Enhanced Transcriptional Signature and Expression of Histone-Modifying Enzymes in Salivary Gland Tumors
Abstract
1. Introduction
2. Materials and Methods
2.1. Public Data Acquisition
2.2. Analyzing RNA Sequencing Data
2.3. Differential Gene Expression Analysis and Functional Enrichment Analysis
2.4. Patients’ Tissues Description
2.5. Immunohistochemistry
2.6. Evaluation of Immunohistochemistry
3. Results
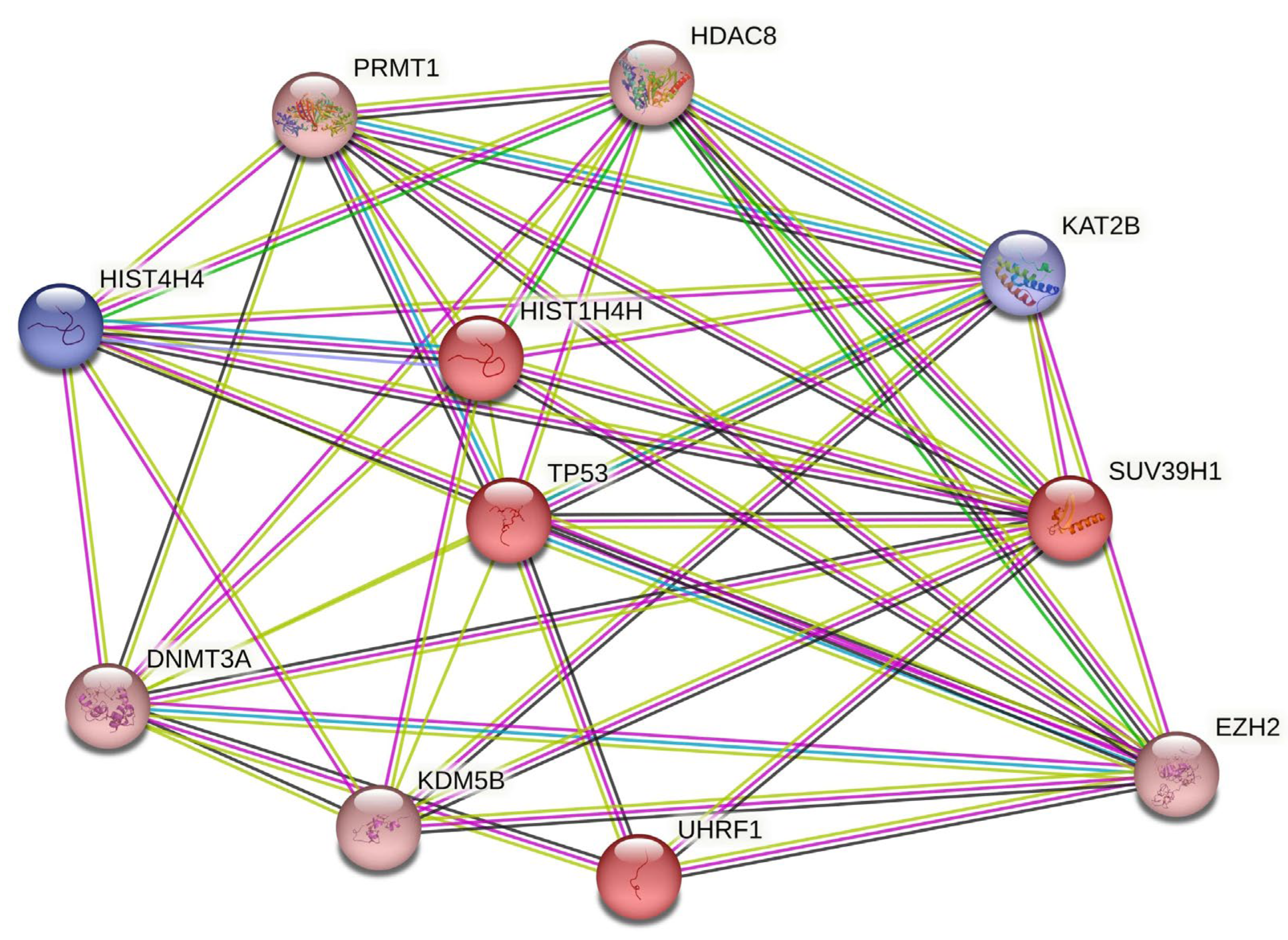
3.1. Increased Transcriptional Signature of Chromatin-modifying Enzymes in ACC Tumors
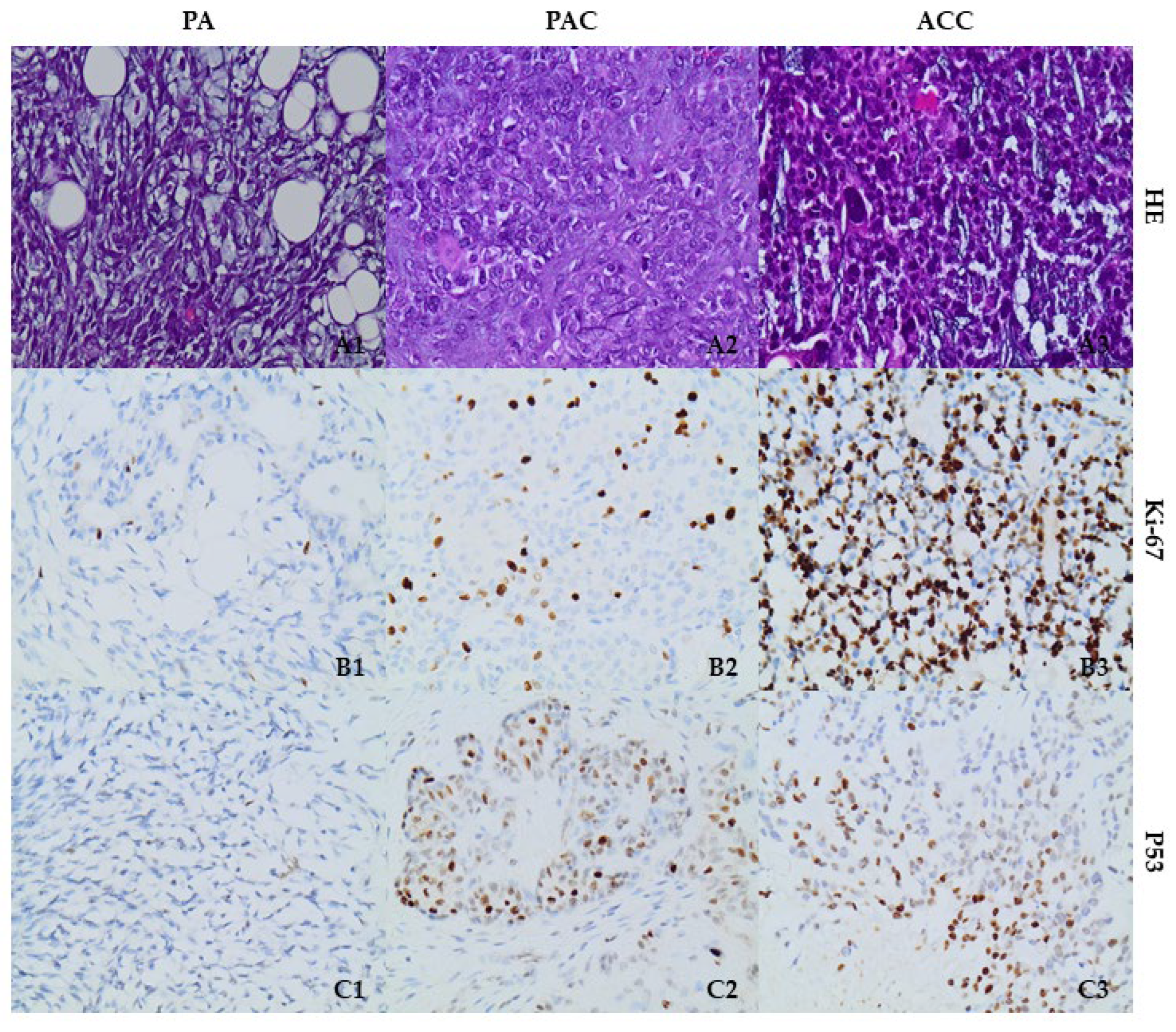
3.2. p53 and Histone-Modifying Enzyme Protein Expression Is Upregulated in SGT Tissues
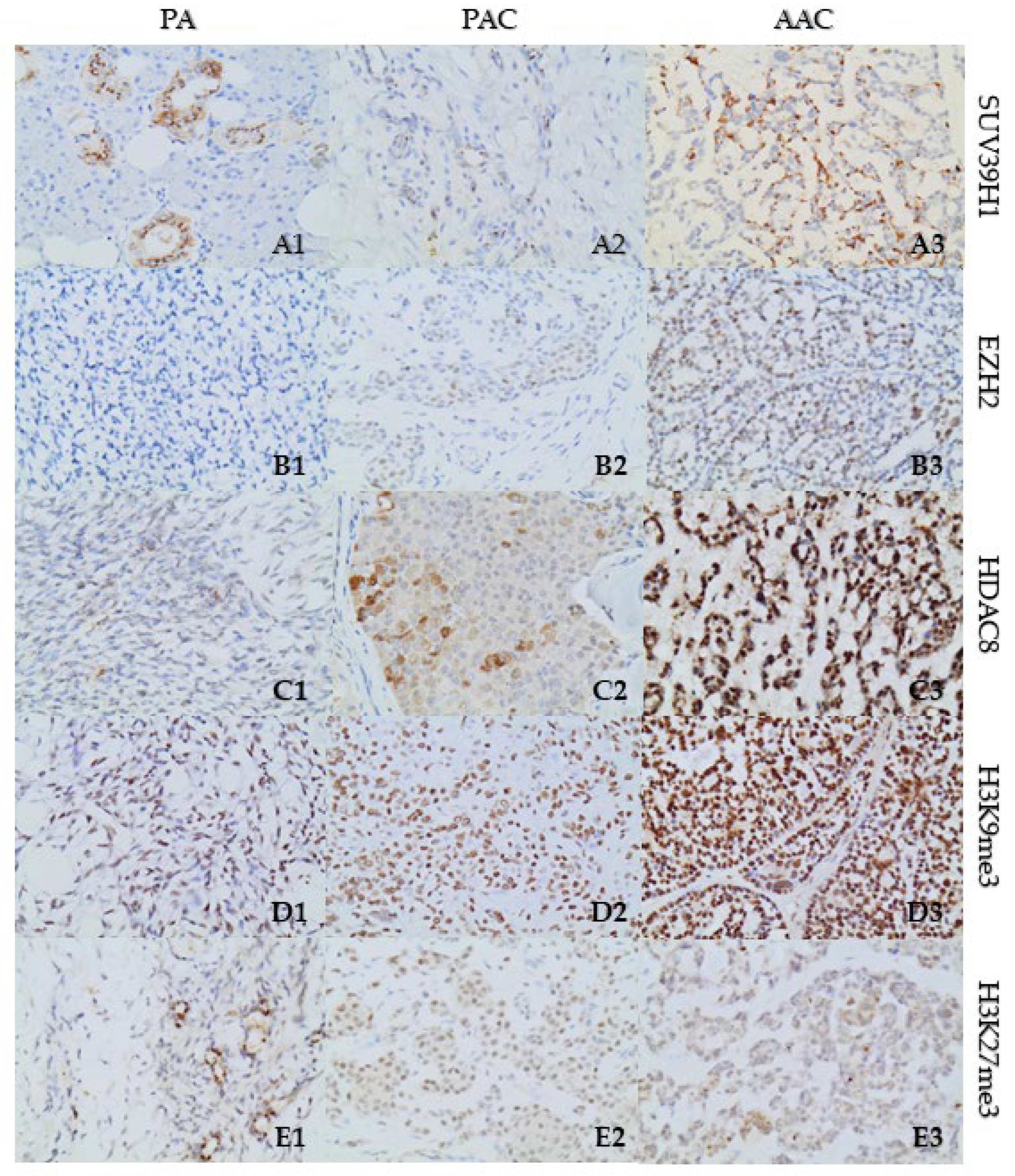
3.3. Increased Expression of Repressive Histone Marks H3K27me3 and H3K9me3 in SGT Tissues
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Da Silva, L.P.; Serpa, M.S.; Viveiros, S.K.; Sena, D.A.C.; de Carvalho Pinho, R.F.; de Abreu Guimaraes, L.D.; de Sousa Andrade, E.S.; Dias Pereira, J.R.; Silveira, M.; Sobral, A.P.V.; et al. Salivary gland tumors in a Brazilian population: A 20-year retrospective and multicentric study of 2292 cases. J. Craniomaxillofac. Surg. 2018, 46, 2227–2233. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Hao, Y.; Huang, M.X.; Ma, D.Q.; Chen, Y.; Luo, H.Y.; Gao, Y.; Cao, Z.Q.; Peng, X.; Yu, G.Y. Salivary gland tumours in a northern Chinese population: A 50-year retrospective study of 7190 cases. Int. J. Oral Maxillofac. Surg. 2017, 46, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Steuer, C.E.; Hanna, G.J.; Viswanathan, K.; Bates, J.E.; Kaka, A.S.; Schmitt, N.C.; Ho, A.L.; Saba, N.F. The evolving landscape of salivary gland tumors. CA Cancer J. Clin. 2023, 1–23. [Google Scholar] [CrossRef]
- Manou, M.; Kanakoglou, D.S.; Loupis, T.; Vrachnos, D.M.; Theocharis, S.; Papavassiliou, A.G.; Piperi, C. Role of Histone Deacetylases in the Pathogenesis of Salivary Gland Tumors and Therapeutic Targeting Options. Int. J. Mol. Sci. 2023, 24, 10038. [Google Scholar] [CrossRef]
- Mat Lazim, N.; Yousaf, A.; Abusalah, M.A.H.; Sulong, S.; Mohd Ismail, Z.I.; Mohamud, R.; Abu-Harirah, H.A.; AlRamadneh, T.N.; Hassan, R.; Abdullah, B. The Epigenesis of Salivary Glands Carcinoma: From Field Cancerization to Carcinogenesis. Cancers 2023, 15, 2111. [Google Scholar] [CrossRef] [PubMed]
- Karpinets, T.V.; Mitani, Y.; Liu, B.; Zhang, J.; Pytynia, K.B.; Sellen, L.D.; Karagiannis, D.T.; Ferrarotto, R.; Futreal, A.P.; El-Naggar, A.K. Whole-Genome Sequencing of Common Salivary Gland Carcinomas: Subtype-Restricted and Shared Genetic Alterations. Clin. Cancer Res. 2021, 27, 3960–3969. [Google Scholar] [CrossRef]
- Mitani, Y.; Li, J.; Rao, P.H.; Zhao, Y.J.; Bell, D.; Lippman, S.M.; Weber, R.S.; Caulin, C.; El-Naggar, A.K. Comprehensive analysis of the MYB-NFIB gene fusion in salivary adenoid cystic carcinoma: Incidence, variability, and clinicopathologic significance. Clin. Cancer Res. 2010, 16, 4722–4731. [Google Scholar] [CrossRef]
- Chau, Y.; Hongyo, T.; Aozasa, K.; Chan, J.K. Dedifferentiation of adenoid cystic carcinoma: Report of a case implicating p53 gene mutation. Hum. Pathol. 2001, 32, 1403–1407. [Google Scholar] [CrossRef]
- Nagao, T.; Gaffey, T.A.; Serizawa, H.; Sugano, I.; Ishida, Y.; Yamazaki, K.; Tokashiki, R.; Yoshida, T.; Minato, H.; Kay, P.A.; et al. Dedifferentiated adenoid cystic carcinoma: A clinicopathologic study of 6 cases. Mod. Pathol. 2003, 16, 1265–1272. [Google Scholar] [CrossRef]
- Cheuk, W.; Chan, J.K.; Ngan, R.K. Dedifferentiation in adenoid cystic carcinoma of salivary gland: An uncommon complication associated with an accelerated clinical course. Am. J. Surg. Pathol. 1999, 23, 465–472. [Google Scholar] [CrossRef]
- Seethala, R.R.; Hunt, J.L.; Baloch, Z.W.; Livolsi, V.A.; Leon Barnes, E. Adenoid cystic carcinoma with high-grade transformation: A report of 11 cases and a review of the literature. Am. J. Surg. Pathol. 2007, 31, 1683–1694. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.F.; Altemani, A.; Vekony, H.; Bloemena, E.; Fresno, F.; Suarez, C.; Llorente, J.L.; Hermsen, M. Genetic profile of adenoid cystic carcinomas (ACC) with high-grade transformation versus solid type. Cell Oncol. 2011, 34, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Saleh, E.; Ukwas, A. Adenoid Cystic Carcinoma of Salivary Glands: A Ten-Year Review and an Assessment of the Current Management, Surgery, Radiotherapy, and Chemotherapy. Int. J. Otolaryngol. 2023, 2023, 7401458. [Google Scholar] [CrossRef] [PubMed]
- Magno Guimaraes, D.; deLucas da Silva Almeida, F.; Moraes Castilho, R.; Eduardo Nor, J.; Daumas Nunes, F. DNA methyltransferase expression is associated with cell proliferation in salivary mucoepidermoid carcinoma. J. Oral. Pathol. Med. 2020, 49, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- Ahn, M.Y.; Yoon, J.H. Histone deacetylase 7 silencing induces apoptosis and autophagy in salivary mucoepidermoid carcinoma cells. J. Oral. Pathol. Med. 2017, 46, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Panaccione, A.; Zhang, Y.; Mi, Y.; Mitani, Y.; Yan, G.; Prasad, M.L.; McDonald, W.H.; El-Naggar, A.K.; Yarbrough, W.G.; Ivanov, S.V. Chromosomal abnormalities and molecular landscape of metastasizing mucinous salivary adenocarcinoma. Oral. Oncol. 2017, 66, 38–45. [Google Scholar] [CrossRef]
- Hajosi-Kalcakosz, S.; Vincze, E.; Dezso, K.; Paku, S.; Rokusz, A.; Sapi, Z.; Toth, E.; Nagy, P. EZH2 is a sensitive marker of malignancy in salivary gland tumors. Diagn. Pathol. 2015, 10, 163. [Google Scholar] [CrossRef]
- Pouloudi, D.; Manou, M.; Sarantis, P.; Tsoukalas, N.; Tsourouflis, G.; Dana, E.; Karamouzis, M.V.; Klijanienko, J.; Theocharis, S. Clinical Significance of Histone Deacetylase (HDAC)-1, -2, -4 and -6 Expression in Salivary Gland Tumors. Diagnostics 2021, 11, 517. [Google Scholar] [CrossRef]
- Daa, T.; Kashima, K.; Kondo, Y.; Yada, N.; Suzuki, M.; Yokoyama, S. Aberrant methylation in promoter regions of cyclin-dependent kinase inhibitor genes in adenoid cystic carcinoma of the salivary gland. APMIS 2008, 116, 21–26. [Google Scholar] [CrossRef]
- Maruya, S.; Kurotaki, H.; Wada, R.; Saku, T.; Shinkawa, H.; Yagihashi, S. Promoter methylation and protein expression of the E-cadherin gene in the clinicopathologic assessment of adenoid cystic carcinoma. Mod. Pathol. 2004, 17, 637–645. [Google Scholar] [CrossRef][Green Version]
- Zhang, C.Y.; Mao, L.; Li, L.; Tian, Z.; Zhou, X.J.; Zhang, Z.Y.; Li, J. Promoter methylation as a common mechanism for inactivating E-cadherin in human salivary gland adenoid cystic carcinoma. Cancer 2007, 110, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P.J.; Davies, H.R.; Mitani, Y.; Van Loo, P.; Shlien, A.; Tarpey, P.S.; Papaemmanuil, E.; Cheverton, A.; Bignell, G.R.; Butler, A.P.; et al. Whole exome sequencing of adenoid cystic carcinoma. J. Clin. Investig. 2013, 123, 2965–2968. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.S.; Kannan, K.; Roy, D.M.; Morris, L.G.; Ganly, I.; Katabi, N.; Ramaswami, D.; Walsh, L.A.; Eng, S.; Huse, J.T.; et al. The mutational landscape of adenoid cystic carcinoma. Nat. Genet. 2013, 45, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Lam-Ubol, A.; Phattarataratip, E. Distinct histone H3 modification profiles correlate with aggressive characteristics of salivary gland neoplasms. Sci. Rep. 2022, 12, 15063. [Google Scholar] [CrossRef]
- Frerich, C.A.; Brayer, K.J.; Painter, B.M.; Kang, H.; Mitani, Y.; El-Naggar, A.K.; Ness, S.A. Transcriptomes define distinct subgroups of salivary gland adenoid cystic carcinoma with different driver mutations and outcomes. Oncotarget 2018, 9, 7341–7358. [Google Scholar] [CrossRef]
- Leinonen, R.; Sugawara, H.; Shumway, M. The sequence read archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef]
- Barrett, T.; Clark, K.; Gevorgyan, R.; Gorelenkov, V.; Gribov, E.; Karsch-Mizrachi, I.; Kimelman, M.; Pruitt, K.D.; Resenchuk, S.; Tatusova, T.; et al. BioProject and BioSample databases at NCBI: Facilitating capture and organization of metadata. Nucleic Acids Res. 2012, 40, D57–D63. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 18 October 2022).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- The Gene Ontology, C. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Ho, A.S.; Ochoa, A.; Jayakumaran, G.; Zehir, A.; Valero Mayor, C.; Tepe, J.; Makarov, V.; Dalin, M.G.; He, J.; Bailey, M.; et al. Genetic hallmarks of recurrent/metastatic adenoid cystic carcinoma. J. Clin. Investig. 2019, 129, 4276–4289. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef]
- Adderley, H.; Rack, S.; Hapuarachi, B.; Feeney, L.; Morgan, D.; Hussell, T.; Wallace, A.J.; Betts, G.; Hodgson, C.; Harrington, K.; et al. The utility of TP53 and PIK3CA mutations as prognostic biomarkers in salivary adenoid cystic carcinoma. Oral. Oncol. 2021, 113, 105095. [Google Scholar] [CrossRef]
- Yang, X.; Jing, D.; Liu, L.; Shen, Z.; Ju, J.; Ma, C.; Sun, M. Downregulation of p53 promotes in vitro perineural invasive activity of human salivary adenoid cystic carcinoma cells through epithelial-mesenchymal transition-like changes. Oncol. Rep. 2015, 33, 1650–1656. [Google Scholar] [CrossRef] [PubMed]
- Weirich, S.; Khella, M.S.; Jeltsch, A. Structure, Activity and Function of the Suv39h1 and Suv39h2 Protein Lysine Methyltransferases. Life 2021, 11, 703. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhang, J.; Ma, Y.; Wu, C.; Cui, W.; Wang, L. Histone methyltransferase and drug resistance in cancers. J. Exp. Clin. Cancer Res. 2020, 39, 173. [Google Scholar] [CrossRef]
- Chen, J.H.; Yeh, K.T.; Yang, Y.M.; Chang, J.G.; Lee, H.E.; Hung, S.Y. High expressions of histone methylation- and phosphorylation-related proteins are associated with prognosis of oral squamous cell carcinoma in male population of Taiwan. Med. Oncol. 2013, 30, 513. [Google Scholar] [CrossRef]
- Xia, R.; Zhou, R.; Tian, Z.; Zhang, C.; Wang, L.; Hu, Y.; Han, J.; Li, J. High expression of H3K9me3 is a strong predictor of poor survival in patients with salivary adenoid cystic carcinoma. Arch. Pathol. Lab. Med. 2013, 137, 1761–1769. [Google Scholar] [CrossRef]
- Bae, W.K.; Hennighausen, L. Canonical and non-canonical roles of the histone methyltransferase EZH2 in mammary development and cancer. Mol. Cell Endocrinol. 2014, 382, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.P.; Ling, K. EZH2 and histone deacetylase inhibitors induce apoptosis in triple negative breast cancer cells by differentially increasing H3 Lys(27) acetylation in the BIM gene promoter and enhancers. Oncol. Lett. 2017, 14, 5735–5742. [Google Scholar] [CrossRef]
- Vekony, H.; Raaphorst, F.M.; Otte, A.P.; van Lohuizen, M.; Leemans, C.R.; van der Waal, I.; Bloemena, E. High expression of Polycomb group protein EZH2 predicts poor survival in salivary gland adenoid cystic carcinoma. J. Clin. Pathol. 2008, 61, 744–749. [Google Scholar] [CrossRef]
- Chen, C.W.; Fu, M.; Du, Z.H.; Zhao, F.; Yang, W.W.; Xu, L.H.; Li, S.L.; Ge, X.Y. Long Noncoding RNA MRPL23-AS1 Promotes Adenoid Cystic Carcinoma Lung Metastasis. Cancer Res. 2020, 80, 2273–2285. [Google Scholar] [CrossRef]
- Auclair, Y.; Richard, S. The role of arginine methylation in the DNA damage response. DNA Repair 2013, 12, 459–465. [Google Scholar] [CrossRef]
- Scorilas, A.; Black, M.H.; Talieri, M.; Diamandis, E.P. Genomic organization, physical mapping, and expression analysis of the human protein arginine methyltransferase 1 gene. Biochem. Biophys. Res. Commun. 2000, 278, 349–359. [Google Scholar] [CrossRef] [PubMed]
- An, W.; Kim, J.; Roeder, R.G. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 2004, 117, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, D.; Lu, J.; Huang, B.; Wang, Y.; Dong, M.; Fan, D.; Li, H.; Gao, Y.; Hou, P.; et al. Methylation of EZH2 by PRMT1 regulates its stability and promotes breast cancer metastasis. Cell Death Differ. 2020, 27, 3226–3242. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Cho, H.; Yoo, J.; Kim, G.W.; Jeon, Y.H.; Lee, S.W.; Kwon, S.H. Pathological Role of HDAC8: Cancer and Beyond. Cells 2022, 11, 3161. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Wong, V.W.; Wong, G.L.; Yang, W.; Sun, H.; Shen, J.; Tong, J.H.; Go, M.Y.; Cheung, Y.S.; Lai, P.B.; et al. Histone Deacetylase HDAC8 Promotes Insulin Resistance and beta-Catenin Activation in NAFLD-Associated Hepatocellular Carcinoma. Cancer Res. 2015, 75, 4803–4816. [Google Scholar] [CrossRef]
- Ahn, M.Y.; Yoon, J.H. Histone deacetylase 8 as a novel therapeutic target in oral squamous cell carcinoma. Oncol. Rep. 2017, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Ahn, M.Y. HDAC inhibitor apicidin suppresses murine oral squamous cell carcinoma cell growth in vitro and in vivo via inhibiting HDAC8 expression. Oncol. Lett. 2018, 16, 6552–6560. [Google Scholar] [CrossRef]
- Hamidi, T.; Singh, A.K.; Chen, T. Genetic alterations of DNA methylation machinery in human diseases. Epigenomics 2015, 7, 247–265. [Google Scholar] [CrossRef]
- Turek-Plewa, J.; Jagodzinski, P.P. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol. Biol. Lett. 2005, 10, 631–647. [Google Scholar]
- Daniel, F.I.; Alves, S.R.; Vieira, D.S.; Biz, M.T.; Daniel, I.W.; Modolo, F. Immunohistochemical expression of DNA methyltransferases 1, 3a, and 3b in actinic cheilitis and lip squamous cell carcinomas. J. Oral. Pathol. Med. 2016, 45, 774–779. [Google Scholar] [CrossRef]
- Adhikari, B.R.; Uehara, O.; Matsuoka, H.; Takai, R.; Harada, F.; Utsunomiya, M.; Chujo, T.; Morikawa, T.; Shakya, M.; Yoshida, K.; et al. Immunohistochemical evaluation of Klotho and DNA methyltransferase 3a in oral squamous cell carcinomas. Med. Mol. Morphol. 2017, 50, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Gazdzicka, J.; Golabek, K.; Strzelczyk, J.K.; Ostrowska, Z. Epigenetic Modifications in Head and Neck Cancer. Biochem. Genet. 2020, 58, 213–244. [Google Scholar] [CrossRef] [PubMed]
- Cavalieri Gomes, C.; da Silveira e Oliveira, C.; Santos Pimenta, L.G.; De Marco, L.; Santiago Gomez, R. Immunolocalization of DNMT1 and DNMT3a in salivary gland neoplasms. Pathobiology 2009, 76, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Li, P.; Fang, L.; Zhu, H.; Xu, L.; Cheng, H.; Zhang, J.; Li, F.; Feng, Y.; Li, Y.; et al. Negative regulation of DNMT3A de novo DNA methylation by frequently overexpressed UHRF family proteins as a mechanism for widespread DNA hypomethylation in cancer. Cell Discov. 2016, 2, 16007. [Google Scholar] [CrossRef]
- Vaughan, R.M.; Dickson, B.M.; Cornett, E.M.; Harrison, J.S.; Kuhlman, B.; Rothbart, S.B. Comparative biochemical analysis of UHRF proteins reveals molecular mechanisms that uncouple UHRF2 from DNA methylation maintenance. Nucleic Acids Res. 2018, 46, 4405–4416. [Google Scholar] [CrossRef]
- Frierson, H.F., Jr.; El-Naggar, A.K.; Welsh, J.B.; Sapinoso, L.M.; Su, A.I.; Cheng, J.; Saku, T.; Moskaluk, C.A.; Hampton, G.M. Large scale molecular analysis identifies genes with altered expression in salivary adenoid cystic carcinoma. Am. J. Pathol. 2002, 161, 1315–1323. [Google Scholar] [CrossRef]
- Chen, H.; Ma, H.; Inuzuka, H.; Diao, J.; Lan, F.; Shi, Y.G.; Wei, W.; Shi, Y. DNA damage regulates UHRF1 stability via the SCF(beta-TrCP) E3 ligase. Mol. Cell Biol. 2013, 33, 1139–1148. [Google Scholar] [CrossRef]
- Knippschild, U.; Kruger, M.; Richter, J.; Xu, P.; Garcia-Reyes, B.; Peifer, C.; Halekotte, J.; Bakulev, V.; Bischof, J. The CK1 Family: Contribution to Cellular Stress Response and Its Role in Carcinogenesis. Front. Oncol. 2014, 4, 96. [Google Scholar] [CrossRef]
- Hervouet, E.; Lalier, L.; Debien, E.; Cheray, M.; Geairon, A.; Rogniaux, H.; Loussouarn, D.; Martin, S.A.; Vallette, F.M.; Cartron, P.F. Disruption of Dnmt1/PCNA/UHRF1 interactions promotes tumorigenesis from human and mice glial cells. PLoS ONE 2010, 5, e11333. [Google Scholar] [CrossRef]
- Pacaud, R.; Brocard, E.; Lalier, L.; Hervouet, E.; Vallette, F.M.; Cartron, P.F. The DNMT1/PCNA/UHRF1 disruption induces tumorigenesis characterized by similar genetic and epigenetic signatures. Sci. Rep. 2014, 4, 4230. [Google Scholar] [CrossRef]
- Barrett, A.; Santangelo, S.; Tan, K.; Catchpole, S.; Roberts, K.; Spencer-Dene, B.; Hall, D.; Scibetta, A.; Burchell, J.; Verdin, E.; et al. Breast cancer associated transcriptional repressor PLU-1/JARID1B interacts directly with histone deacetylases. Int. J. Cancer 2007, 121, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liang, J.; Li, Q. Coordinated regulation of retinoic acid signaling pathway by KDM5B and polycomb repressive complex 2. J. Cell Biochem. 2014, 115, 1528–1538. [Google Scholar] [CrossRef] [PubMed]
- Facompre, N.D.; Harmeyer, K.M.; Sole, X.; Kabraji, S.; Belden, Z.; Sahu, V.; Whelan, K.; Tanaka, K.; Weinstein, G.S.; Montone, K.T.; et al. JARID1B Enables Transit between Distinct States of the Stem-like Cell Population in Oral Cancers. Cancer Res. 2016, 76, 5538–5549. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Ohtani, H.; Chakravarthy, A.; De Carvalho, D.D. Epigenetic therapy in immune-oncology. Nat. Rev. Cancer 2019, 19, 151–161. [Google Scholar] [CrossRef]
- Schlesinger, S.; Meshorer, E. Open Chromatin, Epigenetic Plasticity, and Nuclear Organization in Pluripotency. Dev. Cell 2019, 48, 135–150. [Google Scholar] [CrossRef]
- Wan, J.; Zhan, J.; Li, S.; Ma, J.; Xu, W.; Liu, C.; Xue, X.; Xie, Y.; Fang, W.; Chin, Y.E.; et al. PCAF-primed EZH2 acetylation regulates its stability and promotes lung adenocarcinoma progression. Nucleic Acids Res. 2015, 43, 3591–3604. [Google Scholar] [CrossRef]
- Hamamori, Y.; Sartorelli, V.; Ogryzko, V.; Puri, P.L.; Wu, H.Y.; Wang, J.Y.; Nakatani, Y.; Kedes, L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell 1999, 96, 405–413. [Google Scholar] [CrossRef]
- Qiu, L.; Liu, X.; Xia, H.; Xu, C. Downregulation of P300/CBP-Associated Factor Protects from Vascular Aging via Nrf2 Signal Pathway Activation. Int. J. Mol. Sci. 2022, 23, 12574. [Google Scholar] [CrossRef]
- Okumura, K.; Mendoza, M.; Bachoo, R.M.; DePinho, R.A.; Cavenee, W.K.; Furnari, F.B. PCAF modulates PTEN activity. J. Biol. Chem. 2006, 281, 26562–26568. [Google Scholar] [CrossRef]
- Liu, L.; Scolnick, D.M.; Trievel, R.C.; Zhang, H.B.; Marmorstein, R.; Halazonetis, T.D.; Berger, S.L. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell Biol. 1999, 19, 1202–1209. [Google Scholar] [CrossRef]
- Laptenko, O.; Prives, C. The p53-HAT connection: PCAF rules? Cell Cycle 2012, 11, 2975–2976. [Google Scholar] [CrossRef] [PubMed]
- Love, I.M.; Sekaric, P.; Shi, D.; Grossman, S.R.; Androphy, E.J. The histone acetyltransferase PCAF regulates p21 transcription through stress-induced acetylation of histone H3. Cell Cycle 2012, 11, 2458–2466. [Google Scholar] [CrossRef] [PubMed]
- Koutelou, E.; Farria, A.T.; Dent, S.Y.R. Complex functions of Gcn5 and Pcaf in development and disease. Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194609. [Google Scholar] [CrossRef] [PubMed]
- Kyakumoto, S.; Kito, N.; Sato, N. Expression of cAMP response element binding protein (CREB)-binding protein (CBP) and the implication in retinoic acid-inducible transcription activation in human salivary gland adenocarcinoma cell line HSG. Endocr. Res. 2003, 29, 277–289. [Google Scholar] [CrossRef] [PubMed]
| Primary Antibody | Company | Catalogue No. | Clone | Host | IHC Dilution | Antigen Retrieval | Positive Control |
|---|---|---|---|---|---|---|---|
| Ki-67 | Dako | M7240 | Monoclonal | Mouse | 1:200 | pH = 9 | - |
| p53 | Dako | M7001 | Monoclonal | Mouse | 1:200 | pH = 6 | - |
| SUV39H1 | EMD Millipore | 05-615 | Monoclonal | Mouse | 1:300 | pH = 6 | Human breast cancer |
| Ezh2 | Abcam | Ab283270 | Monoclonal | Mouse | 1:100 | pH = 9 | Human breast cancer |
| HDAC8 | Abcam | Ab217702 | Polyclonal | Rabbit | 1:150 | pH = 6 | Human brain |
| H3K27me3 | Millipore, MA | 07-449 | Polyclonal | Rabbit | 1:50 | pH = 6 | Human normal breast |
| H3K9me3 | BioLegend | 815601 | Monoclonal | Mouse | 1:80 | pH = 6 | Human breast |
| Ensembl ID | Gene Symbol | Adj. p-Value | Log2FC |
|---|---|---|---|
| ENSG00000276043 | UHRF1 | 1.72 × 10−2 | 2.638120288 |
| ENSG00000101945 | SUV39H1 | 3.52 × 10−2 | 2.243544044 |
| ENSG00000141510 | TP53 | 1.78 × 10−2 | 2.1709429 |
| ENSG00000106462 | EZH2 | 5.29 × 10−3 | 1.8144852 |
| ENSG00000126457 | PRMT1 | 9.91 × 10−3 | 1.58807423 |
| ENSG00000119772 | DNMT3A | 1.85 × 10−2 | 1.524719521 |
| ENSG00000117139 | KDM5B | 6.76 × 10−3 | 1.400579007 |
| ENSG00000147099 | HDAC8 | 1.24 × 10−2 | 1.297709246 |
| ENSG00000114166 | KAT2B | 2.49 × 10−2 | −2.244361094 |
| Antibody | SGT Immunoreactivity | ||
|---|---|---|---|
| PA | PAC | ACC | |
| Ki-67 | 2–3% | 30% | 70% |
| p53 | 30%; weak nuclear | 60%; high nuclear | 50%; high nuclear |
| EZH2 | 5%; weak nuclear | 15%; weak nuclear | 70%; high nuclear |
| SUV39H1 | 20%; weak cytoplasmic | 2%; weak cytoplasmic | 40%; high cytoplasmic and paranuclear stain with punctate staining type |
| HDAC8 | 10%; high nuclear and cytoplasmic | 40%; high nuclear and cytoplasmic | 50%; high nuclear and cytoplasmic |
| H3K9me3 | 40%; weak | 95%; high | 98%; high |
| H3K27me3 | 15%; focal, weak | 80%; weak | 20%; regional, weak |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manou, M.; Loupis, T.; Vrachnos, D.M.; Katsoulas, N.; Theocharis, S.; Kanakoglou, D.S.; Basdra, E.K.; Piperi, C.; Papavassiliou, A.G. Enhanced Transcriptional Signature and Expression of Histone-Modifying Enzymes in Salivary Gland Tumors. Cells 2023, 12, 2437. https://doi.org/10.3390/cells12202437
Manou M, Loupis T, Vrachnos DM, Katsoulas N, Theocharis S, Kanakoglou DS, Basdra EK, Piperi C, Papavassiliou AG. Enhanced Transcriptional Signature and Expression of Histone-Modifying Enzymes in Salivary Gland Tumors. Cells. 2023; 12(20):2437. https://doi.org/10.3390/cells12202437
Chicago/Turabian StyleManou, Maria, Theodoros Loupis, Dimitrios M. Vrachnos, Nikolaos Katsoulas, Stamatios Theocharis, Dimitrios S. Kanakoglou, Efthimia K. Basdra, Christina Piperi, and Athanasios G. Papavassiliou. 2023. "Enhanced Transcriptional Signature and Expression of Histone-Modifying Enzymes in Salivary Gland Tumors" Cells 12, no. 20: 2437. https://doi.org/10.3390/cells12202437
APA StyleManou, M., Loupis, T., Vrachnos, D. M., Katsoulas, N., Theocharis, S., Kanakoglou, D. S., Basdra, E. K., Piperi, C., & Papavassiliou, A. G. (2023). Enhanced Transcriptional Signature and Expression of Histone-Modifying Enzymes in Salivary Gland Tumors. Cells, 12(20), 2437. https://doi.org/10.3390/cells12202437











