E.; Cheng, J. LncRNA MALAT1 protects cardiomyocytes from isoproterenol-induced apoptosis through sponging miR-558 to enhance ULK1-mediated protective autophagy. J. Cell Physiol. 2019, 234, 10842–10854. [Google Scholar] [CrossRef]
- Bhan, A.; Mandal, S.S. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta 2015, 1856, 151–164. [Google Scholar] [CrossRef]
- Sorensen, K.P.; Thomassen, M.; Tan, Q.; Bak, M.; Cold, S.; Burton, M.; Larsen, M.J.; Kruse, T.A. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res. Treat. 2013, 142, 529–536. [Google Scholar] [CrossRef]
- Bhan, A.; Hussain, I.; Ansari, K.I.; Kasiri, S.; Bashyal, A.; Mandal, S.S. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J. Mol. Biol. 2013, 425, 3707–3722. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Chiyomaru, T.; Yamamura, S.; Fukuhara, S.; Yoshino, H.; Kinoshita, T.; Majid, S.; Saini, S.; Chang, I.; Tanaka, Y.; Enokida, H.; et al. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PLoS ONE 2013, 8, e70372. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Jutooru, I.; Chadalapaka, G.; Johnson, G.; Frank, J.; Burghardt, R.; Kim, S.; Safe, S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 2013, 32, 1616–1625. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Xu, S.T.; Ren, K. LncRNA HOTAIR promotes myocardial fibrosis by suppressing miR-124. Int. J. Cardiol. 2023, 374, 94. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Wang, K.; Yang, X.; Wang, K.; Wang, N.; Jiang, T.B. LncRNA HOTAIR promotes myocardial fibrosis in atrial fibrillation through binding with PTBP1 to increase the stability of Wnt5a. Int. J. Cardiol. 2022, 369, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.C.; Cui, H.H.; Qiu, C.G. HOTAIR promotes myocardial fibrosis through regulating URI1 expression via Wnt pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6983–6990. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hu, Z.Q. LncRNA HOTAIR aggravates myocardial ischemia-reperfusion injury by sponging microRNA-126 to upregulate SRSF1. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 9046–9054. [Google Scholar] [CrossRef]
- Ni, S.Y.; Xu, W.T.; Liao, G.Y.; Wang, Y.L.; Li, J. LncRNA HOTAIR Promotes LPS-Induced Inflammation and Apoptosis of Cardiomyocytes via Lin28-Mediated PDCD4 Stability. Inflammation 2021, 44, 1452–1463. [Google Scholar] [CrossRef]
- Meng, K.; Jiao, J.; Zhu, R.R.; Wang, B.Y.; Mao, X.B.; Zhong, Y.C.; Zhu, Z.F.; Yu, K.W.; Ding, Y.; Xu, W.B.; et al. The Long Noncoding RNA Hotair Regulates Oxidative Stress and Cardiac Myocyte Apoptosis during Ischemia-Reperfusion Injury. Oxid. Med. Cell Longev. 2020, 2020, 1645249. [Google Scholar] [CrossRef]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal 2010, 3, ra8. [Google Scholar] [CrossRef]
- Williams, G.T.; Mourtada-Maarabouni, M.; Farzaneh, F. A critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytes. Biochem. Soc. Trans. 2011, 39, 482–486. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.Y.; Zhan, H.L.; Li, M.K.; Wu, G.D.; Liu, Z.; Wu, L.F. Long noncoding RNA Gas5 induces cell apoptosis and inhibits tumor growth via activating the CHOP-dependent endoplasmic reticulum stress pathway in human hepatoblastoma HepG2 cells. J. Cell Biochem. 2022, 123, 231–247. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wu, N.; Xia, F.; Liu, S.; Jia, D. Long non-coding RNA GAS5 regulates myocardial ischemia-reperfusion injury through the PI3K/AKT apoptosis pathway by sponging miR-532-5p. Int. J. Mol. Med. 2020, 45, 858–872. [Google Scholar] [CrossRef] [PubMed]
- Jing, W.; Tuxiu, X.; Xiaobing, L.; Guijun, J.; Lulu, K.; Jie, J.; Lu, Y.; Liying, Z.; Xiaoxing, X.; Jingjun, L. LncRNA GAS5/miR-137 Is a Hypoxia-Responsive Axis Involved in Cardiac Arrest and Cardiopulmonary Cerebral Resuscitation. Front. Immunol. 2021, 12, 790750. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, Y.; Han, J.; Su, H. GAS5 knockdown ameliorates apoptosis and inflammatory response by modulating miR-26b-5p/Smad1 axis in cerebral ischaemia/reperfusion injury. Behav. Brain Res. 2020, 379, 112370. [Google Scholar] [CrossRef]
- The Gene Ontology, C. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Sequeira, V.; Nijenkamp, L.L.; Regan, J.A.; van der Velden, J. The physiological role of cardiac cytoskeleton and its alterations in heart failure. Biochim. Biophys. Acta 2014, 1838, 700–722. [Google Scholar] [CrossRef]
- Deswal, A.; Petersen, N.J.; Feldman, A.M.; Young, J.B.; White, B.G.; Mann, D.L. Cytokines and cytokine receptors in advanced heart failure: An analysis of the cytokine database from the Vesnarinone trial (VEST). Circulation 2001, 103, 2055–2059. [Google Scholar] [CrossRef]
- Ramos, G.; Frantz, S. Myocardial Metabolism Under Control of a Cytokine Receptor. J. Am. Heart Assoc. 2017, 6, e006291. [Google Scholar] [CrossRef]
- Shiraishi, M.; Yamaguchi, A.; Suzuki, K. Nrg1/ErbB signaling-mediated regulation of fibrosis after myocardial infarction. FASEB J. 2022, 36, e22150. [Google Scholar] [CrossRef] [PubMed]
- Muslin, A.J. MAPK signalling in cardiovascular health and disease: Molecular mechanisms and therapeutic targets. Clin. Sci. 2008, 115, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Aalikhani, M.; Alikhani, M.; Khajeniazi, S.; Khosravi, A.; Bazi, Z.; Kianmehr, A. Positive effect of miR-2392 on fibroblast to cardiomyocyte-like cell fate transition: An in silico and in vitro study. Gene 2023, 879, 147598. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yang, T.; Duan, J.; Mu, N.; Zhang, T. MALAT1/miR-15b-5p/MAPK1 mediates endothelial progenitor cells autophagy and affects coronary atherosclerotic heart disease via mTOR signaling pathway. Aging 2019, 11, 1089–1109. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; He, Y.; Qiong, A.; Zhang, W. Tanshinone IIA Regulates MAPK/mTOR Signal-Mediated Autophagy to Alleviate Atherosclerosis through the miR-214-3p/ATG16L1 Axis. Int. Heart J. 2023, 64, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008, 456, 980–984. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.F.; Ding, Y.J.; Zhang, Z.X.; Wang, Z.F.; Luo, C.L.; Li, B.X.; Shen, Y.W.; Tao, L.Y.; Zhao, Z.Q. MicroRNA-21 regulation of the progression of viral myocarditis to dilated cardiomyopathy. Mol. Med. Rep. 2014, 10, 161–168. [Google Scholar] [CrossRef]
- Song, X.; Cui, Y.; Zhu, T. MicroRNA-19 upregulation attenuates cardiac fibrosis via targeting connective tissue growth factor. Am. J. Med. Sci. 2023, 365, 375–385. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, X.; Hu, G.; Li, C.; Guo, L.; Zhang, L.; Sun, F.; Xia, Y.; Yan, W.; Cui, Z.; et al. Small Extracellular Vesicles From Brown Adipose Tissue Mediate Exercise Cardioprotection. Circ. Res. 2022, 130, 1490–1506. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samidurai, A.; Olex, A.L.; Ockaili, R.; Kraskauskas, D.; Roh, S.K.; Kukreja, R.C.; Das, A. Integrated Analysis of lncRNA–miRNA–mRNA Regulatory Network in Rapamycin-Induced Cardioprotection against Ischemia/Reperfusion Injury in Diabetic Rabbits. Cells 2023, 12, 2820. https://doi.org/10.3390/cells12242820
Samidurai A, Olex AL, Ockaili R, Kraskauskas D, Roh SK, Kukreja RC, Das A. Integrated Analysis of lncRNA–miRNA–mRNA Regulatory Network in Rapamycin-Induced Cardioprotection against Ischemia/Reperfusion Injury in Diabetic Rabbits. Cells. 2023; 12(24):2820. https://doi.org/10.3390/cells12242820
Chicago/Turabian StyleSamidurai, Arun, Amy L. Olex, Ramzi Ockaili, Donatas Kraskauskas, Sean K. Roh, Rakesh C. Kukreja, and Anindita Das. 2023. "Integrated Analysis of lncRNA–miRNA–mRNA Regulatory Network in Rapamycin-Induced Cardioprotection against Ischemia/Reperfusion Injury in Diabetic Rabbits" Cells 12, no. 24: 2820. https://doi.org/10.3390/cells12242820
APA StyleSamidurai, A., Olex, A. L., Ockaili, R., Kraskauskas, D., Roh, S. K., Kukreja, R. C., & Das, A. (2023). Integrated Analysis of lncRNA–miRNA–mRNA Regulatory Network in Rapamycin-Induced Cardioprotection against Ischemia/Reperfusion Injury in Diabetic Rabbits. Cells, 12(24), 2820. https://doi.org/10.3390/cells12242820