Lysine Demethylase KDM2A Promotes Proteasomal Degradation of TCF/LEF Transcription Factors in a Neddylation-Dependent Manner
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells, Transfection, and Treatment
2.2. Plasmids
2.3. RNA and q-RT-PCR
2.4. Luciferase Reporter Assay
2.5. Antibodies
2.6. Proteins and Western Blot
2.7. Co-Immunoprecipitation and GFP-Trap
3. Results
3.1. KDM2A Promotes Destabilization of TCF/LEF Proteins Independently of Its Demethylase Domain and in a CXXC-Domain-Dependent Manner
3.2. KDM2A Interacts with TCF/LEFs and Promotes Their Proteasomal Degradation in a Neddylation-Dependent Manner
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Anastas, J.N.; Moon, R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 2013, 13, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H.; Nusse, R. Wnt/β-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- Sokol, S.Y. Maintaining embryonic stem cell pluripotency with Wnt signaling. Development 2011, 138, 4341–4350. [Google Scholar] [CrossRef]
- Cadigan, K.M.; Peifer, M. Wnt signaling from development to disease: Insights from model systems. Cold Spring Harb. Perspect. Biol. 2009, 1, a002881. [Google Scholar] [CrossRef]
- Liu, C.; Li, Y.; Semenov, M.; Han, C.; Baeg, G.H.; Tan, Y.; Zhang, Z.; Lin, X.; He, X. Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 2002, 108, 837–847. [Google Scholar] [CrossRef]
- Shen, C.; Wang, D.; Liu, X.; Gu, B.; Du, Y.; Wei, F.Z.; Cao, L.L.; Song, B.; Lu, X.; Yang, Q.; et al. SET7/9 regulates cancer cell proliferation by influencing β-catenin stability. FASEB J. 2015, 29, 4313–4323. [Google Scholar] [CrossRef]
- Lu, L.; Gao, Y.; Zhang, Z.; Cao, Q.; Zhang, X.; Zou, J.; Cao, Y. Kdm2a/b Lysine Demethylases Regulate Canonical Wnt Signaling by Modulating the Stability of Nuclear β-Catenin. Dev. Cell 2015, 33, 660–674. [Google Scholar] [CrossRef]
- Gao, C.; Xiao, G.; Hu, J. Regulation of Wnt/beta-catenin signaling by posttranslational modifications. Cell Biosci. 2014, 4, 13. [Google Scholar] [CrossRef]
- Gay, F.; Calvo, D.; Lo, M.C.; Ceron, J.; Maduro, M.; Lin, R.; Shi, Y. Acetylation regulates subcellular localization of the Wnt signaling nuclear effector POP-1. Genes Dev. 2003, 17, 717–722. [Google Scholar] [CrossRef][Green Version]
- Hikasa, H.; Ezan, J.; Itoh, K.; Li, X.; Klymkowsky, M.W.; Sokol, S.Y. Regulation of TCF3 by Wnt-dependent phosphorylation during vertebrate axis specification. Dev. Cell 2010, 19, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Hikasa, H.; Sokol, S.Y. Phosphorylation of TCF proteins by homeodomain-interacting protein kinase 2. J. Biol. Chem. 2011, 286, 12093–12100. [Google Scholar] [CrossRef] [PubMed]
- Ishitani, T.; Ninomiya-Tsuji, J.; Nagai, S.; Nishita, M.; Meneghini, M.; Barker, N.; Waterman, M.; Bowerman, B.; Clevers, H.; Shibuya, H.; et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature 1999, 399, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Vacik, T.; Ladinovic, D.; Raska, I. KDM2A/B lysine demethylases and their alternative isoforms in development and disease. Nucleus 2018, 9, 431–441. [Google Scholar] [CrossRef]
- Tsukada, Y.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006, 439, 811–816. [Google Scholar] [CrossRef]
- Liu, L.; Liu, J.; Lin, Q. Histone demethylase KDM2A: Biological functions and clinical values (Review). Exp. Ther. Med. 2021, 22, 723. [Google Scholar] [CrossRef]
- Dhar, S.S.; Alam, H.; Li, N.; Wagner, K.W.; Chung, J.; Ahn, Y.W.; Lee, M.G. Transcriptional repression of histone deacetylase 3 by the histone demethylase KDM2A is coupled to tumorigenicity of lung cancer cells. J. Biol. Chem. 2014, 289, 7483–7496. [Google Scholar] [CrossRef]
- Wagner, K.W.; Alam, H.; Dhar, S.S.; Giri, U.; Li, N.; Wei, Y.; Giri, D.; Cascone, T.; Kim, J.H.; Ye, Y.; et al. KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J. Clin. Investig. 2013, 123, 5231–5246. [Google Scholar] [CrossRef]
- Kooistra, S.M.; Helin, K. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 2012, 13, 297–311. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Zhou, J.C.; Tolstorukov, M.Y.; Farcas, A.M.; Park, P.J.; Klose, R.J. CpG islands recruit a histone H3 lysine 36 demethylase. Mol. Cell 2010, 38, 179–190. [Google Scholar] [CrossRef]
- Lu, T.; Jackson, M.W.; Wang, B.; Yang, M.; Chance, M.R.; Miyagi, M.; Gudkov, A.V.; Stark, G.R. Regulation of NF-κB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc. Natl. Acad. Sci. USA 2010, 107, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Borgel, J.; Tyl, M.; Schiller, K.; Pusztai, Z.; Dooley, C.M.; Deng, W.; Wooding, C.; White, R.J.; Warnecke, T.; Leonhardt, H.; et al. KDM2A integrates DNA and histone modification signals through a CXXC/PHD module and direct interaction with HP1. Nucleic Acids Res. 2017, 45, 1114–1129. [Google Scholar] [CrossRef] [PubMed]
- Ladinovic, D.; Novotna, J.; Jaksova, S.; Raska, I.; Vacik, T. A demethylation deficient isoform of the lysine demethylase KDM2A interacts with pericentromeric heterochromatin in an HP1a-dependent manner. Nucleus 2017, 8, 563–572. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Frescas, D.; Guardavaccaro, D.; Kuchay, S.M.; Kato, H.; Poleshko, A.; Basrur, V.; Elenitoba-Johnson, K.S.; Katz, R.A.; Pagano, M. KDM2A represses transcription of centromeric satellite repeats and maintains the heterochromatic state. Cell Cycle 2008, 7, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Bueno, M.T.D.; Baldascini, M.; Richard, S.; Lowndes, N.F. Recruitment of lysine demethylase 2A to DNA double strand breaks and its interaction with 53BP1 ensures genome stability. Oncotarget 2018, 9, 15915–15930. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, J.; Wang, Z.; Meng, J.; Wang, A.; Zhao, X.; Xu, Q.; Cai, Z.; Hu, Z. KDM2A Targets PFKFB3 for Ubiquitylation to Inhibit the Proliferation and Angiogenesis of Multiple Myeloma Cells. Front. Oncol. 2021, 11, 653788. [Google Scholar] [CrossRef]
- Gorelik, M.; Manczyk, N.; Pavlenco, A.; Kurinov, I.; Sidhu, S.S.; Sicheri, F. A Structure-Based Strategy for Engineering Selective Ubiquitin Variant Inhibitors of Skp1-Cul1-F-Box Ubiquitin Ligases. Structure 2018, 26, 1226–1236.e3. [Google Scholar] [CrossRef]
- Zhang, H.; Rong, X.; Wang, C.; Liu, Y.; Lu, L.; Li, Y.; Zhao, C.; Zhou, J. VBP1 modulates Wnt/β-catenin signaling by mediating the stability of the transcription factors TCF/LEFs. J. Biol. Chem. 2020, 295, 16826–16839. [Google Scholar] [CrossRef]
- Yamada, M.; Ohnishi, J.; Ohkawara, B.; Iemura, S.; Satoh, K.; Hyodo-Miura, J.; Kawachi, K.; Natsume, T.; Shibuya, H. NARF, an nemo-like kinase (NLK)-associated ring finger protein regulates the ubiquitylation and degradation of T cell factor/lymphoid enhancer factor (TCF/LEF). J. Biol. Chem. 2006, 281, 20749–20760. [Google Scholar] [CrossRef]
- Hou, P.; Ma, X.; Zhang, Q.; Wu, C.J.; Liao, W.; Li, J.; Wang, H.; Zhao, J.; Zhou, X.; Guan, C.; et al. USP21 deubiquitinase promotes pancreas cancer cell stemness via Wnt pathway activation. Genes Dev. 2019, 33, 1361–1366. [Google Scholar] [CrossRef]
- Han, W.; Lee, H.; Han, J.K. Ubiquitin C-terminal hydrolase37 regulates Tcf7 DNA binding for the activation of Wnt signalling. Sci. Rep. 2017, 7, 42590. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Cai, M.; Wu, L.; Zhen, X.; Chen, Y.; Peng, J.; Han, S.; Zhang, P. Ubiquitin-specific protease 28 deubiquitinates TCF7L2 to govern the action of the Wnt signaling pathway in hepatic carcinoma. Cancer Sci. 2022, 113, 3463–3475. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, L.; Holowatyj, A.; Jiang, Y.; Yang, Z.Q. Integrated genomic and functional analyses of histone demethylases identify oncogenic KDM2A isoform in breast cancer. Mol. Carcinog. 2016, 55, 977–990. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Liu, Y.; Yu, L.; Chen, J.; Hou, J.; Cui, L.; Ma, D.; Lu, W. Histone demethylase KDM2A promotes tumor cell growth and migration in gastric cancer. Tumor Biol. 2015, 36, 271–278. [Google Scholar] [CrossRef]
- Zhu, Q.; Shen, Y.; Chen, X.; He, J.; Liu, J.; Zu, X. Self-Renewal Signalling Pathway Inhibitors: Perspectives on Therapeutic Approaches for Cancer Stem Cells. OncoTargets Ther. 2020, 13, 525–540. [Google Scholar] [CrossRef]
- Fendler, A.; Bauer, D.; Busch, J.; Jung, K.; Wulf-Goldenberg, A.; Kunz, S.; Song, K.; Myszczyszyn, A.; Elezkurtaj, S.; Erguen, B.; et al. Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Nat. Commun. 2020, 11, 929. [Google Scholar] [CrossRef]
- Clara, J.A.; Monge, C.; Yang, Y.; Takebe, N. Targeting signalling pathways and the immune microenvironment of cancer stem cells—A clinical update. Nat. Rev. Clin. Oncol. 2019, 17, 204–232. [Google Scholar] [CrossRef]
- Ladinovic, D.; Pinkas, D.; Sopin, T.; Raska, O.; Liska, F.; Raska, I.; Vacik, T. Alternative isoforms of KDM2A and KDM2B lysine demethylases negatively regulate canonical Wnt signaling. PLoS ONE 2020, 15, e0236612. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Weise, A.; Bruser, K.; Elfert, S.; Wallmen, B.; Wittel, Y.; Wohrle, S.; Hecht, A. Alternative splicing of Tcf7l2 transcripts generates protein variants with differential promoter-binding and transcriptional activation properties at Wnt/β-catenin targets. Nucleic Acids Res. 2010, 38, 1964–1981. [Google Scholar] [CrossRef]
- Sato, N.; Meijer, L.; Skaltsounis, L.; Greengard, P.; Brivanlou, A.H. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 2004, 10, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.; Goedert, M. GSK3 inhibitors: Development and therapeutic potential. Nat. Rev. Drug Discov. 2004, 3, 479–487. [Google Scholar] [CrossRef]
- Li, V.S.; Ng, S.S.; Boersema, P.J.; Low, T.Y.; Karthaus, W.R.; Gerlach, J.P.; Mohammed, S.; Heck, A.J.; Maurice, M.M.; Mahmoudi, T.; et al. Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell 2012, 149, 1245–1256. [Google Scholar] [CrossRef]
- Sarikas, A.; Hartmann, T.; Pan, Z.Q. The cullin protein family. Genome Biol. 2011, 12, 220. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, N.E.; Guven, A.; Gray, K.; Shah, P.; Kasture, G.; Nastke, M.D.; Thakurta, A.; Gesta, S.; Vishnudas, V.K.; Narain, N.R.; et al. The Next Frontier: Translational Development of Ubiquitination, SUMOylation, and NEDDylation in Cancer. Int. J. Mol. Sci. 2022, 23, 3480. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xiang, Y.; Fan, M.; Fang, S.; Hua, Q. The Ubiquitin-Proteasome System in Tumor Metabolism. Cancers 2023, 15, 2385. [Google Scholar] [CrossRef]
- Graham, T.A.; Ferkey, D.M.; Mao, F.; Kimelman, D.; Xu, W. Tcf4 can specifically recognize β-catenin using alternative conformations. Nat. Struct. Biol. 2001, 8, 1048–1052. [Google Scholar] [CrossRef]
- Zuo, Y.; Zhan, L.; Wen, H.; Xue, J.; Tan, Y.; Sun, W.; Xu, E. Stabilization of nuclear β-catenin by inhibiting KDM2A mediates cerebral ischemic tolerance. FASEB J. 2023, 37, e22796. [Google Scholar] [CrossRef]
- Hwang, S.G.; Yu, S.S.; Ryu, J.H.; Jeon, H.B.; Yoo, Y.J.; Eom, S.H.; Chun, J.S. Regulation of beta-catenin signaling and maintenance of chondrocyte differentiation by ubiquitin-independent proteasomal degradation of α-catenin. J. Biol. Chem. 2005, 280, 12758–12765. [Google Scholar] [CrossRef]
- Schwechheimer, C. NEDD8-its role in the regulation of Cullin-RING ligases. Curr. Opin. Plant Biol. 2018, 45, 112–119. [Google Scholar] [CrossRef]
- Wang, B.; Wang, T.; Zhu, H.; Yan, R.; Li, X.; Zhang, C.; Tao, W.; Ke, X.; Hao, P.; Qu, Y. Neddylation is essential for β-catenin degradation in Wnt signaling pathway. Cell Rep. 2022, 38, 110538. [Google Scholar] [CrossRef]
- Zhang, L.; Jing, H.; Li, H.; Chen, W.; Luo, B.; Zhang, H.; Dong, Z.; Li, L.; Su, H.; Xiong, W.C.; et al. Neddylation is critical to cortical development by regulating Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 26448–26459. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, S.; Li, B.; Li, Y.; Xia, K.; Aman, S.; Yang, Y.; Ahmad, B.; Zhao, B.; Wu, H. FBXL10 promotes ERRα protein stability and proliferation of breast cancer cells by enhancing the mono-ubiquitylation of ERRα. Cancer Lett. 2021, 502, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.Z.; Lin, H.Y.; Kuei, C.H.; Lin, C.H.; Lee, H.H.; Lee, H.L.; Lu, H.W.; Su, C.Y.; Chiu, H.W.; Lin, Y.F. NEDD8 promotes radioresistance via triggering autophagy formation and serves as a novel prognostic marker in oral squamous cell carcinoma. Cancer Cell Int. 2023, 23, 41. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Lin, X.; Zhu, J.; Zhang, L.; Chen, S.; Yang, H.; Jia, L.; Chen, B. NEDD8-conjugating enzyme E2s: Critical targets for cancer therapy. Cell Death Discov. 2023, 9, 23. [Google Scholar] [CrossRef]
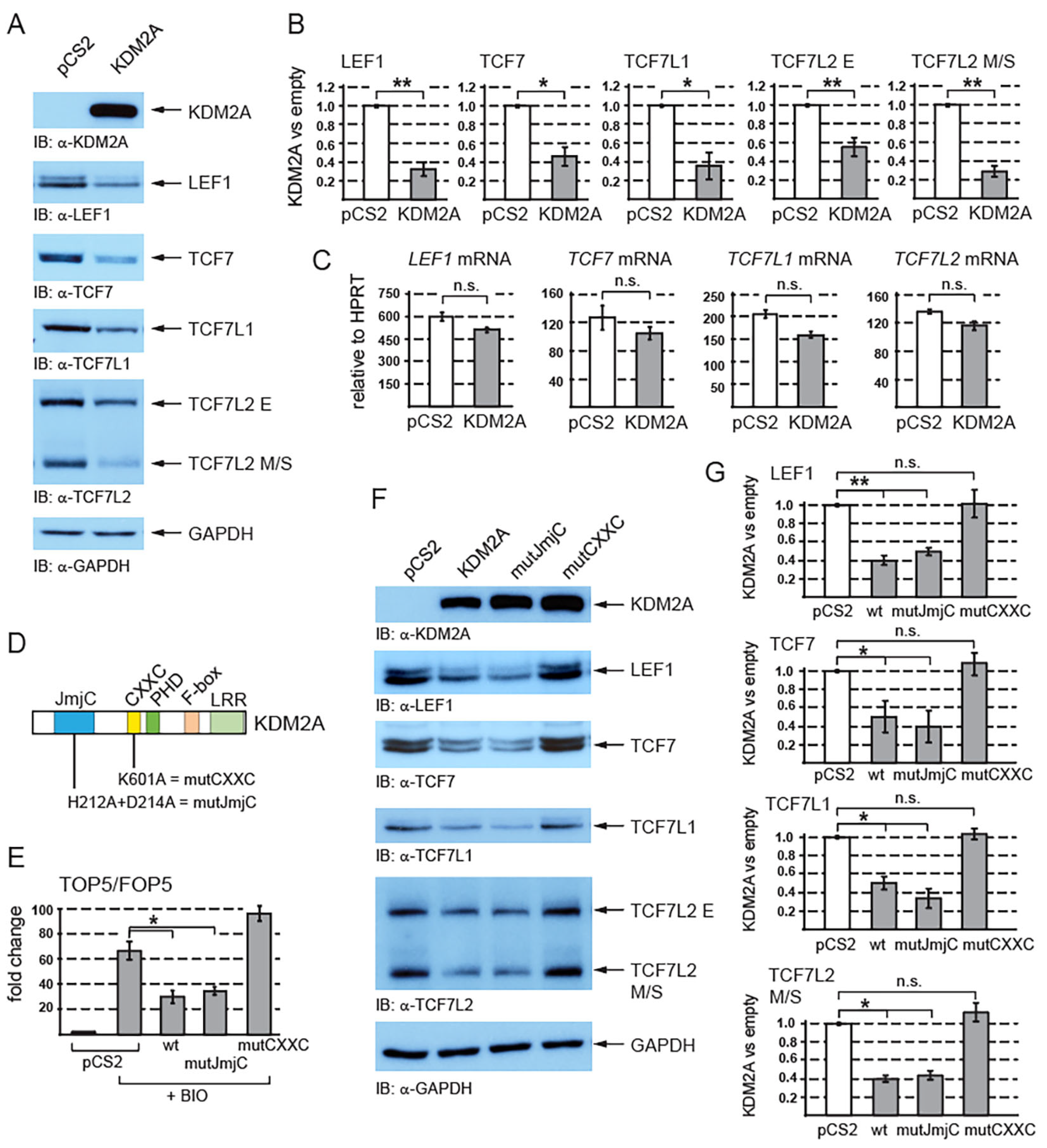

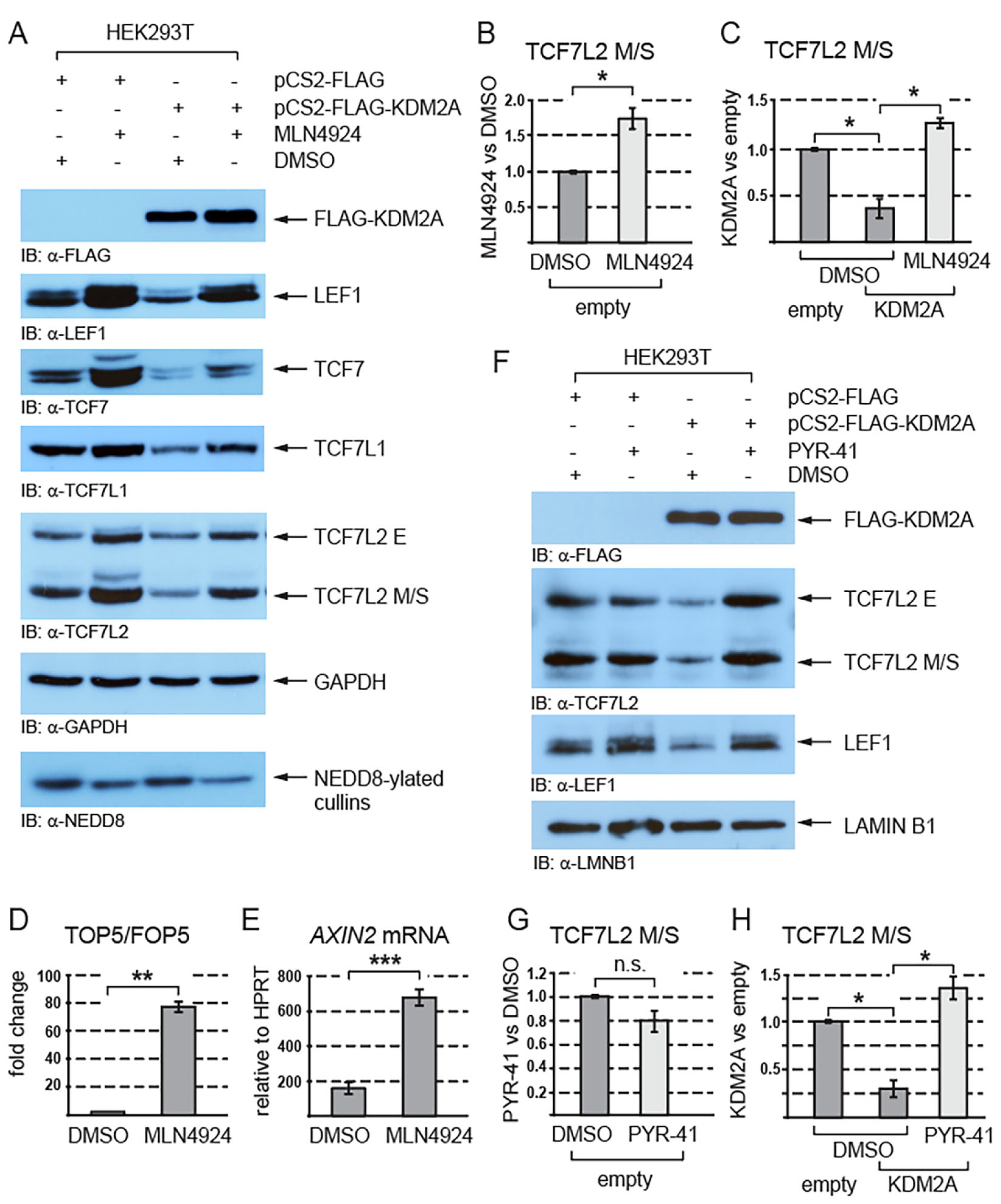
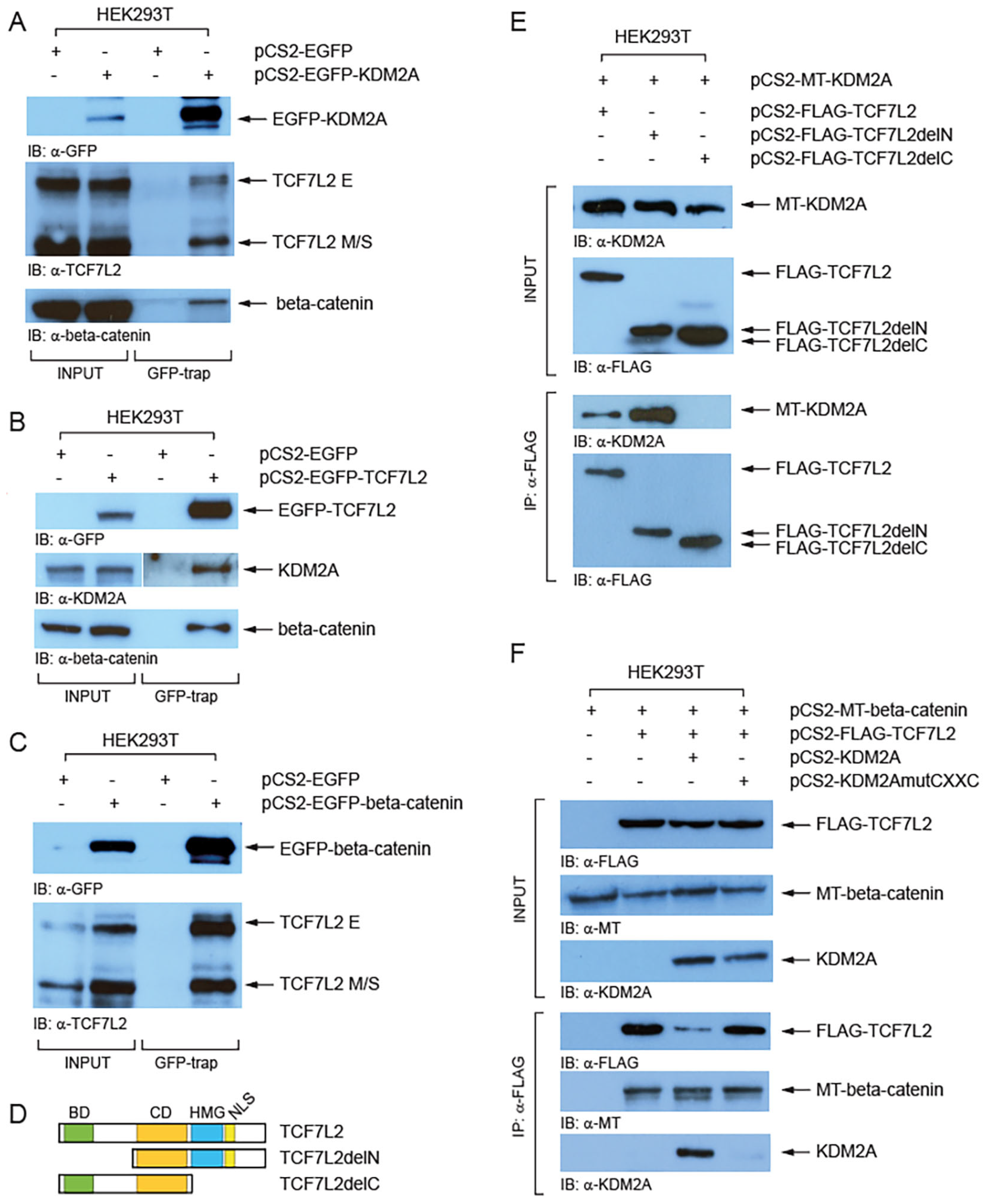
| Gene | Primer | Sequence (5′ to 3′) |
|---|---|---|
| HPRT1 | forward | TCTTTGCTGACCTGCTGGATTAC |
| reverse | GTCTGCATTGTTTTGCCAGTGTC | |
| RPL32 | forward | CTCAGACCCCTTGTGAAGCC |
| reverse | TTGCTTCCATAACCAATGTTGG | |
| LEF1 | forward | GGTGAACGAGTCTGAAATCATCC |
| reverse | TGTTCTCTGGCCTTGTCGTG | |
| TCF7 | forward | ACGAACATTTCAACAGCCCAC |
| reverse | TCAGGGAGTAGAAGCCAGAGAGG | |
| TCF7L1 | forward | TTCCTGATGATCCCGGACC |
| reverse | GAGATGGTGACCTCGTGTCCTT | |
| TCF7L2 | forward | GACAAGCCCTCAAGGATGCC |
| reverse | CGTCGGCTGGTAAGTGTGG | |
| AXIN2 | forward | CTGACGGATGATTCCATGTCC |
| reverse | GGGAAATGAGGTAGAGACACTTGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šopin, T.; Liška, F.; Kučera, T.; Cmarko, D.; Vacík, T. Lysine Demethylase KDM2A Promotes Proteasomal Degradation of TCF/LEF Transcription Factors in a Neddylation-Dependent Manner. Cells 2023, 12, 2620. https://doi.org/10.3390/cells12222620
Šopin T, Liška F, Kučera T, Cmarko D, Vacík T. Lysine Demethylase KDM2A Promotes Proteasomal Degradation of TCF/LEF Transcription Factors in a Neddylation-Dependent Manner. Cells. 2023; 12(22):2620. https://doi.org/10.3390/cells12222620
Chicago/Turabian StyleŠopin, Tijana, František Liška, Tomáš Kučera, Dušan Cmarko, and Tomáš Vacík. 2023. "Lysine Demethylase KDM2A Promotes Proteasomal Degradation of TCF/LEF Transcription Factors in a Neddylation-Dependent Manner" Cells 12, no. 22: 2620. https://doi.org/10.3390/cells12222620
APA StyleŠopin, T., Liška, F., Kučera, T., Cmarko, D., & Vacík, T. (2023). Lysine Demethylase KDM2A Promotes Proteasomal Degradation of TCF/LEF Transcription Factors in a Neddylation-Dependent Manner. Cells, 12(22), 2620. https://doi.org/10.3390/cells12222620






