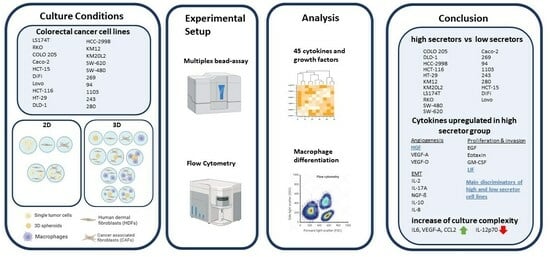
A Subset of Colon Cancer Cell Lines Displays a Cytokine Profile Linked to Angiogenesis, EMT and Invasion Which Is Modulated by the Culture Conditions In Vitro
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Isolation and Differentiation of Human Monocytes
2.3. 2D and 3D Spheroids Cultures
2.4. 3D Matrigel-Collagen Matrix
2.5. Flow Cytometry
2.6. Multiplex Bead Assay Analysis
2.7. Incucyte Software Analysis
3. Results
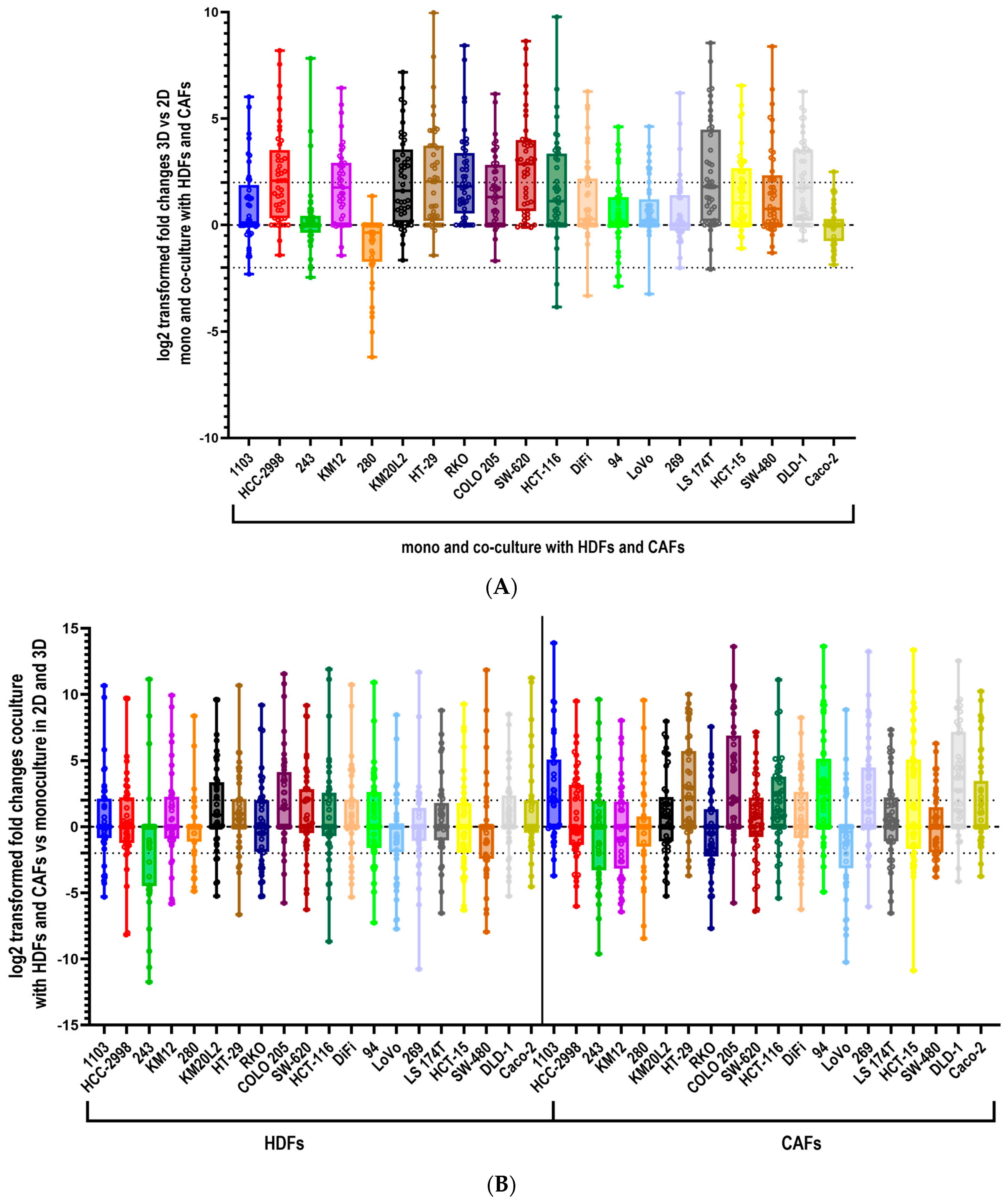
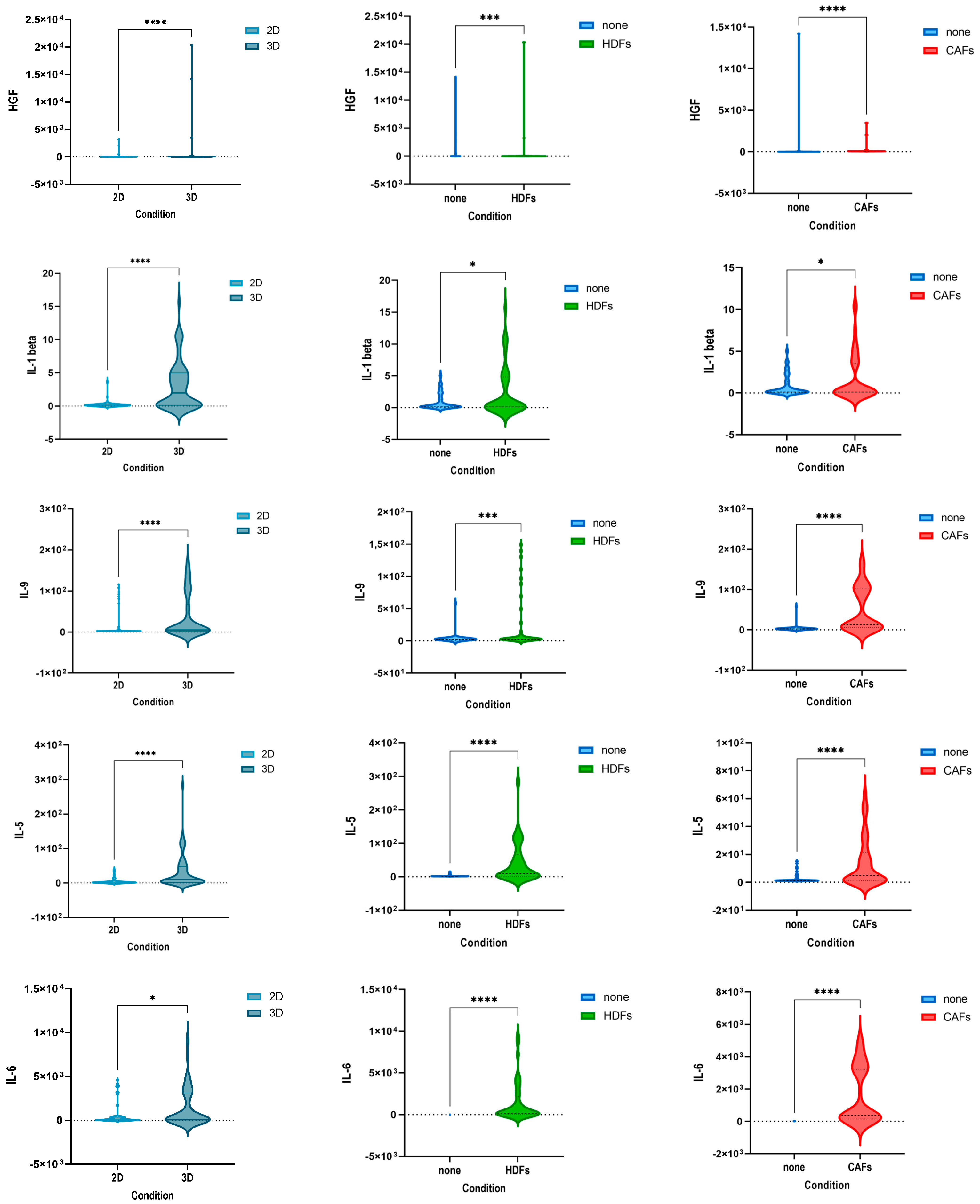
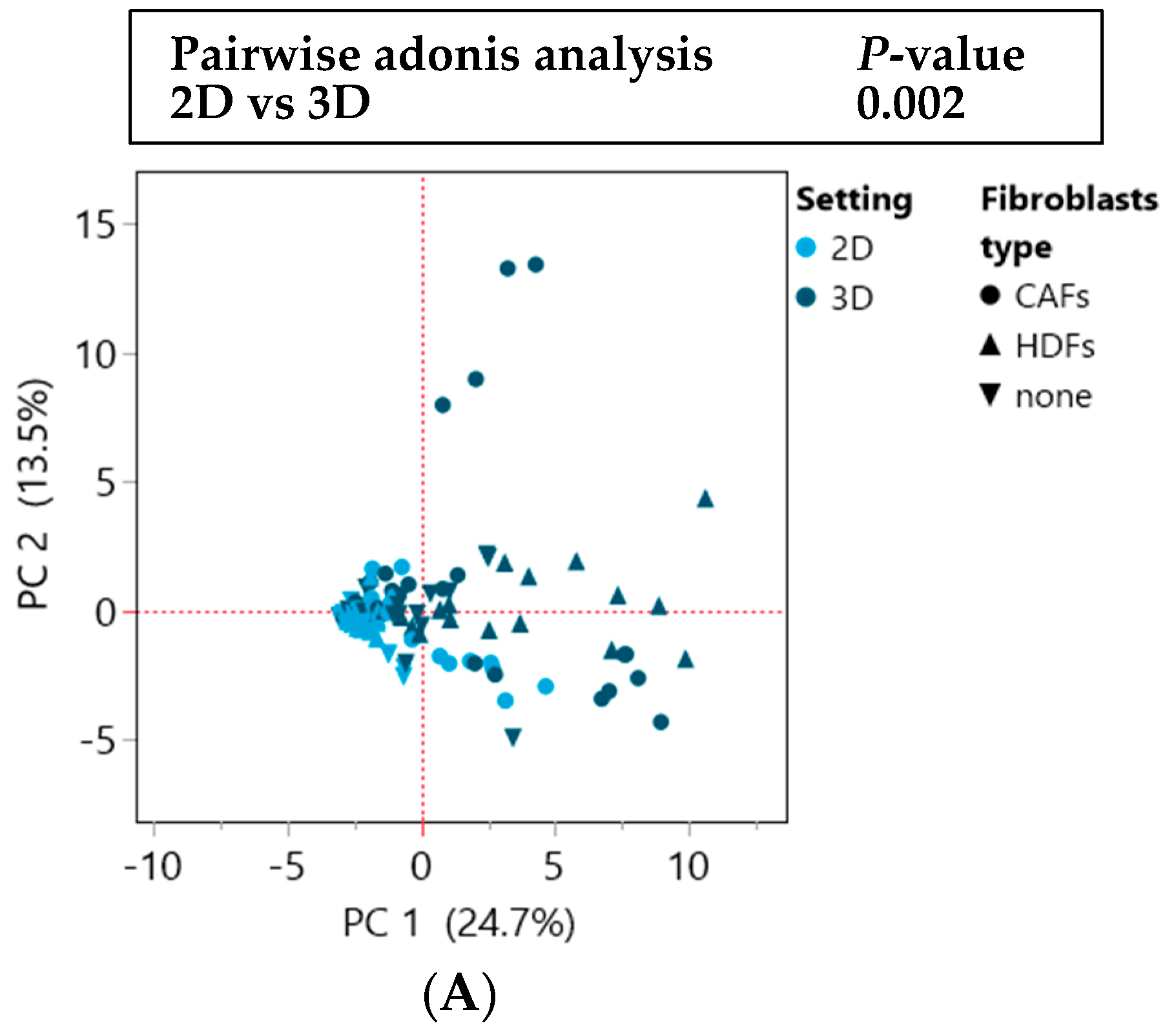
3.1. Culturing Colorectal Cancer Cell Lines in 3D Spheroid Cultures and in Coculture with Fibroblasts Enhances the Secretion of Cytokines
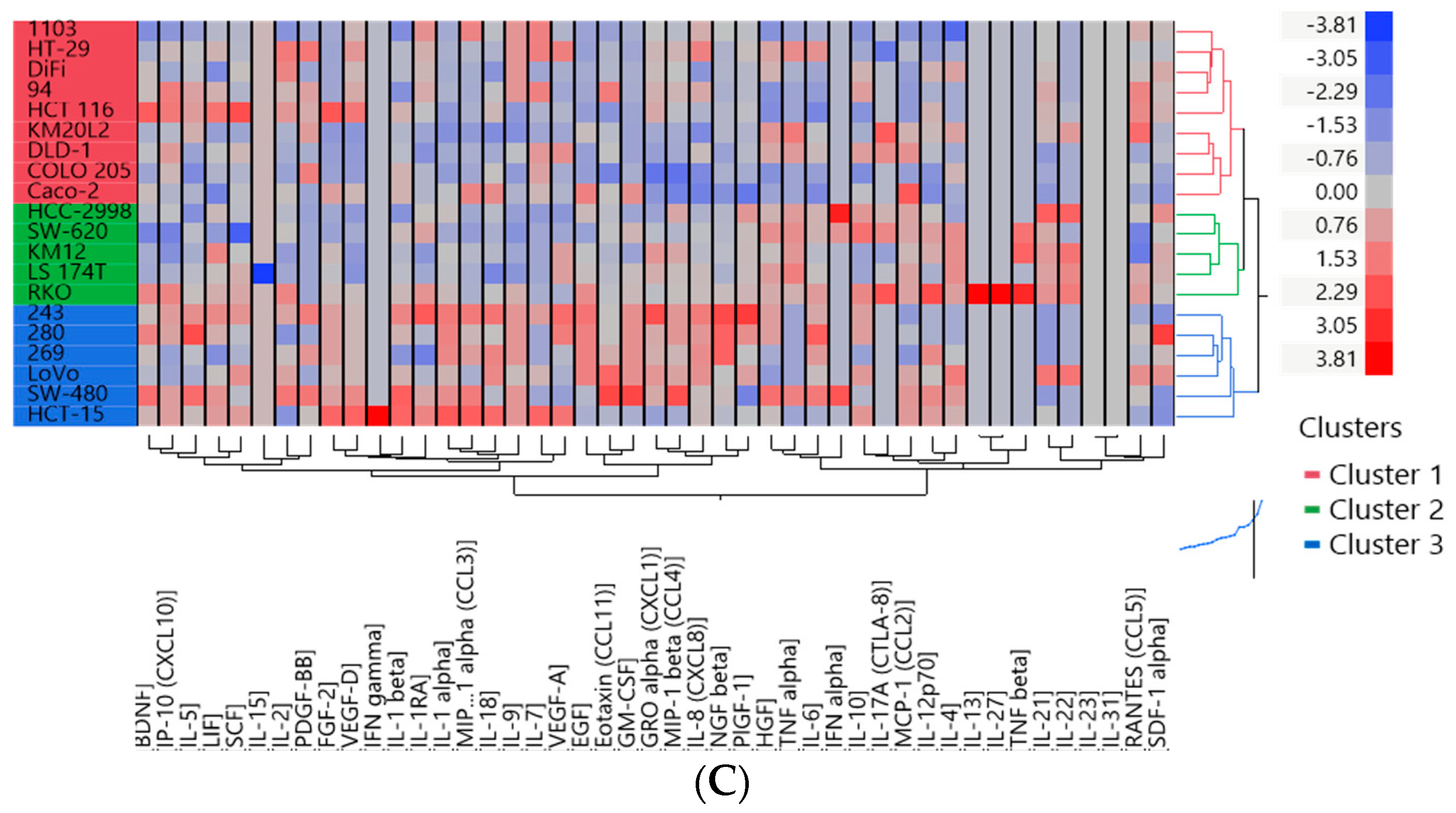
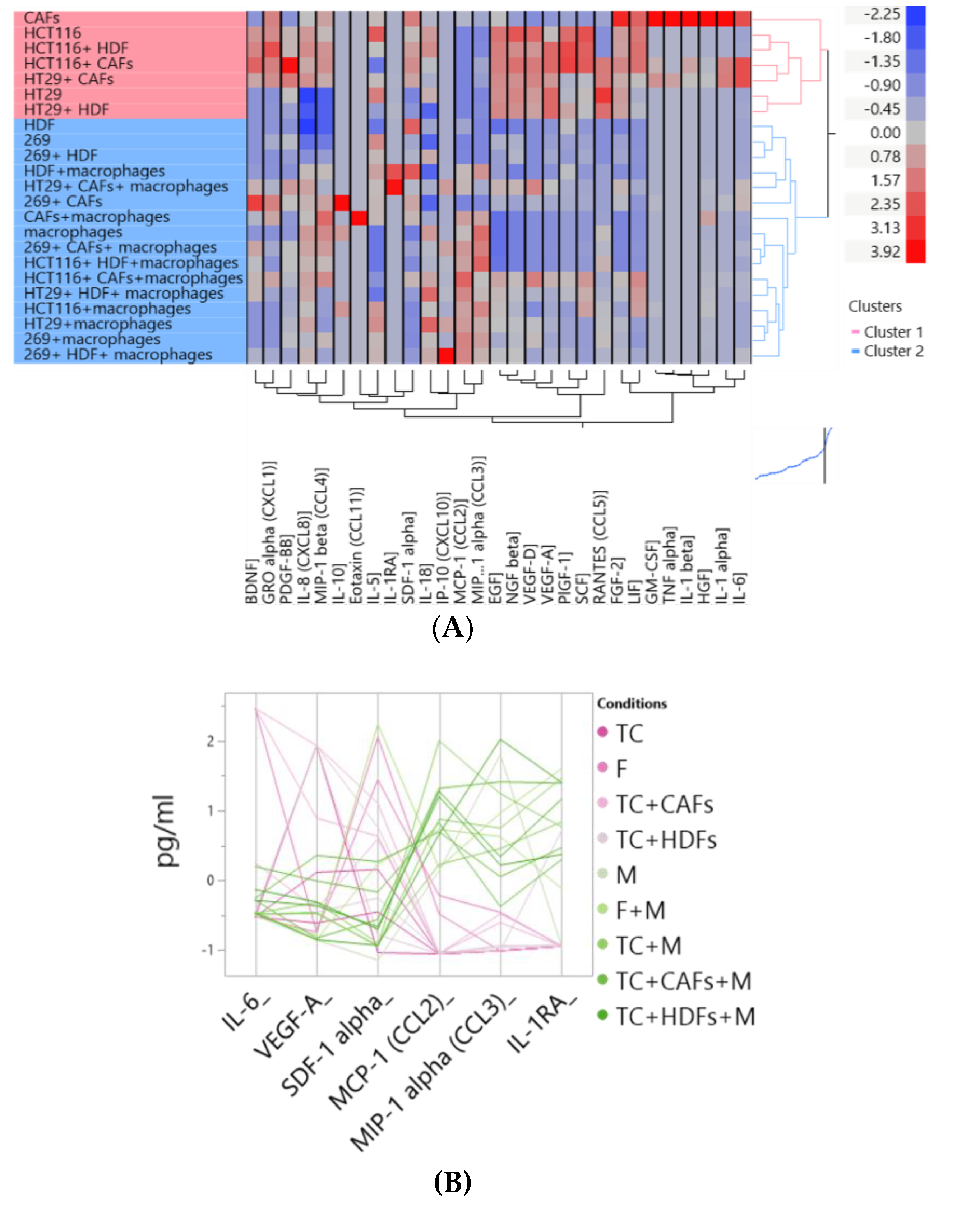
3.2. Two Distinct Subgroups Were Identified Based on the Cytokine Profile In Vitro Displaying Differences in the Secretion of Cytokines Related to Angiogenesis, EMT and Invasion
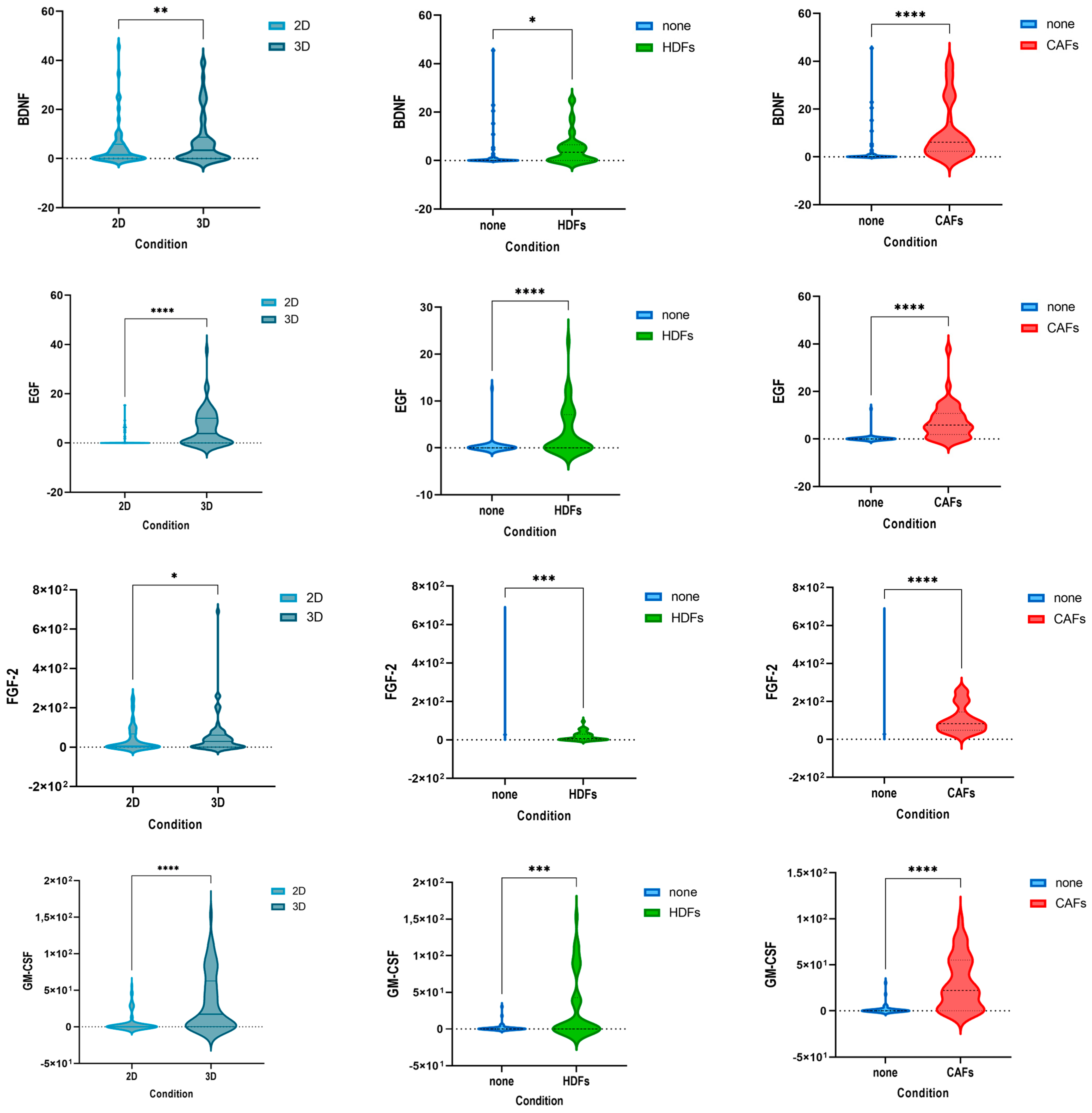
3.3. The Cytokine Expression of CRC Cell Lines Was Significantly Influenced by the Culture Condition and the Presence of Fibroblasts
3.4. The Investigated Tumor-Intrinsic Features of the CRC Lines Had No Major Impact on Their Cytokine Profile
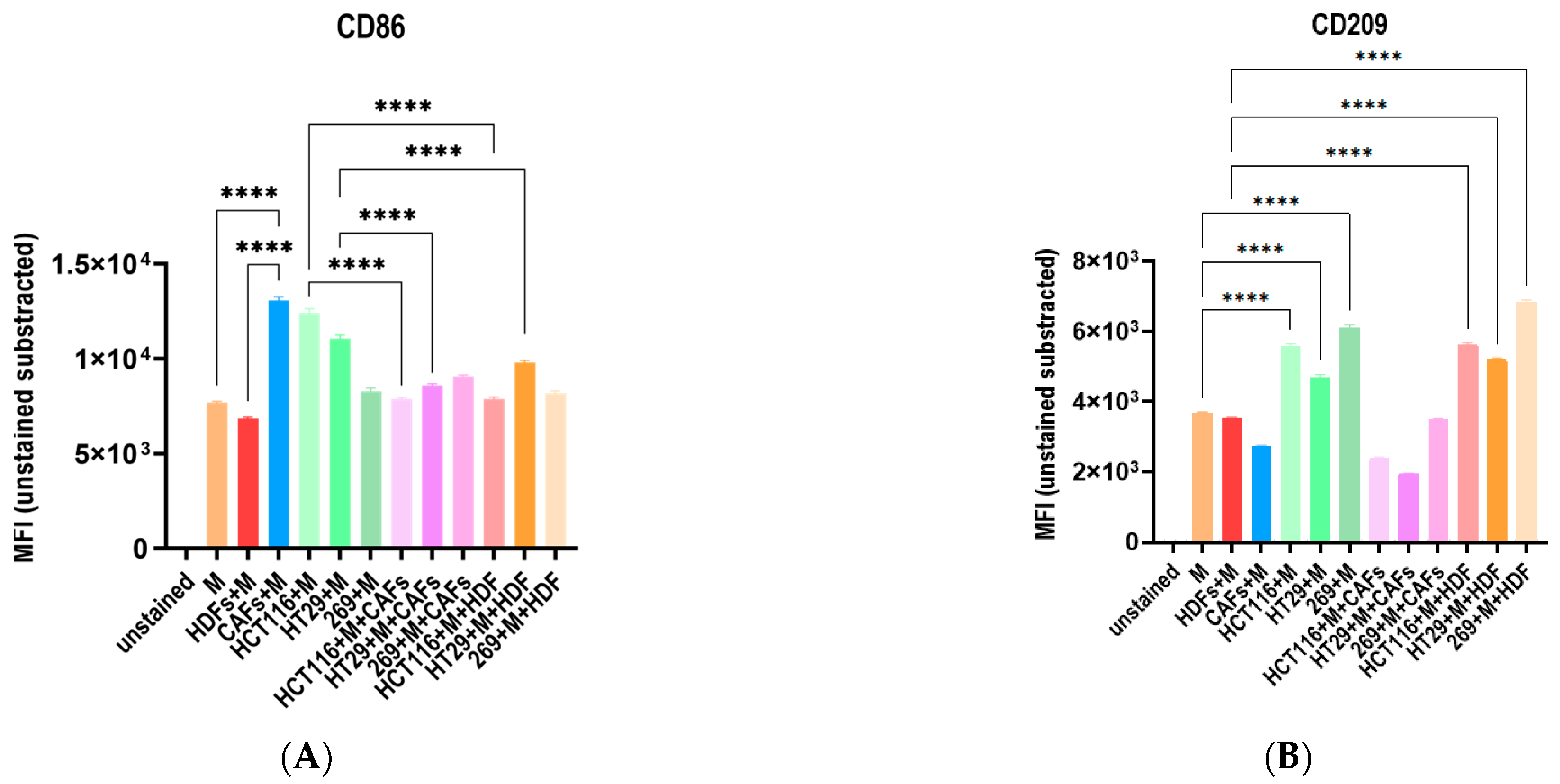
3.5. The Co-Culture with Different Tumor Cell Lines and Fibroblasts Influence the Macrophage Polarization Distinctively
3.6. The Cocultures with Macrophages and Fibroblasts Modulates the Cytike Profile and Confirms the Impact of the Fibrobast Source on Macrophage Polarization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pretzsch, E.; Bösch, F.; Neumann, J.; Ganschow, P.; Bazhin, A.; Guba, M.; Werner, J.; Angele, M. Mechanisms of Metastasis in Colorectal Cancer and Metastatic Organotropism: Hematogenous versus Peritoneal Spread. J. Oncol. 2019, 2019, 7407190. [Google Scholar] [CrossRef] [PubMed]
- Farhan, H.; Rabouille, C. Signalling to and from the secretory pathway. J. Cell Sci. 2011, 124, 171–180. [Google Scholar] [CrossRef] [PubMed]
- La Vecchia, S.; Sebastián, C. Metabolic pathways regulating colorectal cancer initiation and progression. Semin. Cell Dev. Biol. 2020, 98, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Karagiannis, G.S.; Pavlou, M.P.; Diamandis, E.P. Cancer secretomics reveal pathophysiological pathways in cancer molecular oncology. Mol. Oncol. 2010, 4, 496–510. [Google Scholar] [CrossRef]
- Maggio, M.C.; Miniaci, A.; Gallizzi, R.; Civino, A. “Neuroimmunoendocrinology” in Children with Rheumatic Diseases: How Glucocorticoids Are the Orchestra Director. Int. J. Mol. Sci. 2023, 24, 13192. [Google Scholar] [CrossRef]
- da Cunha, B.R.; Domingos, C.; Stefanini, A.C.B.; Henrique, T.; Polachini, G.M.; Castelo-Branco, P.; Tajara, E.H. Cellular Interactions in the Tumor Microenvironment: The Role of Secretome. J. Cancer 2019, 10, 4574–4587. [Google Scholar] [CrossRef]
- Javarsiani, M.H.; Javanmard, S.H.; Colonna, F. Metastatic components in colorectal cancer. J. Res. Med. Sci. 2019, 24, 75. [Google Scholar] [CrossRef] [PubMed]
- Rudisch, A.; Dewhurst, M.R.; Horga, L.G.; Kramer, N.; Harrer, N.; Dong, M.; van der Kuip, H.; Wernitznig, A.; Bernthaler, A.; Dolznig, H.; et al. High EMT Signature Score of Invasive Non-Small Cell Lung Cancer (NSCLC) Cells Correlates with NFκB Driven Colony-Stimulating Factor 2 (CSF2/GM-CSF) Secretion by Neighboring Stromal Fibroblasts. PLoS ONE 2015, 10, e0124283. [Google Scholar] [CrossRef]
- Alkasalias, T.; Moyano-Galceran, L.; Arsenian-Henriksson, M.; Lehti, K. Fibroblasts in the Tumor Microenvironment: Shield or Spear? Int. J. Mol. Sci. 2018, 19, 1532. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Jiang, N.; Zeng, J.; Wang, Y.; Cui, H. The Versatile Roles of Cancer-Associated Fibroblasts in Colorectal Cancer and Therapeutic Implications. Front. Cell Dev. Biol. 2021, 9, 733270. [Google Scholar] [CrossRef]
- Fujimoto, H.; Sangai, T.; Ishii, G.; Ikehara, A.; Nagashima, T.; Miyazaki, M.; Ochiai, A. Stromal MCP-1 in mammary tumors induces tumor-associated macrophage infiltration and contributes to tumor progression. Int. J. Cancer 2009, 125, 1276–1284. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Xu, J.; Lan, H. Tumor-associated macrophages in tumor metastasis: Biological roles and clinical therapeutic applications. J. Hematol. Oncol. 2019, 12, 76. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Seo, Y.; Loke, K.M.; Kim, S.-W.; Oh, S.-M.; Kim, J.-H.; Soh, J.; Kim, H.S.; Lee, H.; Kim, J.; et al. Cancer-Stimulated CAFs Enhance Monocyte Differentiation and Protumoral TAM Activation via IL6 and GM-CSF Secretion. Clin. Cancer Res. 2018, 24, 5407–5421. [Google Scholar] [CrossRef]
- Pinto, M.L.; Rios, E.; Durães, C.; Ribeiro, R.; Machado, J.C.; Mantovani, A.; Barbosa, M.A.; Carneiro, F.; Oliveira, M.J. The Two Faces of Tumor-Associated Macrophages and Their Clinical Significance in Colorectal Cancer. Front. Immunol. 2019, 10, 1875. [Google Scholar] [CrossRef]
- Schneider, M.; Huber, J.; Hadaschik, B.; Siegers, G.M.; Fiebig, H.H.; Schüler, J. Characterization of colon cancer cells: A functional approach characterizing CD133 as a potential stem cell marker. BMC Cancer 2012, 12, 96. [Google Scholar] [CrossRef] [PubMed]
- Néron, S.; Thibault, L.; Dussault, N.; Côté, G.; Ducas, É.; Pineault, N.; Roy, A. Characterization of mononuclear cells remaining in the leukoreduction system chambers of apheresis instruments after routine platelet collection: A new source of viable human blood cells. Transfusion 2007, 47, 1042–1049. [Google Scholar] [CrossRef]
- Brown, W.E.; Hu, J.C.; Athanasiou, K.A. Ammonium-Chloride-Potassium Lysing Buffer Treatment of Fully Differentiated Cells Increases Cell Purity and Resulting Neotissue Functional Properties. Tissue Eng. Part C Methods 2016, 22, 895–903. [Google Scholar] [CrossRef]
- Tjalsma, H.; Bolhuis, A.; Jongbloed, J.D.; Bron, S.; van Dijl, J.M. Signal peptide-dependent protein transport in Bacillus subtilis: A genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 2000, 64, 515–547. [Google Scholar] [CrossRef]
- Cevenini, A.; Orrù, S.; Imperlini, E. Secretome Proteomic Approaches for Biomarker Discovery: An Update on Colorectal Cancer. Medicina 2020, 56, 443. [Google Scholar] [CrossRef]
- Chen, F.; Zhuang, X.; Lin, L.; Yu, P.; Wang, Y.; Shi, Y.; Hu, G.; Sun, Y. New horizons in tumor microenvironment biology: Challenges and opportunities. BMC Med. 2015, 13, 45. [Google Scholar] [CrossRef]
- Xing, F.; Saidou, J.; Watabe, K. Cancer associated fibroblasts (CAFs) in tumor microenvironment. Front. Biosci. 2010, 15, 166–179. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.; Heinrich, M.A.; Teixeira, L.M.; Prakash, J. 3D In Vitro Model (R)evolution: Unveiling Tumor-Stroma Interactions. Trends Cancer 2021, 7, 249–264. [Google Scholar] [CrossRef] [PubMed]
- LeBleu, V.S.; Kalluri, R. A peek into cancer-associated fibroblasts: Origins, functions and translational impact. Dis. Models Mech. 2018, 11, dmm029447. [Google Scholar] [CrossRef]
- Toti, A.; Santi, A.; Pardella, E.; Nesi, I.; Tomasini, R.; Mello, T.; Paoli, P.; Caselli, A.; Cirri, P. Activated fibroblasts enhance cancer cell migration by microvesicles-mediated transfer of Galectin-1. J. Cell Commun. Signal. 2021, 15, 405–419. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.; Sainson, R.C. Regulation of the anti-tumour immune response by cancer-associated fibroblasts. Semin. Cancer Biol. 2014, 25, 69–77. [Google Scholar] [CrossRef]
- Dias Carvalho, P.; Mendonça, S.; Martins, F.; Oliveira, M.J.; Velho, S. Modulation of Fibroblast Phenotype by Colorectal Cancer Cell-Secreted Factors Is Mostly Independent of Oncogenic KRAS. Cells 2022, 11, 2490. [Google Scholar] [CrossRef]
- Yu, H.; Yue, X.; Zhao, Y.; Li, X.; Wu, L.; Zhang, C.; Liu, Z.; Lin, K.; Xu-Monette, Z.Y.; Young, K.H.; et al. LIF negatively regulates tumour-suppressor p53 through Stat3/ID1/MDM2 in colorectal cancers. Nat. Commun. 2014, 5, 5218. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Wang, J.; Hu, W.; Feng, Z. The emerging role of leukemia inhibitory factor in cancer and therapy. Pharmacol. Ther. 2021, 221, 107754. [Google Scholar] [CrossRef]
- Takumi, Y.; Arai, S.; Suzuki, C.; Fukuda, K.; Nishiyama, A.; Takeuchi, S.; Sato, H.; Matsumoto, K.; Sugio, K.; Yano, S. MET kinase inhibitor reverses resistance to entrectinib induced by hepatocyte growth factor in tumors with NTRK1 or ROS1 rearrangements. Cancer Med. 2023, 12, 5809–5820. [Google Scholar] [CrossRef]
- Xiong, M.; Wang, M.; Yan, Y.; Chen, X.; Guo, W.; Xu, M.; Guo, S.; Wang, Y. MACC1 Promotes the Progression and Is a Novel Biomarker for Predicting Immunotherapy Response in Colorectal Cancer. J. Oncol. 2022, 2022, 8326940. [Google Scholar] [CrossRef]
- Grivennikov, S.; Karin, E.; Terzic, J.; Mucida, D.; Yu, G.Y.; Vallabhapurapu, S.; Scheller, J.; Rose-John, S.; Cheroutre, H.; Eckmann, L.; et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 2009, 15, 103–113. [Google Scholar] [CrossRef]
- Nagasaki, T.; Hara, M.; Nakanishi, H.; Takahashi, H.; Sato, M.; Takeyama, H. Interleukin-6 released by colon cancer-associated fibroblasts is critical for tumour angiogenesis: Anti-interleukin-6 receptor antibody suppressed angiogenesis and inhibited tumour-stroma interaction. Br. J. Cancer 2014, 110, 469–478. [Google Scholar] [CrossRef]
- Michielsen, A.J.; Ryan, E.J.; O’Sullivan, J.N. Dendritic cell inhibition correlates with survival of colorectal cancer patients on bevacizumab treatment. Oncoimmunology 2012, 1, 1445–1447. [Google Scholar] [CrossRef][Green Version]
- O’Toole, A.; Michielsen, A.J.; Nolan, B.; Tosetto, M.; Sheahan, K.; Mulcahy, H.E.; Winter, D.C.; Hyland, J.M.; O’Connell, P.R.; Fennelly, D.; et al. Tumour microenvironment of both early- and late-stage colorectal cancer is equally immunosuppressive. Br. J. Cancer 2014, 111, 927–932. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, Z.; Deng, B.; Wu, D.; Quan, Y.; Min, Z. Interleukin 1β/1RA axis in colorectal cancer regulates tumor invasion, proliferation and apoptosis via autophagy. Oncol. Rep. 2020, 43, 908–918. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, L.; Zhao, H.; Yan, Y.; Lu, J. The Role of Interleukins in Colorectal Cancer. Int. J. Biol. Sci. 2020, 16, 2323–2339. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lei, Y.; Sun, J.; Zhang, W.; Li, X.; Chen, S.; Kong, D.; Chen, C.; Bi, K.; Luo, X.; et al. A promising research direction for colorectal cancer immunotherapy: The regulatory mechanism of CCL5 in colorectal cancer. Front. Oncol. 2022, 12, 1020400. [Google Scholar] [CrossRef] [PubMed]
- Chun, E.; Lavoie, S.; Michaud, M.; Gallini, C.A.; Kim, J.; Soucy, G.; Odze, R.; Glickman, J.N.; Garrett, W.S. CCL2 Promotes Colorectal Carcinogenesis by Enhancing Polymorphonuclear Myeloid-Derived Suppressor Cell Population and Function. Cell Rep. 2015, 12, 244–257. [Google Scholar] [CrossRef] [PubMed]
- Stadler, M.; Pudelko, K.; Biermeier, A.; Walterskirchen, N.; Gaigneaux, A.; Weindorfer, C.; Harrer, N.; Klett, H.; Hengstschläger, M.; Schüler, J.; et al. Stromal fibroblasts shape the myeloid phenotype in normal colon and colorectal cancer and induce CD163 and CCL2 expression in macrophages. Cancer Lett. 2021, 520, 184–200. [Google Scholar] [CrossRef]
- Yuan, X.; Liu, W.; Li, Y.; Chen, K.; Li, H.; Tang, H.; Yin, Y.; Song, Z.; Chen, D. CCL3 aggravates intestinal damage in NEC by promoting macrophage chemotaxis and M1 macrophage polarization. Pediatr. Res. 2023, 94, 119–128. [Google Scholar] [CrossRef]
- Pan, B.; Wan, T.; Zhou, Y.; Huang, S.; Yuan, L.; Jiang, Y.; Zheng, X.; Liu, P.; Xiang, H.; Ju, M.; et al. The MYBL2-CCL2 axis promotes tumor progression and resistance to anti-PD-1 therapy in ovarian cancer by inducing immunosuppressive macrophages. Cancer Cell Int. 2023, 23, 248. [Google Scholar] [CrossRef] [PubMed]
- Strasser, K.; Birnleitner, H.; Beer, A.; Pils, D.; Gerner, M.C.; Schmetterer, K.G.; Bachleitner-Hofmann, T.; Stift, A.; Bergmann, M.; Oehler, R. Immunological differences between colorectal cancer and normal mucosa uncover a prognostically relevant immune cell profile. Oncoimmunology 2019, 8, e1537693. [Google Scholar] [CrossRef] [PubMed]








| Cell Lines | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 94 | 243 | 269 | 280 | 1103 | Caco-2 | DiFi | HCT-15 | LoVo | COLO 205 | DLD-1 | HCC-2998 | HCT 116 | HT-29 | KM12 | KM20L2 | LS 174T | RKO | SW-480 | SW-620 | ||
| cluster | 3D monoculture | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 3 | 1 | 2 | 2 | 2 | 2 | 4 | 2 | 2 |
| 3D co-culture | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | |
| 3D mono- vs. co-culture fold change | 1 | 3 | 3 | 3 | 1 | 1 | 1 | 3 | 3 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 2 | 2 | 3 | 2 | |
| group | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| low secretor | high secretor | ||||||||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bersano, J.; Lashuk, K.; Edinger, A.; Schueler, J. A Subset of Colon Cancer Cell Lines Displays a Cytokine Profile Linked to Angiogenesis, EMT and Invasion Which Is Modulated by the Culture Conditions In Vitro. Cells 2023, 12, 2539. https://doi.org/10.3390/cells12212539
Bersano J, Lashuk K, Edinger A, Schueler J. A Subset of Colon Cancer Cell Lines Displays a Cytokine Profile Linked to Angiogenesis, EMT and Invasion Which Is Modulated by the Culture Conditions In Vitro. Cells. 2023; 12(21):2539. https://doi.org/10.3390/cells12212539
Chicago/Turabian StyleBersano, Jacqueline, Kanstantsin Lashuk, Anna Edinger, and Julia Schueler. 2023. "A Subset of Colon Cancer Cell Lines Displays a Cytokine Profile Linked to Angiogenesis, EMT and Invasion Which Is Modulated by the Culture Conditions In Vitro" Cells 12, no. 21: 2539. https://doi.org/10.3390/cells12212539
APA StyleBersano, J., Lashuk, K., Edinger, A., & Schueler, J. (2023). A Subset of Colon Cancer Cell Lines Displays a Cytokine Profile Linked to Angiogenesis, EMT and Invasion Which Is Modulated by the Culture Conditions In Vitro. Cells, 12(21), 2539. https://doi.org/10.3390/cells12212539