The Identification of Nuclear FMRP Isoform Iso6 Partners
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines, Cell Culture, and Drug Treatment
2.2. Antibodies
2.3. Confocal Microscopy
2.4. GST-Pull Down and GFP-Trap Assays
2.5. Stable GFP Transfection
2.6. Affinity Purification and MS Experiments
2.7. Data-Dependent Acquisition MS
2.8. Protein Identification
2.9. Experimental Design and Statistical Rationale for MS Experiments
3. Results
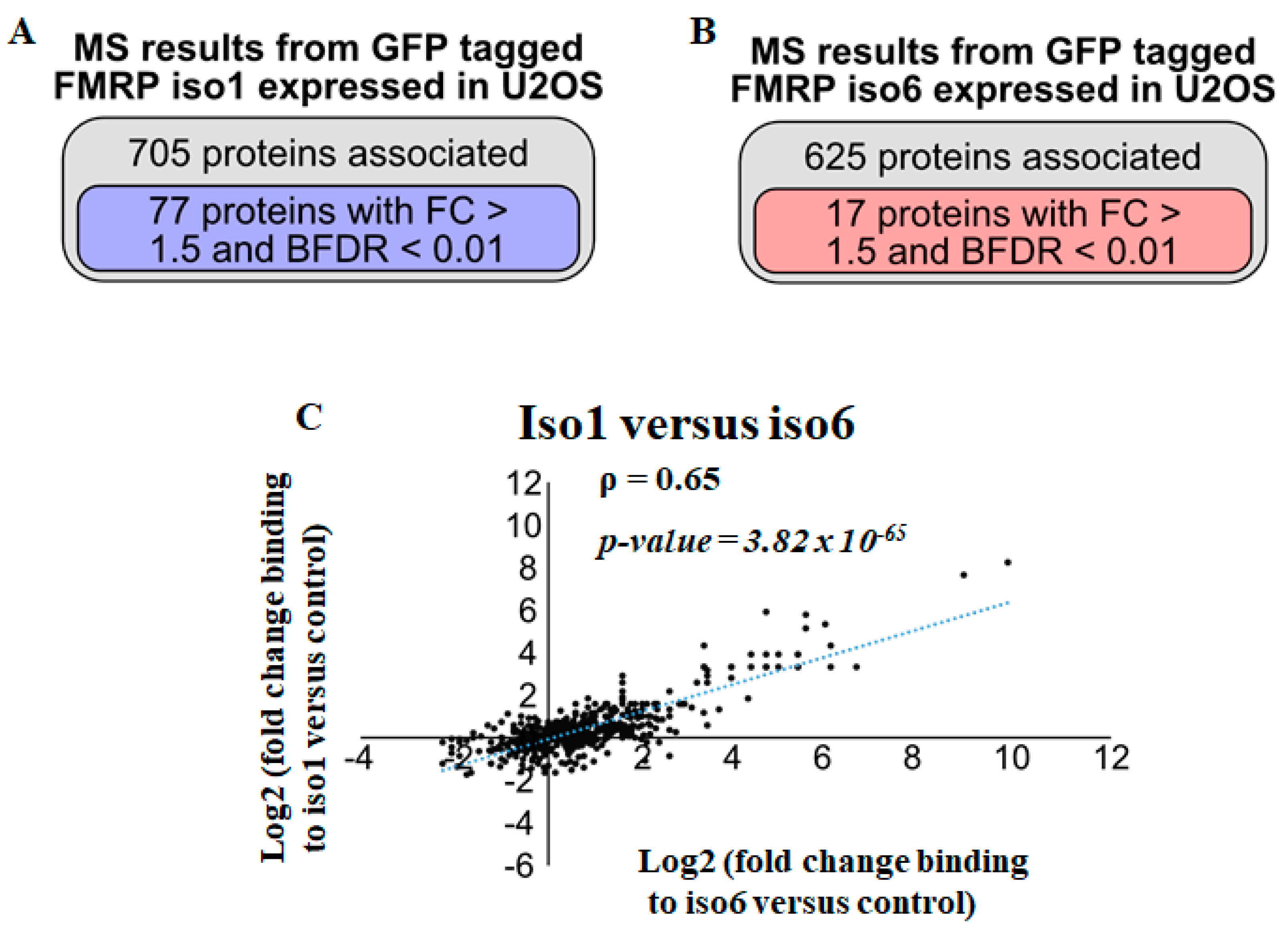
3.1. Identification of nFMRP Partners
3.2. Identification of Potential nFMRP-Specific Partners
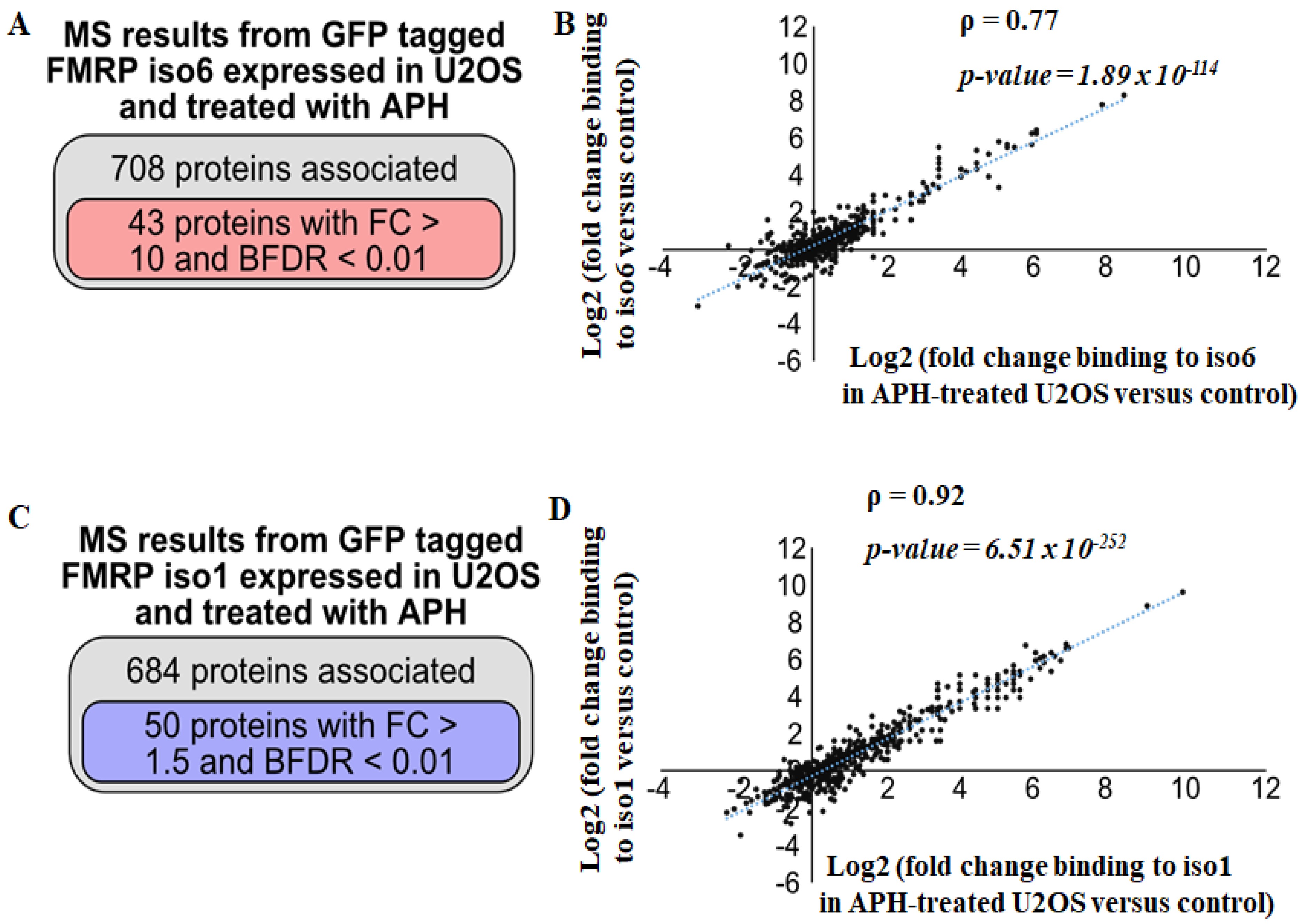
3.3. Regulation of nFMRP Interactions by Replicative Stress
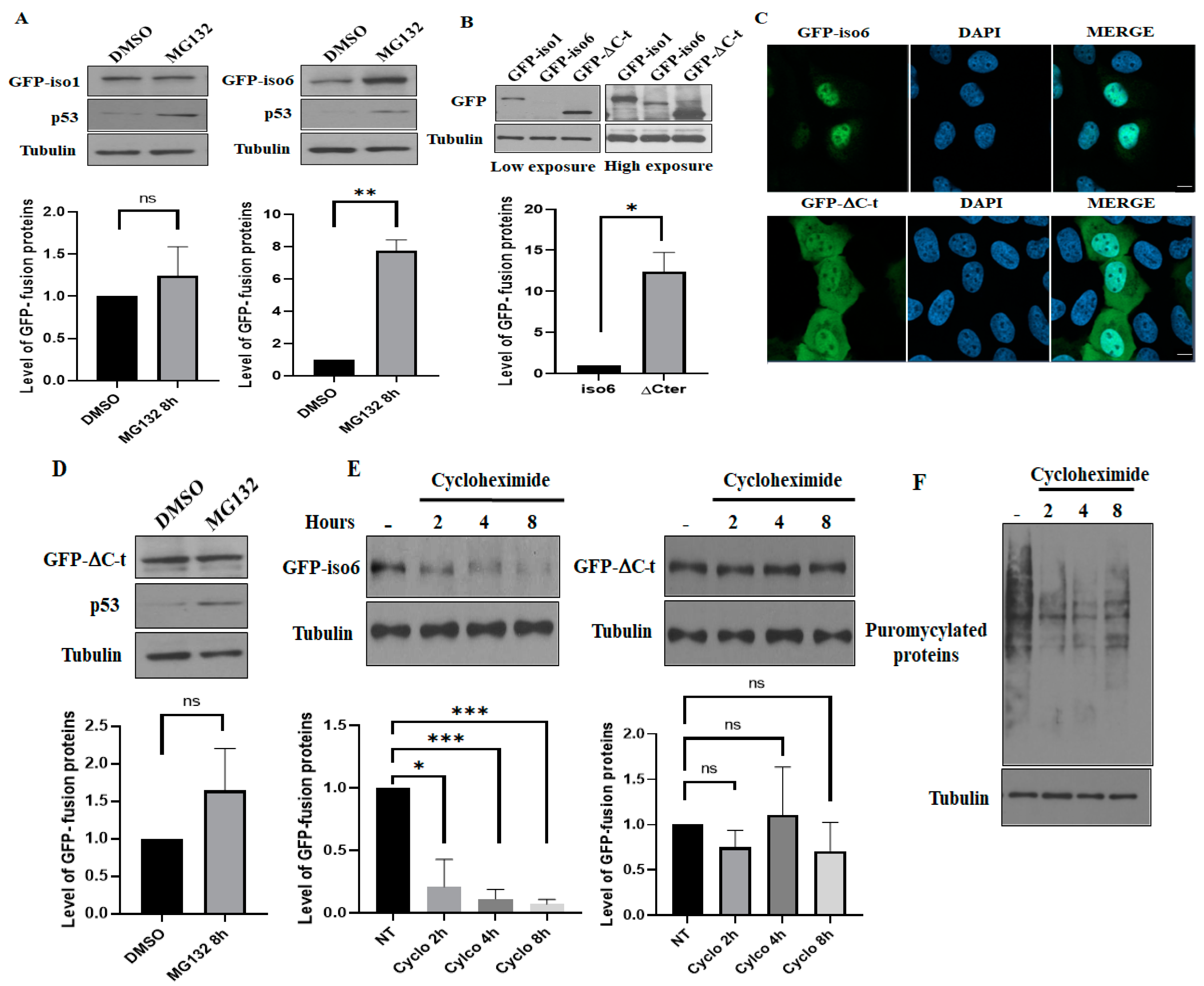
3.4. Iso6 Binds Proteasomal Proteins, While Its Expression Is Regulated by the Proteasome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Banerjee, A.; Ifrim, M.F.; Valdez, A.N.; Raj, N.; Bassell, G.J. Aberrant RNA translation in fragile X syndrome: From FMRP mechanisms to emerging therapeutic strategies. Brain Res. 2018, 1693, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Hagerman, R.J.; Berry-Kravis, E.; Hazlett, H.C.; Bailey, D.B., Jr.; Moine, H.; Kooy, R.F.; Tassone, F.; Gantois, I.; Sonenberg, N.; Mandel, J.L.; et al. Fragile X syndrome. Nat. Rev. Dis. Primers 2017, 3, 17065. [Google Scholar] [CrossRef] [PubMed]
- Dury, A.Y.; El Fatimy, R.; Tremblay, S.; Rose, T.M.; Cote, J.; De Koninck, P.; Khandjian, E.W. Nuclear Fragile X Mental Retardation Protein is localized to Cajal bodies. PLoS Genet. 2013, 9, e1003890. [Google Scholar] [CrossRef]
- Sittler, A.; Devys, D.; Weber, C.; Mandel, J.L. Alternative splicing of exon 14 determines nuclear or cytoplasmic localisation of fmr1 protein isoforms. Hum. Mol. Genet. 1996, 5, 95–102. [Google Scholar] [CrossRef]
- Valverde, R.; Pozdnyakova, I.; Kajander, T.; Venkatraman, J.; Regan, L. Fragile X mental retardation syndrome: Structure of the KH1-KH2 domains of fragile X mental retardation protein. Structure 2007, 15, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Myrick, L.K.; Hashimoto, H.; Cheng, X.; Warren, S.T. Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain. Hum. Mol. Genet. 2015, 24, 1733–1740. [Google Scholar] [CrossRef]
- Eberhart, D.E.; Warren, S.T. The molecular basis of fragile X syndrome. Cold Spring Harb. Symp. Quant. Biol. 1996, 61, 679–687. [Google Scholar]
- Siomi, H.; Siomi, M.C.; Nussbaum, R.L.; Dreyfuss, G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell 1993, 74, 291–298. [Google Scholar] [CrossRef]
- Darnell, J.C.; Jensen, K.B.; Jin, P.; Brown, V.; Warren, S.T.; Darnell, R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 2001, 107, 489–499. [Google Scholar] [CrossRef]
- Ramos, A.; Hollingworth, D.; Pastore, A. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA 2003, 9, 1198–1207. [Google Scholar] [CrossRef]
- Tsang, B.; Arsenault, J.; Vernon, R.M.; Lin, H.; Sonenberg, N.; Wang, L.Y.; Bah, A.; Forman-Kay, J.D. Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proc. Natl. Acad. Sci. USA 2019, 116, 4218–4227. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.C.; Van Driesche, S.J.; Zhang, C.; Hung, K.Y.; Mele, A.; Fraser, C.E.; Stone, E.F.; Chen, C.; Fak, J.J.; Chi, S.W.; et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011, 146, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.D.; Zhao, X. The molecular biology of FMRP: New insights into fragile X syndrome. Nat. Rev. Neurosci. 2021, 22, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Alpatov, R.; Lesch, B.J.; Nakamoto-Kinoshita, M.; Blanco, A.; Chen, S.; Stutzer, A.; Armache, K.J.; Simon, M.D.; Xu, C.; Ali, M.; et al. A chromatin-dependent role of the fragile X mental retardation protein FMRP in the DNA damage response. Cell 2014, 157, 869–881. [Google Scholar] [CrossRef]
- Chakraborty, A.; Jenjaroenpun, P.; Li, J.; El Hilali, S.; McCulley, A.; Haarer, B.; Hoffman, E.A.; Belak, A.; Thorland, A.; Hehnly, H.; et al. Replication Stress Induces Global Chromosome Breakage in the Fragile X Genome. Cell Rep. 2020, 32, 108179. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, Y.; Xiang, Y.; Yadav, T.; Ouyang, J.; Phoon, L.; Zhu, X.; Shi, Y.; Zou, L.; Lan, L. FMRP promotes transcription-coupled homologous recombination via facilitating TET1-mediated m5C RNA modification demethylation. Proc. Natl. Acad. Sci. USA 2022, 119, e2116251119. [Google Scholar] [CrossRef]
- Bardoni, B.; Schenck, A.; Mandel, J.L. A novel RNA-binding nuclear protein that interacts with the fragile X mental retardation (FMR1) protein. Hum. Mol. Genet. 1999, 8, 2557–2566. [Google Scholar] [CrossRef]
- Ledoux, N.; Gauthier-Naud, W.; Lavoie, O.; Watters, V.; Hussein, S.; Adjibade, P.; Mazroui, R. The nuclear isoforms of the Fragile X mental retardation RNA-binding protein associate with genomic DNA bridges. Mol. Biol. Cell 2023, 34, ar36. [Google Scholar] [CrossRef]
- Taha, M.S.; Nouri, K.; Milroy, L.G.; Moll, J.M.; Herrmann, C.; Brunsveld, L.; Piekorz, R.P.; Ahmadian, M.R. Subcellular fractionation and localization studies reveal a direct interaction of the fragile X mental retardation protein (FMRP) with nucleolin. PLoS ONE 2014, 9, e91465. [Google Scholar] [CrossRef]
- Loehr, J.; Kougnassoukou Tchara, P.E.; Gonthier, K.; Noufi, C.; Linteau, N.; Audet-Walsh, E.; Lambert, J.P. A Nutrient-Based Cellular Model to Characterize Acetylation-Dependent Protein-Protein Interactions. Front. Mol. Biosci. 2022, 9, 831758. [Google Scholar] [CrossRef]
- Liu, G.; Knight, J.D.; Zhang, J.P.; Tsou, C.C.; Wang, J.; Lambert, J.P.; Larsen, B.; Tyers, M.; Raught, B.; Bandeira, N.; et al. Data Independent Acquisition analysis in ProHits 4.0. J. Proteomics 2016, 149, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Mendoza, L.; Shteynberg, D.; Slagel, J.; Sun, Z.; Moritz, R.L. Trans-Proteomic Pipeline, a standardized data processing pipeline for large-scale reproducible proteomics informatics. Proteomics Clin. Appl. 2015, 9, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Shteynberg, D.; Deutsch, E.W.; Lam, H.; Eng, J.K.; Sun, Z.; Tasman, N.; Mendoza, L.; Moritz, R.L.; Aebersold, R.; Nesvizhskii, A.I. iProphet: Multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell Proteomics 2011, 10, M111.007690. [Google Scholar] [CrossRef] [PubMed]
- Teo, G.; Liu, G.; Zhang, J.; Nesvizhskii, A.I.; Gingras, A.C.; Choi, H. SAINTexpress: Improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics 2014, 100, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Knight, J.D.R.; Choi, H.; Gupta, G.D.; Pelletier, L.; Raught, B.; Nesvizhskii, A.I.; Gingras, A.C. ProHits-viz: A suite of web tools for visualizing interaction proteomics data. Nat. Methods 2017, 14, 645–646. [Google Scholar] [CrossRef] [PubMed]
- Gareau, C.; Martel, D.; Coudert, L.; Mellaoui, S.; Mazroui, R. Characterization of Fragile X Mental Retardation Protein granules formation and dynamics in Drosophila. Biol. Open 2013, 2, 68–81. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mazroui, R.; Huot, M.E.; Tremblay, S.; Filion, C.; Labelle, Y.; Khandjian, E.W. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 2002, 11, 3007–3017. [Google Scholar] [CrossRef]
- Siomi, M.C.; Zhang, Y.; Siomi, H.; Dreyfuss, G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol. Cell Biol. 1996, 16, 3825–3832. [Google Scholar] [CrossRef]
- Dube, M.; Huot, M.E.; Khandjian, E.W. Muscle specific fragile X related protein 1 isoforms are sequestered in the nucleus of undifferentiated myoblast. BMC Genet. 2000, 1, 4. [Google Scholar] [CrossRef]
- Tamanini, F.; Kirkpatrick, L.L.; Schonkeren, J.; van Unen, L.; Bontekoe, C.; Bakker, C.; Nelson, D.L.; Galjaard, H.; Oostra, B.A.; Hoogeveen, A.T. The fragile X-related proteins FXR1P and FXR2P contain a functional nucleolar-targeting signal equivalent to the HIV-1 regulatory proteins. Hum. Mol. Genet. 2000, 9, 1487–1493. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shimizu, H.; Hohjoh, H. FMRP, FXR1 protein and Dlg4 mRNA, which are associated with fragile X syndrome, are involved in the ubiquitin-proteasome system. Sci. Rep. 2023, 13, 1956. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.L.; Palmai-Pallag, T.; Ying, S.; Hickson, I.D. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 2009, 11, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Casanas, M.; Chan, K.L. The Unresolved Problem of DNA Bridging. Genes 2018, 9, 623. [Google Scholar] [CrossRef] [PubMed]
- Mazouzi, A.; Velimezi, G.; Loizou, J.I. DNA replication stress: Causes, resolution and disease. Exp. Cell Res. 2014, 329, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Steigemann, P.; Wurzenberger, C.; Schmitz, M.H.; Held, M.; Guizetti, J.; Maar, S.; Gerlich, D.W. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell 2009, 136, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Nussenzweig, A.; Takeda, S.; Austin, C. Human topoisomerases and their roles in genome stability and organization. Nat. Rev. Mol. Cell Biol. 2022, 23, 407–427. [Google Scholar] [CrossRef]
- Fanti, L.; Pimpinelli, S. HP1: A functionally multifaceted protein. Curr. Opin. Genet. Dev. 2008, 18, 169–174. [Google Scholar] [CrossRef]
- Maddika, S.; Sy, S.M.; Chen, J. Functional interaction between Chfr and Kif22 controls genomic stability. J. Biol. Chem. 2009, 284, 12998–13003. [Google Scholar] [CrossRef]
- Cao, C.; Han, P.; Liu, L.; Tang, Y.; Tian, S.; Zhang, K.; Shi, L.; Liu, Z.; Zhuo, D.; Ge, W.; et al. Epithelial cell transforming factor ECT2 is an important regulator of DNA double-strand break repair and genome stability. J. Biol. Chem. 2021, 297, 101036. [Google Scholar] [CrossRef]
- Yu, W.; Qiu, Z.; Gao, N.; Wang, L.; Cui, H.; Qian, Y.; Jiang, L.; Luo, J.; Yi, Z.; Lu, H.; et al. PAK1IP1, a ribosomal stress-induced nucleolar protein, regulates cell proliferation via the p53-MDM2 loop. Nucleic Acids Res. 2011, 39, 2234–2248. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hamilton, K.; Tong, L. Recent molecular insights into canonical pre-mRNA 3′-end processing. Transcription 2020, 11, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Boi, D.; Rubini, E.; Breccia, S.; Guarguaglini, G.; Paiardini, A. When Just One Phosphate Is One Too Many: The Multifaceted Interplay between Myc and Kinases. Int. J. Mol. Sci. 2023, 24, 4746. [Google Scholar] [CrossRef]
- Voorhees, P.M.; Orlowski, R.Z. The proteasome and proteasome inhibitors in cancer therapy. Annu. Rev. Pharmacol. Toxicol. 2006, 46, 189–213. [Google Scholar] [CrossRef] [PubMed]
- Fong, K.W.; Li, Y.; Wang, W.; Ma, W.; Li, K.; Qi, R.Z.; Liu, D.; Songyang, Z.; Chen, J. Whole-genome screening identifies proteins localized to distinct nuclear bodies. J. Cell Biol. 2013, 203, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Hebert, M.D.; Poole, A.R. Towards an understanding of regulating Cajal body activity by protein modification. RNA Biol. 2017, 14, 761–778. [Google Scholar] [CrossRef] [PubMed]
- Sahadevan, S.; Perez-Berlanga, M.; Polymenidou, M. Identification of RNA-RBP Interactions in Subcellular Compartments by CLIP-Seq. Methods Mol. Biol. 2022, 2428, 305–323. [Google Scholar]
- Wheeler, E.C.; Van Nostrand, E.L.; Yeo, G.W. Advances and challenges in the detection of transcriptome-wide protein-RNA interactions. Wiley Interdiscip. Rev. RNA 2018, 9, e1436. [Google Scholar] [CrossRef]
- Li, M.; Shin, J.; Risgaard, R.D.; Parries, M.J.; Wang, J.; Chasman, D.; Liu, S.; Roy, S.; Bhattacharyya, A.; Zhao, X. Identification of FMR1-regulated molecular networks in human neurodevelopment. Genome Res. 2020, 30, 361–374. [Google Scholar] [CrossRef]
- Lam, F.C.; Kong, Y.W.; Huang, Q.; Vu Han, T.L.; Maffa, A.D.; Kasper, E.M.; Yaffe, M.B. BRD4 prevents the accumulation of R-loops and protects against transcription-replication collision events and DNA damage. Nat. Commun. 2020, 11, 4083. [Google Scholar] [CrossRef]
- Feng, W.; Jasin, M. BRCA2 suppresses replication stress-induced mitotic and G1 abnormalities through homologous recombination. Nat. Commun. 2017, 8, 525. [Google Scholar] [CrossRef] [PubMed]
- Amaral, N.; Vendrell, A.; Funaya, C.; Idrissi, F.Z.; Maier, M.; Kumar, A.; Neurohr, G.; Colomina, N.; Torres-Rosell, J.; Geli, M.I.; et al. The Aurora-B-dependent NoCut checkpoint prevents damage of anaphase bridges after DNA replication stress. Nat. Cell Biol. 2016, 18, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Gemble, S.; Buhagiar-Labarchede, G.; Onclercq-Delic, R.; Fontaine, G.; Lambert, S.; Amor-Gueret, M. Topoisomerase IIalpha prevents ultrafine anaphase bridges by two mechanisms. Open Biol. 2020, 10, 190259. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.S.; Haghighi, F.; Stefanski, A.; Nakhaei-Rad, S.; Kazemein Jasemi, N.S.; Al Kabbani, M.A.; Gorg, B.; Fujii, M.; Lang, P.A.; Haussinger, D.; et al. Novel FMRP interaction networks linked to cellular stress. FEBS J. 2021, 288, 837–860. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Gutekunst, C.A.; Eberhart, D.E.; Yi, H.; Warren, S.T.; Hersch, S.M. Fragile X mental retardation protein: Nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J. Neurosci. 1997, 17, 1539–1547. [Google Scholar] [CrossRef]
- Kieffer, F.; Hilal, F.; Gay, A.S.; Debayle, D.; Pronot, M.; Poupon, G.; Lacagne, I.; Bardoni, B.; Martin, S.; Gwizdek, C. Combining affinity purification and mass spectrometry to define the network of the nuclear proteins interacting with the N-terminal region of FMRP. Front. Mol. Biosci. 2022, 9, 954087. [Google Scholar] [CrossRef]
- Gumeni, S.; Evangelakou, Z.; Gorgoulis, V.G.; Trougakos, I.P. Proteome Stability as a Key Factor of Genome Integrity. Int. J. Mol. Sci. 2017, 18, 2036. [Google Scholar] [CrossRef]
- Louros, S.R.; Seo, S.S.; Maio, B.; Martinez-Gonzalez, C.; Gonzalez-Lozano, M.A.; Muscas, M.; Verity, N.C.; Wills, J.C.; Li, K.W.; Nolan, M.F.; et al. Excessive proteostasis contributes to pathology in fragile X syndrome. Neuron 2023, 111, 508–525.e507. [Google Scholar] [CrossRef]

| Functions | Genes | Average Spectral Counts | Fold Change | BFDR |
|---|---|---|---|---|
| RNA splicing | SRSF2 | 6 | 20 | 0 |
| SRSF7 | 6 | 60 | 0 | |
| SRSF9 | 6 | 60 | 0 | |
| PRPF3 | 2 | 20 | 0 | |
| PRPF6 | 6 | 6 | 0.01 | |
| PRPF8 | 44.5 | 4.45 | 0 | |
| CPSF1 | 5 | 50 | 0 | |
| RNA modification, RNA structure and stability | FXR1/2 | 90/80 | 900/800 | 0 |
| YTHDF2 | 3.5 | 35 | 0 | |
| DDX50 | 10 | 100 | 0 | |
| DDX52 | 5.5 | 55 | 0 | |
| DDX51 | 3 | 30 | 0 | |
| TRMT1L | 3 | 30 | 0 | |
| Ribosome biogenesis and maturation | WDR3 | 11.5 | 115 | 0 |
| WDR36 | 9.5 | 95 | 0 | |
| WDR46 | 3 | 60 | 0 | |
| MRTO4 | 7.5 | 75 | 0 | |
| NOP9 | 2.5 | 25 | 0 | |
| NOP16 | 7 | 7 | 0.01 | |
| RNA synthesis | POLR2B | 8.5 | 85 | 0 |
| POLR1A | 6.5 | 65 | 0 | |
| POLR1E | 3.5 | 35 | 0 | |
| CEBPZ | 17 | 170 | 0 | |
| SUPT16H | 7 | 70 | 0 | |
| SUPT6H | 3 | 30 | 0 | |
| DNA replication and DNA repair | BRCA2 | 7 | 70 | 0 |
| TOP2A | 25 | 250 | 0 | |
| TOP2B | 10 | 100 | 0 | |
| SMARCA5 | 8.5 | 85 | 0 | |
| SMARCD2 | 2.5 | 25 | 0 | |
| SMARCE1 | 2 | 20 | 0 | |
| Chromatin remodeling and chromosome condensation and segregation | H2BC3 | 33.5 | 335 | 0 |
| H2BC5 | 34.5 | 2.88 | 0.01 | |
| AURKB | 2.5 | 25 | 0 | |
| RCC1 | 7.5 | 75 | 0 | |
| Components of ubiquitin-proteasome system | PSMDs | 4.5–29 | 45–290 | 0–0.01 |
| PSMCs | 4.5–16.5 | 5.5–45 | 0 | |
| UBAP2L | 79.5 | 7.23 | 0 |
| Functions | Genes | Average Spectral Counts | Fold Change | BFDR |
|---|---|---|---|---|
| RNA binding protein | FXR1 | 30 | 300 | 0 |
| FXR2 | 20 | 200 | 0 | |
| Lipoprotein receptor | LRP1 | 6 | 60 | 0 |
| Ubiquitin-proteasome pathway | PSMD2 | 6 | 300 | 0 |
| PSMD1 | 5.5 | 200 | 0 | |
| PSMC4 | 5.5 | 16.5 | 0 | |
| PSMD6 | 3.5 | 35 | 0 | |
| PSMD3 | 3.5 | 35 | 0 | |
| PSMD11 | 3 | 30 | 0.01 | |
| PSMD8 | 2.5 | 25 | 0.01 | |
| PSMD13 | 2.5 | 25 | 0.01 | |
| PSMD14 | 2 | 20 | 0.01 | |
| Regulation of mitotic spindle orientation | ARHGEF2 | 5.5 | 55 | 0 |
| Mitochondrial metabolism | C1QBP | 4 | 40 | 0 |
| Transcription | HP1BP3 | 3.5 | 35 | 0 |
| Biosynthesis of phosphatidylinositol | PI4KA | 3 | 30 | 0.01 |
| Free AA regulation | LPCAT3 | 2 | 20 | 0.01 |
| Genes | Iso6 Fold Change | Iso6 APH Fold Change | Genes | Iso6 Fold Change | Iso6 APH Fold Change |
|---|---|---|---|---|---|
| FXR1 | 300 | 310 | PSMD13 | 25 | NA |
| FXR2 | 200 | 220 | CASK | NA | 25 |
| PSMD2 | 60 | 85 | PSMC6 | NA | 25 |
| LRP1 | 60 | 75 | PSMD4 | NA | 25 |
| PSMD1 | 55 | 75 | NKTR | NA | 25 |
| ECT2 | NA | 60 | PSMD14 | 20 | 25 |
| ARHGEF | 55 | 50 | CPSF1 | NA | 25 |
| PSMD11 | 30 | 55 | ADNP | NA | 25 |
| KIF22 | NA | 50 | NIP7 | NA | 25 |
| PSMD6 | 35 | 50 | MRTO4 | NA | 25 |
| HMGXB | NA | 45 | BRD4 | NA | 25 |
| C1QPB | 40 | 45 | LPCAT3 | 20 | 20 |
| PSMD3 | 35 | 45 | RFC2 | NA | 20 |
| HP1BP3 | 35 | 45 | PCF11 | NA | 20 |
| RPL10A | NA | 40 | NOL7 | NA | 20 |
| PSMD8 | 25 | 35 | YLPM1 | NA | 20 |
| WDR36 | NA | 35 | ITM2B | NA | 20 |
| PI4KA | 30 | NA | TRMT1L | NA | 20 |
| APP | NA | 30 | PLBD2 | NA | 20 |
| PAK1IP1 | NA | 30 | PPA2 | NA | 20 |
| TOP2A | NA | 30 | PSMC4 | 16.5 | 18 |
| WDR3 | NA | 30 | AURKB | NA | 16.5 |
| PSMD7 | NA | 11.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ledoux, N.; Lelong, E.I.J.; Simard, A.; Hussein, S.; Adjibade, P.; Lambert, J.-P.; Mazroui, R. The Identification of Nuclear FMRP Isoform Iso6 Partners. Cells 2023, 12, 2807. https://doi.org/10.3390/cells12242807
Ledoux N, Lelong EIJ, Simard A, Hussein S, Adjibade P, Lambert J-P, Mazroui R. The Identification of Nuclear FMRP Isoform Iso6 Partners. Cells. 2023; 12(24):2807. https://doi.org/10.3390/cells12242807
Chicago/Turabian StyleLedoux, Nassim, Emeline I. J. Lelong, Alexandre Simard, Samer Hussein, Pauline Adjibade, Jean-Philippe Lambert, and Rachid Mazroui. 2023. "The Identification of Nuclear FMRP Isoform Iso6 Partners" Cells 12, no. 24: 2807. https://doi.org/10.3390/cells12242807
APA StyleLedoux, N., Lelong, E. I. J., Simard, A., Hussein, S., Adjibade, P., Lambert, J.-P., & Mazroui, R. (2023). The Identification of Nuclear FMRP Isoform Iso6 Partners. Cells, 12(24), 2807. https://doi.org/10.3390/cells12242807






