Cell Adhesion at the Tight Junctions: New Aspects and New Functions
Abstract
:1. Introduction
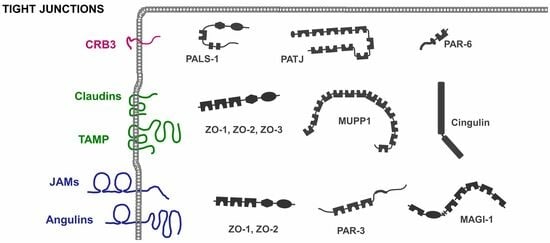
2. Integral Membrane Proteins at the TJs
2.1. CRB3
2.2. Claudins
2.3. TAMPs
2.4. IgSF Proteins
3. Integral Membrane Proteins as Anchors for Cytoskeletal Elements at the TJs
4. Integral Membrane Proteins and the Paracellular Barrier Function of TJs
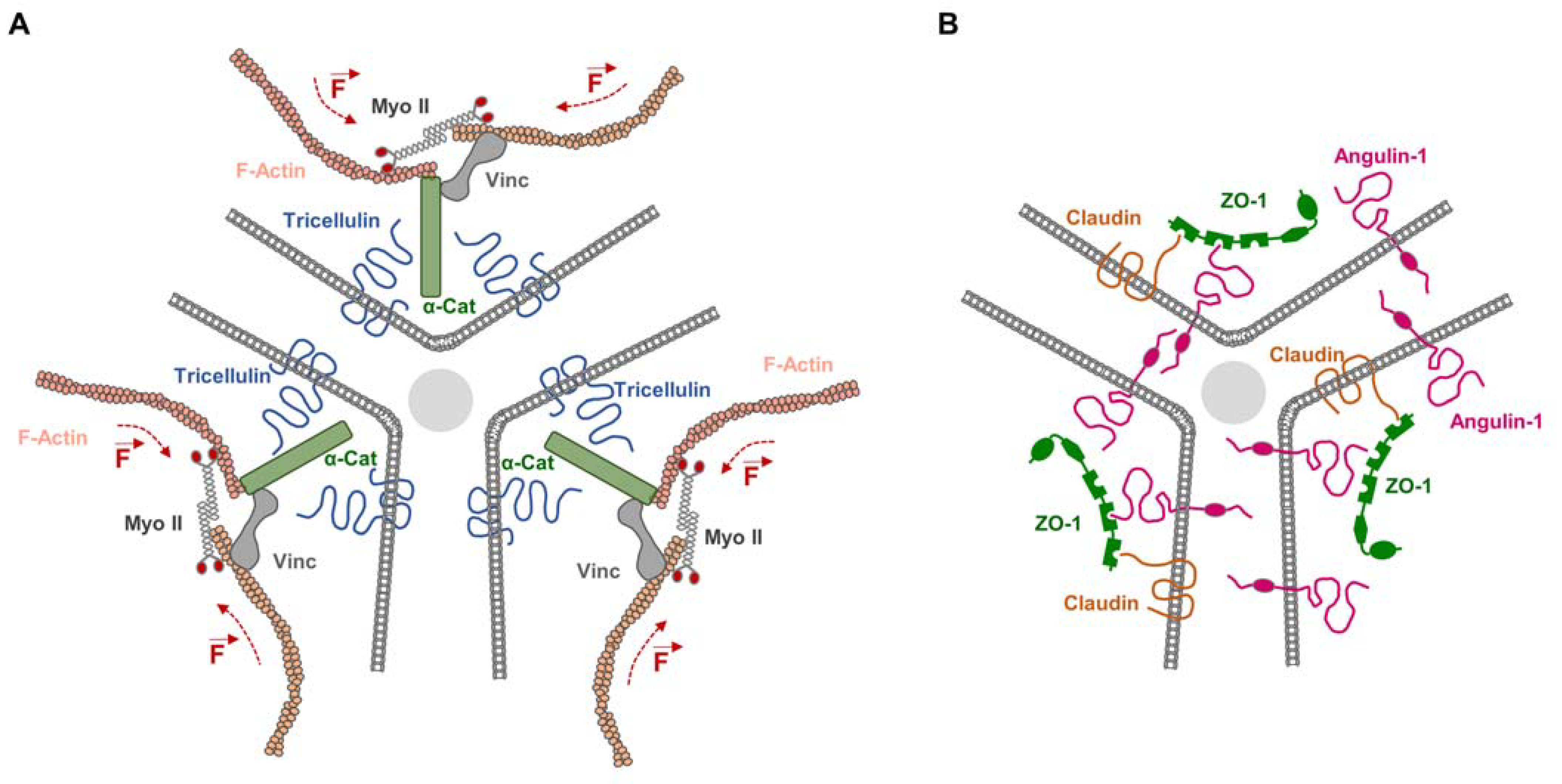
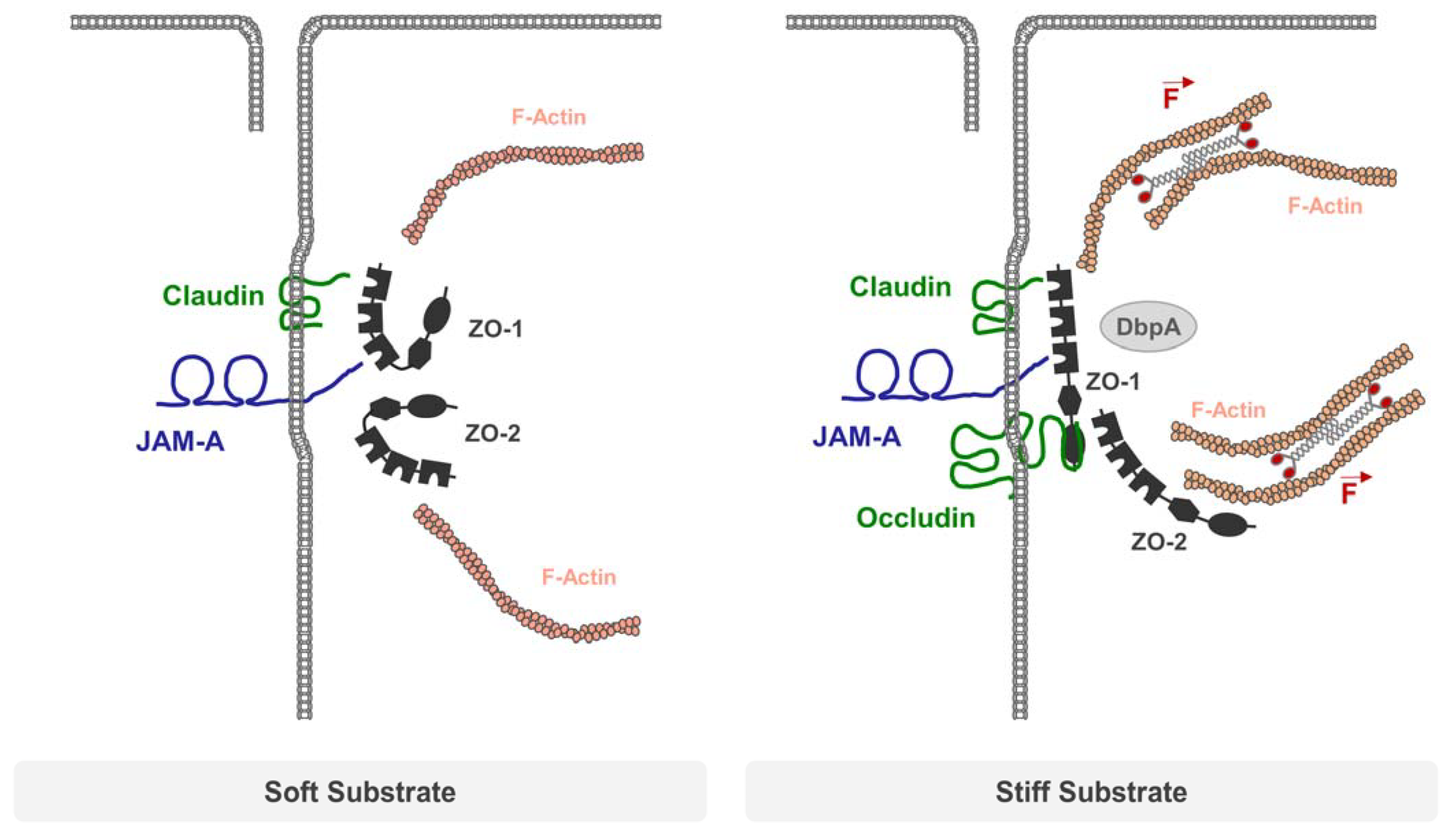
5. Integral Membrane Proteins and the Mechanical Force Load on TJs
6. Integral Membrane Proteins and Phase Separation at the TJs
7. Lipid Modifications of Integral Membrane Proteins at TJs
8. Summary and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Buckley, C.E.; St Johnston, D. Apical-basal polarity and the control of epithelial form and function. Nat. Rev. Mol. Cell Biol. 2022, 23, 559–577. [Google Scholar] [CrossRef] [PubMed]
- Zihni, C.; Mills, C.; Matter, K.; Balda, M.S. Tight junctions: From simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 2016, 17, 564–580. [Google Scholar] [CrossRef] [PubMed]
- Hegazy, M.; Perl, A.L.; Svoboda, S.A.; Green, K.J. Desmosomal Cadherins in Health and Disease. Annu. Rev. Pathol. 2022, 17, 47–72. [Google Scholar] [CrossRef] [PubMed]
- Ladoux, B.; Mege, R.M. Mechanobiology of collective cell behaviours. Nat. Rev. Mol. Cell Biol. 2017, 18, 743–757. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Gonzalez, R.; Peifer, M. Powering morphogenesis: Multiscale challenges at the interface of cell adhesion and the cytoskeleton. Mol. Biol. Cell 2022, 33, pe4. [Google Scholar] [CrossRef] [PubMed]
- Knauf, F.; Brewer, J.R.; Flavell, R.A. Immunity, microbiota and kidney disease. Nat. Rev. Nephrol. 2019, 15, 263–274. [Google Scholar] [CrossRef]
- Lambert, A.W.; Weinberg, R.A. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat. Rev. Cancer 2021, 21, 325–338. [Google Scholar] [CrossRef]
- Martin-Belmonte, F.; Perez-Moreno, M. Epithelial cell polarity, stem cells and cancer. Nat. Rev. Cancer 2011, 12, 23–38. [Google Scholar] [CrossRef]
- Miller, P.W.; Clarke, D.N.; Weis, W.I.; Lowe, C.J.; Nelson, W.J. The evolutionary origin of epithelial cell-cell adhesion mechanisms. Curr. Top. Membr. 2013, 72, 267–311. [Google Scholar] [CrossRef]
- Murray, P.S.; Zaidel-Bar, R. Pre-metazoan origins and evolution of the cadherin adhesome. Biol. Open 2014, 3, 1183–1195. [Google Scholar] [CrossRef]
- Abedin, M.; King, N. Diverse evolutionary paths to cell adhesion. Trends Cell Biol. 2010, 20, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Green, K.J.; Roth-Carter, Q.; Niessen, C.M.; Nichols, S.A. Tracing the Evolutionary Origin of Desmosomes. Curr. Biol. 2020, 30, R535–R543. [Google Scholar] [CrossRef]
- Rice, C.; De, O.; Alhadyian, H.; Hall, S.; Ward, R.E. Expanding the Junction: New Insights into Non-Occluding Roles for Septate Junction Proteins during Development. J. Dev. Biol. 2021, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Takeichi, M. Dynamic contacts: Rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol. 2014, 15, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Vasileva, E.; Citi, S. The role of microtubules in the regulation of epithelial junctions. Tissue Barriers 2018, 6, 1539596. [Google Scholar] [CrossRef] [PubMed]
- Muller, L.; Hatzfeld, M.; Keil, R. Desmosomes as Signaling Hubs in the Regulation of Cell Behavior. Front. Cell Dev. Biol. 2021, 9, 745670. [Google Scholar] [CrossRef]
- Zihni, C.; Balda, M.S.; Matter, K. Signalling at tight junctions during epithelial differentiation and microbial pathogenesis. J. Cell Sci. 2014, 127, 3401–3413. [Google Scholar] [CrossRef]
- Mendonsa, A.M.; Na, T.Y.; Gumbiner, B.M. E-cadherin in contact inhibition and cancer. Oncogene 2018, 37, 4769–4780. [Google Scholar] [CrossRef]
- Broussard, J.A.; Jaiganesh, A.; Zarkoob, H.; Conway, D.E.; Dunn, A.R.; Espinosa, H.D.; Janmey, P.A.; Green, K.J. Scaling up single-cell mechanics to multicellular tissues—The role of the intermediate filament-desmosome network. J. Cell Sci. 2020, 133, jcs228031. [Google Scholar] [CrossRef]
- Heinemann, U.; Schuetz, A. Structural Features of Tight-Junction Proteins. Int. J. Mol. Sci. 2019, 20, 6020. [Google Scholar] [CrossRef]
- Hartmann, C.; Schwietzer, Y.A.; Otani, T.; Furuse, M.; Ebnet, K. Physiological functions of junctional adhesion molecules (JAMs) in tight junctions. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183299. [Google Scholar] [CrossRef]
- Otani, T.; Furuse, M. Tight Junction Structure and Function Revisited. Trends Cell Biol. 2020, 30, 805–817. [Google Scholar] [CrossRef]
- Piontek, J.; Krug, S.M.; Protze, J.; Krause, G.; Fromm, M. Molecular architecture and assembly of the tight junction backbone. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183279. [Google Scholar] [CrossRef]
- Higashi, T.; Chiba, H. Molecular organization, regulation and function of tricellular junctions. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183143. [Google Scholar] [CrossRef]
- Saito, A.C.; Higashi, T.; Chiba, H. Tight-junction strand networks and tightness of the epithelial barrier. Microscopy 2023, 72, 213–225. [Google Scholar] [CrossRef]
- Margolis, B. The Crumbs3 Polarity Protein. Cold Spring Harb. Perspect. Biol. 2018, 10, a027961. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Yatim, S.; Peng, S.; Gunaratne, J.; Hunziker, W.; Ludwig, A. The Mammalian Crumbs Complex Defines a Distinct Polarity Domain Apical of Epithelial Tight Junctions. Curr. Biol. 2020, 30, 2791–2804.e6. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, G.C.; Lucas, E.P.; Brain, R.; Tournier, A.; Thompson, B.J. Positive feedback and mutual antagonism combine to polarize Crumbs in the Drosophila follicle cell epithelium. Curr. Biol. 2012, 22, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Furuse, M.; Tsukita, S. Claudins in occluding junctions of humans and flies. Trends Cell Biol. 2006, 16, 181–188. [Google Scholar] [CrossRef]
- Gunzel, D.; Yu, A.S. Claudins and the modulation of tight junction permeability. Physiol. Rev. 2013, 93, 525–569. [Google Scholar] [CrossRef] [PubMed]
- Kubota, K.; Furuse, M.; Sasaki, H.; Sonoda, N.; Fujita, K.; Nagafuchi, A.; Tsukita, S. Ca(2+)-independent cell-adhesion activity of claudins, a family of integral membrane proteins localized at tight junctions. Curr. Biol. 1999, 9, 1035–1038. [Google Scholar] [CrossRef] [PubMed]
- Furuse, M.; Sasaki, H.; Fujimoto, K.; Tsukita, S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol. 1998, 143, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Furuse, M.; Sasaki, H.; Tsukita, S. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J. Cell Biol. 1999, 147, 891–903. [Google Scholar] [CrossRef]
- Tsukita, S.; Tanaka, H.; Tamura, A. The Claudins: From Tight Junctions to Biological Systems. Trends Biochem. Sci. 2019, 44, 141–152. [Google Scholar] [CrossRef]
- Raleigh, D.R.; Marchiando, A.M.; Zhang, Y.; Shen, L.; Sasaki, H.; Wang, Y.; Long, M.; Turner, J.R. Tight junction-associated MARVEL proteins marveld3, tricellulin, and occludin have distinct but overlapping functions. Mol. Biol. Cell 2010, 21, 1200–1213. [Google Scholar] [CrossRef]
- Sanchez-Pulido, L.; Martin-Belmonte, F.; Valencia, A.; Alonso, M.A. MARVEL: A conserved domain involved in membrane apposition events. Trends Biochem. Sci. 2002, 27, 599–601. [Google Scholar] [CrossRef] [PubMed]
- Ikenouchi, J.; Furuse, M.; Furuse, K.; Sasaki, H.; Tsukita, S.; Tsukita, S. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J. Cell Biol. 2005, 171, 939–945. [Google Scholar] [CrossRef]
- Cording, J.; Berg, J.; Kading, N.; Bellmann, C.; Tscheik, C.; Westphal, J.K.; Milatz, S.; Gunzel, D.; Wolburg, H.; Piontek, J.; et al. In tight junctions, claudins regulate the interactions between occludin, tricellulin and marvelD3, which, inversely, modulate claudin oligomerization. J. Cell Sci. 2013, 126, 554–564. [Google Scholar] [CrossRef]
- Van Itallie, C.M.; Anderson, J.M. Occludin confers adhesiveness when expressed in fibroblasts. J. Cell Sci. 1997, 110 Pt 9, 1113–1121. [Google Scholar] [CrossRef]
- Saitou, M.; Furuse, M.; Sasaki, H.; Schulzke, J.D.; Fromm, M.; Takano, H.; Noda, T.; Tsukita, S. Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol. Biol. Cell 2000, 11, 4131–4142. [Google Scholar] [CrossRef]
- Steed, E.; Rodrigues, N.T.; Balda, M.S.; Matter, K. Identification of MarvelD3 as a tight junction-associated transmembrane protein of the occludin family. BMC Cell Biol. 2009, 10, 95. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, T.; Furuse, K.; Otani, T.; Wakayama, T.; Furuse, M. Angulin-1 seals tricellular contacts independently of tricellulin and claudins. J. Cell Biol. 2021, 220, e202005062. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Haraguchi, D.; Shigetomi, K.; Matsuzawa, K.; Uchida, S.; Ikenouchi, J. Tricellulin secures the epithelial barrier at tricellular junctions by interacting with actomyosin. J. Cell Biol. 2022, 221, e202009037. [Google Scholar] [CrossRef] [PubMed]
- Ebnet, K. Junctional Adhesion Molecules (JAMs): Cell Adhesion Receptors With Pleiotropic Functions in Cell Physiology and Development. Physiol. Rev. 2017, 97, 1529–1554. [Google Scholar] [CrossRef] [PubMed]
- Cohen, C.J.; Shieh, J.T.; Pickles, R.J.; Okegawa, T.; Hsieh, J.T.; Bergelson, J.M. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc. Natl. Acad. Sci. USA 2001, 98, 15191–15196. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, S.; Tajima, M.; Yao, I.; Nishimura, W.; Mori, H.; Hata, Y. JAM4, a junctional cell adhesion molecule interacting with a tight junction protein, MAGI-1. Mol. Cell. Biol. 2003, 23, 4267–4282. [Google Scholar] [CrossRef]
- Raschperger, E.; Engstrom, U.; Pettersson, R.F.; Fuxe, J. CLMP, a novel member of the CTX family and a new component of epithelial tight junctions. J. Biol. Chem. 2004, 279, 796–804. [Google Scholar] [CrossRef]
- Nasdala, I.; Wolburg-Buchholz, K.; Wolburg, H.; Kuhn, A.; Ebnet, K.; Brachtendorf, G.; Samulowitz, U.; Kuster, B.; Engelhardt, B.; Vestweber, D.; et al. A transmembrane tight junction protein selectively expressed on endothelial cells and platelets. J. Biol. Chem. 2002, 277, 16294–16303. [Google Scholar] [CrossRef]
- Hirata, K.; Ishida, T.; Penta, K.; Rezaee, M.; Yang, E.; Wohlgemuth, J.; Quertermous, T. Cloning of an immunoglobulin family adhesion molecule selectively expressed by endothelial cells. J. Biol. Chem. 2001, 276, 16223–16231. [Google Scholar] [CrossRef]
- Furuse, M.; Izumi, Y.; Oda, Y.; Higashi, T.; Iwamoto, N. Molecular organization of tricellular tight junctions. Tissue Barriers 2014, 2, e28960. [Google Scholar] [CrossRef]
- Higashi, T.; Tokuda, S.; Kitajiri, S.; Masuda, S.; Nakamura, H.; Oda, Y.; Furuse, M. Analysis of the ‘angulin’ proteins LSR, ILDR1 and ILDR2--tricellulin recruitment, epithelial barrier function and implication in deafness pathogenesis. J. Cell Sci. 2013, 126, 966–977. [Google Scholar] [CrossRef]
- Masuda, S.; Oda, Y.; Sasaki, H.; Ikenouchi, J.; Higashi, T.; Akashi, M.; Nishi, E.; Furuse, M. LSR defines cell corners for tricellular tight junction formation in epithelial cells. J. Cell Sci. 2011, 124, 548–555. [Google Scholar] [CrossRef]
- Tonikian, R.; Zhang, Y.; Sazinsky, S.L.; Currell, B.; Yeh, J.H.; Reva, B.; Held, H.A.; Appleton, B.A.; Evangelista, M.; Wu, Y.; et al. A specificity map for the PDZ domain family. PLoS Biol. 2008, 6, e239. [Google Scholar] [CrossRef]
- Tateishi, K.; Nishida, T.; Inoue, K.; Tsukita, S. Three-dimensional Organization of Layered Apical Cytoskeletal Networks Associated with Mouse Airway Tissue Development. Sci. Rep. 2017, 7, 43783. [Google Scholar] [CrossRef]
- Rouaud, F.; Sluysmans, S.; Flinois, A.; Shah, J.; Vasileva, E.; Citi, S. Scaffolding proteins of vertebrate apical junctions: Structure, functions and biophysics. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183399. [Google Scholar] [CrossRef] [PubMed]
- Bazzoni, G.; Martinez-Estrada, O.M.; Orsenigo, F.; Cordenonsi, M.; Citi, S.; Dejana, E. Interaction of junctional adhesion molecule with the tight junction components ZO-1, cingulin, and occludin. J. Biol. Chem. 2000, 275, 20520–20526. [Google Scholar] [CrossRef] [PubMed]
- Cordenonsi, M.; D’Atri, F.; Hammar, E.; Parry, D.A.; Kendrick-Jones, J.; Shore, D.; Citi, S. Cingulin contains globular and coiled-coil domains and interacts with ZO-1, ZO-2, ZO-3, and myosin. J. Cell Biol. 1999, 147, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- DiTommaso, T.; Cottle, D.L.; Pearson, H.B.; Schluter, H.; Kaur, P.; Humbert, P.O.; Smyth, I.M. Keratin 76 is required for tight junction function and maintenance of the skin barrier. PLoS Genet. 2014, 10, e1004706. [Google Scholar] [CrossRef]
- Van Itallie, C.M.; Anderson, J.M. Architecture of tight junctions and principles of molecular composition. Semin. Cell Dev. Biol. 2014, 36, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Yano, T.; Kanoh, H.; Tamura, A.; Tsukita, S. Apical cytoskeletons and junctional complexes as a combined system in epithelial cell sheets. Ann. N. Y. Acad. Sci. 2017, 1405, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.I.; Lechuga, S.; Marino-Melendez, A.; Naydenov, N.G. Unique and redundant functions of cytoplasmic actins and nonmuscle myosin II isoforms at epithelial junctions. Ann. N. Y. Acad. Sci. 2022, 1515, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Weber, C.R.; Raleigh, D.R.; Yu, D.; Turner, J.R. Tight junction pore and leak pathways: A dynamic duo. Annu. Rev. Physiol. 2011, 73, 283–309. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, A.; Chanez-Paredes, S.D.; Haest, X.; Turner, J.R. Paracellular permeability and tight junction regulation in gut health and disease. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 1–16. [Google Scholar] [CrossRef]
- Meoli, L.; Gunzel, D. The role of claudins in homeostasis. Nat. Rev. Nephrol. 2023, 19, 587–603. [Google Scholar] [CrossRef]
- Otani, T.; Nguyen, T.P.; Tokuda, S.; Sugihara, K.; Sugawara, T.; Furuse, K.; Miura, T.; Ebnet, K.; Furuse, M. Claudins and JAM-A coordinately regulate tight junction formation and epithelial polarity. J. Cell Biol. 2019, 218, 3372–3396. [Google Scholar] [CrossRef]
- Steinbacher, T.; Kummer, D.; Ebnet, K. Junctional adhesion molecule-A: Functional diversity through molecular promiscuity. Cell Mol. Life Sci. 2018, 75, 1393–1409. [Google Scholar] [CrossRef]
- Martin-Padura, I.; Lostaglio, S.; Schneemann, M.; Williams, L.; Romano, M.; Fruscella, P.; Panzeri, C.; Stoppacciaro, A.; Ruco, L.; Villa, A.; et al. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 1998, 142, 117–127. [Google Scholar] [CrossRef]
- Staehelin, L.A. Further observations on the fine structure of freeze-cleaved tight junctions. J. Cell Sci. 1973, 13, 763–786. [Google Scholar] [CrossRef]
- Wade, J.B.; Karnovsky, M.J. The structure of the zonula occludens. A single fibril model based on freeze-fracture. J. Cell Biol. 1974, 60, 168–180. [Google Scholar] [CrossRef]
- Charras, G.; Yap, A.S. Tensile Forces and Mechanotransduction at Cell-Cell Junctions. Curr. Biol. 2018, 28, R445–R457. [Google Scholar] [CrossRef]
- Hallou, A.; Brunet, T. On growth and force: Mechanical forces in development. Development 2020, 147. [Google Scholar] [CrossRef]
- Fischer, L.S.; Rangarajan, S.; Sadhanasatish, T.; Grashoff, C. Molecular Force Measurement with Tension Sensors. Annu. Rev. Biophys. 2021, 50, 595–616. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, D.; Bellaiche, Y. Mechanical Force-Driven Adherens Junction Remodeling and Epithelial Dynamics. Dev. Cell 2018, 47, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Citi, S. The mechanobiology of tight junctions. Biophys. Rev. 2019, 11, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, R.E.; Higashi, T.; Erofeev, I.S.; Arnold, T.R.; Leda, M.; Goryachev, A.B.; Miller, A.L. Rho Flares Repair Local Tight Junction Leaks. Dev. Cell 2019, 48, 445–459.e5. [Google Scholar] [CrossRef]
- Spadaro, D.; Le, S.; Laroche, T.; Mean, I.; Jond, L.; Yan, J.; Citi, S. Tension-Dependent Stretching Activates ZO-1 to Control the Junctional Localization of Its Interactors. Curr. Biol. 2017, 27, 3783–3795.e8. [Google Scholar] [CrossRef] [PubMed]
- Haas, A.J.; Zihni, C.; Ruppel, A.; Hartmann, C.; Ebnet, K.; Tada, M.; Balda, M.S.; Matter, K. Interplay between Extracellular Matrix Stiffness and JAM-A Regulates Mechanical Load on ZO-1 and Tight Junction Assembly. Cell Rep. 2020, 32, 107924. [Google Scholar] [CrossRef] [PubMed]
- Haas, A.J.; Zihni, C.; Krug, S.M.; Maraspini, R.; Otani, T.; Furuse, M.; Honigmann, A.; Balda, M.S.; Matter, K. ZO-1 Guides Tight Junction Assembly and Epithelial Morphogenesis via Cytoskeletal Tension-Dependent and -Independent Functions. Cells 2022, 11, 3775. [Google Scholar] [CrossRef]
- Hatte, G.; Prigent, C.; Tassan, J.P. Tight junctions negatively regulate mechanical forces applied to adherens junctions in vertebrate epithelial tissue. J. Cell Sci. 2018, 131, jcs208736. [Google Scholar] [CrossRef]
- Ebnet, K.; Schulz, C.U.; Meyer Zu Brickwedde, M.K.; Pendl, G.G.; Vestweber, D. Junctional adhesion molecule interacts with the PDZ domain-containing proteins AF-6 and ZO-1. J. Biol. Chem. 2000, 275, 27979–27988. [Google Scholar] [CrossRef]
- Nomme, J.; Fanning, A.S.; Caffrey, M.; Lye, M.F.; Anderson, J.M.; Lavie, A. The Src homology 3 domain is required for junctional adhesion molecule binding to the third PDZ domain of the scaffolding protein ZO-1. J. Biol. Chem. 2011, 286, 43352–43360. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, A.C.; Sumagin, R.; Rankin, C.R.; Leoni, G.; Mina, M.J.; Reiter, D.M.; Stehle, T.; Dermody, T.S.; Schaefer, S.A.; Hall, R.A.; et al. JAM-A associates with ZO-2, afadin, and PDZ-GEF1 to activate Rap2c and regulate epithelial barrier function. Mol. Biol. Cell 2013, 24, 2849–2860. [Google Scholar] [CrossRef]
- Scott, D.W.; Tolbert, C.E.; Burridge, K. Tension on JAM-A activates RhoA via GEF-H1 and p115 RhoGEF. Mol. Biol. Cell 2016, 27, 1420–1430. [Google Scholar] [CrossRef]
- Vachharajani, V.T.; DeJong, M.P.; Dunn, A.R. PDZ Domains from the Junctional Proteins Afadin and ZO-1 Act as Mechanosensors. bioRxiv 2023. [Google Scholar] [CrossRef]
- Liu, Y.; Nusrat, A.; Schnell, F.J.; Reaves, T.A.; Walsh, S.; Pochet, M.; Parkos, C.A. Human junction adhesion molecule regulates tight junction resealing in epithelia. J. Cell Sci. 2000, 113 Pt 13, 2363–2374. [Google Scholar] [CrossRef] [PubMed]
- Iden, S.; Misselwitz, S.; Peddibhotla, S.S.; Tuncay, H.; Rehder, D.; Gerke, V.; Robenek, H.; Suzuki, A.; Ebnet, K. aPKC phosphorylates JAM-A at Ser285 to promote cell contact maturation and tight junction formation. J. Cell Biol. 2012, 196, 623–639. [Google Scholar] [CrossRef] [PubMed]
- Campbell, H.K.; Maiers, J.L.; DeMali, K.A. Interplay between tight junctions & adherens junctions. Exp. Cell Res. 2017, 358, 39–44. [Google Scholar] [CrossRef]
- Ebnet, K. Organization of multiprotein complexes at cell-cell junctions. Histochem. Cell Biol. 2008, 130, 1–20. [Google Scholar] [CrossRef]
- Pawson, T.; Nash, P. Assembly of cell regulatory systems through protein interaction domains. Science 2003, 300, 445–452. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Case, L.B.; Ditlev, J.A.; Rosen, M.K. Regulation of Transmembrane Signaling by Phase Separation. Annu. Rev. Biophys. 2019, 48, 465–494. [Google Scholar] [CrossRef]
- Mayer, B.J.; Yu, J. Protein Clusters in Phosphotyrosine Signal Transduction. J. Mol. Biol. 2018, 430, 4547–4556. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; LuValle-Burke, I.; Pombo-Garcia, K.; Honigmann, A. Biomolecular condensates in epithelial junctions. Curr. Opin. Cell Biol. 2022, 77, 102089. [Google Scholar] [CrossRef] [PubMed]
- Beutel, O.; Maraspini, R.; Pombo-Garcia, K.; Martin-Lemaitre, C.; Honigmann, A. Phase Separation of Zonula Occludens Proteins Drives Formation of Tight Junctions. Cell 2019, 179, 923–936.e11. [Google Scholar] [CrossRef] [PubMed]
- Schwayer, C.; Shamipour, S.; Pranjic-Ferscha, K.; Schauer, A.; Balda, M.; Tada, M.; Matter, K.; Heisenberg, C.P. Mechanosensation of Tight Junctions Depends on ZO-1 Phase Separation and Flow. Cell 2019, 179, 937–952.e18. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, N.; Yamamoto, T.S.; Yasue, N.; Takagi, C.; Fujimori, T.; Ueno, N. Force-dependent remodeling of cytoplasmic ZO-1 condensates contributes to cell-cell adhesion through enhancing tight junctions. iScience 2022, 25, 103846. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zahao, X.; Wiegand, T.; Bartolucci, G.; Martin-Lemaitre, C.; Grill, S.W.; Hyman, A.A.; Weber, C.; Honigmann, A. Assembly of tight junction belts by surface condensation and actin elongation. bioRxiv 2023. [Google Scholar] [CrossRef]
- Sasaki, H.; Matsui, C.; Furuse, K.; Mimori-Kiyosue, Y.; Furuse, M.; Tsukita, S. Dynamic behavior of paired claudin strands within apposing plasma membranes. Proc. Natl. Acad. Sci. USA 2003, 100, 3971–3976. [Google Scholar] [CrossRef]
- Shen, L.; Weber, C.R.; Turner, J.R. The tight junction protein complex undergoes rapid and continuous molecular remodeling at steady state. J. Cell Biol. 2008, 181, 683–695. [Google Scholar] [CrossRef]
- Varadarajan, S.; Stephenson, R.E.; Miller, A.L. Multiscale dynamics of tight junction remodeling. J. Cell Sci. 2019, 132, jcs229286. [Google Scholar] [CrossRef]
- Shigetomi, K.; Ikenouchi, J. Regulation of the epithelial barrier by post-translational modifications of tight junction membrane proteins. J. Biochem. 2018, 163, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Reiche, J.; Huber, O. Post-translational modifications of tight junction transmembrane proteins and their direct effect on barrier function. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183330. [Google Scholar] [CrossRef]
- Blaskovic, S.; Blanc, M.; van der Goot, F.G. What does S-palmitoylation do to membrane proteins? FEBS J. 2013, 280, 2766–2774. [Google Scholar] [CrossRef]
- Lingwood, D.; Simons, K. Lipid rafts as a membrane-organizing principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Levental, I.; Lingwood, D.; Grzybek, M.; Coskun, U.; Simons, K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 22050–22054. [Google Scholar] [CrossRef] [PubMed]
- Nusrat, A.; Parkos, C.A.; Verkade, P.; Foley, C.S.; Liang, T.W.; Innis-Whitehouse, W.; Eastburn, K.K.; Madara, J.L. Tight junctions are membrane microdomains. J. Cell Sci. 2000, 113 Pt 10, 1771–1781. [Google Scholar] [CrossRef]
- Shigetomi, K.; Ono, Y.; Inai, T.; Ikenouchi, J. Adherens junctions influence tight junction formation via changes in membrane lipid composition. J. Cell Biol. 2018, 217, 2373–2381. [Google Scholar] [CrossRef]
- Van Itallie, C.M.; Gambling, T.M.; Carson, J.L.; Anderson, J.M. Palmitoylation of claudins is required for efficient tight-junction localization. J. Cell Sci. 2005, 118, 1427–1436. [Google Scholar] [CrossRef]
- Heiler, S.; Mu, W.; Zoller, M.; Thuma, F. The importance of claudin-7 palmitoylation on membrane subdomain localization and metastasis-promoting activities. Cell Commun. Signal 2015, 13, 29. [Google Scholar] [CrossRef]
- Rodenburg, R.N.P.; Snijder, J.; van de Waterbeemd, M.; Schouten, A.; Granneman, J.; Heck, A.J.R.; Gros, P. Stochastic palmitoylation of accessible cysteines in membrane proteins revealed by native mass spectrometry. Nat. Commun. 2017, 8, 1280. [Google Scholar] [CrossRef]
- Rajagopal, N.; Irudayanathan, F.J.; Nangia, S. Palmitoylation of Claudin-5 Proteins Influences Their Lipid Domain Affinity and Tight Junction Assembly at the Blood-Brain Barrier Interface. J. Phys. Chem. B 2019, 123, 983–993. [Google Scholar] [CrossRef]
- Yuan, M.; Chen, X.; Sun, Y.; Jiang, L.; Xia, Z.; Ye, K.; Jiang, H.; Yang, B.; Ying, M.; Cao, J.; et al. ZDHHC12-mediated claudin-3 S-palmitoylation determines ovarian cancer progression. Acta Pharm. Sin. B 2020, 10, 1426–1439. [Google Scholar] [CrossRef] [PubMed]
- Shigetomi, K.; Ono, Y.; Matsuzawa, K.; Ikenouchi, J. Cholesterol-rich domain formation mediated by ZO proteins is essential for tight junction formation. Proc. Natl. Acad. Sci. USA 2023, 120, e2217561120. [Google Scholar] [CrossRef] [PubMed]
- Aramsangtienchai, P.; Spiegelman, N.A.; Cao, J.; Lin, H. S-Palmitoylation of Junctional Adhesion Molecule C Regulates Its Tight Junction Localization and Cell Migration. J. Biol. Chem. 2017, 292, 5325–5334. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Sugawara, T.; Fukata, Y.; Izumi, Y.; Otani, T.; Higashi, T.; Fukata, M.; Furuse, M. The extracellular domain of angulin-1 and palmitoylation of its cytoplasmic region are required for angulin-1 assembly at tricellular contacts. J. Biol. Chem. 2020, 295, 4289–4302. [Google Scholar] [CrossRef]
- Itoh, M.; Furuse, M.; Morita, K.; Kubota, K.; Saitou, M.; Tsukita, S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J. Cell Biol. 1999, 147, 1351–1363. [Google Scholar] [CrossRef]
- Umeda, K.; Ikenouchi, J.; Katahira-Tayama, S.; Furuse, K.; Sasaki, H.; Nakayama, M.; Matsui, T.; Tsukita, S.; Furuse, M.; Tsukita, S. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell 2006, 126, 741–754. [Google Scholar] [CrossRef]

| Integral Membrane Protein | Size (AA) | Cytoplasmic Region (AA) | COOH-Terminal Residues (PBM) |
|---|---|---|---|
| CRB3 | 120 | 40 | - EERLI (+) |
| JAM-A | 299 | 40 | - SSFLV (+) |
| JAM-C | 310 | 48 | - SSFVI (+) |
| CAR | 365 | 107 | - DGSIV (+) |
| JAM4 | 407 | 122 | - NTTVV (+) |
| ESAM | 390 | 121 | - AGSLV (+) |
| Angulin-1/LSR | 649 | 369 | - ESLVV (+) |
| Angulin-2/ILDR1 | 546 | 358 | - RSVVI (+) |
| Angulin-3/ILDR2 | 639 | 432 | - MSLVV (+) |
| Claudins (1–26) | 207–305 | 27–66 | - XXXYF (+) |
| Occludin | 522 | 257 | - DRQKT (−) |
| Tricellulin | 558 | 196 | - VQGYS (−) |
| MarvelD3 | 401 | 20 | - EMFEF (+) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wibbe, N.; Ebnet, K. Cell Adhesion at the Tight Junctions: New Aspects and New Functions. Cells 2023, 12, 2701. https://doi.org/10.3390/cells12232701
Wibbe N, Ebnet K. Cell Adhesion at the Tight Junctions: New Aspects and New Functions. Cells. 2023; 12(23):2701. https://doi.org/10.3390/cells12232701
Chicago/Turabian StyleWibbe, Nicolina, and Klaus Ebnet. 2023. "Cell Adhesion at the Tight Junctions: New Aspects and New Functions" Cells 12, no. 23: 2701. https://doi.org/10.3390/cells12232701
APA StyleWibbe, N., & Ebnet, K. (2023). Cell Adhesion at the Tight Junctions: New Aspects and New Functions. Cells, 12(23), 2701. https://doi.org/10.3390/cells12232701