Mutant p53 Gain-of-Function Induces Migration and Invasion through Overexpression of miR-182-5p in Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatments
2.2. Plasmids and Transfections
2.3. RNA Extraction
2.4. Reverse Transcription
2.5. Human miRNome PCR Array
2.6. PCR Array Analysis
2.7. Bioinformatic Analysis and Visualization
2.8. Validation of miRNA Expression by RT-qPCR
2.9. Expression Analysis of miR-182-5p Target Genes via RT-qPCR
2.10. Western Blot
2.11. Cell Migration and Invasion Assays
2.12. Statistical Analysis
3. Results
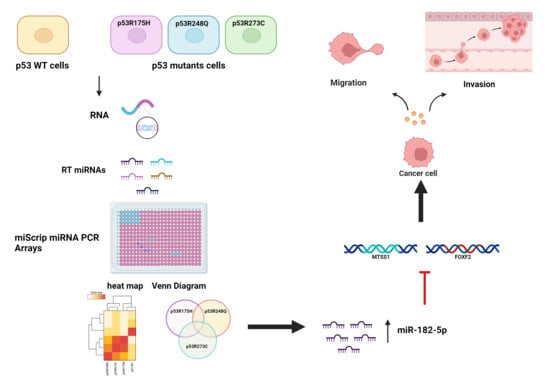
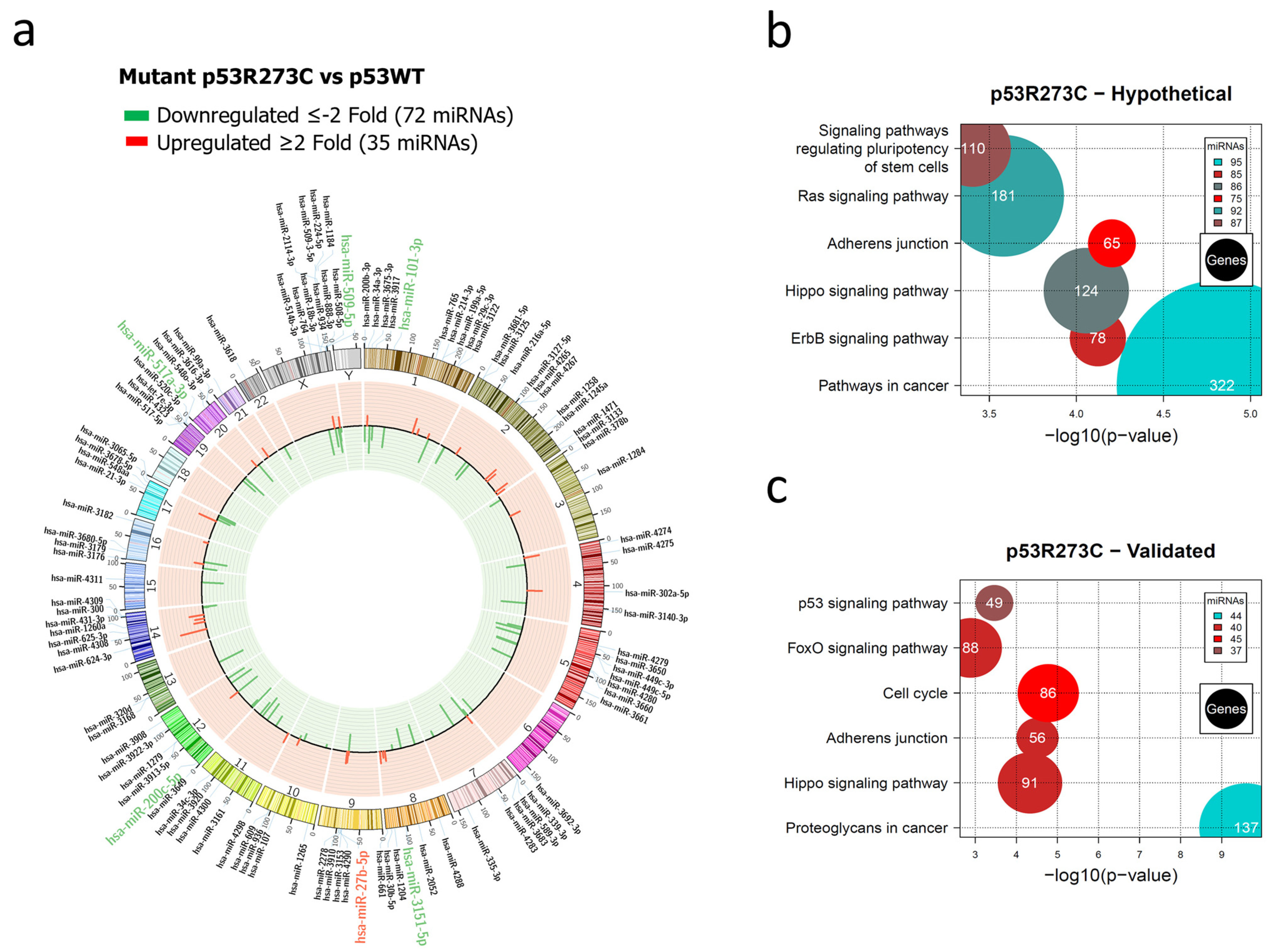
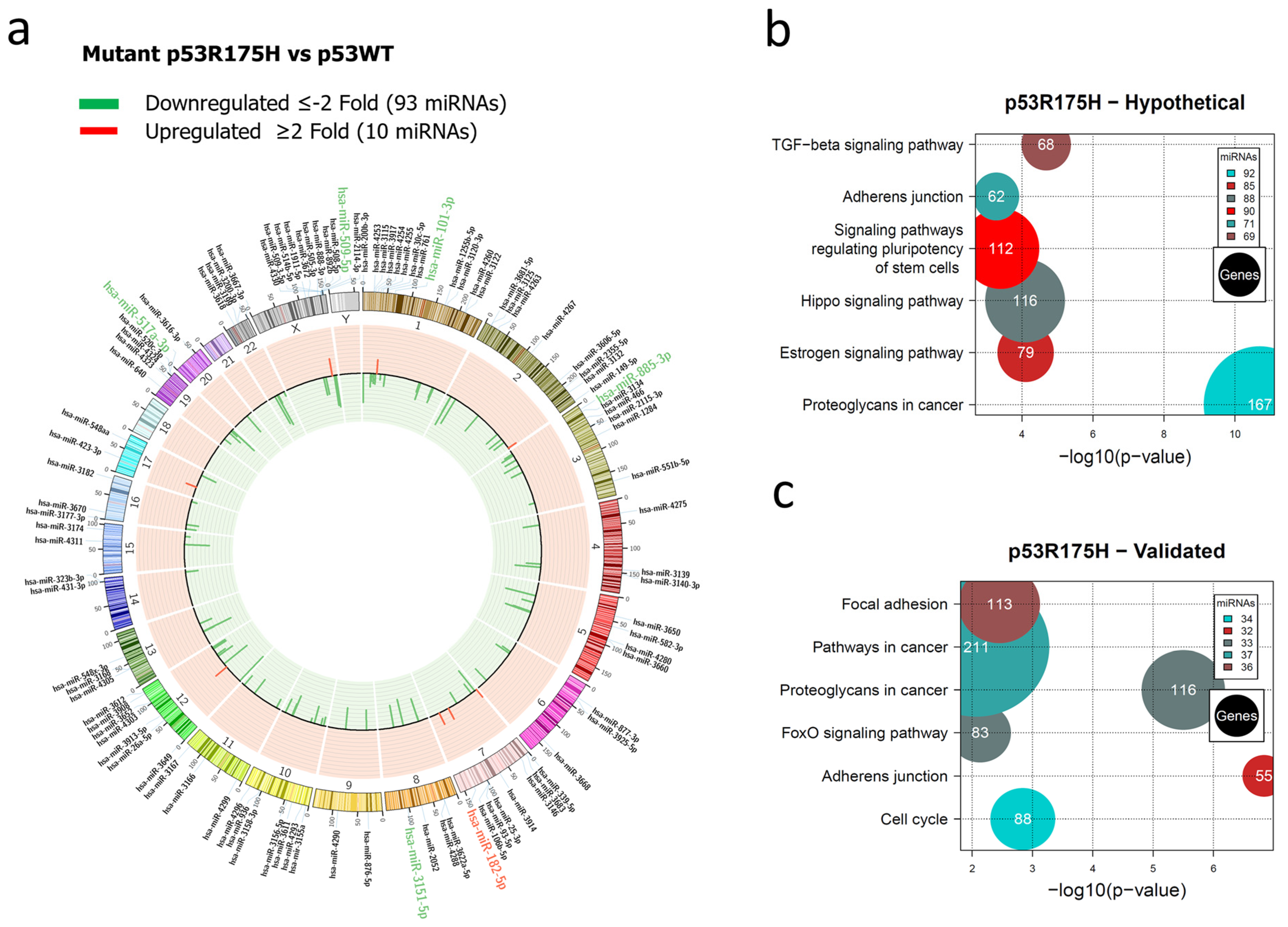
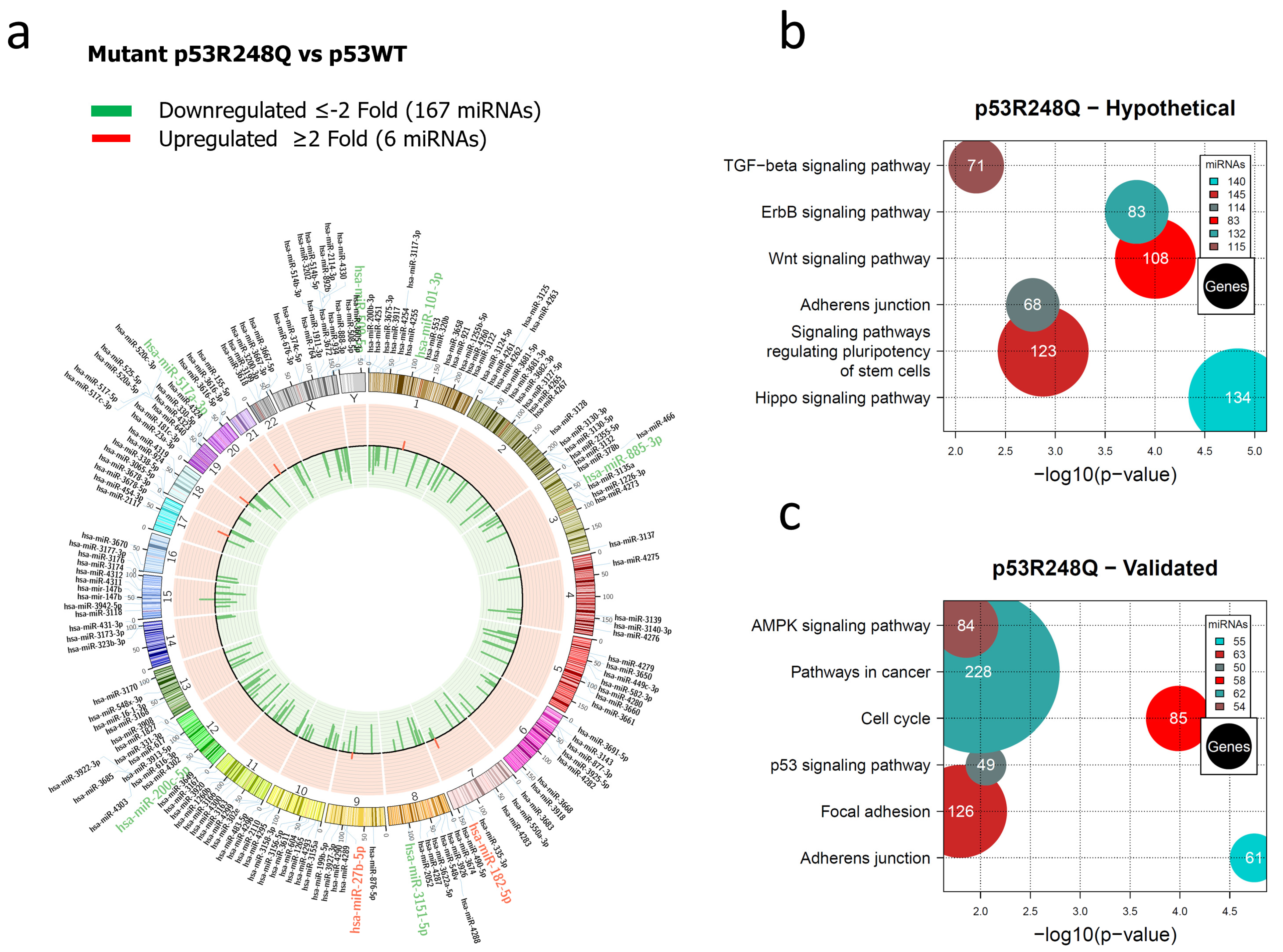
3.1. Global Expression Profiling of miRNAs in Saos-2 Cells Expressing p53R175H, p53R273C, or p53R248Q, as Well as Signaling Pathway Enrichment (KEGG) Analysis
3.2. Heat Map and Venn Diagram of Differentially Expressed miRNAs in p53R273C, p53R248Q, and p53R175H Mutants
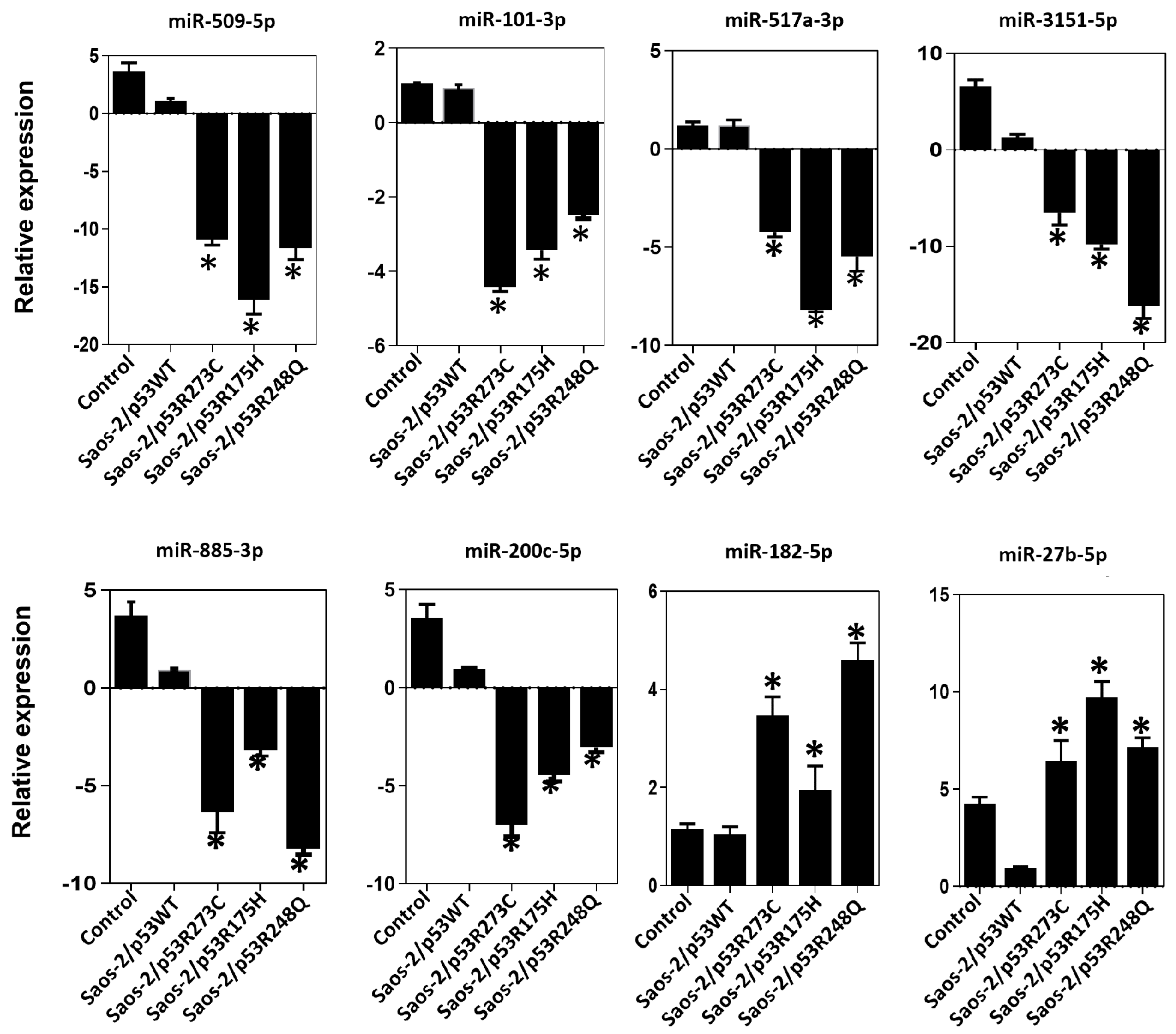
3.3. Validation of Differentially Expressed miRNAs in the Presence of the p53R273C, p53R248Q, and p53R175H Mutants
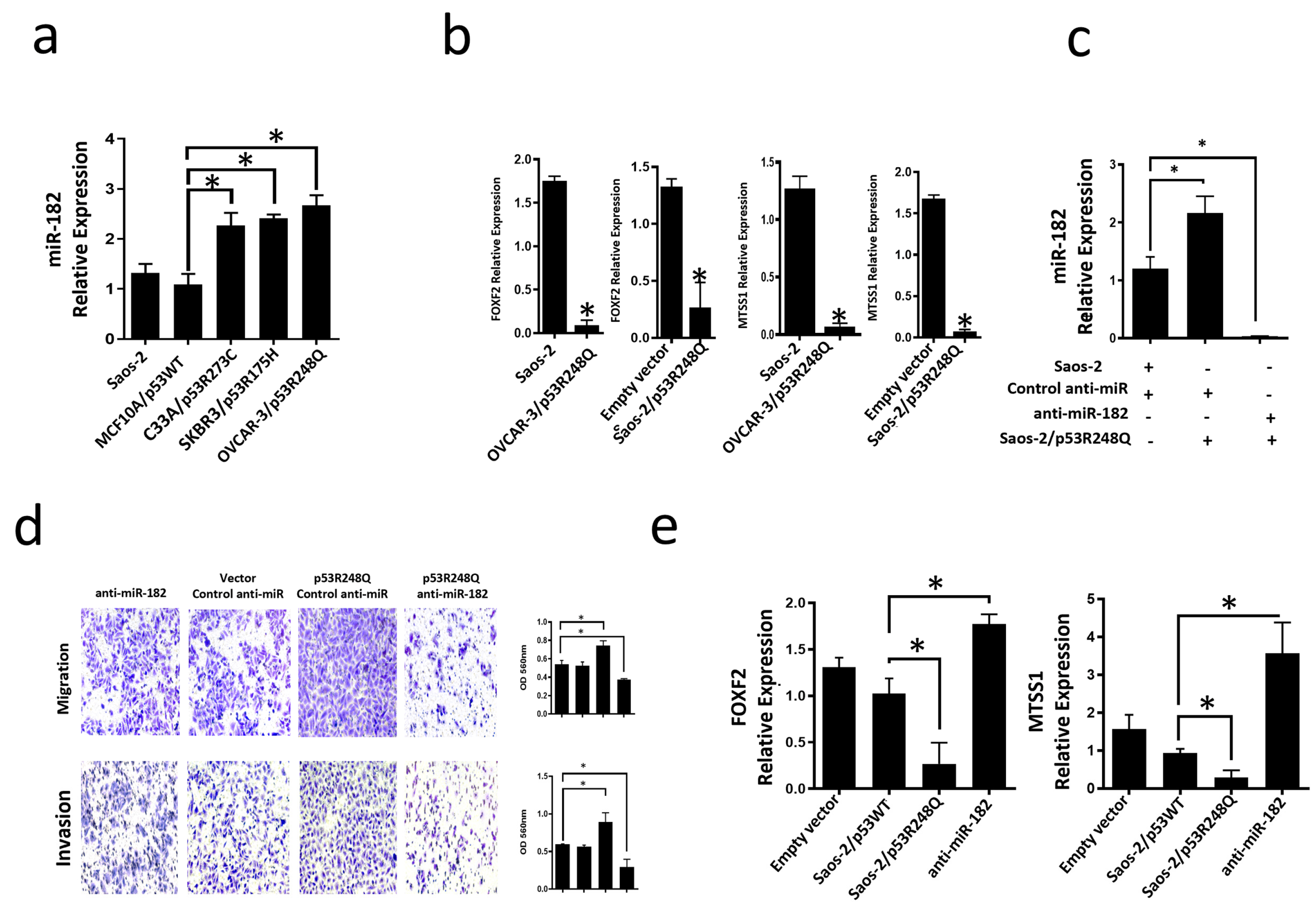
3.4. Mutant p53R248Q Is Associated with Overexpression of miR-185-5p and Downregulation of FOXF2 and MTSS1
3.5. The p53R248Q Mutant Stimulates Invasion and Migration by Overexpressing miR-182-5p in Saos-2 Cells
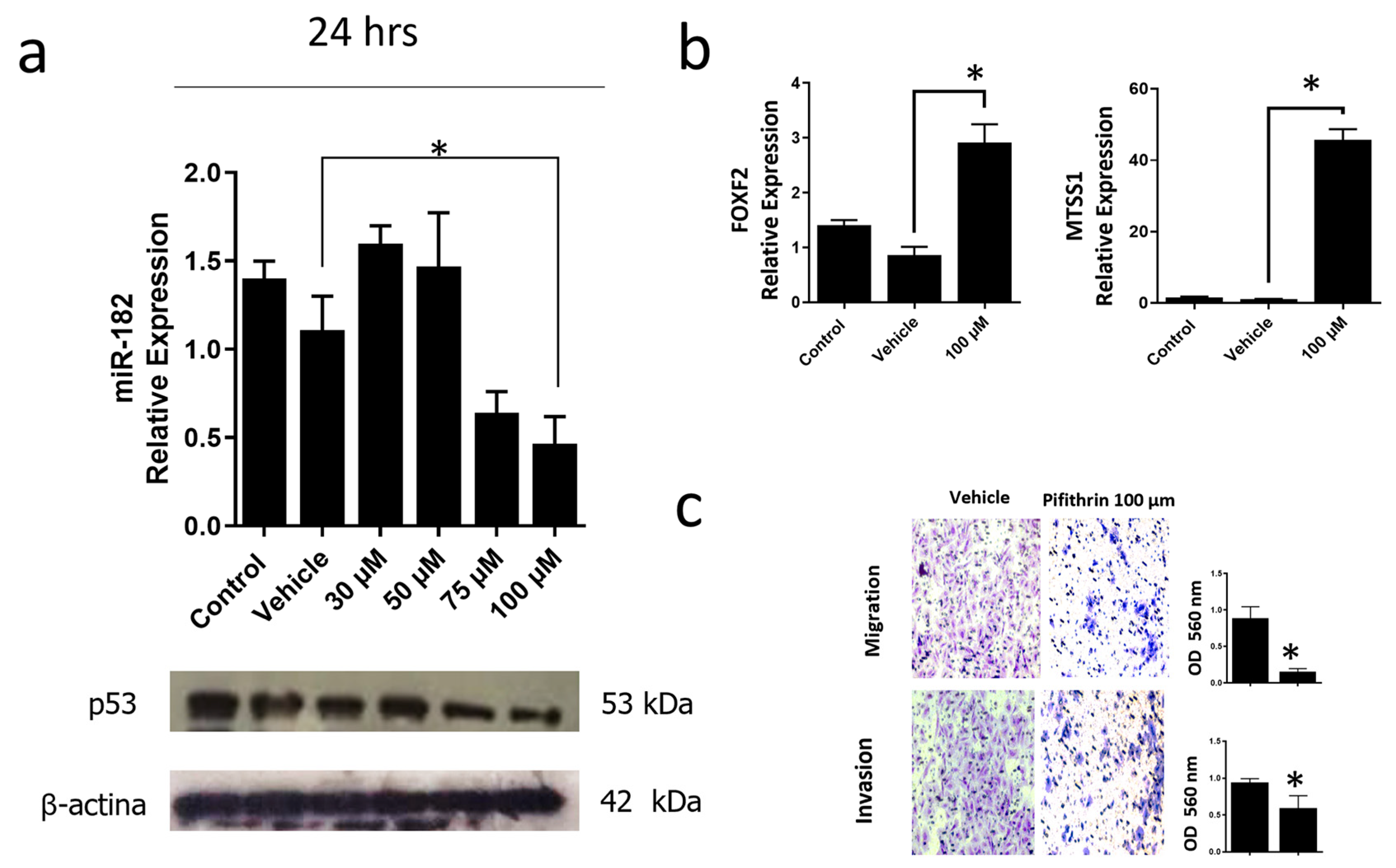
3.6. Inhibition of the p53R248Q Mutant in the OVCAR-3 Cell Line Induces Decreased Cell Migration and Invasion through miR-182-5p
4. Discussion
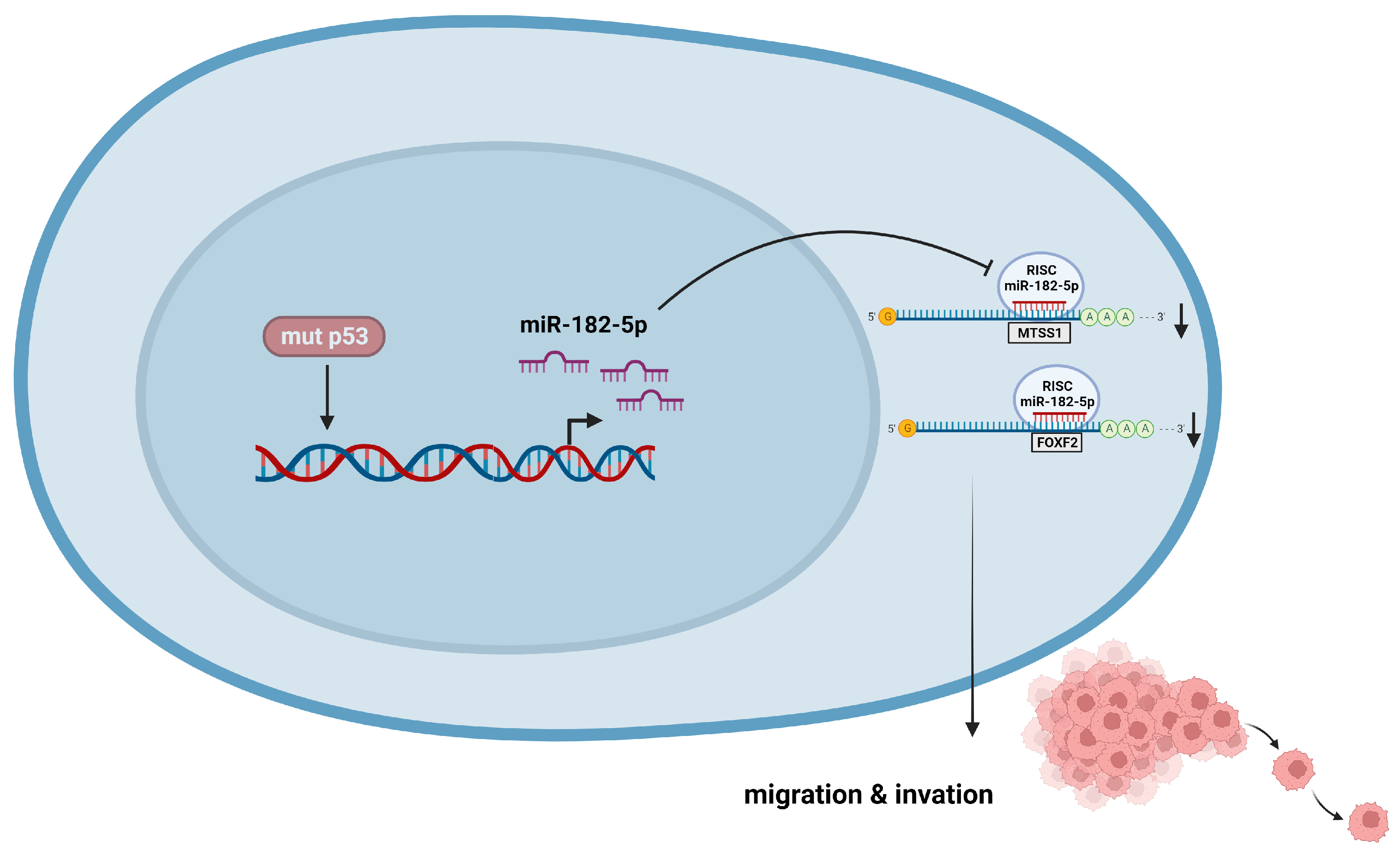
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pfister, N.T.; Prives, C. Transcriptional Regulation by Wild-Type and Cancer-Related Mutant Forms of P53. Cold Spring Harb. Perspect. Med. 2017, 7, a026054. [Google Scholar] [CrossRef] [PubMed]
- Joerger, A.C.; Fersht, A.R. Structural Biology of the Tumor Suppressor P53. Annu. Rev. Biochem. 2008, 77, 557–582. [Google Scholar] [CrossRef] [PubMed]
- Klimovich, B.; Merle, N.; Neumann, M.; Elmshäuser, S.; Nist, A.; Mernberger, M.; Kazdal, D.; Stenzinger, A.; Timofeev, O.; Stiewe, T. P53 Partial Loss-of-Function Mutations Sensitize to Chemotherapy. Oncogene 2022, 41, 1011–1023. [Google Scholar] [CrossRef] [PubMed]
- Freed-Pastor, W.A.; Prives, C. Mutant P53: One Name, Many Proteins. Genes Dev. 2012, 26, 1268–1286. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 Mutations in Human Cancers: Origins, Consequences, and Clinical Use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.A.J.; Vousden, K.H. Mutant P53 in Cancer: New Functions and Therapeutic Opportunities. Cancer Cell 2014, 25, 304–317. [Google Scholar] [CrossRef]
- Blagosklonny, M.V. P53 from Complexity to Simplicity: Mutant P53 Stabilization, Gain-of-Function, and Dominant-Negative Effect. FASEB J. 2000, 14, 1901–1907. [Google Scholar] [CrossRef]
- Sabapathy, K.; Lane, D.P. Therapeutic Targeting of P53: All Mutants Are Equal, but Some Mutants Are More Equal than Others. Nat. Rev. Clin. Oncol. 2018, 15, 13–30. [Google Scholar] [CrossRef]
- Kim, M.P.; Lozano, G. Mutant P53 Partners in Crime. Cell Death Differ. 2018, 25, 161–168. [Google Scholar] [CrossRef]
- Stein, Y.; Rotter, V.; Aloni-Grinstein, R. Gain-of-Function Mutant P53: All the Roads Lead to Tumorigenesis. Int. J. Mol. Sci. 2019, 20, 6197. [Google Scholar] [CrossRef]
- Xiong, S.; Chachad, D.; Zhang, Y.; Gencel-Augusto, J.; Sirito, M.; Pant, V.; Yang, P.; Sun, C.; Chau, G.; Qi, Y.; et al. Differential Gain-of-Function Activity of Three P53 Hotspot Mutants In Vivo. Cancer Res. 2022, 82, 1926–1936. [Google Scholar] [CrossRef] [PubMed]
- Annese, T.; Tamma, R.; De Giorgis, M.; Ribatti, D. MicroRNAs Biogenesis, Functions and Role in Tumor Angiogenesis. Front. Oncol. 2020, 10, 581007. [Google Scholar] [CrossRef] [PubMed]
- Madrigal, T.; Hernández-Monge, J.; Herrera, L.A.; González-De la Rosa, C.H.; Domínguez-Gómez, G.; Candelaria, M.; Luna-Maldonado, F.; Calderón González, K.G.; Díaz-Chávez, J. Regulation of MiRNAs Expression by Mutant P53 Gain of Function in Cancer. Front. Cell Dev. Biol. 2021, 9, 695723. [Google Scholar] [CrossRef] [PubMed]
- Cooks, T.; Pateras, I.S.; Jenkins, L.M.; Patel, K.M.; Robles, A.I.; Morris, J.; Forshew, T.; Appella, E.; Gorgoulis, V.G.; Harris, C.C. Mutant P53 Cancers Reprogram Macrophages to Tumor Supporting Macrophages via Exosomal MiR-1246. Nat. Commun. 2018, 9, 771. [Google Scholar] [CrossRef]
- Chan, K.-T.; Lung, M.L. Mutant P53 Expression Enhances Drug Resistance in a Hepatocellular Carcinoma Cell Line. Cancer Chemother. Pharmacol. 2004, 53, 519–526. [Google Scholar] [CrossRef]
- Baker, S.J.; Markowitz, S.; Fearon, E.R.; Willson, J.K.; Vogelstein, B. Suppression of Human Colorectal Carcinoma Cell Growth by Wild-Type P53. Science 1990, 249, 912–915. [Google Scholar] [CrossRef]
- Loughery, J.; Cox, M.; Smith, L.M.; Meek, D.W. Critical Role for P53-Serine 15 Phosphorylation in Stimulating Transactivation at P53-Responsive Promoters. Nucleic Acids Res. 2014, 42, 7666–7680. [Google Scholar] [CrossRef]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. Gplots: Various R Programming Tools for Plotting Data; R team: Vienna, Austria, 2022. [Google Scholar]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open Software Development for Computational Biology and Bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 7 July 2022).
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Venny 2.1.0. Available online: https://bioinfogp.cnb.csic.es/tools/venny/ (accessed on 7 July 2022).
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-MiRPath v3.0: Deciphering MicroRNA Function with Experimental Support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.-J.; Nong, X.-Y.; Lv, Y.-R.; Sun, H.-H.; An, P.-P.; Wang, F.; Li, X.; Liu, M.; Tang, H. Mir-509-5p Joins the Mdm2/P53 Feedback Loop and Regulates Cancer Cell Growth. Cell Death Dis. 2014, 5, e1387. [Google Scholar] [CrossRef] [PubMed]
- Hiramoto, H.; Muramatsu, T.; Ichikawa, D.; Tanimoto, K.; Yasukawa, S.; Otsuji, E.; Inazawa, J. MiR-509-5p and MiR-1243 Increase the Sensitivity to Gemcitabine by Inhibiting Epithelial-Mesenchymal Transition in Pancreatic Cancer. Sci. Rep. 2017, 7, 4002. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Bai, W.; Zhang, J. MiR-200c-5p Suppresses Proliferation and Metastasis of Human Hepatocellular Carcinoma (HCC) via Suppressing MAD2L1. Biomed. Pharmacother. 2017, 92, 1038–1044. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Liang, H.; Zhou, J.; Wang, Y.; Liu, S.; Wang, X.; Su, L.; Kang, X. LncRNA Small Nucleolar RNA Host Gene 12 Promotes Renal Cell Carcinoma Progression by Modulating the MiR-200c-5p/Collagen Type XI A1 Chain Pathway. Mol. Med. Rep. 2020, 22, 3677–3686. [Google Scholar] [CrossRef]
- Wang, L.Q.; Wong, K.Y.; Rosèn, A.; Chim, C.S. Epigenetic Silencing of Tumor Suppressor MiR-3151 Contributes to Chinese Chronic Lymphocytic Leukemia by Constitutive Activation of MADD/ERK and PIK3R2/AKT Signaling Pathways. Oncotarget 2015, 6, 44422–44436. [Google Scholar] [CrossRef]
- Xiao, F.; Qiu, H.; Cui, H.; Ni, X.; Li, J.; Liao, W.; Lu, L.; Ding, K. MicroRNA-885-3p Inhibits the Growth of HT-29 Colon Cancer Cell Xenografts by Disrupting Angiogenesis via Targeting BMPR1A and Blocking BMP/Smad/Id1 Signaling. Oncogene 2015, 34, 1968–1978. [Google Scholar] [CrossRef]
- Huang, Y.; Chuang, A.Y.; Ratovitski, E.A. Phospho-ΔNp63α/MiR-885-3p Axis in Tumor Cell Life and Cell Death upon Cisplatin Exposure. Cell Cycle 2011, 10, 3938–3947. [Google Scholar] [CrossRef]
- Vijayakumaran, R.; Tan, K.H.; Miranda, P.J.; Haupt, S.; Haupt, Y. Regulation of Mutant P53 Protein Expression. Front. Oncol. 2015, 5, 284. [Google Scholar] [CrossRef]
- Yoshitomi, T.; Kawakami, K.; Enokida, H.; Chiyomaru, T.; Kagara, I.; Tatarano, S.; Yoshino, H.; Arimura, H.; Nishiyama, K.; Seki, N.; et al. Restoration of MiR-517a Expression Induces Cell Apoptosis in Bladder Cancer Cell Lines. Oncol. Rep. 2011, 25, 1661–1668. [Google Scholar] [CrossRef]
- Liu, R.-F.; Xu, X.; Huang, J.; Fei, Q.-L.; Chen, F.; Li, Y.-D.; Han, Z.-G. Down-Regulation of MiR-517a and MiR-517c Promotes Proliferation of Hepatocellular Carcinoma Cells via Targeting Pyk2. Cancer Lett. 2013, 329, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Garibaldi, F.; Falcone, E.; Trisciuoglio, D.; Colombo, T.; Lisek, K.; Walerych, D.; Del Sal, G.; Paci, P.; Bossi, G.; Piaggio, G.; et al. Mutant P53 Inhibits MiRNA Biogenesis by Interfering with the Microprocessor Complex. Oncogene 2016, 35, 3760–3770. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Ye, F.; Xie, X.; Li, X.; Tang, H.; Li, S.; Huang, X.; Song, C.; Wei, W.; Xie, X. Mir-101-3p Is a Key Regulator of Tumor Metabolism in Triple Negative Breast Cancer Targeting AMPK. Oncotarget 2016, 7, 35188–35198. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-Y.; Liu, Z.-J.; He, H.; Zhang, C.; Wang, Y.-L. MicroRNA-101-3p Suppresses Cell Proliferation, Invasion and Enhances Chemotherapeutic Sensitivity in Salivary Gland Adenoid Cystic Carcinoma by Targeting Pim-1. Am. J. Cancer Res. 2015, 5, 3015–3029. [Google Scholar]
- Hou, Y.; Li, L.; Ju, Y.; Lu, Y.; Chang, L.; Xiang, X. MiR-101-3p Regulates the Viability of Lung Squamous Carcinoma Cells via Targeting EZH2. J. Cell Biochem. 2017, 118, 3142–3149. [Google Scholar] [CrossRef]
- Kundu, S.T.; Byers, L.A.; Peng, D.H.; Roybal, J.D.; Diao, L.; Wang, J.; Tong, P.; Creighton, C.J.; Gibbons, D.L. The MiR-200 Family and the MiR-183~96~182 Cluster Target Foxf2 to Inhibit Invasion and Metastasis in Lung Cancers. Oncogene 2016, 35, 173–186. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Shen, J.; Wang, C.; Yang, L.; Zhang, X. MicroRNA-182 Downregulates Metastasis Suppressor 1 and Contributes to Metastasis of Hepatocellular Carcinoma. BMC Cancer 2012, 12, 227. [Google Scholar] [CrossRef]
- Lei, R.; Tang, J.; Zhuang, X.; Deng, R.; Li, G.; Yu, J.; Liang, Y.; Xiao, J.; Wang, H.-Y.; Yang, Q.; et al. Suppression of MIM by MicroRNA-182 Activates RhoA and Promotes Breast Cancer Metastasis. Oncogene 2014, 33, 1287–1296. [Google Scholar] [CrossRef]
- Xue, J.; Zhou, A.; Wu, Y.; Morris, S.-A.; Lin, K.; Amin, S.; Verhaak, R.; Fuller, G.; Xie, K.; Heimberger, A.B.; et al. MiR-182-5p Induced by STAT3 Activation Promotes Glioma Tumorigenesis. Cancer Res. 2016, 76, 4293–4304. [Google Scholar] [CrossRef]
- Xiao, Y.; Huang, W.; Huang, H.; Wang, L.; Wang, M.; Zhang, T.; Fang, X.; Xia, X. MiR-182-5p and MiR-96-5p Target PIAS1 and Mediate the Negative Feedback Regulatory Loop between PIAS1 and STAT3 in Endometrial Cancer. DNA Cell Biol. 2021, 40, 618–628. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, J.; Segura, M.F.; Shao, C.; Lee, P.; Gong, Y.; Hernando, E.; Wei, J.-J. MiR-182 Overexpression in Tumourigenesis of High-Grade Serous Ovarian Carcinoma. J. Pathol. 2012, 228, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Rathinam, R.; Walch, A.; Alahari, S.K. ST14 (Suppression of Tumorigenicity 14) Gene Is a Target for MiR-27b, and the Inhibitory Effect of ST14 on Cell Growth Is Independent of MiR-27b Regulation. J. Biol. Chem. 2009, 284, 23094–23106. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, S.; Zhao, Z.; Mao, X.; Huang, J.; Wu, Z.; Zheng, L.; Wang, Q. MicroRNA-27b up-Regulated by Human Papillomavirus 16 E7 Promotes Proliferation and Suppresses Apoptosis by Targeting Polo-like Kinase2 in Cervical Cancer. Oncotarget 2016, 7, 19666–19679. [Google Scholar] [CrossRef]
- Eastlack, S.C.; Dong, S.; Ivan, C.; Alahari, S.K. Suppression of PDHX by MicroRNA-27b Deregulates Cell Metabolism and Promotes Growth in Breast Cancer. Mol. Cancer 2018, 17, 100. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Y.; Fang, J.-Y.; Xu, J. Gain-of-Function MiRNA Signature by Mutant P53 Associates with Poor Cancer Outcome. Oncotarget 2016, 7, 11056–11066. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, W.; Chen, X. Mutant P53 Disrupts MCF-10A Cell Polarity in Three-Dimensional Culture via Epithelial-to-Mesenchymal Transitions. J. Biol. Chem. 2011, 286, 16218–16228. [Google Scholar] [CrossRef] [PubMed]
- Neilsen, P.M.; Noll, J.E.; Mattiske, S.; Bracken, C.P.; Gregory, P.A.; Schulz, R.B.; Lim, S.P.; Kumar, R.; Suetani, R.J.; Goodall, G.J.; et al. Mutant P53 Drives Invasion in Breast Tumors through Up-Regulation of MiR-155. Oncogene 2013, 32, 2992–3000. [Google Scholar] [CrossRef]
- Schulz-Heddergott, R.; Stark, N.; Edmunds, S.J.; Li, J.; Conradi, L.-C.; Bohnenberger, H.; Ceteci, F.; Greten, F.R.; Dobbelstein, M.; Moll, U.M. Therapeutic Ablation of Gain-of-Function Mutant P53 in Colorectal Cancer Inhibits Stat3-Mediated Tumor Growth and Invasion. Cancer Cell 2018, 34, 298–314.e7. [Google Scholar] [CrossRef]
- Klemke, L.; Fehlau, C.F.; Winkler, N.; Toboll, F.; Singh, S.K.; Moll, U.M.; Schulz-Heddergott, R. The Gain-of-Function P53 R248W Mutant Promotes Migration by STAT3 Deregulation in Human Pancreatic Cancer Cells. Front. Oncol. 2021, 11, 642603. [Google Scholar] [CrossRef]
- Zhang, W.; Qian, P.; Zhang, X.; Zhang, M.; Wang, H.; Wu, M.; Kong, X.; Tan, S.; Ding, K.; Perry, J.K.; et al. Autocrine/Paracrine Human Growth Hormone-Stimulated MicroRNA 96-182-183 Cluster Promotes Epithelial-Mesenchymal Transition and Invasion in Breast Cancer. J. Biol. Chem. 2015, 290, 13812–13829. [Google Scholar] [CrossRef]
- Fu, M.; Hu, Y.; Lan, T.; Guan, K.-L.; Luo, T.; Luo, M. The Hippo Signalling Pathway and Its Implications in Human Health and Diseases. Signal Transduct. Target Ther. 2022, 7, 376. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Liu, J.; Feng, J.; Liu, Z.; Liu, S.; Zhang, M.; Zhang, Y.; Hou, Y.; Wu, D.; Li, C.; et al. Variation in the Life History Strategy Underlies Functional Diversity of Tumors. Natl. Sci. Rev. 2021, 8, nwaa124. [Google Scholar] [CrossRef] [PubMed]
- Di Agostino, S.; Sorrentino, G.; Ingallina, E.; Valenti, F.; Ferraiuolo, M.; Bicciato, S.; Piazza, S.; Strano, S.; Del Sal, G.; Blandino, G. YAP Enhances the Pro-Proliferative Transcriptional Activity of Mutant P53 Proteins. EMBO Rep. 2016, 17, 188–201. [Google Scholar] [CrossRef] [PubMed]
- Ferraiuolo, M.; Verduci, L.; Blandino, G.; Strano, S. Mutant P53 Protein and the Hippo Transducers YAP and TAZ: A Critical Oncogenic Node in Human Cancers. Int. J. Mol. Sci. 2017, 18, 961. [Google Scholar] [CrossRef] [PubMed]
- Elbendary, A.A.; Cirisano, F.D.; Evans, A.C.; Davis, P.L.; Iglehart, J.D.; Marks, J.R.; Berchuck, A. Relationship between P21 Expression and Mutation of the P53 Tumor Suppressor Gene in Normal and Malignant Ovarian Epithelial Cells. Clin. Cancer Res 1996, 2, 1571–1575. [Google Scholar]
- Jin, Q.; He, W.; Chen, L.; Yang, Y.; Shi, K.; You, Z. MicroRNA-101-3p Inhibits Proliferation in Retinoblastoma Cells by Targeting EZH2 and HDAC9. Exp. Ther. Med. 2018, 16, 1663–1670. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, D.; Xia, F.; Xian, H.; Zhu, X.; Cui, H.; Huang, Z. Downregulation of HDAC9 Inhibits Cell Proliferation and Tumor Formation by Inducing Cell Cycle Arrest in Retinoblastoma. Biochem. Biophys. Res. Commun. 2016, 473, 600–606. [Google Scholar] [CrossRef]
- Chang, C.-J.; Hung, M.-C. The Role of EZH2 in Tumour Progression. Br. J. Cancer 2012, 106, 243–247. [Google Scholar] [CrossRef]
- Zenz, T.; Eichhorst, B.; Busch, R.; Denzel, T.; Häbe, S.; Winkler, D.; Bühler, A.; Edelmann, J.; Bergmann, M.; Hopfinger, G.; et al. TP53 Mutation and Survival in Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2010, 28, 4473–4479. [Google Scholar] [CrossRef]
- Ratovitski, E.A.; Patturajan, M.; Hibi, K.; Trink, B.; Yamaguchi, K.; Sidransky, D. P53 Associates with and Targets Delta Np63 into a Protein Degradation Pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 1817–1822. [Google Scholar] [CrossRef]
- Jeng, K.S.; Sheen, I.S.; Chen, B.F.; Wu, J.Y. Is the P53 Gene Mutation of Prognostic Value in Hepatocellular Carcinoma after Resection? Arch. Surg. 2000, 135, 1329–1333. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Sbisà, E.; Tullo, A.; Papeo, P.A.; Saccone, C.; Poole, S.; Pignatelli, M.; Mitry, R.R.; Ding, S.; Isla, A.; et al. P53 Mutation Is a Poor Prognostic Indicator for Survival in Patients with Hepatocellular Carcinoma Undergoing Surgical Tumour Ablation. Br. J. Cancer 1998, 77, 776–782. [Google Scholar] [CrossRef] [PubMed]
- Heinze, T.; Jonas, S.; Kärsten, A.; Neuhaus, P. Determination of the Oncogenes P53 and C-Erb B2 in the Tumour Cytosols of Advanced Hepatocellular Carcinoma (HCC) and Correlation to Survival Time. Anticancer Res. 1999, 19, 2501–2503. [Google Scholar] [PubMed]
- Yu, J.; Shen, W.; Gao, B.; Zhao, H.; Xu, J.; Gong, B. MicroRNA-182 Targets FOXF2 to Promote the Development of Triple-Negative Breast Cancer. Neoplasma 2017, 64, 209–215. [Google Scholar] [CrossRef]
- Wang, Q.-S.; Kong, P.-Z.; Li, X.-Q.; Yang, F.; Feng, Y.-M. FOXF2 Deficiency Promotes Epithelial-Mesenchymal Transition and Metastasis of Basal-like Breast Cancer. Breast Cancer Res. 2015, 17, 30. [Google Scholar] [CrossRef]
- Xu, X.; Ayub, B.; Liu, Z.; Serna, V.A.; Qiang, W.; Liu, Y.; Hernando, E.; Zabludoff, S.; Kurita, T.; Kong, B.; et al. Anti-MiR182 Reduces Ovarian Cancer Burden, Invasion, and Metastasis: An in Vivo Study in Orthotopic Xenografts of Nude Mice. Mol. Cancer Ther. 2014, 13, 1729–1739. [Google Scholar] [CrossRef]
- Saarikangas, J.; Mattila, P.K.; Varjosalo, M.; Bovellan, M.; Hakanen, J.; Calzada-Wack, J.; Tost, M.; Jennen, L.; Rathkolb, B.; Hans, W.; et al. Missing-in-Metastasis MIM/MTSS1 Promotes Actin Assembly at Intercellular Junctions and Is Required for Integrity of Kidney Epithelia. J. Cell Sci. 2011, 124, 1245–1255. [Google Scholar] [CrossRef]
- Wang, L.; Jiang, H.; Li, W.; Jia, C.; Zhang, H.; Sun, Y.; Chen, X.; Song, X. Overexpression of TP53 Mutation-Associated MicroRNA-182 Promotes Tumor Cell Proliferation and Migration in Head and Neck Squamous Cell Carcinoma. Arch. Oral. Biol. 2017, 73, 105–112. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madrigal, T.; Ortega-Bernal, D.; Herrera, L.A.; González-De la Rosa, C.H.; Domínguez-Gómez, G.; Aréchaga-Ocampo, E.; Díaz-Chávez, J. Mutant p53 Gain-of-Function Induces Migration and Invasion through Overexpression of miR-182-5p in Cancer Cells. Cells 2023, 12, 2506. https://doi.org/10.3390/cells12202506
Madrigal T, Ortega-Bernal D, Herrera LA, González-De la Rosa CH, Domínguez-Gómez G, Aréchaga-Ocampo E, Díaz-Chávez J. Mutant p53 Gain-of-Function Induces Migration and Invasion through Overexpression of miR-182-5p in Cancer Cells. Cells. 2023; 12(20):2506. https://doi.org/10.3390/cells12202506
Chicago/Turabian StyleMadrigal, Tzitzijanik, Daniel Ortega-Bernal, Luis A. Herrera, Claudia Haydée González-De la Rosa, Guadalupe Domínguez-Gómez, Elena Aréchaga-Ocampo, and José Díaz-Chávez. 2023. "Mutant p53 Gain-of-Function Induces Migration and Invasion through Overexpression of miR-182-5p in Cancer Cells" Cells 12, no. 20: 2506. https://doi.org/10.3390/cells12202506
APA StyleMadrigal, T., Ortega-Bernal, D., Herrera, L. A., González-De la Rosa, C. H., Domínguez-Gómez, G., Aréchaga-Ocampo, E., & Díaz-Chávez, J. (2023). Mutant p53 Gain-of-Function Induces Migration and Invasion through Overexpression of miR-182-5p in Cancer Cells. Cells, 12(20), 2506. https://doi.org/10.3390/cells12202506