Interaction of the C9orf72-Amyotrophic Lateral Sclerosis-Related Proline–Arginine Dipeptide Repeat Protein with the RNA-Binding Protein NOVA1 Causes Decreased Expression of UNC13A Due to Enhanced Inclusion of Cryptic Exons, Which Is Reversed by Betulin Treatment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals, Media, and Neuron-like Cell Line Maintenance
2.2. Processing of Small RNA Interference in SH-SY5Y and SK-N-DZ Cells
2.3. Western Blotting of SH-SY5Y and SK-N-DZ Cells
2.4. Total RNA Extraction and Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR) of SH-SY5Y and SK-N-DZ Cells
2.5. Recombinant Plasmid Construction and Transfection
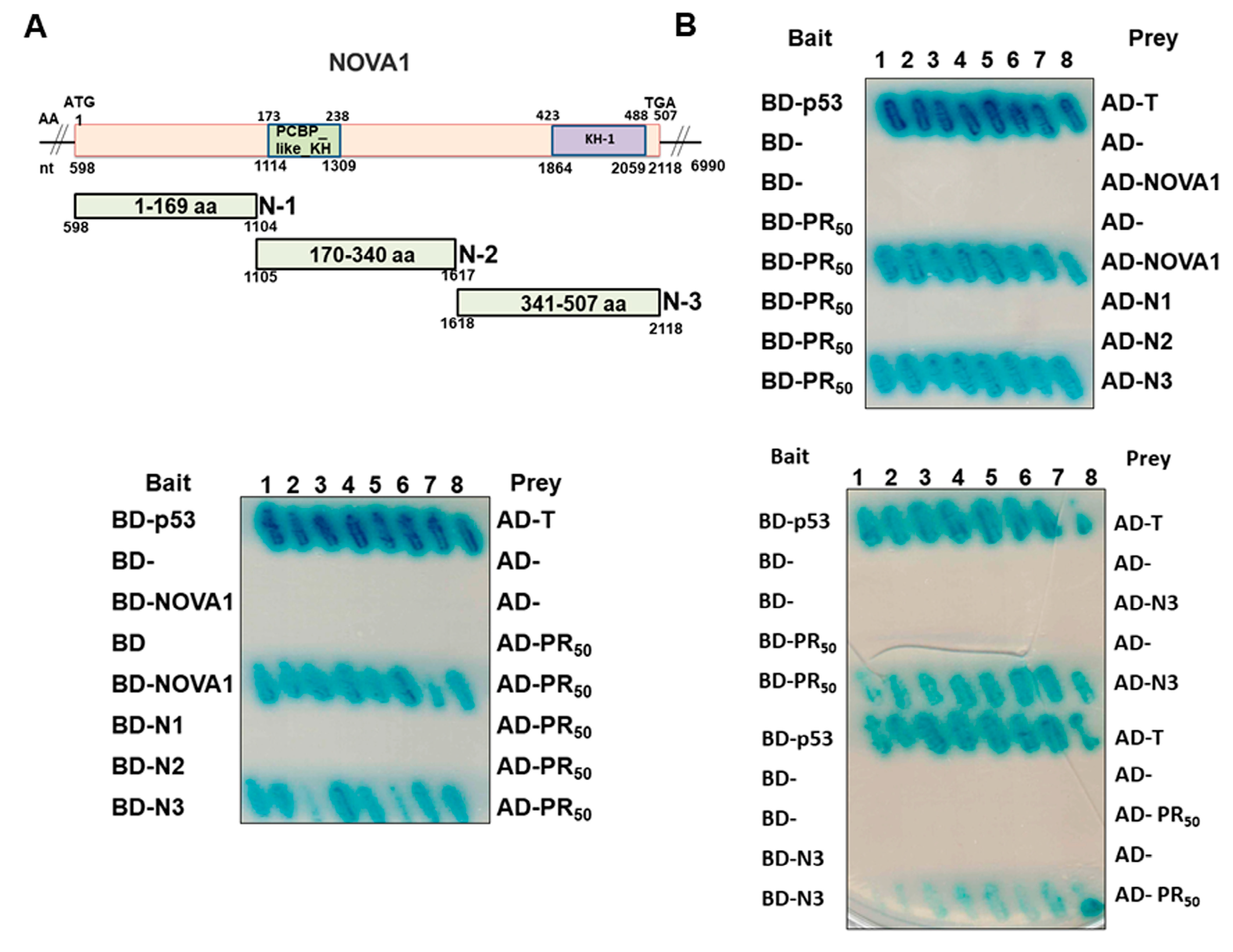
2.6. Yeast Two-Hybrid Screening of PR50-Associated Protein
2.7. Yeast Two-Hybrid Assay for PR50 and NOVA1
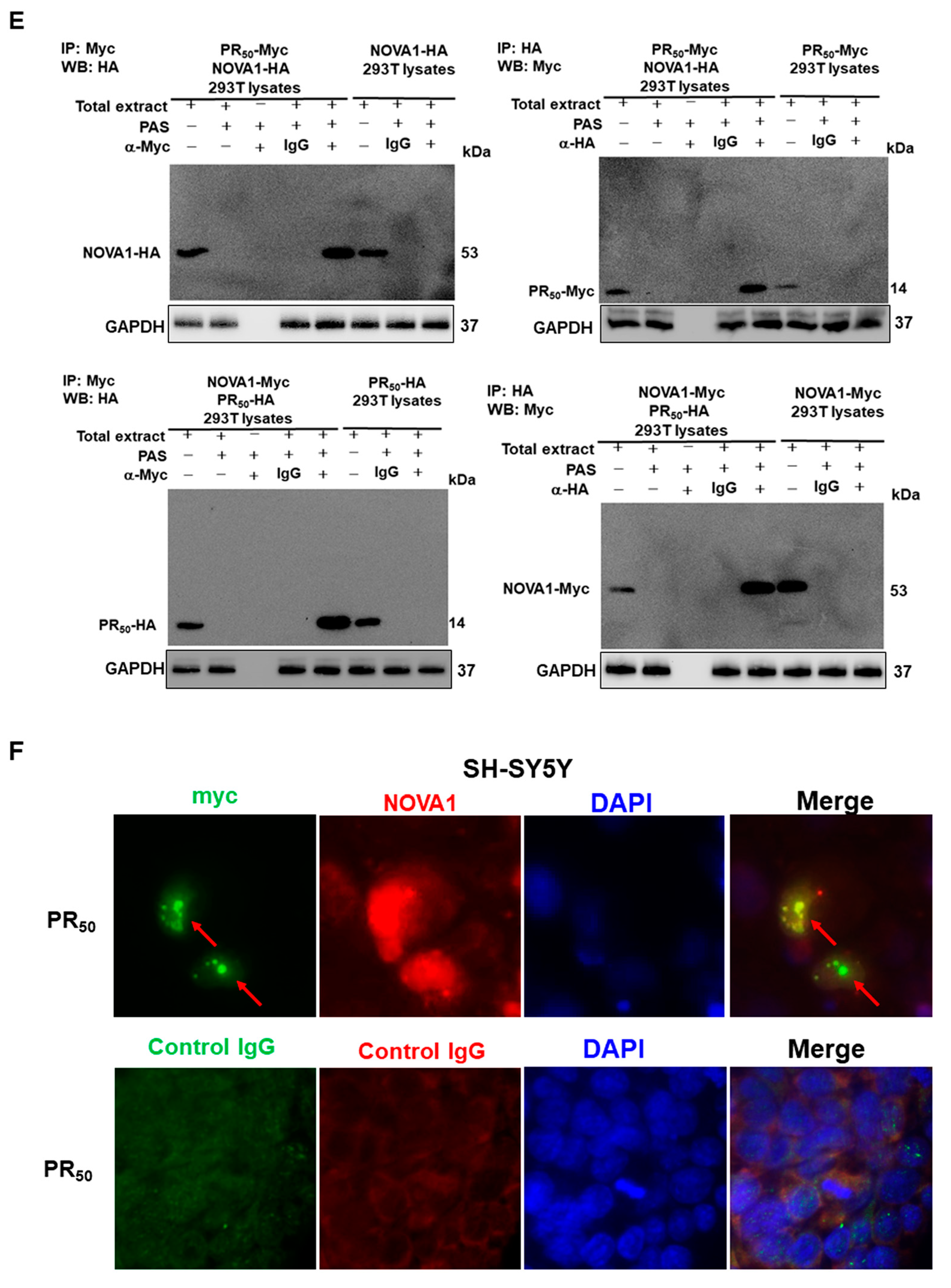
2.8. Co-Immunoprecipitation Analysis of 293T Cells
2.9. Immunofluorescence Staining of PR50-Expressing SH-SY5Y Cells
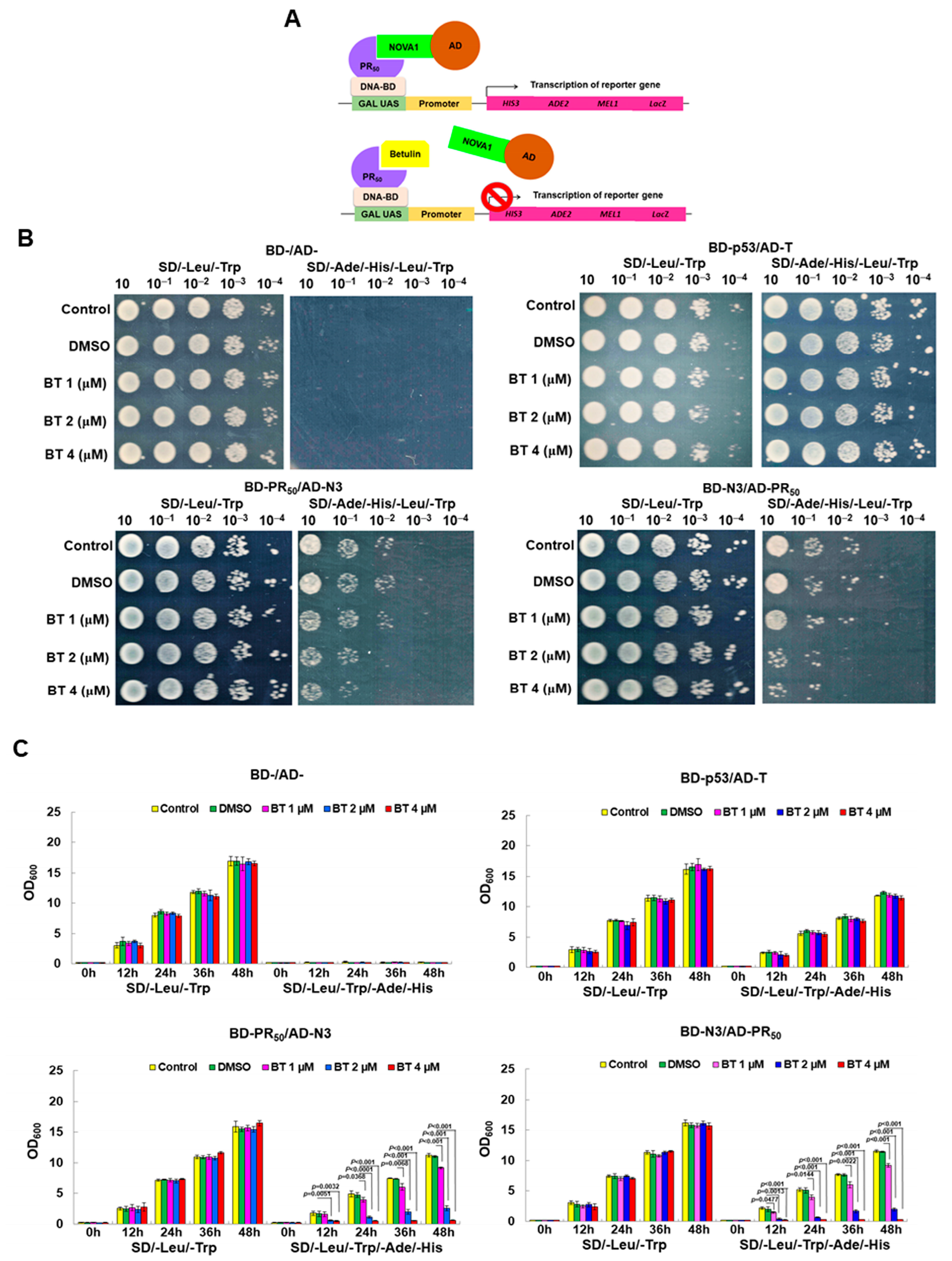
2.10. Screening Inhibitors of the Interaction between GA50 and NOVA1 Using a Yeast Two-Hybrid-Based Growth Assay
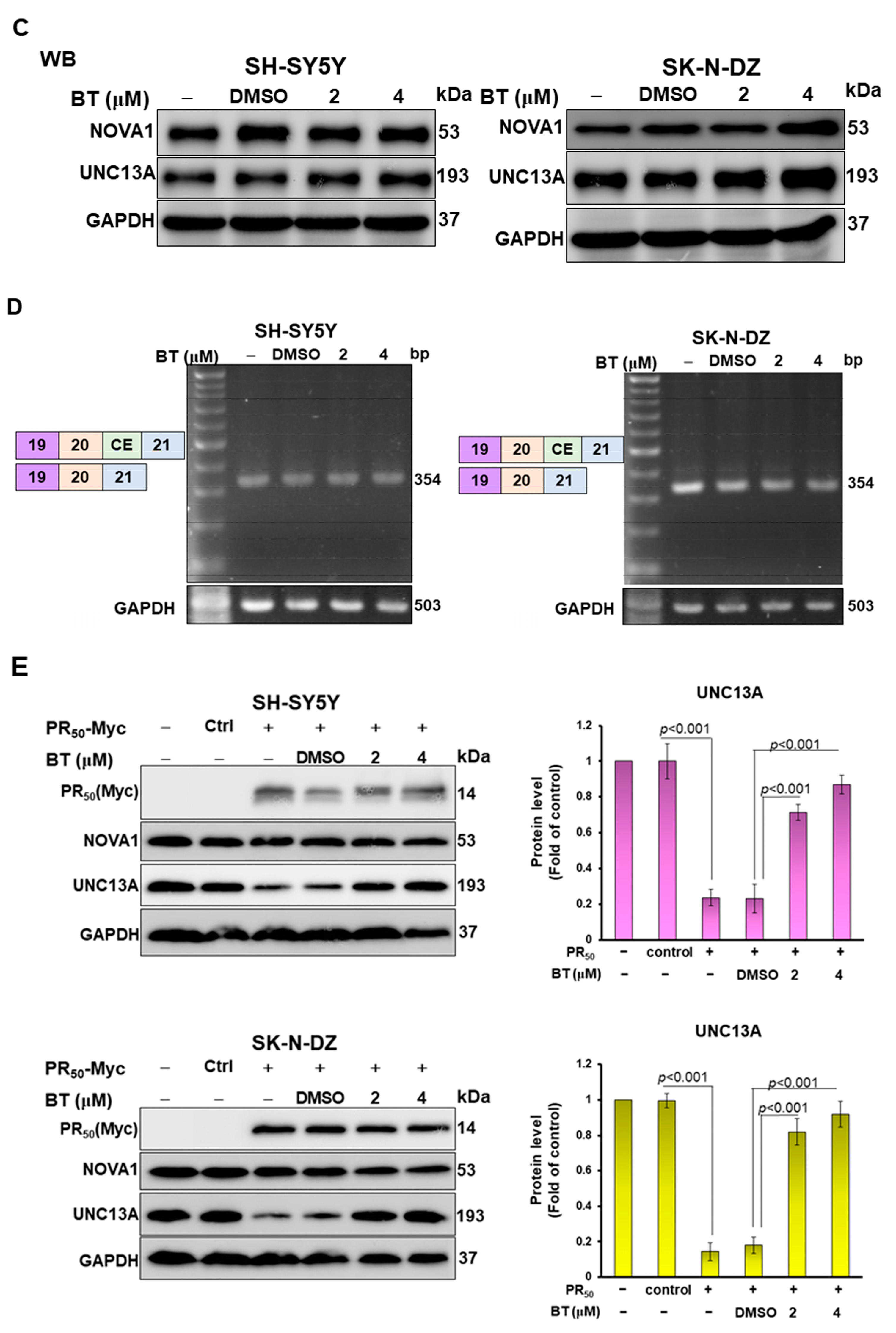
2.11. Treatment with Betulin and Cell Viability Analysis of SH-SY5Y and SK-N-DZ Cells
2.12. Statistical Methods Used in This Study
3. Results
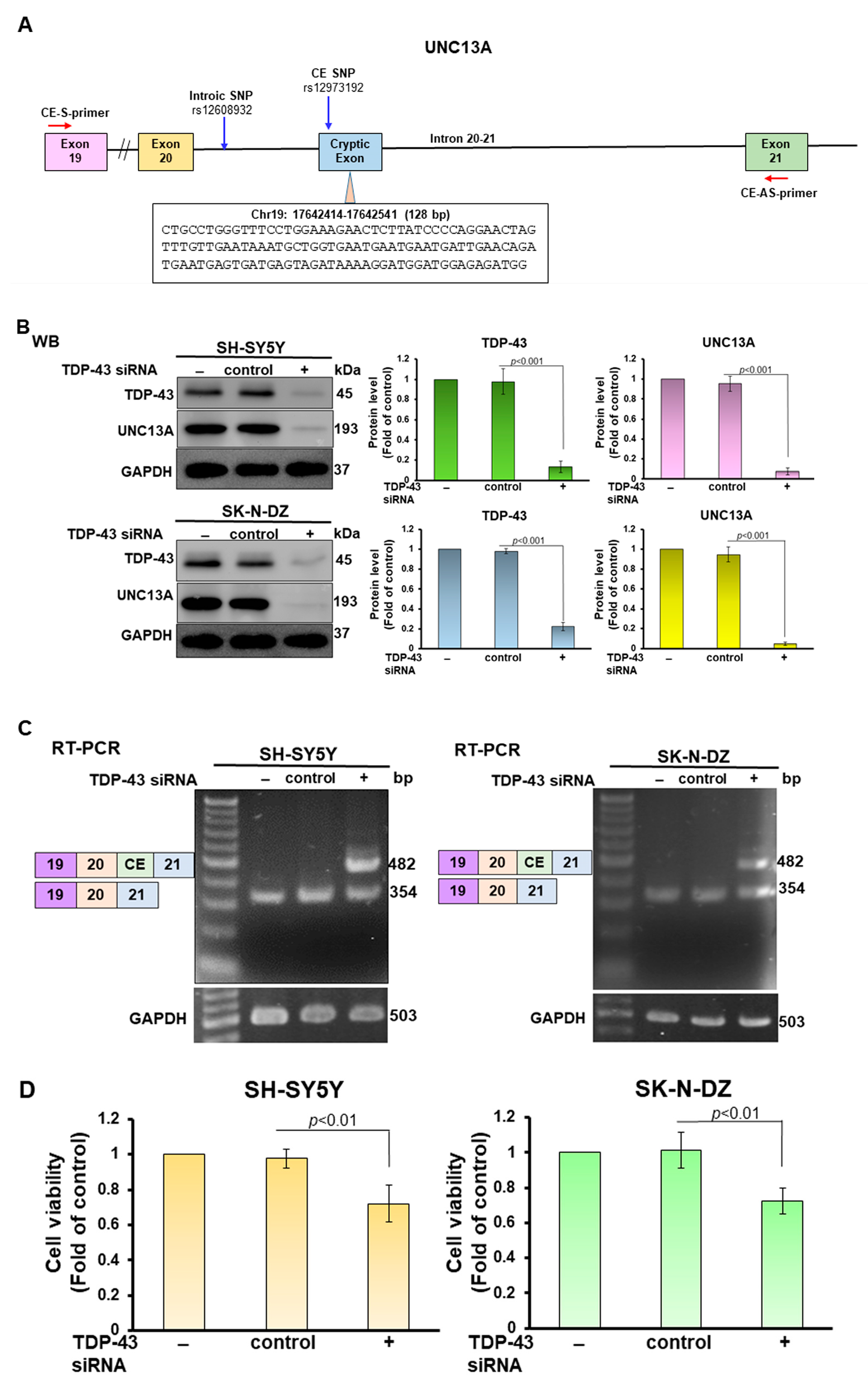
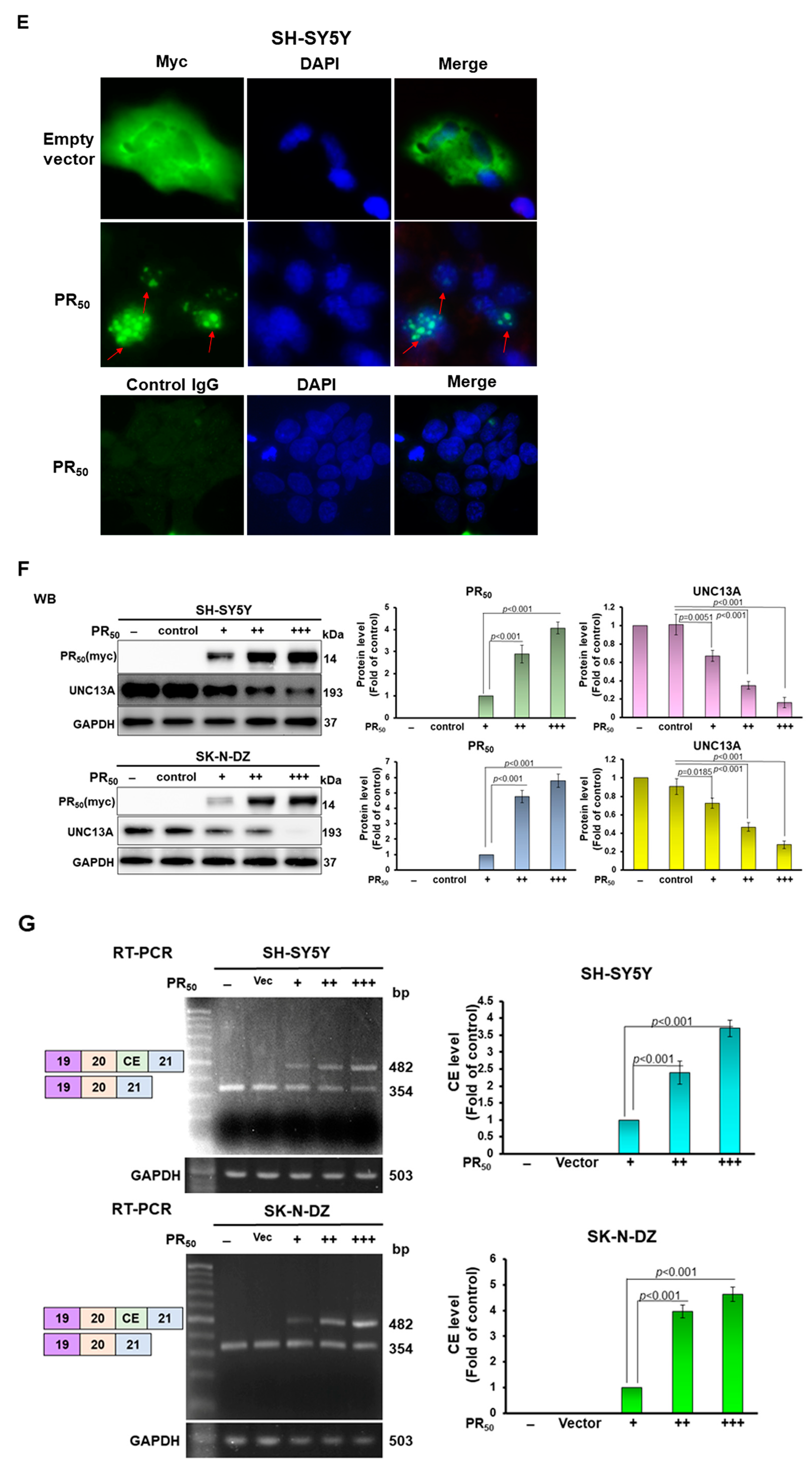
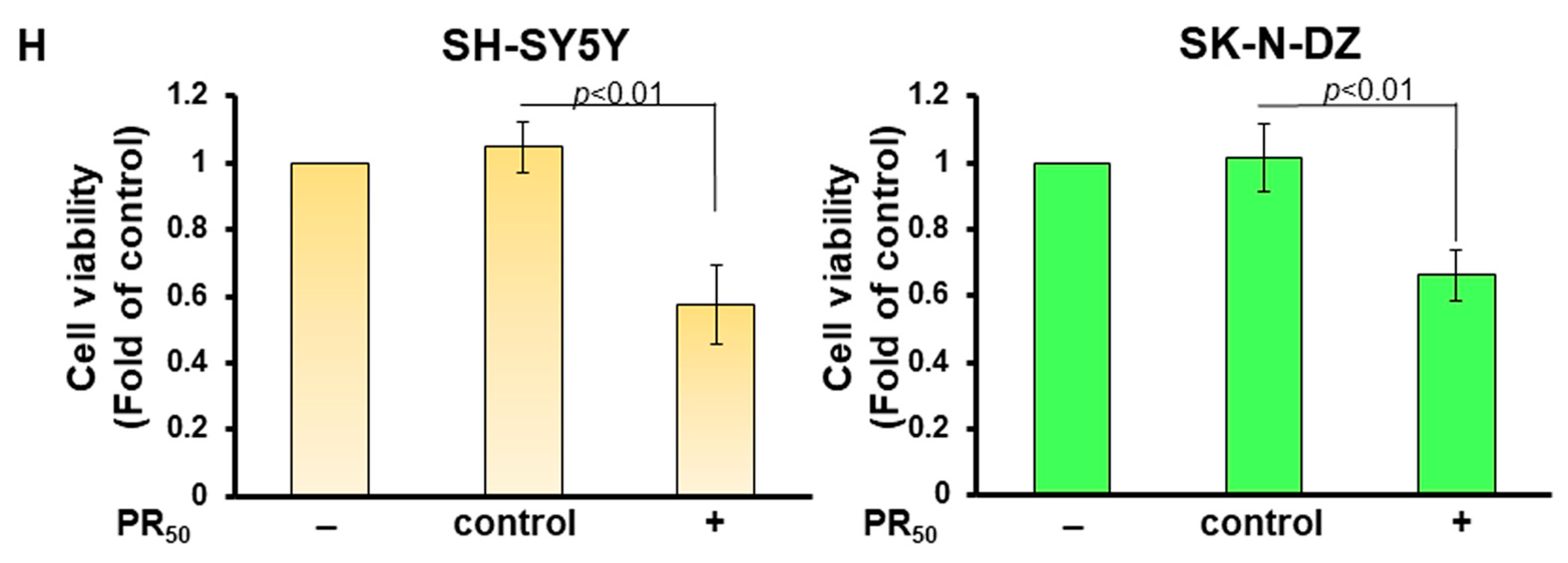
3.1. PR-DPR Expression Promotes the Inclusion of Cryptic Exon (CE) in UNC13A mRNA in Neuronal Cell Lines, Resulting in a Decrease in the Expression of UNC13A Protein
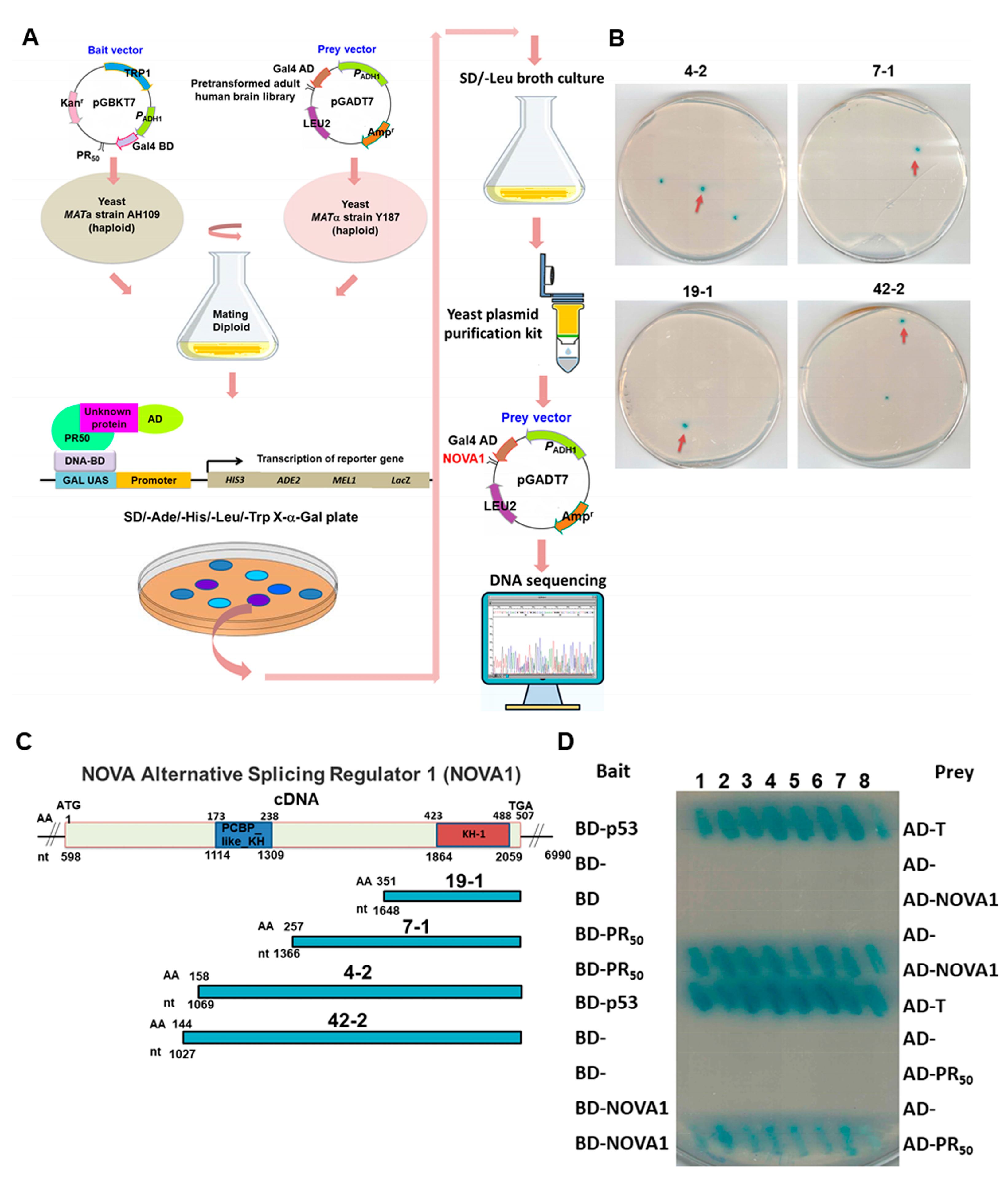
3.2. RNA-Binding Protein NOVA1 Is a Specific Interaction Partner of PR-DPR
3.3. PR-DPR Interacts with the Amino Acid Fragment at the C Terminus of NOVA1
3.4. CE Inclusion of UNC13A mRNA in Neuronal Cell Lines Will Be Significantly Increased by Knockdown of NOVA1, Resulting in Downregulation of UNC13A Protein Expression
3.5. Betulin (BT) Can Directly Interfere with the Interaction between PR-DPR and NOVA1 in the Yeast Two-Hybrid Model
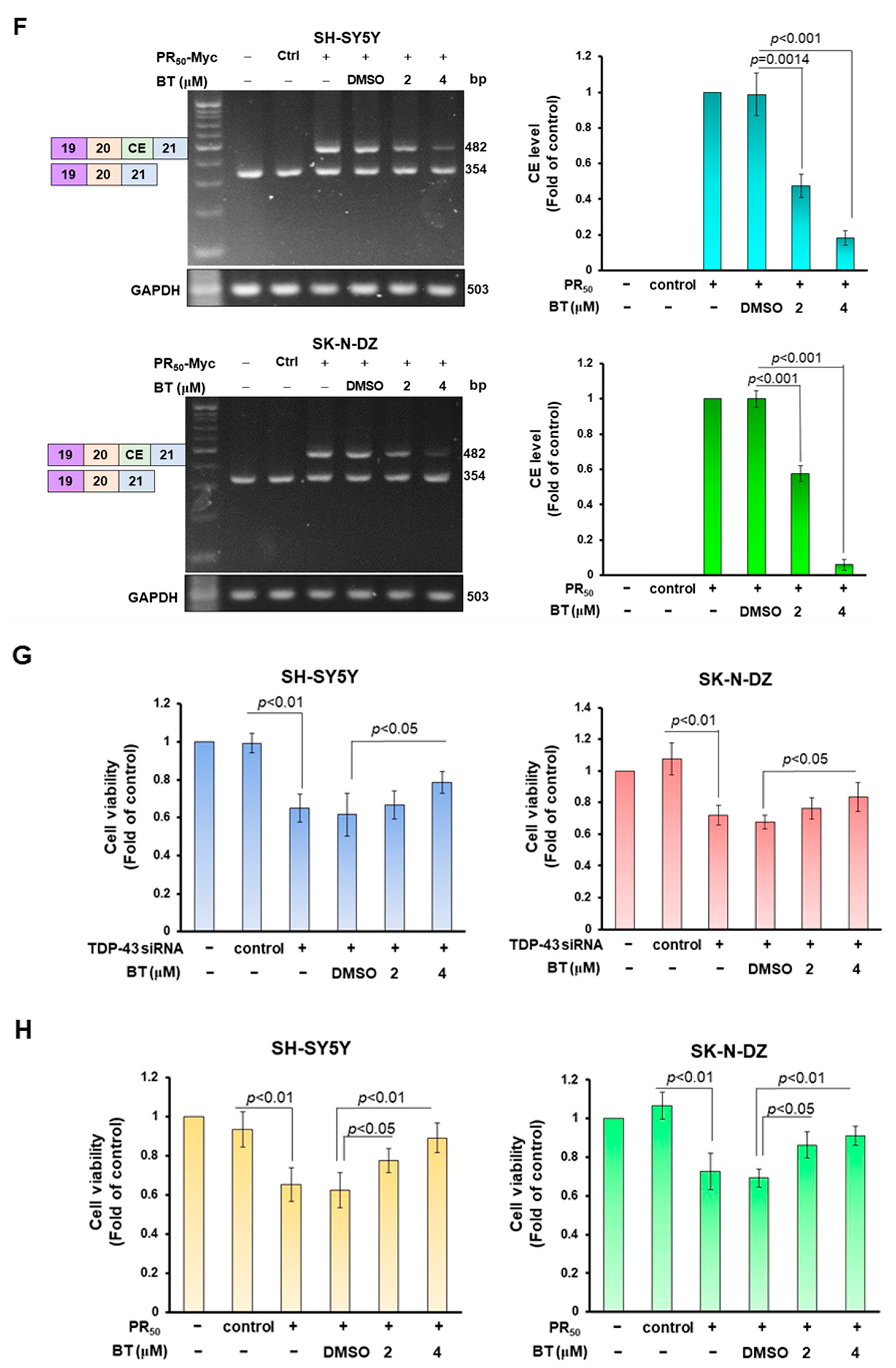
3.6. BT treatment Can Significantly Inhibit the CE Inclusion of UNC13 A mRNA in Neuronal Cell Lines to Avoid Downregulation of UNC13 A Protein Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mead, R.J.; Shan, N.; Reiser, H.J.; Marshall, F.; Shaw, P.J. Amyotrophic lateral sclerosis: A neurodegenerative disorder poised for successful therapeutic translation. Nat. Rev. Drug Discov. 2023, 22, 185–212. [Google Scholar] [CrossRef]
- Talbott, E.O.; Malek, A.M.; Lacomis, D. The epidemiology of amyotrophic lateral sclerosis. Handb. Clin. Neurol. 2016, 138, 225–238. [Google Scholar]
- Lepine, S.; Castellanos-Montiel, M.J.; Durcan, T.M. TDP-43 dysregulation and neuromuscular junction disruption in amyotrophic lateral sclerosis. Transl. Neurodegener. 2022, 11, 56. [Google Scholar] [CrossRef]
- Zampatti, S.; Peconi, C.; Campopiano, R.; Gambardella, S.; Caltagirone, C.; Giardina, E. C9orf72-Related Neurodegenerative Diseases: From Clinical Diagnosis to Therapeutic Strategies. Front. Aging Neurosci. 2022, 14, 907122. [Google Scholar] [CrossRef]
- Schmitz, A.; Pinheiro Marques, J.; Oertig, I.; Maharjan, N.; Saxena, S. Emerging Perspectives on Dipeptide Repeat Proteins in C9ORF72 ALS/FTD. Front. Cell. Neurosci. 2021, 15, 637548. [Google Scholar] [CrossRef]
- Fu, R.H.; Tsai, C.W.; Chiu, S.C.; Liu, S.P.; Chiang, Y.T.; Kuo, Y.H.; Shyu, W.C.; Lin, S.Z. C9-ALS-Associated Proline-Arginine Dipeptide Repeat Protein Induces Activation of NLRP3 Inflammasome of HMC3 Microglia Cells by Binding of Complement Component 1 Q Subcomponent-Binding Protein (C1QBP), and Syringin Prevents This Effect. Cells 2022, 11, 3128. [Google Scholar] [CrossRef]
- Babu, M.; Favretto, F.; de Opakua, A.I.; Rankovic, M.; Becker, S.; Zweckstetter, M. Proline/arginine dipeptide repeat polymers derail protein folding in amyotrophic lateral sclerosis. Nat. Commun. 2021, 12, 3396. [Google Scholar] [CrossRef]
- Gupta, R.; Lan, M.; Mojsilovic-Petrovic, J.; Choi, W.H.; Safren, N.; Barmada, S.; Lee, M.J.; Kalb, R. The Proline/Arginine Dipeptide from Hexanucleotide Repeat Expanded C9ORF72 Inhibits the Proteasome. eNeuro 2017, 4, ENEURO.0249-16.2017. [Google Scholar] [CrossRef]
- Maor-Nof, M.; Shipony, Z.; Lopez-Gonzalez, R.; Nakayama, L.; Zhang, Y.J.; Couthouis, J.; Blum, J.A.; Castruita, P.A.; Linares, G.R.; Ruan, K.; et al. p53 is a central regulator driving neurodegeneration caused by C9orf72 poly(PR). Cell 2021, 184, 689–708.e20. [Google Scholar] [CrossRef]
- Jafarinia, H.; Van der Giessen, E.; Onck, P.R. Molecular basis of C9orf72 poly-PR interference with the beta-karyopherin family of nuclear transport receptors. Sci. Rep. 2022, 12, 21324. [Google Scholar] [CrossRef]
- Tg, S.; Chase, K.J.; Liu, F.; Lloyd, T.E.; Rossoll, W.; Zhang, K. c-Jun N-terminal Kinase Promotes Stress Granule Assembly and Neurodegeneration in C9orf72-mediated ALS and FTD. J. Neurosci. 2023, 43, 3186–3197. [Google Scholar]
- Loveland, A.B.; Svidritskiy, E.; Susorov, D.; Lee, S.; Park, A.; Zvornicanin, S.; Demo, G.; Gao, F.B.; Korostelev, A.A. Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM. Nat. Commun. 2022, 13, 2776. [Google Scholar] [CrossRef]
- Shiota, T.; Nagata, R.; Kikuchi, S.; Nanaura, H.; Matsubayashi, M.; Nakanishi, M.; Kobashigawa, S.; Isozumi, N.; Kiriyama, T.; Nagayama, K.; et al. C9orf72-Derived Proline:Arginine Poly-Dipeptides Modulate Cytoskeleton and Mechanical Stress Response. Front. Cell Dev. Biol. 2022, 10, 750829. [Google Scholar] [CrossRef]
- Jo, Y.; Lee, J.; Lee, S.Y.; Kwon, I.; Cho, H. Poly-dipeptides produced from C9orf72 hexanucleotide repeats cause selective motor neuron hyperexcitability in ALS. Proc. Natl. Acad. Sci. USA 2022, 119, e2113813119. [Google Scholar] [CrossRef]
- Scotter, E.L.; Chen, H.J.; Shaw, C.E. TDP-43 Proteinopathy and ALS: Insights into Disease Mechanisms and Therapeutic Targets. Neurotherapeutics 2015, 12, 352–363. [Google Scholar] [CrossRef]
- Donde, A.; Sun, M.; Ling, J.P.; Braunstein, K.E.; Pang, B.; Wen, X.; Cheng, X.; Chen, L.; Wong, P.C. Splicing repression is a major function of TDP-43 in motor neurons. Acta Neuropathol. 2019, 138, 813–826. [Google Scholar] [CrossRef]
- Melamed, Z.; Lopez-Erauskin, J.; Baughn, M.W.; Zhang, O.; Drenner, K.; Sun, Y.; Freyermuth, F.; McMahon, M.A.; Beccari, M.S.; Artates, J.W.; et al. Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat. Neurosci. 2019, 22, 180–190. [Google Scholar] [CrossRef]
- Klim, J.R.; Williams, L.A.; Limone, F.; Guerra San Juan, I.; Davis-Dusenbery, B.N.; Mordes, D.A.; Burberry, A.; Steinbaugh, M.J.; Gamage, K.K.; Kirchner, R.; et al. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat. Neurosci. 2019, 22, 167–179. [Google Scholar] [CrossRef]
- Brown, A.L.; Wilkins, O.G.; Keuss, M.J.; Hill, S.E.; Zanovello, M.; Lee, W.C.; Bampton, A.; Lee, F.C.Y.; Masino, L.; Qi, Y.A.; et al. TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature 2022, 603, 131–137. [Google Scholar] [CrossRef]
- Manini, A.; Casiraghi, V.; Brusati, A.; Maranzano, A.; Gentile, F.; Colombo, E.; Bonetti, R.; Peverelli, S.; Invernizzi, S.; Gentilini, D.; et al. Association of the risk factor UNC13A with survival and upper motor neuron involvement in amyotrophic lateral sclerosis. Front. Aging Neurosci. 2023, 15, 1067954. [Google Scholar] [CrossRef]
- Tan, H.H.G.; Westeneng, H.J.; van der Burgh, H.K.; van Es, M.A.; Bakker, L.A.; van Veenhuijzen, K.; van Eijk, K.R.; van Eijk, R.P.A.; Veldink, J.H.; van den Berg, L.H. The Distinct Traits of the UNC13A Polymorphism in Amyotrophic Lateral Sclerosis. Ann. Neurol. 2020, 88, 796–806. [Google Scholar] [CrossRef]
- Dittman, J.S. Unc13: A multifunctional synaptic marvel. Curr. Opin. Neurobiol. 2019, 57, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Augustin, I.; Rosenmund, C.; Sudhof, T.C.; Brose, N. Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature 1999, 400, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Varoqueaux, F.; Sigler, A.; Rhee, J.S.; Brose, N.; Enk, C.; Reim, K.; Rosenmund, C. Total arrest of spontaneous and evoked synaptic transmission but normal synaptogenesis in the absence of Munc13-mediated vesicle priming. Proc. Natl. Acad. Sci. USA 2002, 99, 9037–9042. [Google Scholar] [CrossRef]
- van Es, M.A.; Veldink, J.H.; Saris, C.G.; Blauw, H.M.; van Vught, P.W.; Birve, A.; Lemmens, R.; Schelhaas, H.J.; Groen, E.J.; Huisman, M.H.; et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat. Genet. 2009, 41, 1083–1087. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.R.; Prudencio, M.; Koike, Y.; Vatsavayai, S.C.; Kim, G.; Harbinski, F.; Briner, A.; Rodriguez, C.M.; Guo, C.; Akiyama, T.; et al. TDP-43 represses cryptic exon inclusion in the FTD-ALS gene UNC13A. Nature 2022, 603, 124–130. [Google Scholar] [CrossRef]
- Calvo, A.; Canosa, A.; Moglia, C.; Manera, U.; Grassano, M.; Vasta, R.; Palumbo, F.; Cugnasco, P.; Gallone, S.; Brunetti, M.; et al. Clinical and Metabolic Signature of UNC13A rs12608932 Variant in Amyotrophic Lateral Sclerosis. Neurol. Genet. 2022, 8, e200033. [Google Scholar] [CrossRef]
- Koike, Y.; Pickles, S.; Estades Ayuso, V.; Jansen-West, K.; Qi, Y.A.; Li, Z.; Daughrity, L.M.; Yue, M.; Zhang, Y.J.; Cook, C.N.; et al. TDP-43 and other hnRNPs regulate cryptic exon inclusion of a key ALS/FTD risk gene, UNC13A. PLoS Biol. 2023, 21, e3002028. [Google Scholar]
- Meldolesi, J. Alternative Splicing by NOVA Factors: From Gene Expression to Cell Physiology and Pathology. Int. J. Mol. Sci. 2020, 21, 3941. [Google Scholar] [CrossRef]
- Krach, F.; Wheeler, E.C.; Regensburger, M.; Boerstler, T.; Wend, H.; Vu, A.Q.; Wang, R.; Reischl, S.; Boldt, K.; Batra, R.; et al. Aberrant NOVA1 function disrupts alternative splicing in early stages of amyotrophic lateral sclerosis. Acta Neuropathol. 2022, 144, 413–435. [Google Scholar] [CrossRef]
- Suzuki, H.; Shibagaki, Y.; Hattori, S.; Matsuoka, M. C9-ALS/FTD-linked proline-arginine dipeptide repeat protein associates with paraspeckle components and increases paraspeckle formation. Cell Death Dis. 2019, 10, 746. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.; Rollinson, S.; Hobbs, E.; Pickering-Brown, S. C9orf72 dipeptides disrupt the nucleocytoplasmic transport machinery and cause TDP-43 mislocalisation to the cytoplasm. Sci. Rep. 2022, 12, 4799. [Google Scholar] [CrossRef] [PubMed]
- Perrone, B.; La Cognata, V.; Sprovieri, T.; Ungaro, C.; Conforti, F.L.; Ando, S.; Cavallaro, S. Alternative Splicing of ALS Genes: Misregulation and Potential Therapies. Cell. Mol. Neurobiol. 2020, 40, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Nikom, D.; Zheng, S. Alternative splicing in neurodegenerative disease and the promise of RNA therapies. Nat. Rev. Neurosci. 2023, 24, 457–473. [Google Scholar] [CrossRef]
- Tassinari, V.; La Rosa, P.; Guida, E.; Colopi, A.; Caratelli, S.; De Paolis, F.; Gallo, A.; Cenciarelli, C.; Sconocchia, G.; Dolci, S.; et al. Contribution of A-to-I RNA editing, M6A RNA Methylation, and Alternative Splicing to physiological brain aging and neurodegenerative diseases. Mech. Ageing Dev. 2023, 212, 111807. [Google Scholar] [CrossRef]
- Shao, W.; Todd, T.W.; Wu, Y.; Jones, C.Y.; Tong, J.; Jansen-West, K.; Daughrity, L.M.; Park, J.; Koike, Y.; Kurti, A.; et al. Two FTD-ALS genes converge on the endosomal pathway to induce TDP-43 pathology and degeneration. Science 2022, 378, 94–99. [Google Scholar] [CrossRef]
- Yang, Z.; Dong, P.; Cao, J.; Lin, N.; Ma, S.; Cao, R.; Cai, L.; Wang, L.; Cao, C.; Xue, Y.; et al. NOVA1 prevents overactivation of the unfolded protein response and facilitates chromatin access during human white adipogenesis. Nucleic Acids Res. 2023, 51, 6981–6998. [Google Scholar] [CrossRef]
- Jensen, K.B.; Dredge, B.K.; Stefani, G.; Zhong, R.; Buckanovich, R.J.; Okano, H.J.; Yang, Y.Y.; Darnell, R.B. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 2000, 25, 359–371. [Google Scholar] [CrossRef]
- Du, L.L.; Sun, J.J.; Chen, Z.H.; Shao, Y.X.; Wu, L.C. NOVA1 promotes SMN2 exon 7 splicing by binding the UCAC motif and increases SMN protein expression. Neural Regen. Res. 2022, 17, 2530–2536. [Google Scholar]
- Eom, T.; Zhang, C.; Wang, H.; Lay, K.; Fak, J.; Noebels, J.L.; Darnell, R.B. NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure. eLife 2013, 2, e00178. [Google Scholar] [CrossRef]
- Ule, J.; Ule, A.; Spencer, J.; Williams, A.; Hu, J.S.; Cline, M.; Wang, H.; Clark, T.; Fraser, C.; Ruggiu, M.; et al. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 2005, 37, 844–852. [Google Scholar] [CrossRef] [PubMed]
- Paz, I.; Kosti, I.; Ares, M., Jr.; Cline, M.; Mandel-Gutfreund, Y. RBPmap: A web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014, 42, W361–W367. [Google Scholar] [CrossRef] [PubMed]
- Adepoju, F.O.; Duru, K.C.; Li, E.; Kovaleva, E.G.; Tsurkan, M.V. Pharmacological Potential of Betulin as a Multitarget Compound. Biomolecules 2023, 13, 1105. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, R.-H.; Chen, H.-J.; Hong, S.-Y. Interaction of the C9orf72-Amyotrophic Lateral Sclerosis-Related Proline–Arginine Dipeptide Repeat Protein with the RNA-Binding Protein NOVA1 Causes Decreased Expression of UNC13A Due to Enhanced Inclusion of Cryptic Exons, Which Is Reversed by Betulin Treatment. Cells 2023, 12, 2476. https://doi.org/10.3390/cells12202476
Fu R-H, Chen H-J, Hong S-Y. Interaction of the C9orf72-Amyotrophic Lateral Sclerosis-Related Proline–Arginine Dipeptide Repeat Protein with the RNA-Binding Protein NOVA1 Causes Decreased Expression of UNC13A Due to Enhanced Inclusion of Cryptic Exons, Which Is Reversed by Betulin Treatment. Cells. 2023; 12(20):2476. https://doi.org/10.3390/cells12202476
Chicago/Turabian StyleFu, Ru-Huei, Hui-Jye Chen, and Syuan-Yu Hong. 2023. "Interaction of the C9orf72-Amyotrophic Lateral Sclerosis-Related Proline–Arginine Dipeptide Repeat Protein with the RNA-Binding Protein NOVA1 Causes Decreased Expression of UNC13A Due to Enhanced Inclusion of Cryptic Exons, Which Is Reversed by Betulin Treatment" Cells 12, no. 20: 2476. https://doi.org/10.3390/cells12202476
APA StyleFu, R.-H., Chen, H.-J., & Hong, S.-Y. (2023). Interaction of the C9orf72-Amyotrophic Lateral Sclerosis-Related Proline–Arginine Dipeptide Repeat Protein with the RNA-Binding Protein NOVA1 Causes Decreased Expression of UNC13A Due to Enhanced Inclusion of Cryptic Exons, Which Is Reversed by Betulin Treatment. Cells, 12(20), 2476. https://doi.org/10.3390/cells12202476






