SGC-CAMKK2-1: A Chemical Probe for CAMKK2
Abstract
1. Introduction
2. Materials and Methods
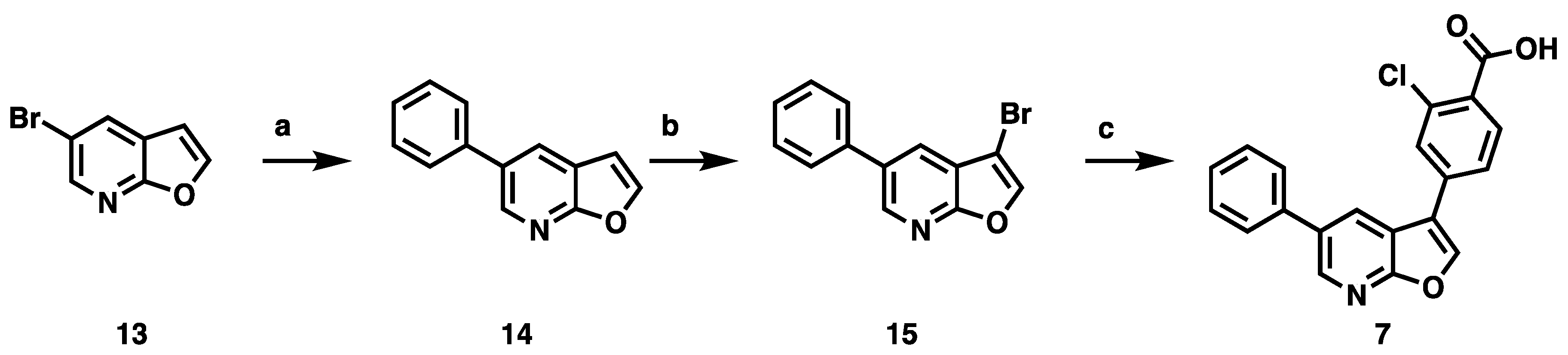
2.1. Synthesis
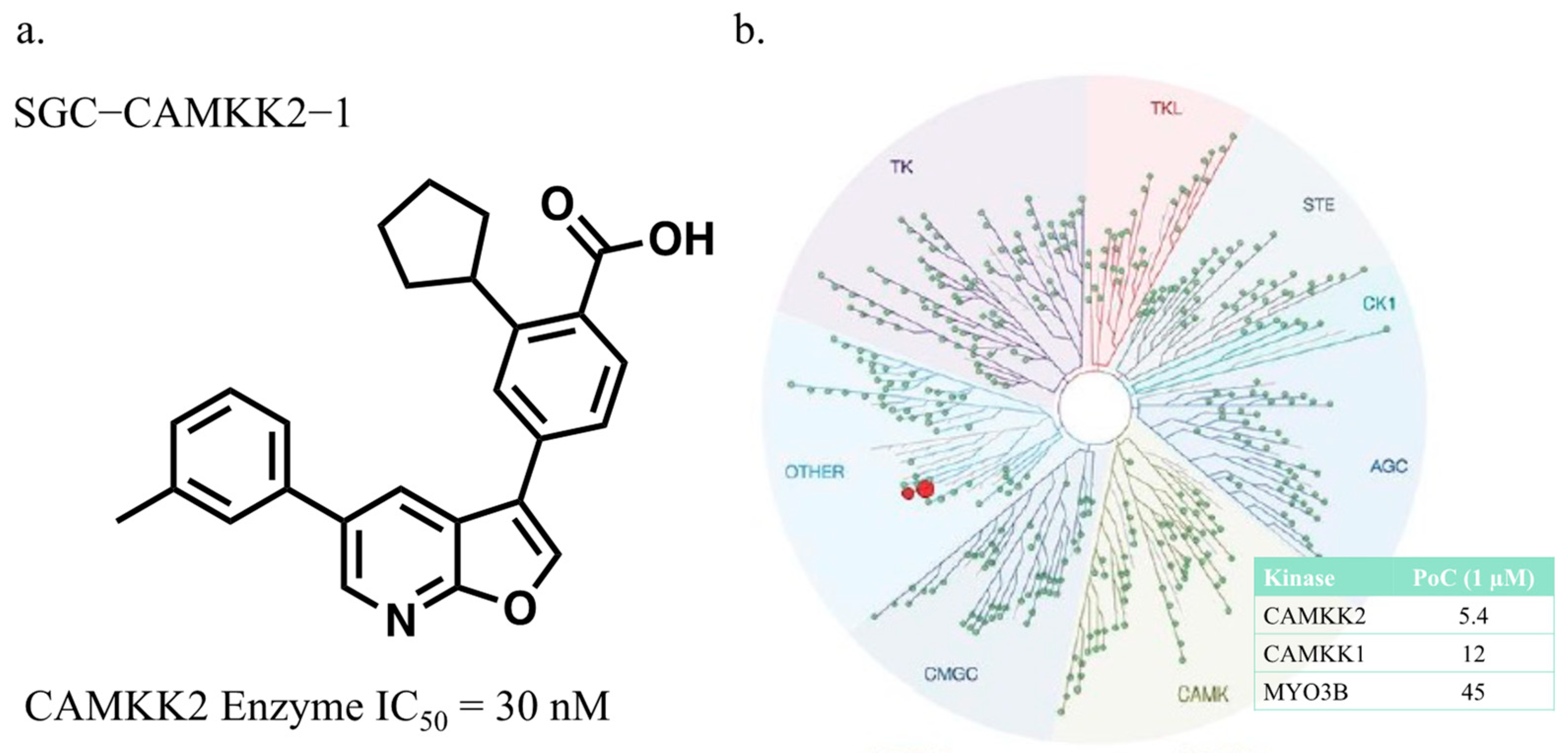
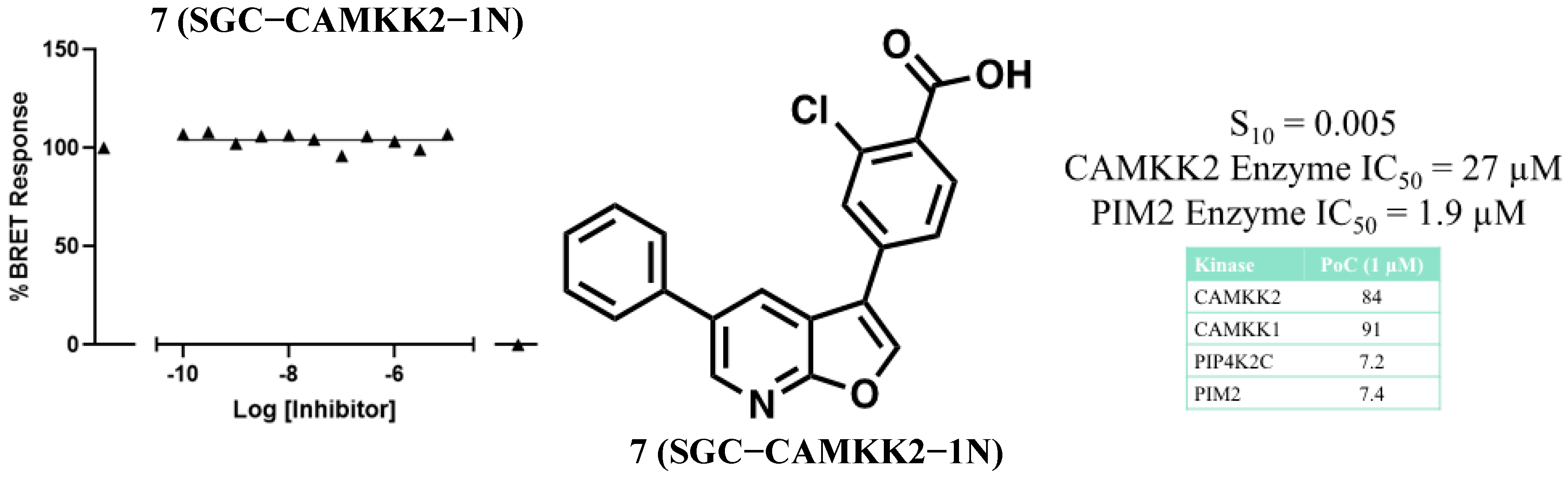
2.2. Eurofins DiscoverX Broad Kinome Profiling (KINOMEscan™)
2.3. CAMKK2 Enzyme Assay
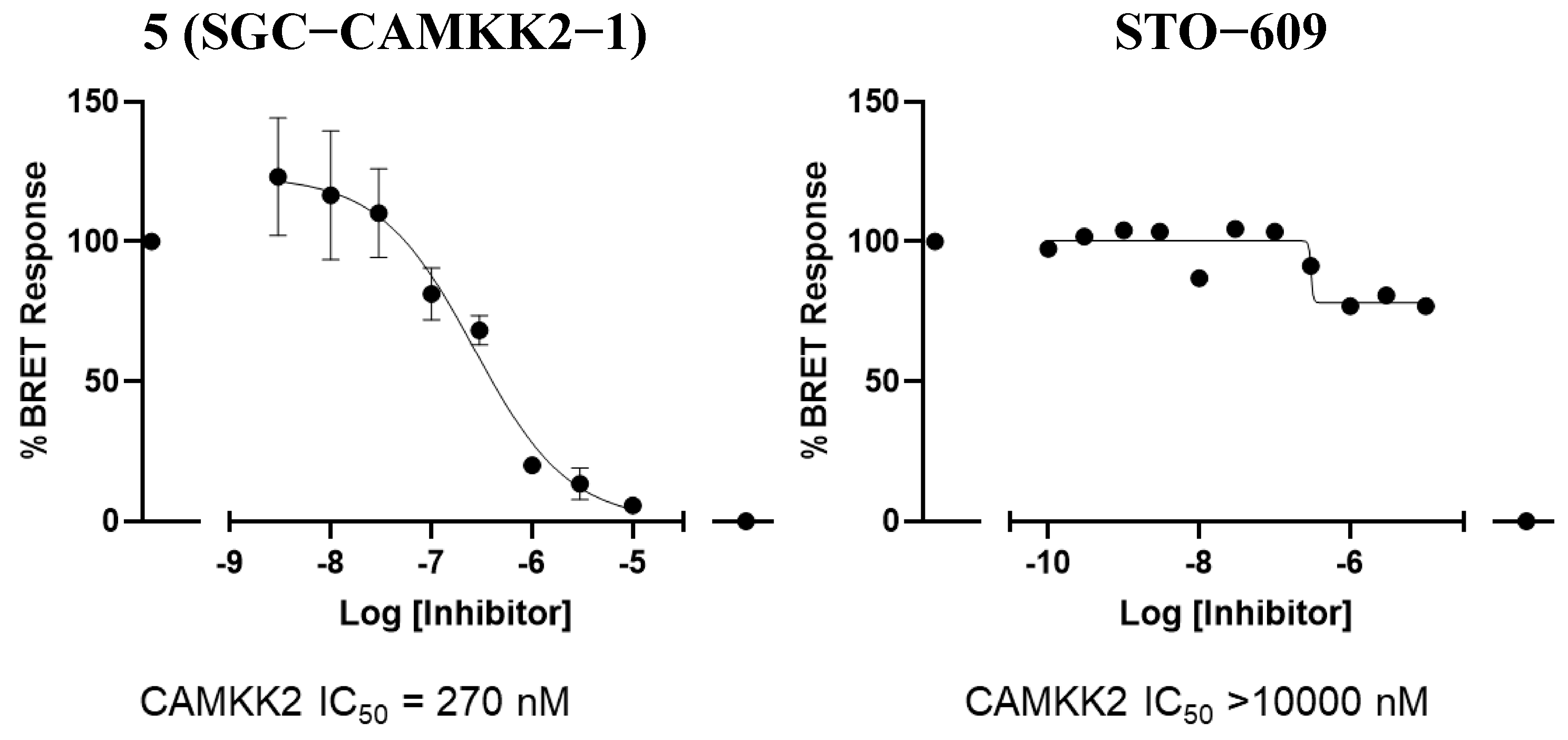
2.4. NanoBRET Cellular Target Engagement
2.5. In Silico Docking of SGC-CAMKK2-1
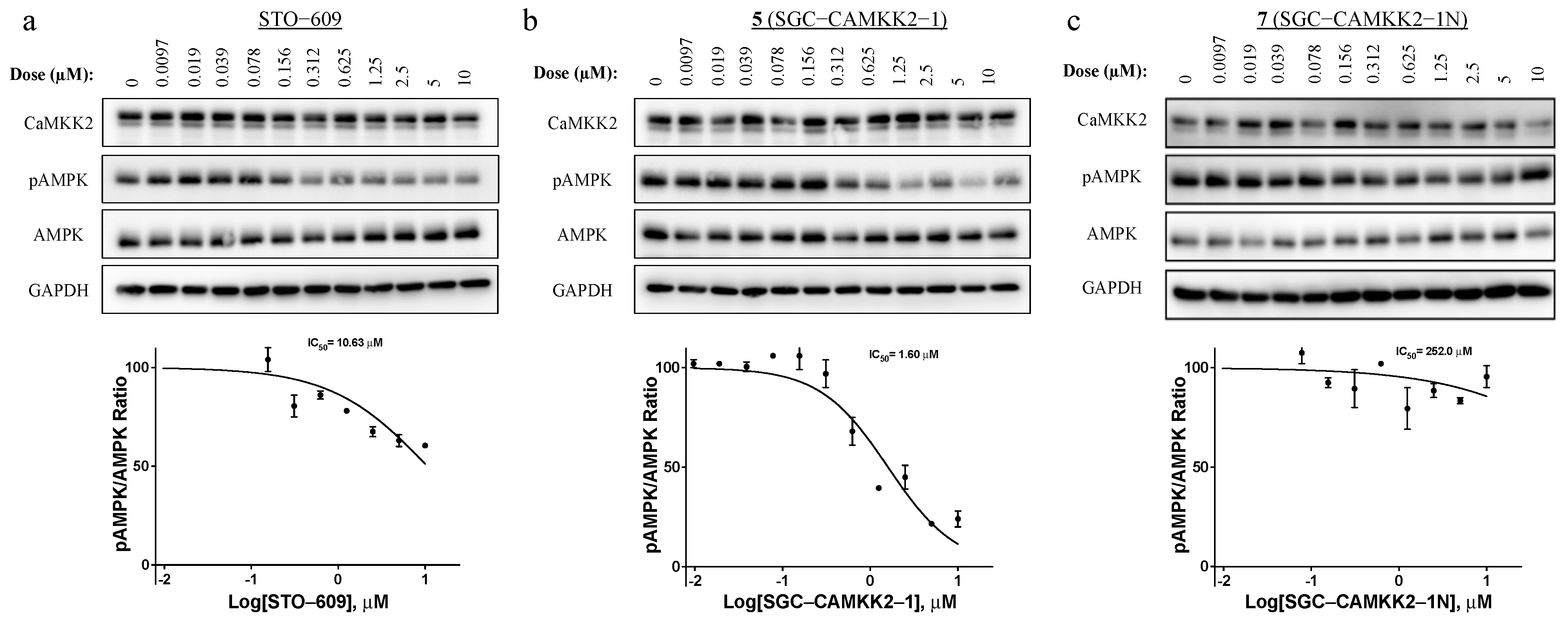
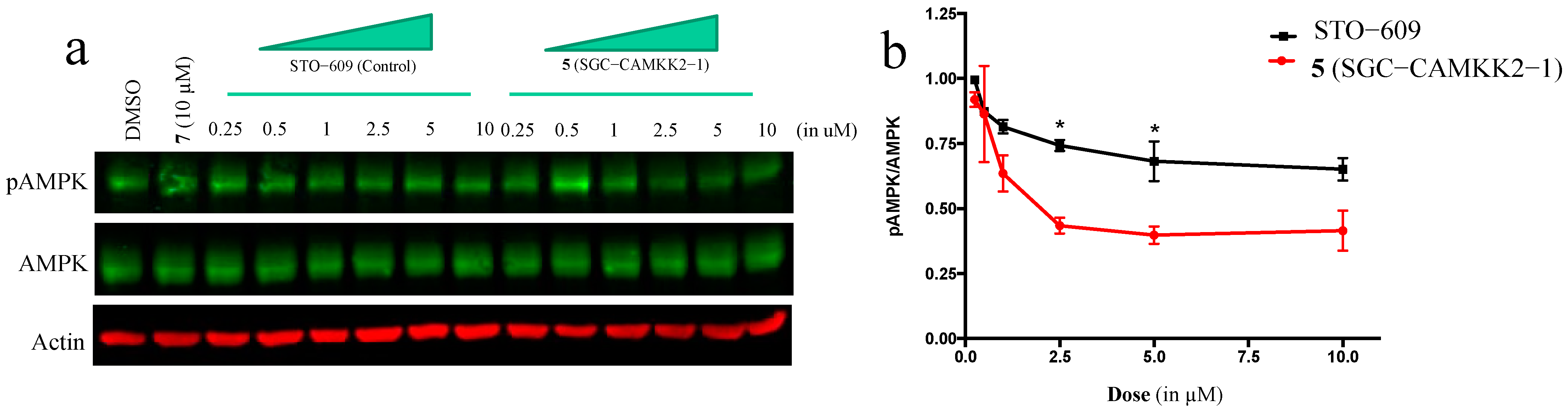
2.6. Western Blot Analysis
2.6.1. Prostate Cancer Cellular Screening
2.6.2. Breast Cancer Cellular Screening
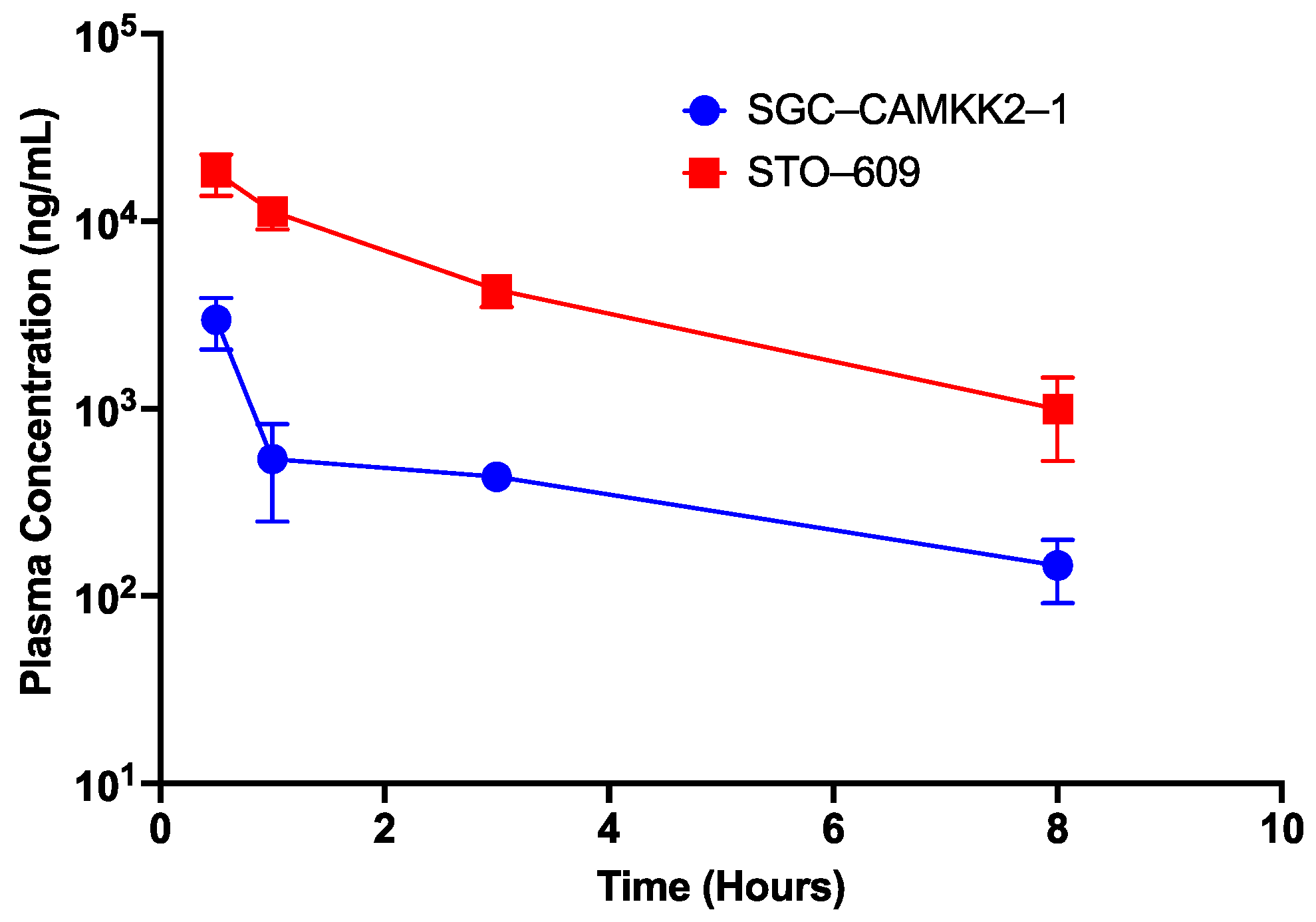
2.7. Pharmacokinetic Analysis
2.7.1. Animals
2.7.2. Animal Experiments
2.7.3. Pharmacokinetic (PK) Study of Compound 5 and STO-609 in Mice
3. Results
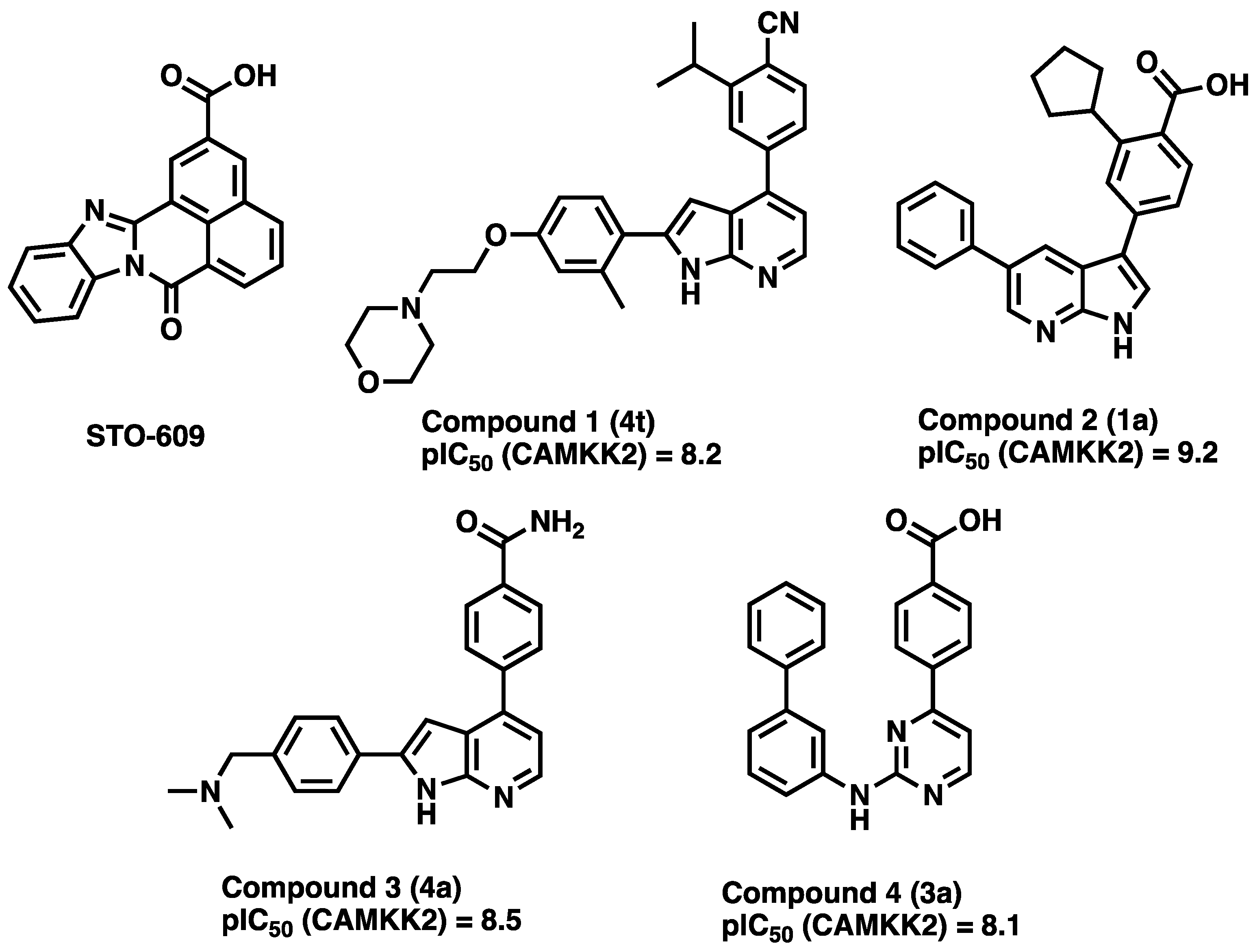
3.1. CAMKK2 Probe Development Synthesis and Profiling
3.2. Development of a Structurally Related Negative Control
3.3. Molecular Basis for Ligand Binding
3.4. CAMKK2 NanoBRET in Cell Target Engagement
3.5. On-Target Cellular Effect (Western Blot)
3.6. Pharmacokinetic Studies
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anderson, K.A.; Ribar, T.J.; Lin, F.; Noeldner, P.K.; Green, M.F.; Muehlbauer, M.J.; Witters, L.A.; Kemp, B.E.; Means, A.R. Hypothalamic CaMKK2 contributes to the regulation of energy balance. Cell Metab. 2008, 7, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, Z.J.; Cary, R.L.; Yang, C.; Novack, D.V.; Voor, M.J.; Sankar, U. Inhibition of CaMKK2 reverses age-associated decline in bone mass. Bone 2015, 75, 120–127. [Google Scholar] [CrossRef]
- Cary, R.L.; Waddell, S.; Racioppi, L.; Long, F.; Novack, D.V.; Voor, M.J.; Sankar, U. Inhibition of Ca(2)(+)/calmodulin-dependent protein kinase kinase 2 stimulates osteoblast formation and inhibits osteoclast differentiation. J. Bone Min. Res. 2013, 28, 1599–1610. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.N.; Kambrath, A.V.; Patel, R.B.; Kang, K.S.; Mevel, E.; Li, Y.; Cheng, Y.H.; Pucylowski, A.J.; Hassert, M.A.; Voor, M.J.; et al. Inhibition of CaMKK2 Enhances Fracture Healing by Stimulating Indian Hedgehog Signaling and Accelerating Endochondral Ossification. J. Bone Min. Res. 2018, 33, 930–944. [Google Scholar] [CrossRef] [PubMed]
- Broxmeyer, H.E.; Ropa, J.; Capitano, M.L.; Cooper, S.; Racioppi, L.; Sankar, U. CaMKK2 Knockout Bone Marrow Cells Collected/Processed in Low Oxygen (Physioxia) Suggests CaMKK2 as a Hematopoietic Stem to Progenitor Differentiation Fate Determinant. Stem Cell Rev. Rep. 2022, 18, 2513–2521. [Google Scholar] [CrossRef]
- Racioppi, L.; Lento, W.; Huang, W.; Arvai, S.; Doan, P.L.; Harris, J.R.; Marcon, F.; Nakaya, H.I.; Liu, Y.; Chao, N. Calcium/calmodulin-dependent kinase kinase 2 regulates hematopoietic stem and progenitor cell regeneration. Cell Death Dis. 2017, 8, e3076. [Google Scholar] [CrossRef]
- Racioppi, L.; Nelson, E.R.; Huang, W.; Mukherjee, D.; Lawrence, S.A.; Lento, W.; Masci, A.M.; Jiao, Y.; Park, S.; York, B.; et al. CaMKK2 in myeloid cells is a key regulator of the immune-suppressive microenvironment in breast cancer. Nat. Commun. 2019, 10, 2450. [Google Scholar] [CrossRef]
- Huang, W.; Liu, Y.; Luz, A.; Berrong, M.; Meyer, J.N.; Zou, Y.; Swann, E.; Sundaramoorthy, P.; Kang, Y.; Jauhari, S.; et al. Calcium/Calmodulin Dependent Protein Kinase Kinase 2 Regulates the Expansion of Tumor-Induced Myeloid-Derived Suppressor Cells. Front. Immunol. 2021, 12, 754083. [Google Scholar] [CrossRef]
- Casciano, J.C.; Perry, C.; Cohen-Nowak, A.J.; Miller, K.D.; Vande Voorde, J.; Zhang, Q.; Chalmers, S.; Sandison, M.E.; Liu, Q.; Hedley, A.; et al. MYC regulates fatty acid metabolism through a multigenic program in claudin-low triple negative breast cancer. Br. J. Cancer 2020, 122, 868–884. [Google Scholar] [CrossRef]
- Jauhari, S.; Volkheimer, A.D.; Barak, I.; Guadalupe, E.; Weinberg, J.B.; Chao, N.J.A.; Li, Z.; Racioppi, L. Expression and prognostic relevance of calcium calmodulin-dependent protein kinase kinase 2 (CaMKK2) in chronic lymphocytic leukemia (CLL). J. Clin. Oncol. 2019, 37, e19002. [Google Scholar] [CrossRef]
- Liu, D.M.; Wang, H.J.; Han, B.; Meng, X.Q.; Chen, M.H.; Yang, D.B.; Sun, Y.; Li, Y.L.; Jiang, C.L. CAMKK2, Regulated by Promoter Methylation, is a Prognostic Marker in Diffuse Gliomas. CNS Neurosci. 2016, 22, 518–524. [Google Scholar] [CrossRef]
- Subbannayya, Y.; Syed, N.; Barbhuiya, M.A.; Raja, R.; Marimuthu, A.; Sahasrabuddhe, N.; Pinto, S.M.; Manda, S.S.; Renuse, S.; Manju, H.C.; et al. Calcium calmodulin dependent kinase kinase 2—A novel therapeutic target for gastric adenocarcinoma. Cancer Biol. 2015, 16, 336–345. [Google Scholar] [CrossRef]
- Lin, F.; Marcelo, K.L.; Rajapakshe, K.; Coarfa, C.; Dean, A.; Wilganowski, N.; Robinson, H.; Sevick, E.; Bissig, K.D.; Goldie, L.C.; et al. The camKK2/camKIV relay is an essential regulator of hepatic cancer. Hepatology 2015, 62, 505–520. [Google Scholar] [CrossRef]
- Jin, L.; Chun, J.; Pan, C.; Kumar, A.; Zhang, G.; Ha, Y.; Li, D.; Alesi, G.N.; Kang, Y.; Zhou, L.; et al. The PLAG1-GDH1 Axis Promotes Anoikis Resistance and Tumor Metastasis through CamKK2-AMPK Signaling in LKB1-Deficient Lung Cancer. Mol. Cell 2018, 69, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Gocher, A.M.; Azabdaftari, G.; Euscher, L.M.; Dai, S.; Karacosta, L.G.; Franke, T.F.; Edelman, A.M. Akt activation by Ca(2+)/calmodulin-dependent protein kinase kinase 2 (CaMKK2) in ovarian cancer cells. J. Biol. Chem. 2017, 292, 14188–14204. [Google Scholar] [CrossRef] [PubMed]
- Penfold, L.; Woods, A.; Muckett, P.; Nikitin, A.Y.; Kent, T.R.; Zhang, S.; Graham, R.; Pollard, A.; Carling, D. CAMKK2 Promotes Prostate Cancer Independently of AMPK via Increased Lipogenesis. Cancer Res. 2018, 78, 6747–6761. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Blessing, A.M.; Pulliam, T.L.; Shi, Y.; Wilkenfeld, S.R.; Han, J.J.; Murray, M.M.; Pham, A.H.; Duong, K.; Brun, S.N.; et al. Inhibition of CAMKK2 impairs autophagy and castration-resistant prostate cancer via suppression of AMPK-ULK1 signaling. Oncogene 2021, 40, 1690–1705. [Google Scholar] [CrossRef]
- Saxena, N.; Beraldi, E.; Fazli, L.; Somasekharan, S.P.; Adomat, H.; Zhang, F.; Molokwu, C.; Gleave, A.; Nappi, L.; Nguyen, K.; et al. Androgen receptor (AR) antagonism triggers acute succinate-mediated adaptive responses to reactivate AR signaling. EMBO Mol. Med. 2021, 13, e13427. [Google Scholar] [CrossRef]
- Pulliam, T.L.; Goli, P.; Awad, D.; Lin, C.; Wilkenfeld, S.R.; Frigo, D.E. Regulation and role of CAMKK2 in prostate cancer. Nat. Rev. Urol. 2022, 19, 367–380. [Google Scholar] [CrossRef]
- Pulliam, T.L.; Awad, D.; Han, J.J.; Murray, M.M.; Ackroyd, J.J.; Goli, P.; Oakhill, J.S.; Scott, J.W.; Ittmann, M.M.; Frigo, D.E. Systemic Ablation of Camkk2 Impairs Metastatic Colonization and Improves Insulin Sensitivity in TRAMP Mice: Evidence for Cancer Cell-Extrinsic CAMKK2 Functions in Prostate Cancer. Cells 2022, 11, 1890. [Google Scholar] [CrossRef]
- Zhang, Y.; Recouvreux, M.V.; Jung, M.; Galenkamp, K.M.O.; Li, Y.; Zagnitko, O.; Scott, D.A.; Lowy, A.M.; Commisso, C. Macropinocytosis in Cancer-Associated Fibroblasts Is Dependent on CaMKK2/ARHGEF2 Signaling and Functions to Support Tumor and Stromal Cell Fitness. Cancer Discov. 2021, 11, 1808–1825. [Google Scholar] [CrossRef] [PubMed]
- Najar, M.A.; Rex, D.A.B.; Modi, P.K.; Agarwal, N.; Dagamajalu, S.; Karthikkeyan, G.; Vijayakumar, M.; Chatterjee, A.; Sankar, U.; Prasad, T.S.K. A complete map of the Calcium/calmodulin-dependent protein kinase kinase 2 (CAMKK2) signaling pathway. J. Cell Commun Signal. 2021, 15, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Racioppi, L.; Noeldner, P.K.; Lin, F.; Arvai, S.; Means, A.R. Calcium/calmodulin-dependent protein kinase kinase 2 regulates macrophage-mediated inflammatory responses. J. Biol. Chem. 2012, 287, 11579–11591. [Google Scholar] [CrossRef] [PubMed]
- Teng, E.C.; Racioppi, L.; Means, A.R. A cell-intrinsic role for CaMKK2 in granulocyte lineage commitment and differentiation. J. Leukoc. Biol. 2011, 90, 897–909. [Google Scholar] [CrossRef]
- Saito, M.; Saito, M.; Das, B.C. Involvement of AMP-activated protein kinase in neuroinflammation and neurodegeneration in the adult and developing brain. Int, J. Dev. Neurosci 2019, 77, 48–59. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, Y.; Wang, Y.; Wang, Y.; He, L.; Jiang, Z.; Huang, Z.; Liao, H.; Li, J.; Saavedra, J.M.; et al. Telmisartan prevention of LPS-induced microglia activation involves M2 microglia polarization via CaMKKbeta-dependent AMPK activation. Brain Behav. Immun. 2015, 50, 298–313. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, N.; Ding, Y.; Zhang, Y.; Li, Q.; Flores, J.; Haghighiabyaneh, M.; Doycheva, D.; Tang, J.; Zhang, J.H. Chemerin suppresses neuroinflammation and improves neurological recovery via CaMKK2/AMPK/Nrf2 pathway after germinal matrix hemorrhage in neonatal rats. Brain Behav. Immun. 2018, 70, 179–193. [Google Scholar] [CrossRef]
- Silwal, P.; Kim, J.K.; Yuk, J.M.; Jo, E.K. AMP-Activated Protein Kinase and Host Defense against Infection. Int. J. Mol. Sci. 2018, 19, 3495. [Google Scholar] [CrossRef]
- Williams, J.N.; Sankar, U. CaMKK2 Signaling in Metabolism and Skeletal Disease: A New Axis with Therapeutic Potential. Curr. Osteoporos Rep. 2019, 17, 169–177. [Google Scholar] [CrossRef]
- Lin, F.; Ribar, T.J.; Means, A.R. The Ca2+/calmodulin-dependent protein kinase kinase, CaMKK2, inhibits preadipocyte differentiation. Endocrinology 2011, 152, 3668–3679. [Google Scholar] [CrossRef]
- York, B.; Li, F.; Lin, F.; Marcelo, K.L.; Mao, J.; Dean, A.; Gonzales, N.; Gooden, D.; Maity, S.; Coarfa, C.; et al. Pharmacological inhibition of CaMKK2 with the selective antagonist STO-609 regresses NAFLD. Sci. Rep. 2017, 7, 11793. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Chen, Z.; Zhang, F.; Cui, X.; Sun, W.; Geary, G.G.; Wang, Y.; Johnson, D.A.; Zhu, Y.; Chien, S.; et al. Ca2+/calmodulin-dependent protein kinase kinase beta phosphorylation of Sirtuin 1 in endothelium is atheroprotective. Proc. Natl. Acad. Sci. USA 2013, 110, E2420–E2427. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yu, J.; Li, D.; Yu, S.; Ke, J.; Wang, L.; Wang, Y.; Qiu, Y.; Gao, X.; Zhang, J.; et al. Store-operated calcium entry-activated autophagy protects EPC proliferation via the CAMKK2-MTOR pathway in ox-LDL exposure. Autophagy 2017, 13, 82–98. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Zhang, D.; Zhao, L.; Li, Y.; Yao, X.; Wang, H.; Zhang, S.; Liu, W.; Cao, H.; Yu, S.; et al. CaMKK2 Suppresses Muscle Regeneration through the Inhibition of Myoblast Proliferation and Differentiation. Int J. Mol. Sci. 2016, 17, 1695. [Google Scholar] [CrossRef]
- Mairet-Coello, G.; Courchet, J.; Pieraut, S.; Courchet, V.; Maximov, A.; Polleux, F. The CAMKK2-AMPK kinase pathway mediates the synaptotoxic effects of Abeta oligomers through Tau phosphorylation. Neuron 2013, 78, 94–108. [Google Scholar] [CrossRef]
- Peters, M.; Mizuno, K.; Ris, L.; Angelo, M.; Godaux, E.; Giese, K.P. Loss of Ca2+/calmodulin kinase kinase beta affects the formation of some, but not all, types of hippocampus-dependent long-term memory. J. Neurosci. 2003, 23, 9752–9760. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Antunes-Martins, A.; Ris, L.; Peters, M.; Godaux, E.; Giese, K. Calcium/calmodulin kinase kinase β has a male-specific role in memory formation. Neuroscience 2007, 145, 393–402. [Google Scholar] [CrossRef]
- Scott, J.W.; Park, E.; Rodriguiz, R.M.; Oakhill, J.S.; Issa, S.; O’Brien, M.T.; Dite, T.A.; Langendorf, C.G.; Wetsel, W.C.; Means, A.R. Autophosphorylation of CaMKK2 generates autonomous activity that is disrupted by a T85S mutation linked to anxiety and bipolar disorder. Sci. Rep. 2015, 5, 1–10. [Google Scholar] [CrossRef]
- Erhardt, A.; Lucae, S.; Unschuld, P.G.; Ising, M.; Kern, N.; Salyakina, D.; Lieb, R.; Uhr, M.; Binder, E.B.; Keck, M.E.; et al. Association of polymorphisms in P2RX7 and CaMKKb with anxiety disorders. J. Affect. Disord. 2007, 101, 159–168. [Google Scholar] [CrossRef]
- Luo, X.J.; Li, M.; Huang, L.; Steinberg, S.; Mattheisen, M.; Liang, G.; Donohoe, G.; Shi, Y.; Chen, C.; Yue, W.; et al. Convergent lines of evidence support CAMKK2 as a schizophrenia susceptibility gene. Mol. Psychiatry 2014, 19, 774–783. [Google Scholar] [CrossRef]
- Sayad, A.; Ranjbaran, F.; Ghafouri-Fard, S.; Arsang-Jang, S.; Taheri, M. Expression Analysis of CYFIP1 and CAMKK2 Genes in the Blood of Epileptic and Schizophrenic Patients. J. Mol. Neurosci. 2018, 65, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; McCullough, L.; Li, J. Genetic deletion of calcium/calmodulin-dependent protein kinase kinase beta (CaMKK beta) or CaMK IV exacerbates stroke outcomes in ovariectomized (OVXed) female mice. BMC Neurosci. 2014, 15, 118. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Bu, F.; Min, J.W.; Munshi, Y.; Howe, M.D.; Liu, L.; Koellhoffer, E.C.; Qi, L.; McCullough, L.D.; Li, J. Inhibition of calcium/calmodulin-dependent protein kinase kinase (CaMKK) exacerbates impairment of endothelial cell and blood-brain barrier after stroke. Eur J. Neurosci. 2019, 49, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Tokumitsu, H.; Inuzuka, H.; Ishikawa, Y.; Ikeda, M.; Saji, I.; Kobayashi, R. STO-609, a specific inhibitor of the Ca(2+)/calmodulin-dependent protein kinase kinase. J. Biol. Chem. 2002, 277, 15813–15818. [Google Scholar] [CrossRef]
- O’Byrne, S.N.; Scott, J.W.; Pilotte, J.R.; Santiago, A.d.S.; Langendorf, C.G.; Oakhill, J.S.; Eduful, B.J.; Couñago, R.M.; Wells, C.I.; Zuercher, W.J.; et al. In Depth Analysis of Kinase Cross Screening Data to Identify CAMKK2 Inhibitory Scaffolds. Molecules 2020, 25, 325. [Google Scholar] [CrossRef]
- Edwards, A.M.; Bountra, C.; Kerr, D.J.; Willson, T.M. Open access chemical and clinical probes to support drug discovery. Nat. Chem. Biol. 2009, 5, 436–440. [Google Scholar] [CrossRef]
- Price, D.J.; Drewry, D.H.; Schaller, L.T.; Thompson, B.D.; Reid, P.R.; Maloney, P.R.; Liang, X.; Banker, P.; Buckholz, R.G.; Selley, P.K.; et al. An orally available, brain-penetrant CAMKK2 inhibitor reduces food intake in rodent model. Bioorg. Med. Chem. Lett. 2018, 28, 1958–1963. [Google Scholar] [CrossRef]
- Fabian, M.A.; Biggs, W.H., 3rd; Treiber, D.K.; Atteridge, C.E.; Azimioara, M.D.; Benedetti, M.G.; Carter, T.A.; Ciceri, P.; Edeen, P.T.; Floyd, M.; et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 2005, 23, 329–336. [Google Scholar] [CrossRef]
- Wodicka, L.M.; Ciceri, P.; Davis, M.I.; Hunt, J.P.; Floyd, M.; Salerno, S.; Hua, X.H.; Ford, J.M.; Armstrong, R.C.; Zarrinkar, P.P.; et al. Activation state-dependent binding of small molecule kinase inhibitors: Structural insights from biochemistry. Chem. Biol. 2010, 17, 1241–1249. [Google Scholar] [CrossRef]
- O’Brien, M.T.; Oakhill, J.S.; Ling, N.X.; Langendorf, C.G.; Hoque, A.; Dite, T.A.; Means, A.R.; Kemp, B.E.; Scott, J.W. Impact of Genetic Variation on Human CaMKK2 Regulation by Ca(2+)-Calmodulin and Multisite Phosphorylation. Sci. Rep. 2017, 7, 43264. [Google Scholar] [CrossRef]
- Vasta, J.D.; Corona, C.R.; Wilkinson, J.; Zimprich, C.A.; Hartnett, J.R.; Ingold, M.R.; Zimmerman, K.; Machleidt, T.; Kirkland, T.A.; Huwiler, K.G.; et al. Quantitative, Wide-Spectrum Kinase Profiling in Live Cells for Assessing the Effect of Cellular ATP on Target Engagement. Cell Chem. Biol. 2018, 25, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Eduful, B.J.; O’Byrne, S.N.; Temme, L.; Asquith, C.R.M.; Liang, Y.; Picado, A.; Pilotte, J.R.; Hossain, M.A.; Wells, C.I.; Zuercher, W.J.; et al. Hinge Binder Scaffold Hopping Identifies Potent Calcium/Calmodulin-Dependent Protein Kinase Kinase 2 (CAMKK2) Inhibitor Chemotypes. J. Med. Chem. 2021, 64, 10849–10877. [Google Scholar] [CrossRef]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided. Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Olsson, M.H.; Sondergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- O’Byrne, S.N.; Eduful, B.J.; Willson, T.M.; Drewry, D.H. Concise, gram-scale synthesis of furo[2,3-b]pyridines with functional handles for chemoselective cross-coupling. Tetrahedron Lett 2020, 61, 152353. [Google Scholar] [CrossRef]
- Bunnage, M.E.; Chekler, E.L.; Jones, L.H. Target validation using chemical probes. Nat. Chem. Biol. 2013, 9, 195–199. [Google Scholar] [CrossRef]
- Poli, A.; Zaurito, A.E.; Abdul-Hamid, S.; Fiume, R.; Faenza, I.; Divecha, N. Phosphatidylinositol 5 Phosphate (PI5P): From Behind the Scenes to the Front (Nuclear) Stage. Int J. Mol. Sci. 2019, 20, 2080. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.H.; Irvine, R.F. Evolutionarily conserved structural changes in phosphatidylinositol 5-phosphate 4-kinase (PI5P4K) isoforms are responsible for differences in enzyme activity and localization. Biochem. J. 2013, 454, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.H.; Giudici, M.L.; Burke, J.E.; Williams, R.L.; Maloney, D.J.; Marugan, J.; Irvine, R.F. The function of phosphatidylinositol 5-phosphate 4-kinase gamma (PI5P4Kgamma) explored using a specific inhibitor that targets the PI5P-binding site. Biochem. J. 2015, 466, 359–367. [Google Scholar] [CrossRef]
- Saitoh, M.; Kunitomo, J.; Kimura, E.; Iwashita, H.; Uno, Y.; Onishi, T.; Uchiyama, N.; Kawamoto, T.; Tanaka, T.; Mol, C.D.; et al. 2-3-[4-(Alkylsulfinyl)phenyl]-1-benzofuran-5-yl-5-methyl-1,3,4-oxadiazole derivatives as novel inhibitors of glycogen synthase kinase-3beta with good brain permeability. J. Med. Chem. 2009, 52, 6270–6286. [Google Scholar] [CrossRef] [PubMed]
- Ghose, A.K.; Herbertz, T.; Pippin, D.A.; Salvino, J.M.; Mallamo, J.P. Knowledge based prediction of ligand binding modes and rational inhibitor design for kinase drug discovery. J. Med. Chem. 2008, 51, 5149–5171. [Google Scholar] [CrossRef]
- Davis, M.I.; Hunt, J.P.; Herrgard, S.; Ciceri, P.; Wodicka, L.M.; Pallares, G.; Hocker, M.; Treiber, D.K.; Zarrinkar, P.P. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol 2011, 29, 1046–1051. [Google Scholar] [CrossRef]
- Wu, F.; Hill, K.; Fang, Q.; He, Z.; Zheng, H.; Wang, X.; Xiong, H.; Sha, S.H. Traumatic-noise-induced hair cell death and hearing loss is mediated by activation of CaMKKbeta. Cell Mol. Life Sci 2022, 79, 249. [Google Scholar] [CrossRef] [PubMed]
- McArdle, J.; Schafer, X.L.; Munger, J. Inhibition of calmodulin-dependent kinase kinase blocks human cytomegalovirus-induced glycolytic activation and severely attenuates production of viral progeny. J. Virol. 2011, 85, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Stewart, L.M.; Gerner, L.; Rettel, M.; Stein, F.; Burrows, J.F.; Mills, I.G.; Evergren, E. CaMKK2 facilitates Golgi-associated vesicle trafficking to sustain cancer cell proliferation. Cell Death Dis. 2021, 12, 1040. [Google Scholar] [CrossRef]
- Chen, Z.; Sun, X.; Xia, Z.; Wang, J.; Guo, N.; Zhang, Y. CaMKK2 Promotes the Progression of Ovarian Carcinoma through the PI3K/PDK1/Akt Axis. Comput Math. Methods Med. 2022, 2022, 7187940. [Google Scholar] [CrossRef]
- Najar, M.A.; Aravind, A.; Dagamajalu, S.; Sidransky, D.; Ashktorab, H.; Smoot, D.T.; Gowda, H.; Prasad, T.S.K.; Modi, P.K.; Chatterjee, A. Hyperactivation of MEK/ERK pathway by Ca(2+) /calmodulin-dependent protein kinase kinase 2 promotes cellular proliferation by activating cyclin-dependent kinases and minichromosome maintenance protein in gastric cancer cells. Mol. Carcinog. 2021, 60, 769–783. [Google Scholar] [CrossRef]
- Huang, Y.K.; Su, Y.F.; Lieu, A.S.; Loh, J.K.; Li, C.Y.; Wu, C.H.; Kuo, K.L.; Lin, C.L. MiR-1271 regulates glioblastoma cell proliferation and invasion by directly targeting the CAMKK2 gene. Neurosci. Lett. 2020, 737, 135289. [Google Scholar] [CrossRef]


| Disease | Role of CAMKK2 | Pathway | References |
|---|---|---|---|
| Bone | |||
| Age-associated bone degeneration (ex. osteoporosis) | CAMKK2 inhibition promotes trabecular bone formation | Decrease in CAMKK2 causes an increase in osteoblast/osteoclast ratio 1 | [2,3] |
| Fracture Healing | CAMKK2 inhibition accelerates bone healing | Decrease in CAMKK2 causes an increase in Indian Hedgehog Signaling | [4] |
| Hematopoiesis | |||
| Hematopoietic stem cells (HSC) | CaMKK2 regulates stress response to oxygen | HSC functions | [5] |
| Acute radiation syn-drome | CaMKK2 inhibition promote hematopoietic recovery | AMPK/p53 | [6] |
| Cancer | |||
| Breast cancer | Driver | 1. Myeloid functions | [7] |
| 2. CAMKK2-AMPK | [8] | ||
| 3. CAMKK2-FAO | [9] | ||
| Chronic lymphocytic leukemia | Prognostic marker | Not determined | [10] |
| Diffuse gliomas | Driver and prognostic marker | Promoter hypomethylation-↑CAMKK2 | [11] |
| Gastric cancer | Driver | Not determined | [12] |
| Hepatic cancer | Driver | CAMKK2-CaMKIV-mTOR | [13] |
| Lung cancer | Driver | PLAG-GDH1-aKG-CAMKK2-AMPK | [14] |
| Ovarian cancer | Driver | CAMKK2-AKT | [15] |
| Prostate cancer | Driver and prognostic marker | 1. AR-CAMKK2-AMPK 2. Triptolide-CAMKK2-AMPK 3. AR-CAMKK2-Lipogenesis | [16,17,18,19,20] |
| Pancreatic cancer | CaMKK2/ARHGEF2 | [21] | |
| Infectious Disease | |||
| Immune response | Immune suppression: 1. macrophage maturation 2. granulocyte differentiation | CAMKK2-AMPK | [8,22,23,24] |
| Neuroinflammation | CAMKK2 activation of AMPK reduces LPS mediated neuroinflammation | 1. 3C-CAMKK2-AMPK- PGC1α 2. Telmisartan-CAMKK2- AMPK 3. Chemerin-CAMKK2- AMPK-Nrf2 | [25,26,27] |
| Viral infection | Promotes viral replication via diversion of host autophagy | CAMKK2-AMPK-autophagy | [28] |
| Metabolic Disease | |||
| Diabetes (I and II) | Promotes global glucose intolerance Negative impact on pancreatic β cells | CAMKK2-AMPK | [29] |
| Diet-induced obesity | CAMKK2 inhibition decreases appetite and weight gain | CAMKK2-AMPK | [30] |
| Nonalcoholic fatty liver disease (NFALD) | CAMKK2 inhibition regresses NFALD | Not determined | [31] |
| Heart Disease | |||
| Atherosclerosis | CAMKK2 phosphorylation of SIRT1 is atheroprotective | CAMKK2-SIRT1 | [32] |
| Hypercholesterolemia (precedes atherosclerosis) | CAMKK2-mediated autophagy protects EPC proliferation | ox-LDL exposure-stored Ca2+ release-CAMKK2-mTOR- autophagy | [33] |
| Muscle | |||
| Duchenne muscular dystrophy and general muscular injury | CAMKK2 loss promotes muscle regeneration through C2C12 myoblast proliferation and differentiation | 1. CAMKK2-AMPK-cdc2-Tyr15-cell cycle arrest | [34] |
| 2. CAMKK2-AMPK-PGC1α-repression of differentiation | [34] | ||
| Neurological Disorders | |||
| Alzheimer’s | Promotes Aβ oligomer synaptotoxicity | CAMKK2-AMPK | [35] |
| Memory loss | Involved in long-term memory formation | CAMKK2-CREB | [36,37] |
| Anxiety and bipolar disorder | Loss of CAMKK2 activity | CAMKK2 autonomous activity | [38,39] |
| Schizophrenia | Loss of CAMKK2 activity | Not determined | [40] |
| Epilepsy | Loss of CAMKK2 activity | Not determined | [41] |
| Stroke | CAMKK2 loss exacerbates stroke outcomes | 1. CAMKK2-CaMKIV- CREB/Bcl-2 | [42] |
| 2. CaMKK2-CaMKIV-reduced inflammatory response | [42,43] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wells, C.; Liang, Y.; Pulliam, T.L.; Lin, C.; Awad, D.; Eduful, B.; O’Byrne, S.; Hossain, M.A.; Catta-Preta, C.M.C.; Ramos, P.Z.; et al. SGC-CAMKK2-1: A Chemical Probe for CAMKK2. Cells 2023, 12, 287. https://doi.org/10.3390/cells12020287
Wells C, Liang Y, Pulliam TL, Lin C, Awad D, Eduful B, O’Byrne S, Hossain MA, Catta-Preta CMC, Ramos PZ, et al. SGC-CAMKK2-1: A Chemical Probe for CAMKK2. Cells. 2023; 12(2):287. https://doi.org/10.3390/cells12020287
Chicago/Turabian StyleWells, Carrow, Yi Liang, Thomas L. Pulliam, Chenchu Lin, Dominik Awad, Benjamin Eduful, Sean O’Byrne, Mohammad Anwar Hossain, Carolina Moura Costa Catta-Preta, Priscila Zonzini Ramos, and et al. 2023. "SGC-CAMKK2-1: A Chemical Probe for CAMKK2" Cells 12, no. 2: 287. https://doi.org/10.3390/cells12020287
APA StyleWells, C., Liang, Y., Pulliam, T. L., Lin, C., Awad, D., Eduful, B., O’Byrne, S., Hossain, M. A., Catta-Preta, C. M. C., Ramos, P. Z., Gileadi, O., Gileadi, C., Couñago, R. M., Stork, B., Langendorf, C. G., Nay, K., Oakhill, J. S., Mukherjee, D., Racioppi, L., ... Drewry, D. H. (2023). SGC-CAMKK2-1: A Chemical Probe for CAMKK2. Cells, 12(2), 287. https://doi.org/10.3390/cells12020287









