The Histone H3K27 Demethylase REF6 Is a Positive Regulator of Light-Initiated Seed Germination in Arabidopsis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. phyB-Dependent Germination Assays
2.3. RNA Isolation and Quantitative RT-PCR (qRT-PCR) Analysis
2.4. RNA-Sequencing (RNA-seq) Analysis
2.5. ChIP Assays
2.6. Statistical Analysis
3. Results
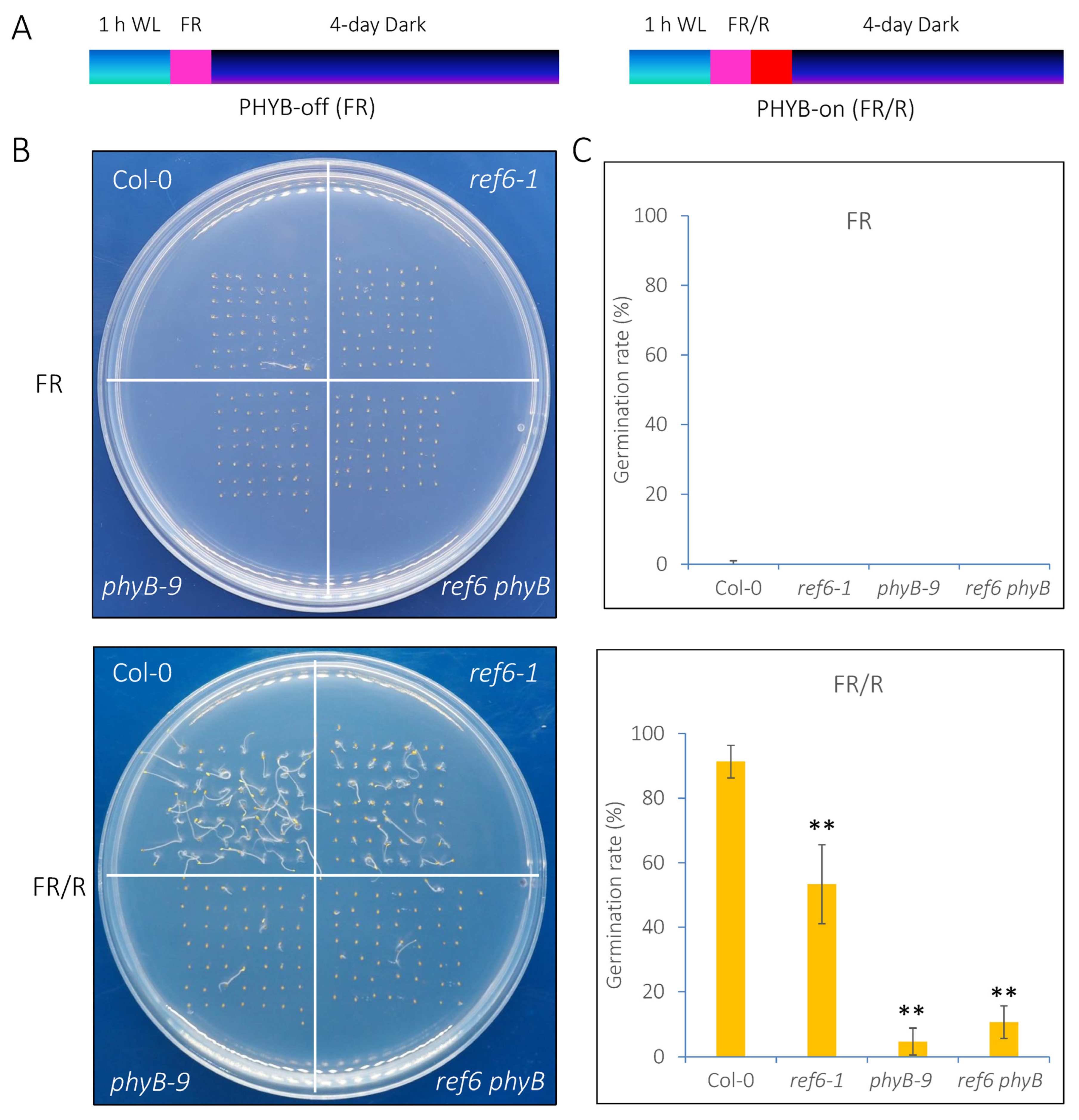
3.1. REF6 Is a Positive Regulator in phyB-Mediated Seed Germination
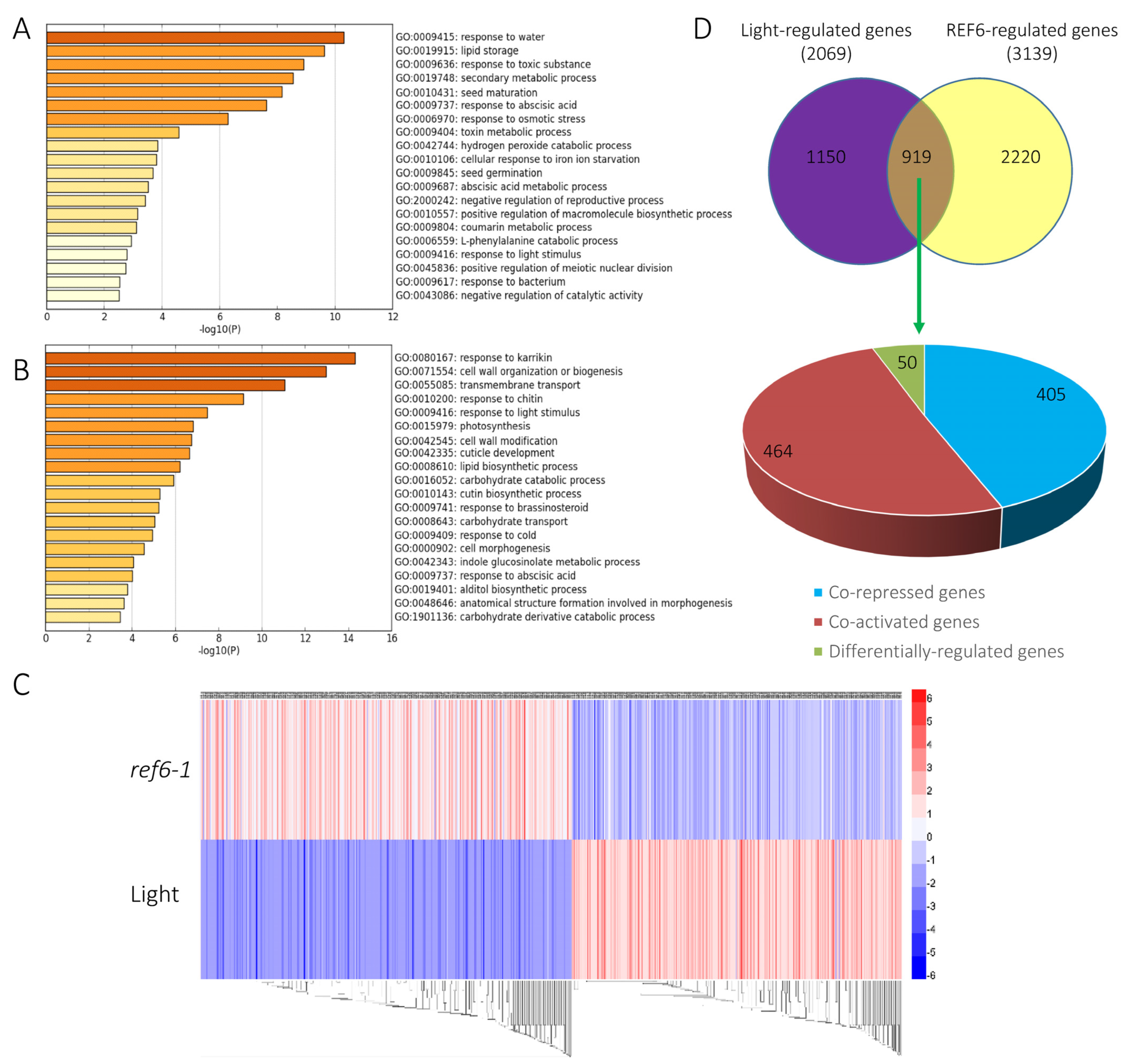
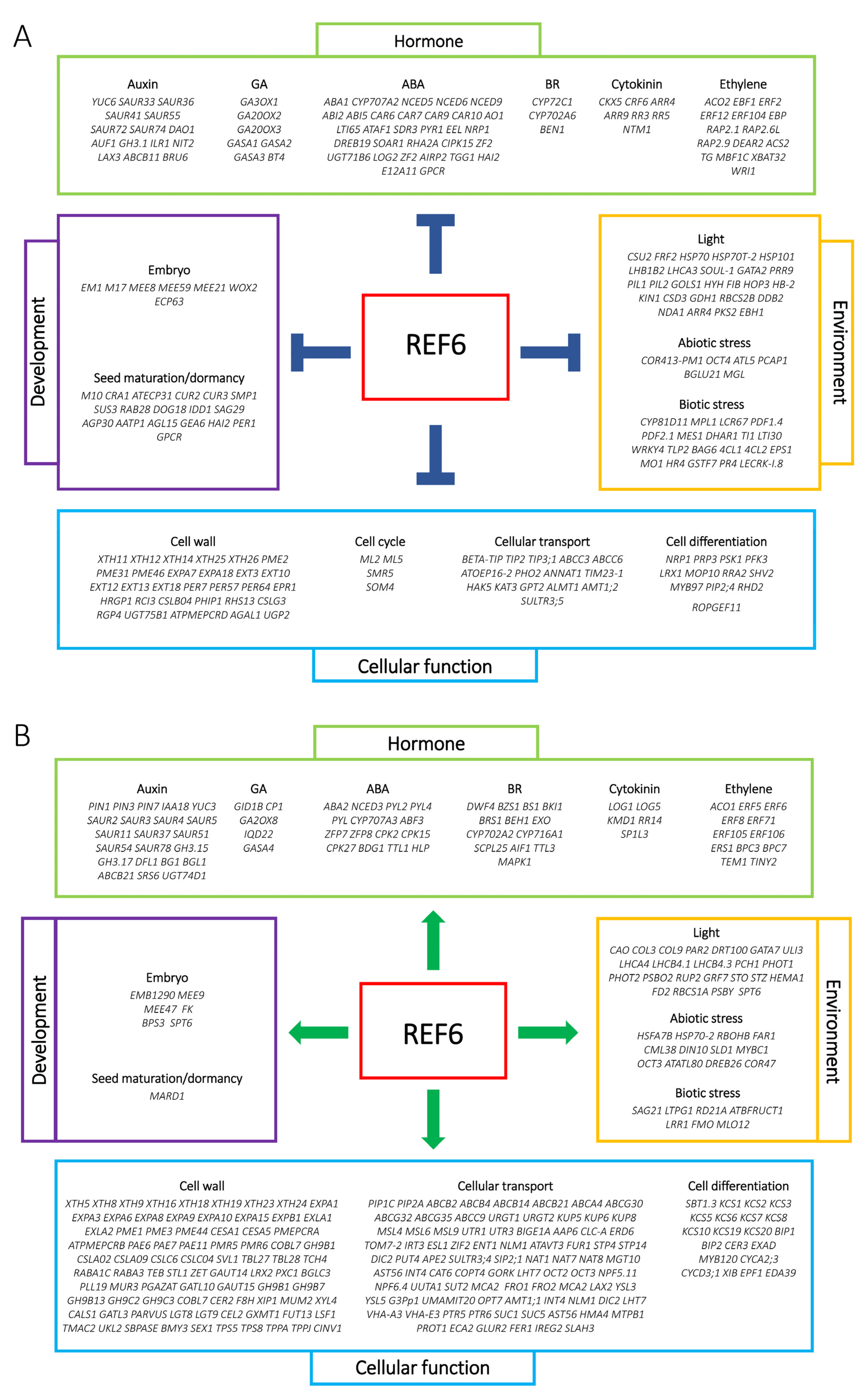
3.2. Genome-Wide Analysis of REF6-Regulated Transcriptome during Seed Germination
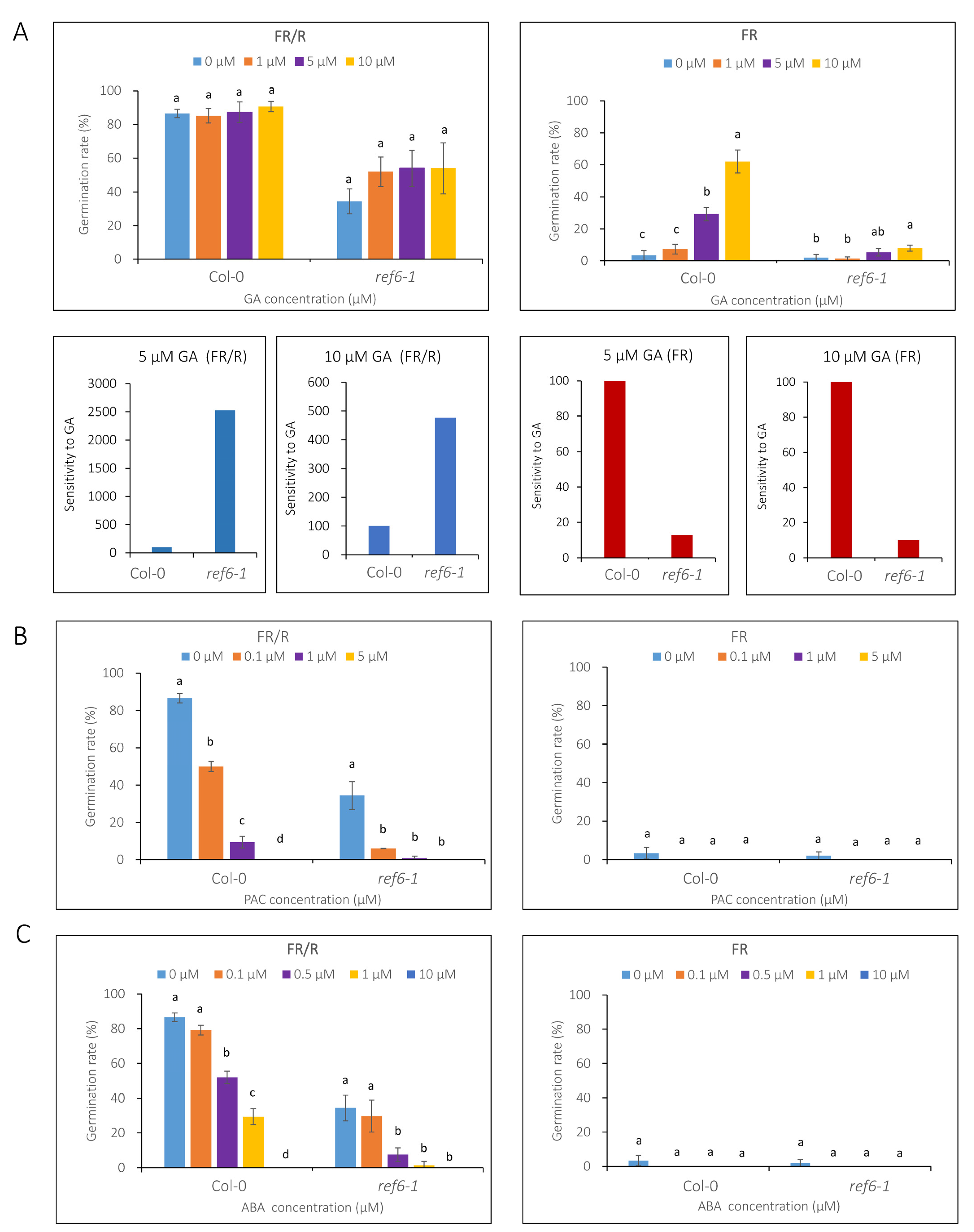
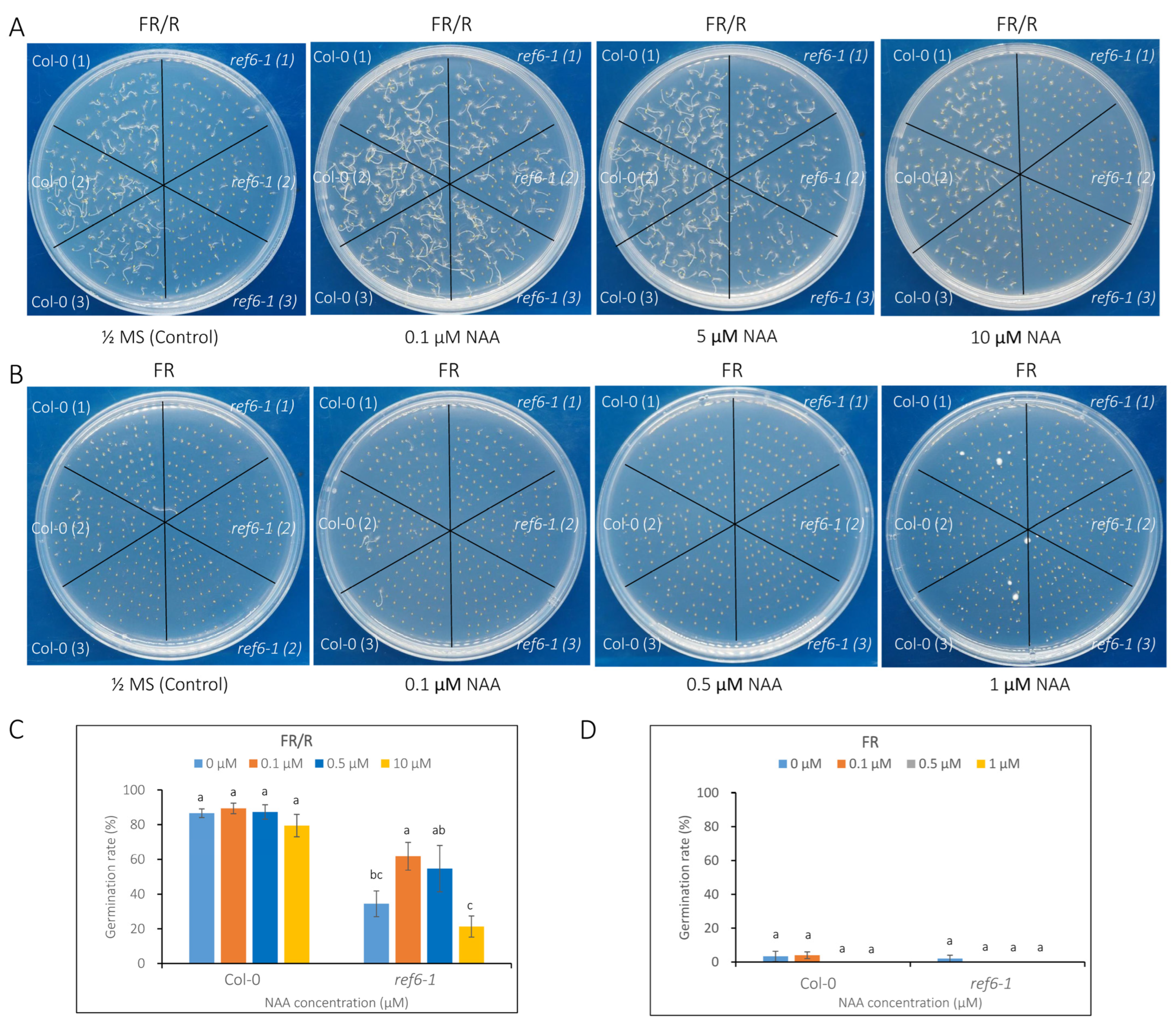
3.3. Seed Germination Phenotype of Ref6 Mutant in Response to ABA, GA and Auxin
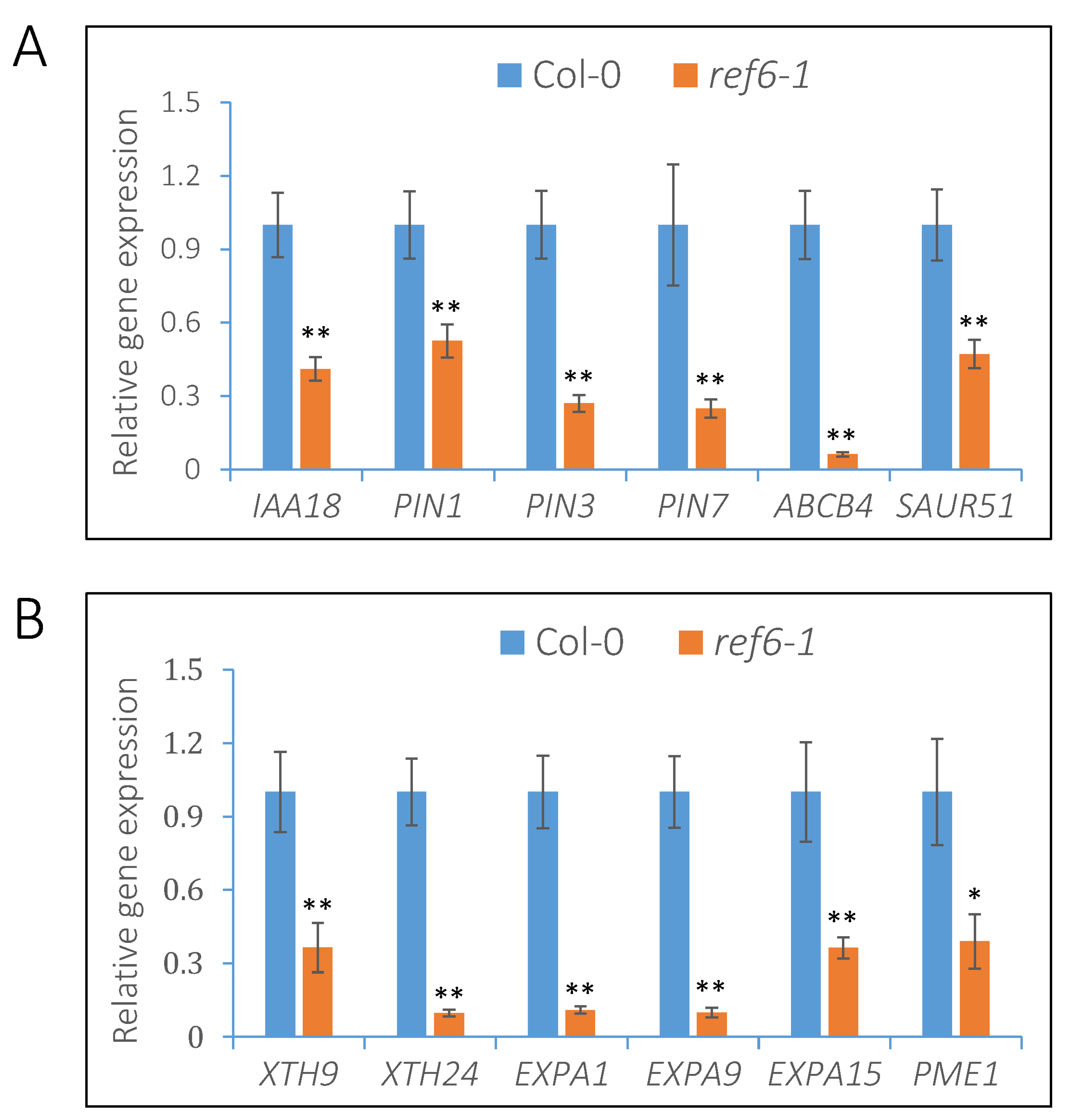
3.4. REF6 Activates the Expression of Auxin-Signaling- and Cell-Wall-Loosening-Related Genes
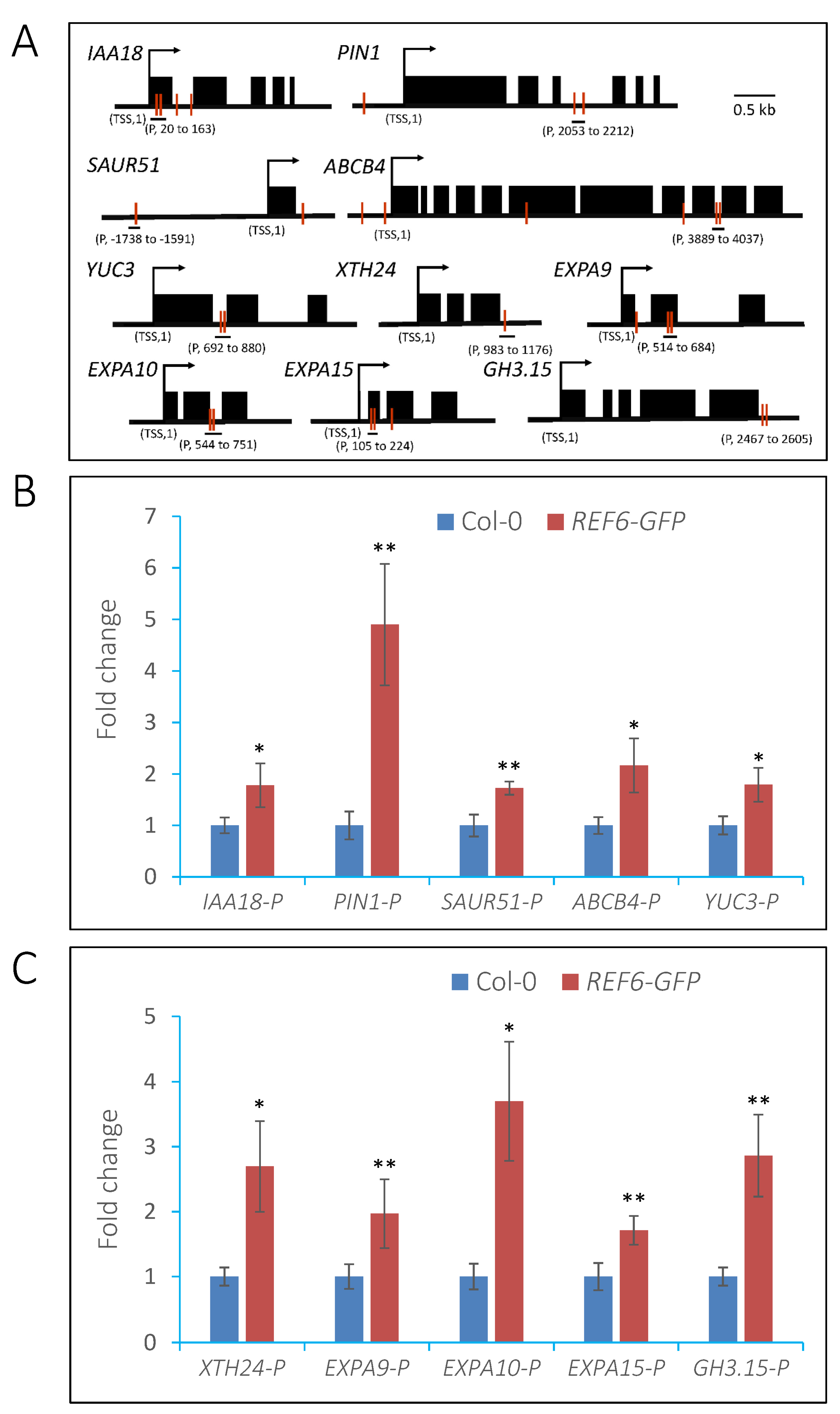
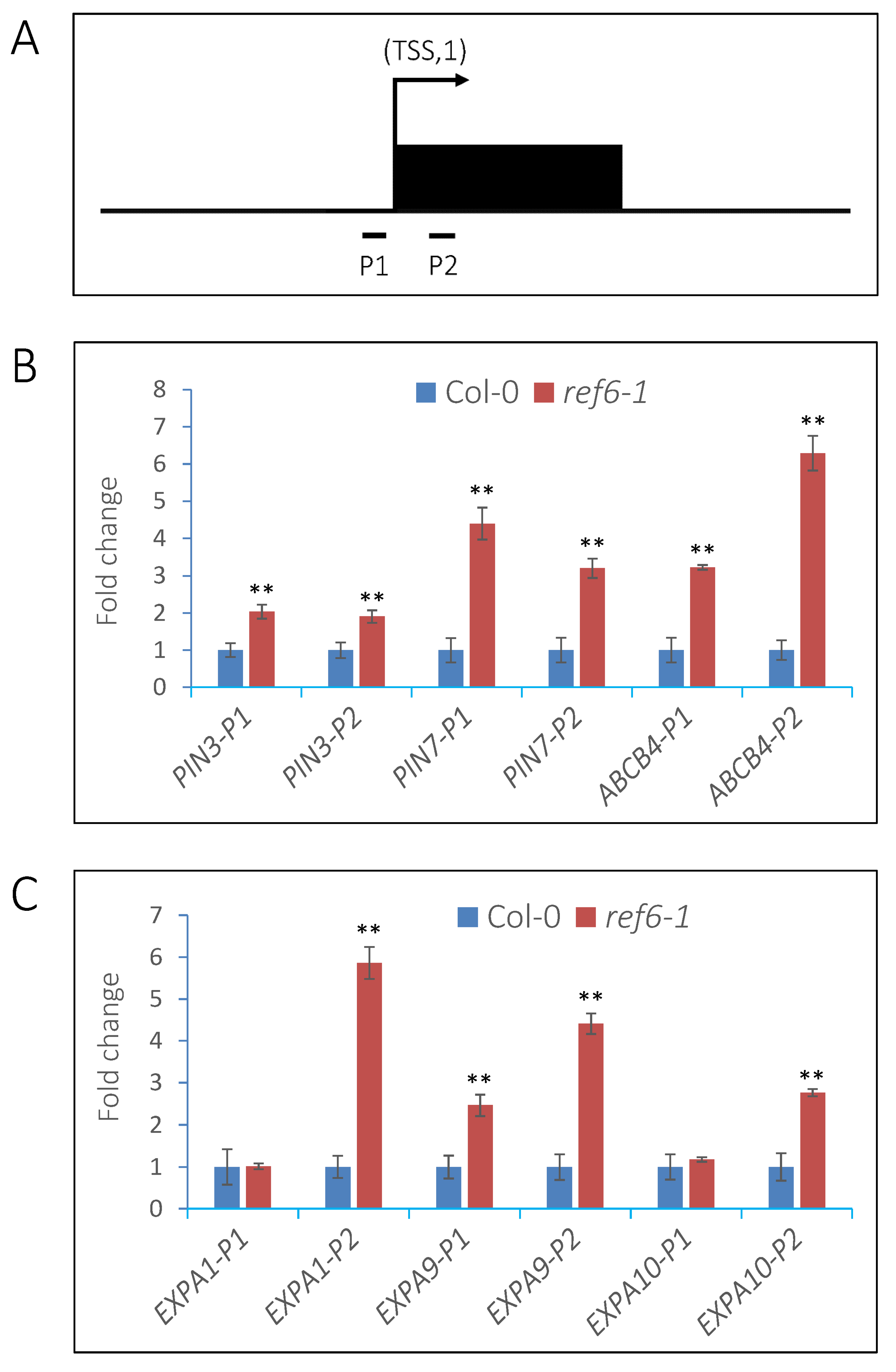
3.5. REF6 Directly Targets Auxin-Signaling- and Cell-Wall-Loosening-Related Genes
3.6. REF6 Activates Target Genes Expression via Removal of Histone H3K27me3
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holdsworth, M.J.; Bentsink, L.; Soppe, W.J.J. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008, 179, 33–54. [Google Scholar] [CrossRef] [PubMed]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef] [PubMed]
- Weitbrecht, K.; Muller, K.; Leubner-Metzger, G. First off the mark: Early seed germination. J. Exp. Bot. 2011, 62, 3289–3309. [Google Scholar] [CrossRef] [PubMed]
- De Wit, M.; Galvao, V.C.; Fankhauser, C. Light-Mediated Hormonal Regulation of Plant Growth and Development. Annu. Rev. Plant Biol. 2016, 67, 513–537. [Google Scholar] [CrossRef]
- Shu, K.; Liu, X.D.; Xie, Q.; He, Z.H. Two Faces of One Seed: Hormonal Regulation of Dormancy and Germination. Mol. Plant. 2016, 9, 34–45. [Google Scholar] [CrossRef]
- Jiao, Y.; Lau, O.S.; Deng, X.W. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 2007, 8, 217–230. [Google Scholar] [CrossRef]
- Borthwick, H.A.; Hendricks, S.B.; Parker, M.W.; Toole, E.H.; Toole, V.K. A Reversible Photoreaction Controlling Seed Germination. Proc. Natl. Acad. Sci. USA 1952, 38, 662–666. [Google Scholar] [CrossRef]
- Lee, K.P.; Piskurewicz, U.; Tureckova, V.; Carat, S.; Chappuis, R.; Strnad, M.; Fankhauser, C.; Lopez-Molina, L. Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev. 2012, 26, 1984–1996. [Google Scholar] [CrossRef]
- Quail, P.H.; Boylan, M.T.; Parks, B.M.; Short, T.W.; Xu, Y.; Wagner, D. Phytochromes: Photosensory perception and signal transduction. Science 1995, 268, 675–680. [Google Scholar] [CrossRef]
- Burgie, E.S.; Vierstra, R.D. Phytochromes: An atomic perspective on photoactivation and signaling. Plant Cell 2014, 26, 4568–4583. [Google Scholar] [CrossRef]
- Castillon, A.; Shen, H.; Huq, E. Phytochrome Interacting Factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007, 12, 514–521. [Google Scholar] [CrossRef]
- Shinomura, T.; Nagatani, A.; Hanzawa, H.; Kubota, M.; Watanabe, M.; Furuya, M. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1996, 93, 8129–8133. [Google Scholar] [CrossRef]
- Shinomura, T.; Nagatani, A.; Chory, J.; Furuya, M. The Induction of Seed Germination in Arabidopsis thaliana Is Regulated Principally by Phytochrome B and Secondarily by Phytochrome A. Plant. Physiol. 1994, 104, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Nambara, E.; Choi, G.; Yamaguchi, S. Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 2009, 69, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.W.; Liu, S.R.; Lin, R.C. The role of light in regulating seed dormancy and germination. J. Integr. Plant Biol. 2020, 62, 1310–1326. [Google Scholar] [CrossRef]
- Hsueh, Y.L.; Lou, C.H. Effects of 2,4-D on Seed Germination and Respiration. Science 1947, 105, 283–285. [Google Scholar] [CrossRef]
- Shi, H.; Zhong, S.W.; Mo, X.R.; Liu, N.; Nezames, C.D.; Deng, X.W. HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 2013, 25, 3770–3784. [Google Scholar] [CrossRef]
- Ma, L.; Li, J.; Qu, L.; Hager, J.; Chen, Z.; Zhao, H.; Deng, X.W. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 2001, 13, 2589–2607. [Google Scholar] [CrossRef]
- Jiao, Y.; Ma, L.; Strickland, E.; Deng, X.W. Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 2005, 17, 3239–3256. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, C.Y.; Wang, K.C.; Luo, M.; Tai, R.; Yuan, L.; Zhao, M.; Yang, S.; Tian, G.; Cui, Y.; et al. PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 2013, 25, 1258–1273. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Chen, C.Y.; Zhao, M.; Zhao, L.; Duan, X.; Duan, J.; Wu, K.; Liu, X. Identification of HDA15-PIF1 as a key repression module directing the transcriptional network of seed germination in the dark. Nucleic Acids Res. 2017, 45, 7137–7150. [Google Scholar] [CrossRef]
- Zhao, L.; Peng, T.; Chen, C.Y.; Ji, R.; Gu, D.; Li, T.; Zhang, D.; Tu, Y.T.; Wu, K.; Liu, X. HY5 Interacts with the Histone Deacetylase HDA15 to Repress Hypocotyl Cell Elongation in Photomorphogenesis. Plant Physiol. 2019, 180, 1450–1466. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.L. The complex language of chromatin regulation during transcription. Nature 2007, 447, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lu, F.; Cui, X.; Cao, X. Histone methylation in higher plants. Annu. Rev. Plant Biol. 2010, 61, 395–420. [Google Scholar] [CrossRef]
- Lee, N.; Kang, H.; Lee, D.; Choi, G. A histone methyltransferase inhibits seed germination by increasing PIF1 mRNA expression in imbibed seeds. Plant J. 2014, 78, 282–293. [Google Scholar] [CrossRef]
- Gu, D.; Ji, R.; He, C.; Peng, T.; Zhang, M.; Duan, J.; Xiong, C.; Liu, X. Arabidopsis Histone Methyltransferase SUVH5 Is a Positive Regulator of Light-Mediated Seed Germination. Front. Plant Sci. 2019, 10, 841. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.N.; Ryu, J.Y.; Jeong, Y.M.; Park, J.; Song, J.J.; Amasino, R.M.; Noh, B.; Noh, Y.S. Control of seed germination by light-induced histone arginine demethylation activity. Dev. Cell 2012, 22, 736–748. [Google Scholar] [CrossRef]
- Noh, B.; Lee, S.H.; Kim, H.J.; Yi, G.; Shin, E.A.; Lee, M.; Jung, K.J.; Doyle, M.R.; Amasino, R.M.; Noh, Y.S. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 2004, 16, 2601–2613. [Google Scholar] [CrossRef]
- Reed, J.W.; Nagpal, P.; Poole, D.S.; Furuya, M.; Chory, J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 1993, 5, 147–157. [Google Scholar] [CrossRef]
- Oh, E.; Yamaguchi, S.; Hu, J.; Yusuke, J.; Jung, B.; Paik, I.; Lee, H.S.; Sun, T.P.; Kamiya, Y.; Choi, G. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 2007, 19, 1192–1208. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef]
- Gendrel, A.V.; Lippman, Z.; Martienssen, R.; Colot, V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2005, 2, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Yamaguchi, S.; Kamiya, Y.; Bae, G.; Chung, W.I.; Choi, G. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006, 47, 124–139. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Lu, F.; Qiu, Q.; Zhou, B.; Gu, L.; Zhang, S.; Kang, Y.; Cui, X.; Ma, X.; Yao, Q.; et al. REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat. Genet. 2016, 48, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Cui, X.; Zhang, S.; Jenuwein, T.; Cao, X. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 2011, 43, 715–719. [Google Scholar] [CrossRef]
- Xu, P.; Cai, X.T.; Wang, Y.; Xing, L.; Chen, Q.; Xiang, C.B. HDG11 upregulates cell-wall-loosening protein genes to promote root elongation in Arabidopsis. J. Exp. Bot. 2014, 65, 4285–4295. [Google Scholar] [CrossRef]
- Li, C.L.; Gu, L.F.; Gao, L.; Chen, C.; Wei, C.Q.; Qiu, Q.; Chien, C.W.; Wang, S.K.; Jiang, L.H.; Ai, L.F.; et al. Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 2016, 48, 687. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Clarenz, O.; Cokus, S.; Bernatavichute, Y.V.; Pellegrini, M.; Goodrich, J.; Jacobsen, S.E. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PloS Biol. 2007, 5, 1026–1035. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.C.; Yang, S.G.; Zhao, M.L.; Luo, M.; Yu, C.W.; Chen, C.Y.; Tai, R.; Wu, K.Q. Transcriptional Repression by Histone Deacetylases in Plants. Mol. Plant 2014, 7, 764–772. [Google Scholar] [CrossRef]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Tong, J.; Fu, W.; Liang, Z.; Ruan, J.; Yu, Y.; Song, X.; Yuan, L.; Xiao, L.; Liu, J.; et al. The H3K27me3 Demethylase RELATIVE OF EARLY FLOWERING6 Suppresses Seed Dormancy by Inducing Abscisic Acid Catabolism. Plant Physiol. 2020, 184, 1969–1978. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Santos-Gonzalez, J.; Kohler, C. Combinations of maternal-specific repressive epigenetic marks in the endosperm control seed dormancy. Elife 2021, 10, e64593. [Google Scholar] [CrossRef]
- Pan, J.; Zhang, H.; Zhan, Z.; Zhao, T.; Jiang, D. A REF6-dependent H3K27me3-depleted state facilitates gene activation during germination in Arabidopsis. J. Genet Genom. 2022; in press. [Google Scholar] [CrossRef]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef]
- Xiao, J.; Wagner, D. Polycomb repression in the regulation of growth and development in Arabidopsis. Curr. Opin. Plant Biol. 2015, 23, 15–24. [Google Scholar] [CrossRef]
- Kim, J.; Bordiya, Y.; Kathare, P.K.; Zhao, B.; Zong, W.; Huq, E.; Sung, S. Phytochrome B triggers light-dependent chromatin remodelling through the PRC2-associated PHD finger protein VIL1. Nat. Plants 2021, 7, 1213–1219. [Google Scholar] [CrossRef]
- Yan, W.; Chen, D.; Smaczniak, C.; Engelhorn, J.; Liu, H.; Yang, W.; Graf, A.; Carles, C.C.; Zhou, D.X.; Kaufmann, K. Dynamic and spatial restriction of Polycomb activity by plant histone demethylases. Nat. Plants 2018, 4, 681–689. [Google Scholar] [CrossRef]
- Crevillen, P.; Yang, H.C.; Cui, X.; Greeff, C.; Trick, M.; Qiu, Q.; Cao, X.F.; Dean, C. Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 2014, 515, 587. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Mei, H.; Zhu, J.; Qiu, Q.; Cao, X.; Deng, X. The histone H3K27 demethylase REF6/JMJ12 promotes thermomorphogenesis in Arabidopsis. Natl. Sci. Rev. 2022, 9, nwab213. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Gu, D.; Deng, L.; He, C.; Zheng, F.; Liu, X. The Histone H3K27 Demethylase REF6 Is a Positive Regulator of Light-Initiated Seed Germination in Arabidopsis. Cells 2023, 12, 295. https://doi.org/10.3390/cells12020295
Wang Y, Gu D, Deng L, He C, Zheng F, Liu X. The Histone H3K27 Demethylase REF6 Is a Positive Regulator of Light-Initiated Seed Germination in Arabidopsis. Cells. 2023; 12(2):295. https://doi.org/10.3390/cells12020295
Chicago/Turabian StyleWang, Yahan, Dachuan Gu, Ling Deng, Chunmei He, Feng Zheng, and Xuncheng Liu. 2023. "The Histone H3K27 Demethylase REF6 Is a Positive Regulator of Light-Initiated Seed Germination in Arabidopsis" Cells 12, no. 2: 295. https://doi.org/10.3390/cells12020295
APA StyleWang, Y., Gu, D., Deng, L., He, C., Zheng, F., & Liu, X. (2023). The Histone H3K27 Demethylase REF6 Is a Positive Regulator of Light-Initiated Seed Germination in Arabidopsis. Cells, 12(2), 295. https://doi.org/10.3390/cells12020295






