Multiple Roles of the RUNX Gene Family in Hepatocellular Carcinoma and Their Potential Clinical Implications
Abstract
1. Introduction
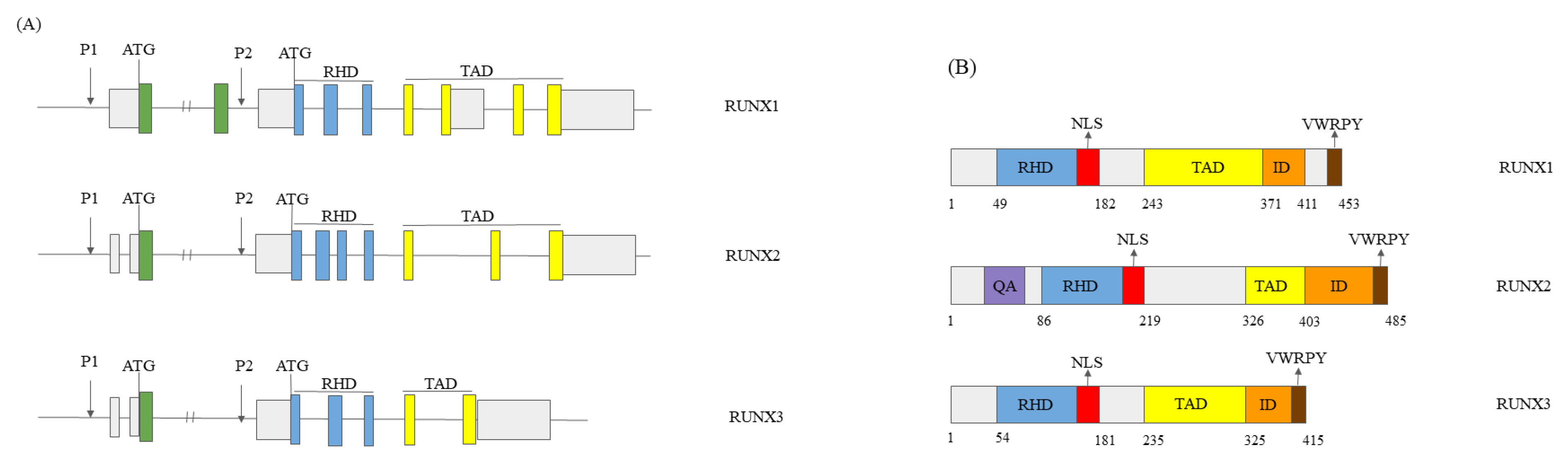
1.1. RUNX Genes’ and Proteins’ Structure
1.2. The Role of the RUNX Genes in Normal Development
1.3. The Role of RUNX Genes in Cancer
1.4. The Mechanisms of Action of RUNX Proteins
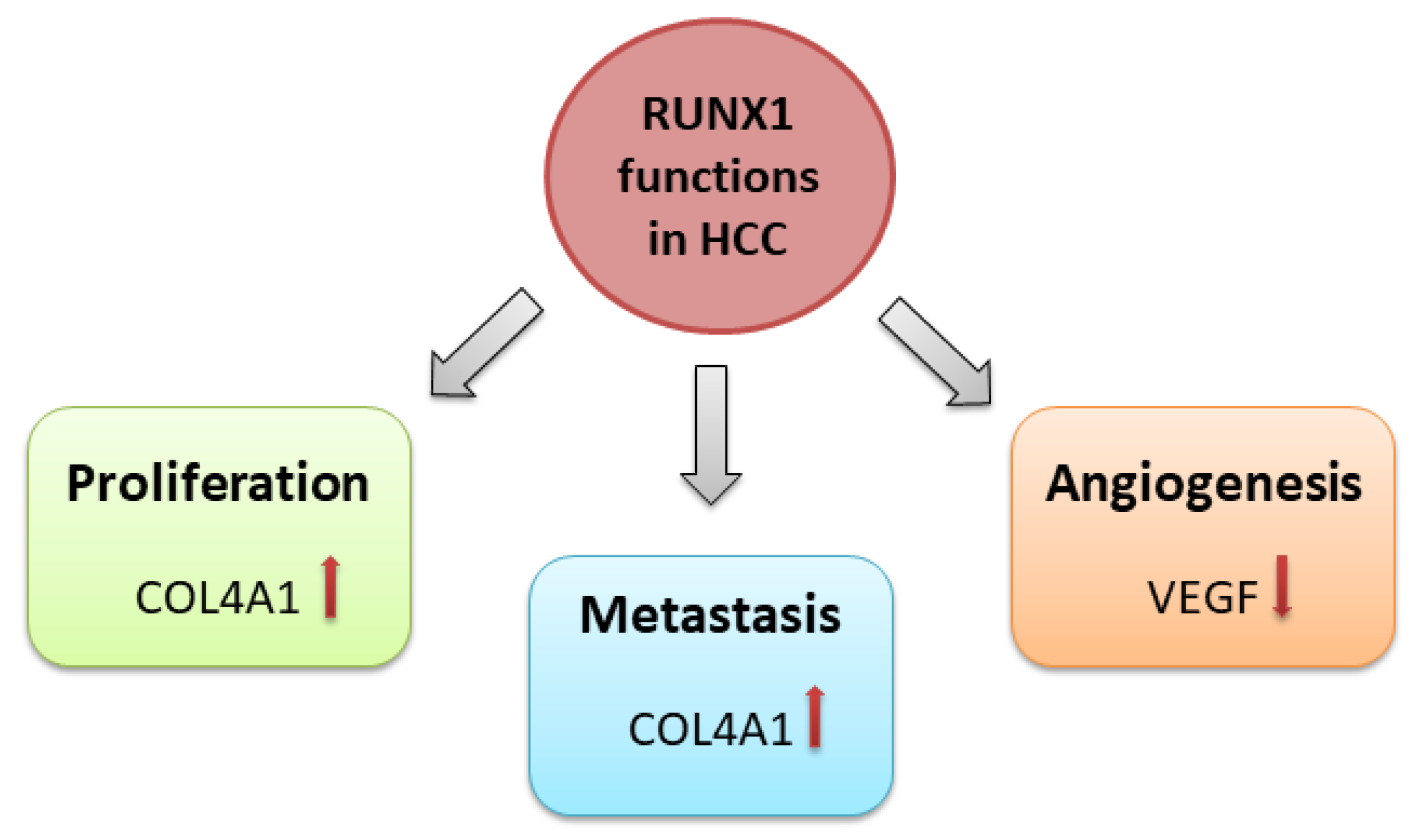
2. RUNX1 in HCC
2.1. RUNX1 Role in HCC
2.2. RUNX1 and miRNAs
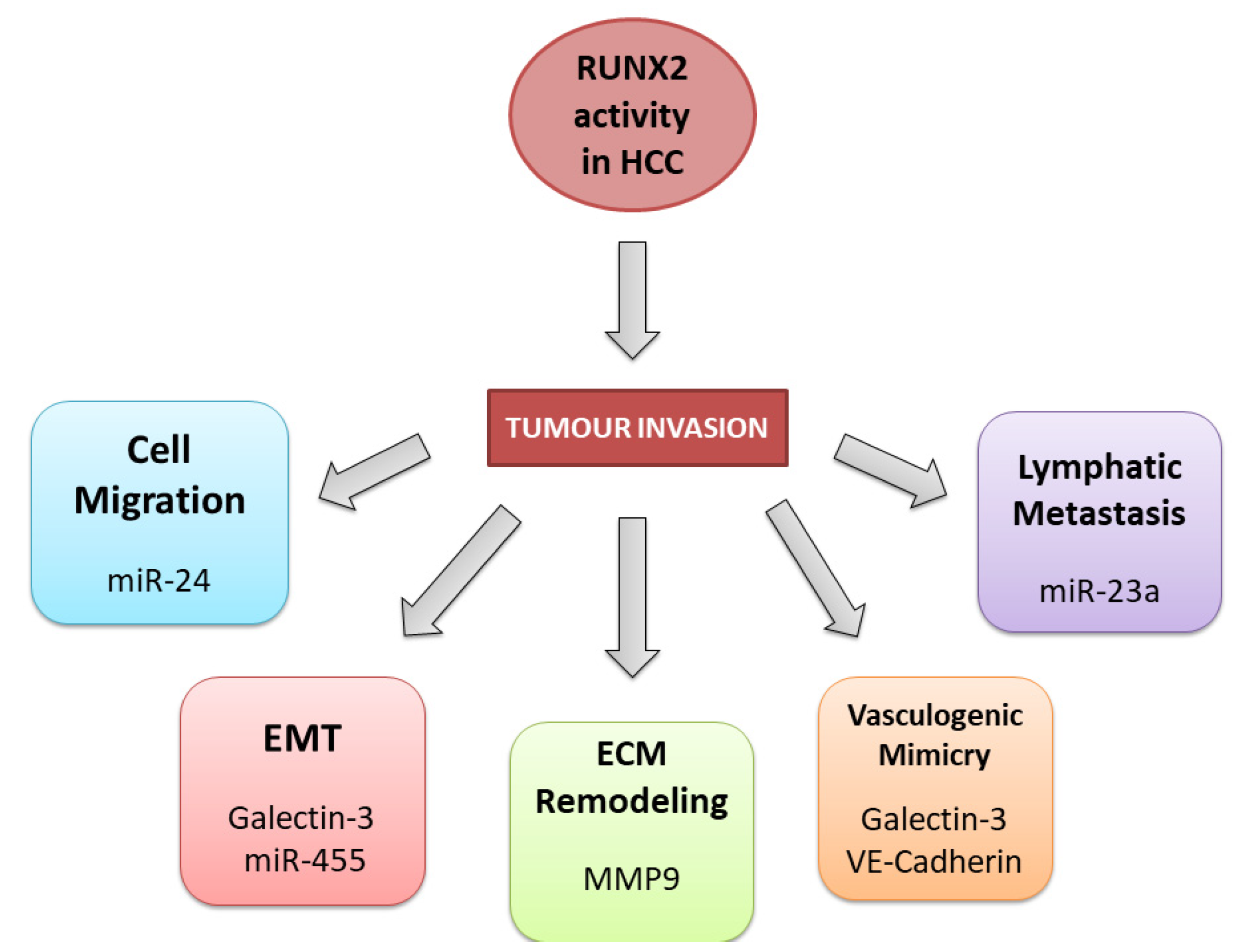
3. RUNX2 in HCC
3.1. General Role of RUNX2 in HCC
3.2. RUNX2 Tumour Invasion Activity in HCC
3.3. RUNX2 and Non-Coding RNAs in HCC
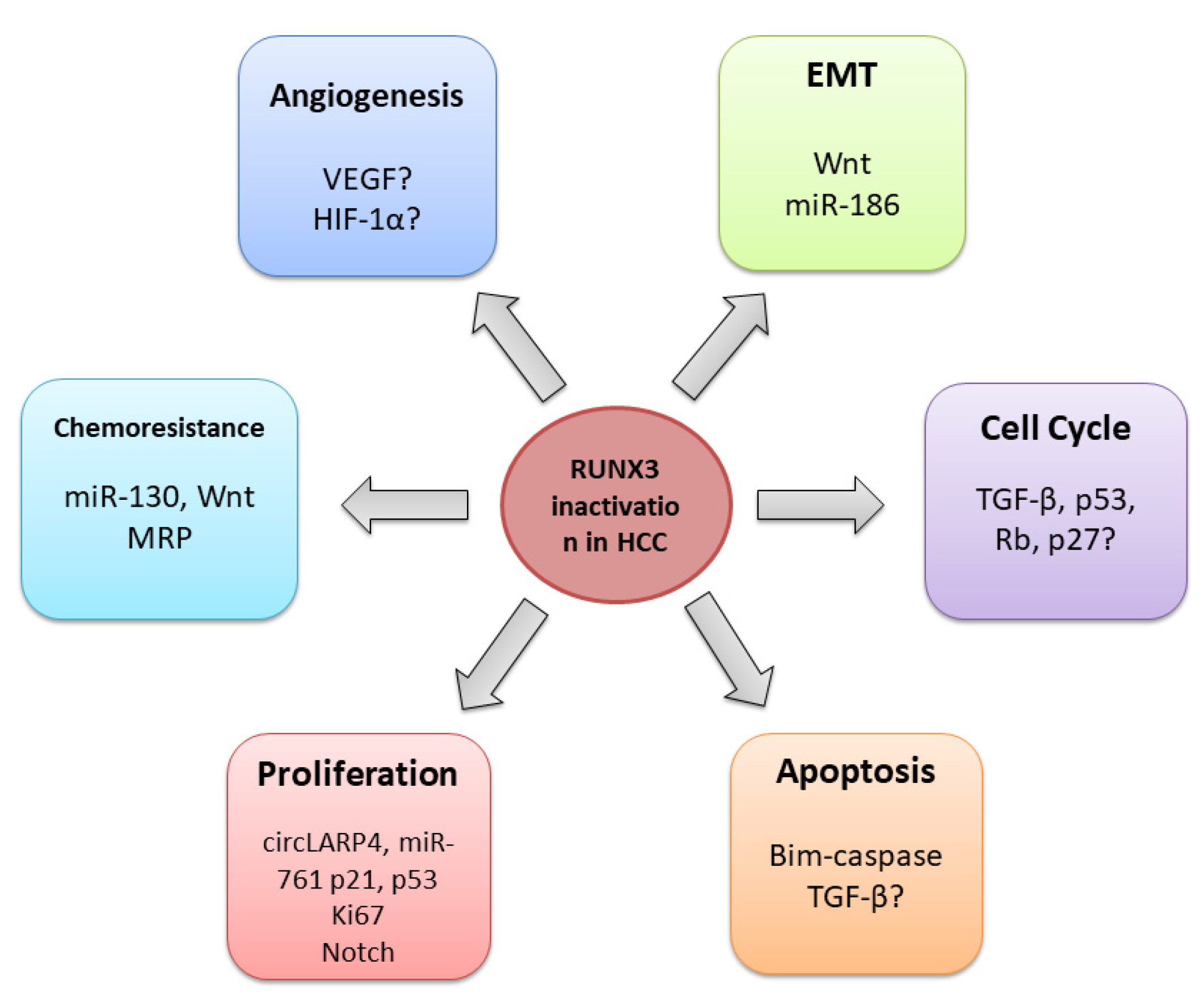
4. RUNX3 in HCC
4.1. General Role of RUNX3 in HCC
4.2. RUNX3 Regulates Cell Cycle, Proliferation, and Apoptosis
4.3. RUNX3 in the Angiogenesis Regulation
4.4. RUNX3 and Epithelial-Mesenchymal Transition
4.5. RUNX3 and Chemoresistance
5. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HCC | Hepatocellular carcinoma; |
| RUNX | Runt-related transcription factors; |
| HBV | Hepatitis B Virus; |
| HCV | Hepatitis C Virus; |
| RHD | Runt homology domain; |
| miRNA | Micro ribonucleic acids; |
| lncRNAs | Long non-coding ribonucleic acids |
References
- Asafo-Agyei, K.O.; Samant, H. Hepatocellular Carcinoma. In StatPearls; StatPearls Publishing: Treasure Islan, FL, USA, 2023. [Google Scholar]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular Carcinoma. Nat. Rev. Dis. Prim. 2021, 7, 6. [Google Scholar] [CrossRef]
- Balogh, J.; Victor, D.; Asham, E.H.; Burroughs, S.G.; Boktour, M.; Saharia, A.; Li, X.; Ghobrial, M.; Monsour, H. Hepatocellular Carcinoma: A Review. J. Hepatocell. Carcinoma 2016, 3, 41–53. [Google Scholar] [CrossRef]
- Sim, H.-W.; Knox, J. Hepatocellular Carcinoma in the Era of Immunotherapy. Curr. Probl. Cancer 2018, 42, 40–48. [Google Scholar] [CrossRef]
- Suresh, D.; Srinivas, A.N.; Prashant, A.; Harikumar, K.B.; Kumar, D.P. Therapeutic Options in Hepatocellular Carcinoma: A Comprehensive Review. Clin. Exp. Med. 2023. [Google Scholar] [CrossRef] [PubMed]
- Chuang, L.S.H.; Ito, K.; Ito, Y. RUNX Family: Regulation and Diversification of Roles through Interacting Proteins. Int. J. Cancer 2013, 132, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Otálora-Otálora, B.A.; Henríquez, B.; López-Kleine, L.; Rojas, A. RUNX Family: Oncogenes or Tumor Suppressors (Review). Oncol. Rep. 2019, 42, 3–19. [Google Scholar] [CrossRef]
- Ito, Y.; Bae, S.-C.; Chuang, L.S.H. The RUNX Family: Developmental Regulators in Cancer. Nat. Rev. Cancer 2015, 15, 81–95. [Google Scholar] [CrossRef]
- Levanon, D.; Groner, Y. Structure and Regulated Expression of Mammalian RUNX Genes. Oncogene 2004, 23, 4211–4219. [Google Scholar] [CrossRef]
- Yi, H.; He, Y.; Zhu, Q.; Fang, L. RUNX Proteins as Epigenetic Modulators in Cancer. Cells 2022, 11, 3687. [Google Scholar] [CrossRef] [PubMed]
- Mevel, R.; Draper, J.E.; Lie-a-Ling, M.; Kouskoff, V.; Lacaud, G. RUNX Transcription Factors: Orchestrators of Development. Development 2019, 146, dev148296. [Google Scholar] [CrossRef]
- Darnell, J.E. Transcription Factors as Targets for Cancer Therapy. Nat. Rev. Cancer 2002, 2, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Sood, R.; Kamikubo, Y.; Liu, P. Role of RUNX1 in Hematological Malignancies. Blood 2017, 129, 2070–2082. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y. Oncogenic Potential of the RUNX Gene Family: ‘Overview’. Oncogene 2004, 23, 4198–4208. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M. RUNX Family in Hypoxic Microenvironment and Angiogenesis in Cancers. Cells 2022, 11, 3098. [Google Scholar] [CrossRef]
- Yamagata, T.; Maki, K.; Mitani, K. Runx1/AML1 in Normal and Abnormal Hematopoiesis. Int. J. Hematol. 2005, 82, 1–8. [Google Scholar] [CrossRef]
- Dalle Carbonare, L.; Innamorati, G.; Valenti, M.T. Transcription Factor Runx2 and Its Application to Bone Tissue Engineering. Stem Cell Rev. Rep. 2012, 8, 891–897. [Google Scholar] [CrossRef]
- Lee, J.-M.; Lee, D.-J.; Bae, S.-C.; Jung, H.-S. Abnormal Liver Differentiation and Excessive Angiogenesis in Mice Lacking Runx3. Histochem. Cell Biol. 2013, 139, 751–758. [Google Scholar] [CrossRef]
- Wang, J.W.; Stifani, S. Roles of Runx Genes in Nervous System Development. In RUNX Proteins in Development and Cancer; Groner, Y., Ito, Y., Liu, P., Neil, J.C., Speck, N.A., Van Wijnen, A., Eds.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2017; Volume 962, pp. 103–116. ISBN 978-981-10-3231-8. [Google Scholar]
- Guilliams, M.; Bonnardel, J.; Haest, B.; Vanderborght, B.; Wagner, C.; Remmerie, A.; Bujko, A.; Martens, L.; Thoné, T.; Browaeys, R.; et al. Spatial Proteogenomics Reveals Distinct and Evolutionarily Conserved Hepatic Macrophage Niches. Cell 2022, 185, 379–396.e38. [Google Scholar] [CrossRef]
- Human All Liver Cells. Available online: https://www.livercellatlas.org/umap-humanAll.php (accessed on 11 March 2023).
- Pan, S.; Sun, S.; Liu, B.; Hou, Y. Pan-Cancer Landscape of the RUNX Protein Family Reveals Their Potential as Carcinogenic Biomarkers and the Mechanisms Underlying Their Action. J. Transl. Intern. Med. 2022, 10, 156–174. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef]
- Kellaway, S.G.; Keane, P.; Edginton-White, B.; Regha, K.; Kennett, E.; Bonifer, C. Different Mutant RUNX1 Oncoproteins Program Alternate Haematopoietic Differentiation Trajectories. Life Sci. Alliance 2021, 4, e202000864. [Google Scholar] [CrossRef] [PubMed]
- Osato, M. Point Mutations in the RUNX1/AML1 Gene: Another Actor in RUNX Leukemia. Oncogene 2004, 23, 4284–4296. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and Management of AML in Adults: 2017 ELN Recommendations from an International Expert Panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Gaidzik, V.I.; Teleanu, V.; Papaemmanuil, E.; Weber, D.; Paschka, P.; Hahn, J.; Wallrabenstein, T.; Kolbinger, B.; Köhne, C.H.; Horst, H.A.; et al. RUNX1 Mutations in Acute Myeloid Leukemia Are Associated with Distinct Clinico-Pathologic and Genetic Features. Leukemia 2016, 30, 2160–2168. [Google Scholar] [CrossRef] [PubMed]
- Romana, S.; Mauchauffe, M.; Le Coniat, M.; Chumakov, I.; Le Paslier, D.; Berger, R.; Bernard, O. The t(12;21) of Acute Lymphoblastic Leukemia Results in a Tel-AML1 Gene Fusion. Blood 1995, 85, 3662–3670. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, H.; Kozu, T.; Shimizu, K.; Enomoto, K.; Maseki, N.; Kaneko, Y.; Kamada, N.; Ohki, M. The t(8;21) Translocation in Acute Myeloid Leukemia Results in Production of an AML1-MTG8 Fusion Transcript. EMBO J. 1993, 12, 2715–2721. [Google Scholar] [CrossRef]
- Mitani, K.; Ogawa, S.; Tanaka, T.; Miyoshi, H.; Kurokawa, M.; Mano, H.; Yazaki, Y.; Ohki, M.; Hirai, H. Generation of the AML1-EVI-1 Fusion Gene in the t(3;21)(Q26;Q22) Causes Blastic Crisis in Chronic Myelocytic Leukemia. EMBO J. 1994, 13, 504–510. [Google Scholar] [CrossRef]
- Keita, M.; Bachvarova, M.; Morin, C.; Plante, M.; Gregoire, J.; Renaud, M.-C.; Sebastianelli, A.; Trinh, X.B.; Bachvarov, D. The RUNX1 Transcription Factor Is Expressed in Serous Epithelial Ovarian Carcinoma and Contributes to Cell Proliferation, Migration and Invasion. Cell Cycle 2013, 12, 972–986. [Google Scholar] [CrossRef]
- Liu, C.; Xu, D.; Xue, B.; Liu, B.; Li, J.; Huang, J. Upregulation of RUNX1 Suppresses Proliferation and Migration through Repressing VEGFA Expression in Hepatocellular Carcinoma. Pathol. Oncol. Res. 2020, 26, 1301–1311. [Google Scholar] [CrossRef]
- Li, Q.; Lai, Q.; He, C.; Fang, Y.; Yan, Q.; Zhang, Y.; Wang, X.; Gu, C.; Wang, Y.; Ye, L.; et al. RUNX1 Promotes Tumour Metastasis by Activating the Wnt/β-Catenin Signalling Pathway and EMT in Colorectal Cancer. J. Exp. Clin. Cancer Res. 2019, 38, 334. [Google Scholar] [CrossRef]
- Lu, C.; Yang, Z.; Yu, D.; Lin, J.; Cai, W. RUNX1 Regulates TGF-β Induced Migration and EMT in Colorectal Cancer. Pathol. Res. Pract. 2020, 216, 153142. [Google Scholar] [CrossRef]
- Sangpairoj, K.; Vivithanaporn, P.; Apisawetakan, S.; Chongthammakun, S.; Sobhon, P.; Chaithirayanon, K. RUNX1 Regulates Migration, Invasion, and Angiogenesis via P38 MAPK Pathway in Human Glioblastoma. Cell. Mol. Neurobiol. 2017, 37, 1243–1255. [Google Scholar] [CrossRef]
- Fritz, A.J.; Hong, D.; Boyd, J.; Kost, J.; Finstaad, K.H.; Fitzgerald, M.P.; Hanna, S.; Abuarqoub, A.H.; Malik, M.; Bushweller, J.; et al. RUNX1 and RUNX2 Transcription Factors Function in Opposing Roles to Regulate Breast Cancer Stem Cells. J. Cell. Physiol. 2020, 235, 7261–7272. [Google Scholar] [CrossRef]
- Lin, T.-C. RUNX2 and Cancer. Int. J. Mol. Sci. 2023, 24, 7001. [Google Scholar] [CrossRef]
- Nie, J.-H.; Yang, T.; Li, H.; Ye, H.-S.; Zhong, G.-Q.; Li, T.-T.; Zhang, C.; Huang, W.-H.; Xiao, J.; Li, Z.; et al. Identification of GPC3 Mutation and Upregulation in a Multidrug Resistant Osteosarcoma and Its Spheroids as Therapeutic Target. J. Bone Oncol. 2021, 30, 100391. [Google Scholar] [CrossRef] [PubMed]
- Lake, S.L.; Jmor, F.; Dopierala, J.; Taktak, A.F.G.; Coupland, S.E.; Damato, B.E. Multiplex Ligation-Dependent Probe Amplification of Conjunctival Melanoma Reveals Common BRAF V600E Gene Mutation and Gene Copy Number Changes. Invest. Ophthalmol. Vis. Sci. 2011, 52, 5598. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-S.; Lee, J.-W.; Jang, J.-W.; Chi, X.-Z.; Kim, J.-H.; Li, Y.-H.; Kim, M.-K.; Kim, D.-M.; Choi, B.-S.; Kim, E.-G.; et al. Runx3 Inactivation Is a Crucial Early Event in the Development of Lung Adenocarcinoma. Cancer Cell 2013, 24, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.-L.; Ito, K.; Sakakura, C.; Fukamachi, H.; Inoue, K.; Chi, X.-Z.; Lee, K.-Y.; Nomura, S.; Lee, C.-W.; Han, S.-B.; et al. Causal Relationship between the Loss of RUNX3 Expression and Gastric Cancer. Cell 2002, 109, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.; Arnold, C.N.; Tassone, P.; Chang, D.K.; Niedzwiecki, D.; Dowell, J.M.; Wasserman, L.; Compton, C.; Mayer, R.J.; Bertagnolli, M.M.; et al. Epigenetic Inactivation OfRUNX3 in Microsatellite Unstable Sporadic Colon Cancers. Int. J. Cancer 2004, 112, 754–759. [Google Scholar] [CrossRef]
- Kang, G.H.; Lee, S.; Lee, H.J.; Hwang, K.S. Aberrant CpG Island Hypermethylation of Multiple Genes in Prostate Cancer and Prostatic Intraepithelial Neoplasia: CpG Island Methylation in Prostate Cancer and PIN. J. Pathol. 2004, 202, 233–240. [Google Scholar] [CrossRef]
- Lau, Q.C.; Raja, E.; Salto-Tellez, M.; Liu, Q.; Ito, K.; Inoue, M.; Putti, T.C.; Loh, M.; Ko, T.K.; Huang, C.; et al. RUNX3 Is Frequently Inactivated by Dual Mechanisms of Protein Mislocalization and Promoter Hypermethylation in Breast Cancer. Cancer Res. 2006, 66, 6512–6520. [Google Scholar] [CrossRef] [PubMed]
- Tsunematsu, T.; Kudo, Y.; Iizuka, S.; Ogawa, I.; Fujita, T.; Kurihara, H.; Abiko, Y.; Takata, T. RUNX3 Has an Oncogenic Role in Head and Neck Cancer. PLoS ONE 2009, 4, e5892. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, G.; Cheng, Y.; Martinka, M.; Li, G. Prognostic Significance of RUNX3 Expression in Human Melanoma: RUNX3 in Melanoma Prognosis. Cancer 2011, 117, 2719–2727. [Google Scholar] [CrossRef]
- Nevadunsky, N.S.; Barbieri, J.S.; Kwong, J.; Merritt, M.A.; Welch, W.R.; Berkowitz, R.S.; Mok, S.C. RUNX3 Protein Is Overexpressed in Human Epithelial Ovarian Cancer. Gynecol. Oncol. 2009, 112, 325–330. [Google Scholar] [CrossRef]
- Kim, W.-J.; Kim, E.-J.; Jeong, P.; Quan, C.; Kim, J.; Li, Q.-L.; Yang, J.-O.; Ito, Y.; Bae, S.-C. RUNX3 Inactivation by Point Mutations and Aberrant DNA Methylation in Bladder Tumors. Cancer Res. 2005, 65, 9347–9354. [Google Scholar] [CrossRef]
- Lund, A.H.; Van Lohuizen, M. RUNX: A Trilogy of Cancer Genes. Cancer Cell 2002, 1, 213–215. [Google Scholar] [CrossRef]
- Nakanishi, Y.; Shiraha, H.; Nishina, S.; Tanaka, S.; Matsubara, M.; Horiguchi, S.; Iwamuro, M.; Takaoka, N.; Uemura, M.; Kuwaki, K.; et al. Loss of Runt-Related Transcription Factor 3 Expression Leads Hepatocellular Carcinoma Cells to Escape Apoptosis. BMC Cancer 2011, 11, 3. [Google Scholar] [CrossRef]
- Mori, T.; Nomoto, S.; Koshikawa, K.; Fujii, T.; Sakai, M.; Nishikawa, Y.; Inoue, S.; Takeda, S.; Kaneko, T.; Nakao, A. Decreased Expression and Frequent Allelic Inactivation of the RUNX3 Gene at 1p36 in Human Hepatocellular Carcinoma. Liver Int. 2005, 25, 380–388. [Google Scholar] [CrossRef]
- Steinhart, Z.; Angers, S. Wnt Signaling in Development and Tissue Homeostasis. Development 2018, 145, dev146589. [Google Scholar] [CrossRef]
- Ito, K.; Lim, A.C.-B.; Salto-Tellez, M.; Motoda, L.; Osato, M.; Chuang, L.S.H.; Lee, C.W.L.; Voon, D.C.-C.; Koo, J.K.W.; Wang, H.; et al. RUNX3 Attenuates β-Catenin/T Cell Factors in Intestinal Tumorigenesis. Cancer Cell 2008, 14, 226–237. [Google Scholar] [CrossRef]
- Lee, Y.M. Control of RUNX3 by Histone Methyltransferases. J. Cell. Biochem. 2011, 112, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kim, J.; Kim, W.-H.; Lee, Y.M. Hypoxic Silencing of Tumor Suppressor RUNX3 by Histone Modification in Gastric Cancer Cells. Oncogene 2009, 28, 184–194. [Google Scholar] [CrossRef] [PubMed]
- cBioPortal For Cancer Genomics. Available online: https://www.cbioportal.org/ (accessed on 22 August 2022).
- Song, W.-J.; Sullivan, M.G.; Legare, R.D.; Hutchings, S.; Tan, X.; Kufrin, D.; Ratajczak, J.; Resende, I.C.; Haworth, C.; Hock, R.; et al. Haploinsufficiency of CBFA2 Causes Familial Thrombocytopenia with Propensity to Develop Acute Myelogenous Leukaemia. Nat. Genet. 1999, 23, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Mendler, J.H.; Maharry, K.; Radmacher, M.D.; Mrózek, K.; Becker, H.; Metzeler, K.H.; Schwind, S.; Whitman, S.P.; Khalife, J.; Kohlschmidt, J.; et al. RUNX1 Mutations Are Associated with Poor Outcome in Younger and Older Patients with Cytogenetically Normal Acute Myeloid Leukemia and with Distinct Gene and MicroRNA Expression Signatures. J. Clin. Oncol. 2012, 30, 3109–3118. [Google Scholar] [CrossRef]
- Wu, C.-Y.; Li, L.; Chen, S.-L.; Yang, X.; Zhang, C.Z.; Cao, Y. A Zic2/Runx2/NOLC1 Signaling Axis Mediates Tumor Growth and Metastasis in Clear Cell Renal Cell Carcinoma. Cell Death Dis. 2021, 12, 319. [Google Scholar] [CrossRef]
- Wang, C.; Shi, Z.; Zhang, Y.; Li, M.; Zhu, J.; Huang, Z.; Zhang, J.; Chen, J. CBFβ Promotes Colorectal Cancer Progression through Transcriptionally Activating OPN, FAM129A, and UPP1 in a RUNX2-Dependent Manner. Cell Death Differ. 2021, 28, 3176–3192. [Google Scholar] [CrossRef]
- Liu, B.; Pan, S.; Liu, J.; Kong, C. Cancer-Associated Fibroblasts and the Related Runt-Related Transcription Factor 2 (RUNX2) Promote Bladder Cancer Progression. Gene 2021, 775, 145451. [Google Scholar] [CrossRef]
- Yang, D.-P.; Huang, W.-Y.; Chen, G.; Chen, S.-W.; Yang, J.; He, R.-Q.; Huang, S.-N.; Gan, T.-Q.; Ma, J.; Yang, L.-J.; et al. Clinical Significance of Transcription Factor RUNX2 in Lung Adenocarcinoma and Its Latent Transcriptional Regulating Mechanism. Comput. Biol. Chem. 2020, 89, 107383. [Google Scholar] [CrossRef]
- Ito, K.; Chuang, L.S.H.; Ito, T.; Chang, T.L.; Fukamachi, H.; Salto–Tellez, M.; Ito, Y. Loss of Runx3 Is a Key Event in Inducing Precancerous State of the Stomach. Gastroenterology 2011, 140, 1536–1546. [Google Scholar] [CrossRef]
- Xiao, W.-H.; Liu, W.-W. Hemizygous Deletion and Hypermethylation of RUNX3 Gene in Hepatocellular Carcinoma. World J. Gastroenterol. 2004, 10, 376. [Google Scholar] [CrossRef]
- Zaret, K.S.; Carroll, J.S. Pioneer Transcription Factors: Establishing Competence for Gene Expression. Genes Dev. 2011, 25, 2227–2241. [Google Scholar] [CrossRef]
- Lichtinger, M.; Ingram, R.; Hannah, R.; Müller, D.; Clarke, D.; Assi, S.A.; Lie-A-Ling, M.; Noailles, L.; Vijayabaskar, M.S.; Wu, M.; et al. RUNX1 Reshapes the Epigenetic Landscape at the Onset of Haematopoiesis: RUNX1 Shifts Transcription Factor Binding Patterns. EMBO J. 2012, 31, 4318–4333. [Google Scholar] [CrossRef] [PubMed]
- Hoogenkamp, M.; Lichtinger, M.; Krysinska, H.; Lancrin, C.; Clarke, D.; Williamson, A.; Mazzarella, L.; Ingram, R.; Jorgensen, H.; Fisher, A.; et al. Early Chromatin Unfolding by RUNX1: A Molecular Explanation for Differential Requirements during Specification versus Maintenance of the Hematopoietic Gene Expression Program. Blood 2009, 114, 299–309. [Google Scholar] [CrossRef]
- Kitabayashi, I. Activation of AML1-Mediated Transcription by MOZ and Inhibition by the MOZ-CBP Fusion Protein. EMBO J. 2001, 20, 7184–7196. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Zhao, X.; Wang, L.; Elf, S.; Xu, H.; Zhao, X.; Sashida, G.; Zhang, Y.; Liu, Y.; Lee, J.; et al. The Ability of MLL to Bind RUNX1 and Methylate H3K4 at PU.1 Regulatory Regions Is Impaired by MDS/AML-Associated RUNX1/AML1 Mutations. Blood 2011, 118, 6544–6552. [Google Scholar] [CrossRef] [PubMed]
- Westendorf, J.J.; Zaidi, S.K.; Cascino, J.E.; Kahler, R.; Van Wijnen, A.J.; Lian, J.B.; Yoshida, M.; Stein, G.S.; Li, X. Runx2 (Cbfa1, AML-3) Interacts with Histone Deacetylase 6 and Represses the P21CIP1/WAF1 Promoter. Mol. Cell. Biol. 2002, 22, 7982–7992. [Google Scholar] [CrossRef]
- Yi, H.; Li, G.; Long, Y.; Liang, W.; Cui, H.; Zhang, B.; Tan, Y.; Li, Y.; Shen, L.; Deng, D.; et al. Integrative Multi-Omics Analysis of a Colon Cancer Cell Line with Heterogeneous Wnt Activity Revealed RUNX2 as an Epigenetic Regulator of EMT. Oncogene 2020, 39, 5152–5164. [Google Scholar] [CrossRef]
- Lee, J.-W.; Kim, D.-M.; Jang, J.-W.; Park, T.-G.; Song, S.-H.; Lee, Y.-S.; Chi, X.-Z.; Park, I.Y.; Hyun, J.-W.; Ito, Y.; et al. RUNX3 Regulates Cell Cycle-Dependent Chromatin Dynamics by Functioning as a Pioneer Factor of the Restriction-Point. Nat. Commun. 2019, 10, 1897. [Google Scholar] [CrossRef]
- National Library of Medicine. RUNX1 RUNX Family Transcription Factor 1 [Homo sapiens (Human)]. Available online: https://www.ncbi.nlm.nih.gov/gene/861 (accessed on 18 March 2023).
- Miyoshi, H.; Shimizu, K.; Kozu, T.; Maseki, N.; Kaneko, Y.; Ohki, M. T(8;21) Breakpoints on Chromosome 21 in Acute Myeloid Leukemia Are Clustered within a Limited Region of a Single Gene, AML1. Proc. Natl. Acad. Sci. USA 1991, 88, 10431–10434. [Google Scholar] [CrossRef]
- Martinez, M.; Hinojosa, M.; Trombly, D.; Morin, V.; Stein, J.; Stein, G.; Javed, A.; Gutierrez, S.E. Transcriptional Auto-Regulation of RUNX1 P1 Promoter. PLoS ONE 2016, 11, e0149119. [Google Scholar] [CrossRef]
- Lin, T.-C. RUNX1 and Cancer. Biochim. Biophys. Acta (BBA) Rev. Cancer 2022, 1877, 188715. [Google Scholar] [CrossRef] [PubMed]
- TIMER2.0. Available online: http://timer.cistrome.org/ (accessed on 3 April 2023).
- GENT2. Available online: http://gent2.appex.kr/gent2/ (accessed on 3 April 2023).
- Zhu, J.; Sanborn, J.Z.; Benz, S.; Szeto, C.; Hsu, F.; Kuhn, R.M.; Karolchik, D.; Archie, J.; Lenburg, M.E.; Esserman, L.J.; et al. The UCSC Cancer Genomics Browser. Nat. Methods 2009, 6, 239–240. [Google Scholar] [CrossRef] [PubMed]
- UCSC Xena. Available online: http://xena.ucsc.edu/welcome-to-ucsc-xena/ (accessed on 3 April 2023).
- Miyagawa, K.; Sakakura, C.; Nakashima, S.; Yoshikawa, T.; Kin, S.; Nakase, Y.; Ito, K.; Yamagishi, H.; Ida, H.; Yazumi, S.; et al. Down-Regulation of RUNX1, RUNX3 and CBFbeta in Hepatocellular Carcinomas in an Early Stage of Hepatocarcinogenesis. Anticancer Res. 2006, 26, 3633–3643. [Google Scholar] [PubMed]
- Wang, T.; Jin, H.; Hu, J.; Li, X.; Ruan, H.; Xu, H.; Wei, L.; Dong, W.; Teng, F.; Gu, J.; et al. COL4A1 Promotes the Growth and Metastasis of Hepatocellular Carcinoma Cells by Activating FAK-Src Signaling. J. Exp. Clin. Cancer Res. 2020, 39, 148. [Google Scholar] [CrossRef]
- Takakura, N.; Watanabe, T.; Suenobu, S.; Yamada, Y.; Noda, T.; Ito, Y.; Satake, M.; Suda, T. A Role for Hematopoietic Stem Cells in Promoting Angiogenesis. Cell 2000, 102, 199–209. [Google Scholar] [CrossRef]
- Iwatsuki, K.; Tanaka, K.; Kaneko, T.; Kazama, R.; Okamoto, S.; Nakayama, Y.; Ito, Y.; Satake, M.; Takahashi, S.-I.; Miyajima, A.; et al. Runx1 Promotes Angiogenesis by Downregulation of Insulin-like Growth Factor-Binding Protein-3. Oncogene 2005, 24, 1129–1137. [Google Scholar] [CrossRef]
- Ter Elst, A.; Ma, B.; Scherpen, F.J.G.; De Jonge, H.J.M.; Douwes, J.; Wierenga, A.T.J.; Schuringa, J.J.; Kamps, W.A.; De Bont, E.S.J.M. Repression of Vascular Endothelial Growth Factor Expression by the Runt-Related Transcription Factor 1 in Acute Myeloid Leukemia. Cancer Res. 2011, 71, 2761–2771. [Google Scholar] [CrossRef]
- Tuo, Z.; Zhang, Y.; Wang, X.; Dai, S.; Liu, K.; Xia, D.; Wang, J.; Bi, L. RUNX1 Is a Promising Prognostic Biomarker and Related to Immune Infiltrates of Cancer-Associated Fibroblasts in Human Cancers. BMC Cancer 2022, 22, 523. [Google Scholar] [CrossRef]
- National Library of Medicine. RUNX1-IT1 RUNX1 Intronic Transcript 1 [Homo sapiens (Human)]. Available online: https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=80215 (accessed on 18 March 2023).
- Yan, P.-H.; Wang, L.; Chen, H.; Yu, F.-Q.; Guo, L.; Liu, Y.; Zhang, W.-J.; Bai, Y.-L. LncRNA RUNX1-IT1 Inhibits Proliferation and Promotes Apoptosis of Hepatocellular Carcinoma by Regulating MAPK Pathways. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8287–8294. [Google Scholar] [CrossRef]
- Sun, L.; Wang, L.; Chen, T.; Shi, Y.; Yao, B.; Liu, Z.; Wang, Y.; Li, Q.; Liu, R.; Niu, Y.; et al. LncRNA RUNX1-IT1 Which Is Downregulated by Hypoxia-Driven Histone Deacetylase 3 Represses Proliferation and Cancer Stem-like Properties in Hepatocellular Carcinoma Cells. Cell Death Dis. 2020, 11, 95. [Google Scholar] [CrossRef]
- Vivacqua, A.; De Marco, P.; Santolla, M.F.; Cirillo, F.; Pellegrino, M.; Panno, M.L.; Abonante, S.; Maggiolini, M. Estrogenic Gper Signaling Regulates Mir144 Expression in Cancer Cells and Cancer-Associated Fibroblasts (Cafs). Oncotarget 2015, 6, 16573–16587. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Zhang, Y.; Lin, J.; Shentu, Y.; Xie, X. MicroRNA-455 Regulates Migration and Invasion of Human Hepatocellular Carcinoma by Targeting Runx2. Oncol. Rep. 2016, 36, 3325–3332. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-Y.; Chen, C.-L.; Hu, Y.-C.; Chi, Y.; Huang, Y.-H.; Su, C.-W.; Jeng, W.-J.; Liang, Y.-J.; Wu, J.-C. High Expression of MicroRNA-196a Is Associated with Progression of Hepatocellular Carcinoma in Younger Patients. Cancers 2019, 11, 1549. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Y.; Zhang, C.; Chen, X.; Huang, H.; Li, W.; Zhang, J.; Liu, Y. Upregulation of OGT by Caveolin-1 Promotes Hepatocellular Carcinoma Cell Migration and Invasion. Cell Biol. Int. 2021, 45, 2251–2263. [Google Scholar] [CrossRef]
- Huang, H.; Liu, Y.; Yu, P.; Qu, J.; Guo, Y.; Li, W.; Wang, S.; Zhang, J. MiR-23a Transcriptional Activated by Runx2 Increases Metastatic Potential of Mouse Hepatoma Cell via Directly Targeting Mgat3. Sci. Rep. 2018, 8, 7366. [Google Scholar] [CrossRef]
- Jing, G.; Zheng, X.; Ji, X. LncRNA HAND2-AS1 Overexpression Inhibits Cancer Cell Proliferation in Hepatocellular Carcinoma by Downregulating RUNX2 Expression. J. Clin. Lab. Anal. 2021, 35, e23717. [Google Scholar] [CrossRef]
- Yu, W.; Qiao, Y.; Tang, X.; Ma, L.; Wang, Y.; Zhang, X.; Weng, W.; Pan, Q.; Yu, Y.; Sun, F.; et al. Tumor Suppressor Long Non-Coding RNA, MT1DP Is Negatively Regulated by YAP and Runx2 to Inhibit FoxA1 in Liver Cancer Cells. Cell. Signal. 2014, 26, 2961–2968. [Google Scholar] [CrossRef]
- Chen, Z.; Zuo, X.; Pu, L.; Zhang, Y.; Han, G.; Zhang, L.; Wu, J.; Wang, X. Circ LARP 4 Induces Cellular Senescence through Regulating MiR-761/RUNX 3/P53/P21 Signaling in Hepatocellular Carcinoma. Cancer Sci. 2019, 110, 568–581. [Google Scholar] [CrossRef]
- Gou, Y.; Zhai, F.; Zhang, L.; Cui, L. RUNX3 Regulates Hepatocellular Carcinoma Cell Metastasis via Targeting MiR-186/E-Cadherin/EMT Pathway. Oncotarget 2017, 8, 61475–61486. [Google Scholar] [CrossRef]
- Li, S.; Li, F.; Xu, L.; Liu, X.; Zhu, X.; Gao, W.; Shen, X. TLR2 Agonist Promotes Myeloid-Derived Suppressor Cell Polarization via Runx1 in Hepatocellular Carcinoma. Int. Immunopharmacol. 2022, 111, 109168. [Google Scholar] [CrossRef]
- Hill, T.P.; Später, D.; Taketo, M.M.; Birchmeier, W.; Hartmann, C. Canonical Wnt/β-Catenin Signaling Prevents Osteoblasts from Differentiating into Chondrocytes. Dev. Cell 2005, 8, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-W.; Yasui, N.; Ito, K.; Huang, G.; Fujii, M.; Hanai, J.; Nogami, H.; Ochi, T.; Miyazono, K.; Ito, Y. A RUNX2/PEBP2 AA/CBFA1 Mutation Displaying Impaired Transactivation and Smad Interaction in Cleidocranial Dysplasia. Proc. Natl. Acad. Sci. USA 2000, 97, 10549–10554. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Yang, H.; Chai, J.; Xing, L. RUNX2 as a Promising Therapeutic Target for Malignant Tumors. Cancer Manag. Res. 2021, 13, 2539–2548. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, W.; Huang, T.; Zhu, Y.; Huang, C. RUNX2 Promotes Hepatocellular Carcinoma Cell Migration and Invasion by Upregulating MMP9 Expression. Oncol. Rep. 2016, 36, 2777–2784. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Sun, B.; Zhao, X.; Zhang, Y.; Gu, Q.; Liang, X.; Dong, X.; Zhao, N. Correction: Cao, Z.; et al. The Expression and Functional Significance of Runx2 in Hepatocellular Carcinoma: Its Role in Vasculogenic Mimicry and Epithelial—Mesenchymal Transition. Int. J. Mol. Sci. 2020, 18, 500. [Google Scholar] [CrossRef]
- Emma, M.R.; Iovanna, J.L.; Bachvarov, D.; Puleio, R.; Loria, G.R.; Augello, G.; Candido, S.; Libra, M.; Gulino, A.; Cancila, V.; et al. NUPR1, a New Target in Liver Cancer: Implication in Controlling Cell Growth, Migration, Invasion and Sorafenib Resistance. Cell Death Dis. 2016, 7, e2269. [Google Scholar] [CrossRef]
- Wang, X.; Li, L.; Wu, Y.; Zhang, R.; Zhang, M.; Liao, D.; Wang, G.; Qin, G.; Xu, R.; Kang, T. CBX4 Suppresses Metastasis via Recruitment of HDAC3 to the Runx2 Promoter in Colorectal Carcinoma. Cancer Res. 2016, 76, 7277–7289. [Google Scholar] [CrossRef]
- Sase, T.; Suzuki, T.; Miura, K.; Shiiba, K.; Sato, I.; Nakamura, Y.; Takagi, K.; Onodera, Y.; Miki, Y.; Watanabe, M.; et al. Runt-Related Transcription Factor 2 in Human Colon Carcinoma: A Potent Prognostic Factor Associated with Estrogen Receptor. Int. J. Cancer 2012, 131, 2284–2293. [Google Scholar] [CrossRef]
- Komori, T. Runx2, A Multifunctional Transcription Factor in Skeletal Development. J. Cell. Biochem. 2002, 87, 1–8. [Google Scholar] [CrossRef]
- Pratap, J.; Javed, A.; Languino, L.R.; Van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. The Runx2 Osteogenic Transcription Factor Regulates Matrix Metalloproteinase 9 in Bone Metastatic Cancer Cells and Controls Cell Invasion. Mol. Cell. Biol. 2005, 25, 8581–8591. [Google Scholar] [CrossRef]
- Boregowda, R.K.; Olabisi, O.O.; Abushahba, W.; Jeong, B.-S.; Haenssen, K.K.; Chen, W.; Chekmareva, M.; Lasfar, A.; Foran, D.J.; Goydos, J.S.; et al. RUNX2 Is Overexpressed in Melanoma Cells and Mediates Their Migration and Invasion. Cancer Lett. 2014, 348, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Q.; Du, X.; Li, D.-M.; Kong, P.-Z.; Sun, Y.; Liu, P.-F.; Wang, Q.-S.; Feng, Y.-M. ITGBL1 Is a Runx2 Transcriptional Target and Promotes Breast Cancer Bone Metastasis by Activating the TGFβ Signaling Pathway. Cancer Res. 2015, 75, 3302–3313. [Google Scholar] [CrossRef] [PubMed]
- El-Gendi, S.M.; Mostafa, M.F. Runx2 Expression as a Potential Prognostic Marker in Invasive Ductal Breast Carcinoma. Pathol. Oncol. Res. 2016, 22, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Baniwal, S.K.; Khalid, O.; Gabet, Y.; Shah, R.R.; Purcell, D.J.; Mav, D.; Kohn-Gabet, A.E.; Shi, Y.; Coetzee, G.A.; Frenkel, B. Runx2 Transcriptome of Prostate Cancer Cells: Insights into Invasiveness and Bone Metastasis. Mol. Cancer 2010, 9, 258. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Sun, B.; Zhao, X.; Zhang, D.; Zhao, X.; Gu, Q.; Dong, X.; Zhao, N.; Liu, P.; Liu, Y. Doxycycline as an Inhibitor of the Epithelial-to-Mesenchymal Transition and Vasculogenic Mimicry in Hepatocellular Carcinoma. Mol. Cancer Ther. 2014, 13, 3107–3122. [Google Scholar] [CrossRef]
- Sun, T.; Zhao, N.; Zhao, X.; Gu, Q.; Zhang, S.; Che, N.; Wang, X.; Du, J.; Liu, Y.; Sun, B. Expression and Functional Significance of Twist1 in Hepatocellular Carcinoma: Its Role in Vasculogenic Mimicry. Hepatology 2010, 51, 545–556. [Google Scholar] [CrossRef]
- Funasaka, T.; Raz, A.; Nangia-Makker, P. Galectin-3 in Angiogenesis and Metastasis. Glycobiology 2014, 24, 886–891. [Google Scholar] [CrossRef]
- Vestweber, D. VE-Cadherin: The Major Endothelial Adhesion Molecule Controlling Cellular Junctions and Blood Vessel Formation. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 223–232. [Google Scholar] [CrossRef]
- Yang, N.; Wang, L.; Chen, T.; Liu, R.; Liu, Z.; Zhang, L. ZNF521 Which Is Downregulated by MiR-802 Suppresses Malignant Progression of Hepatocellular Carcinoma through Regulating Runx2 Expression. J. Cancer 2020, 11, 5831–5839. [Google Scholar] [CrossRef]
- Li, Y.-J.; Ping, C.; Tang, J.; Zhang, W. MicroRNA-455 Suppresses Non-Small Cell Lung Cancer through Targeting ZEB1: The Role of MiRNA-455 in NSCLC. Cell Biol. Int. 2016, 40, 621–628. [Google Scholar] [CrossRef]
- Chai, J.; Wang, S.; Han, D.; Dong, W.; Xie, C.; Guo, H. MicroRNA-455 Inhibits Proliferation and Invasion of Colorectal Cancer by Targeting RAF Proto-Oncogene Serine/Threonine-Protein Kinase. Tumor Biol. 2015, 36, 1313–1321. [Google Scholar] [CrossRef]
- Chou, C.-H.; Shrestha, S.; Yang, C.-D.; Chang, N.-W.; Lin, Y.-L.; Liao, K.-W.; Huang, W.-C.; Sun, T.-H.; Tu, S.-J.; Lee, W.-H.; et al. MiRTarBase Update 2018: A Resource for Experimentally Validated MicroRNA-Target Interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, S.; Wang, B.; Huang, J.; Lu, W.W.; Chen, D. Runx2 and MicroRNA Regulation in Bone and Cartilage Diseases: Runx2 and MiRNAs in Bone and Cartilage. Ann. N. Y. Acad. Sci. 2016, 1383, 80–87. [Google Scholar] [CrossRef]
- Wai, P.Y.; Kuo, P.C. The Role of Osteopontin in Tumor Metastasis. J. Surg. Res. 2004, 121, 228–241. [Google Scholar] [CrossRef]
- Whittle, M.C.; Izeradjene, K.; Rani, P.G.; Feng, L.; Carlson, M.A.; DelGiorno, K.E.; Wood, L.D.; Goggins, M.; Hruban, R.H.; Chang, A.E.; et al. RUNX3 Controls a Metastatic Switch in Pancreatic Ductal Adenocarcinoma. Cell 2015, 161, 1345–1360. [Google Scholar] [CrossRef]
- Kumar, A.; Sundaram, S.; Rayala, S.K.; Venkatraman, G. UnPAKing RUNX3 Functions–Both Sides of the Coin. Small GTPases 2017, 10, 264–270. [Google Scholar] [CrossRef]
- Zhang, X.; He, H.; Zhang, X.; Guo, W.; Wang, Y. RUNX3 Promoter Methylation Is Associated with Hepatocellular Carcinoma Risk: A Meta-Analysis. Cancer Investig. 2015, 33, 121–125. [Google Scholar] [CrossRef]
- Yang, Y.; Ye, Z.; Zou, Z.; Xiao, G.; Luo, G.; Yang, H. Clinicopathological Significance of RUNX3 Gene Hypermethylation in Hepatocellular Carcinoma. Tumor Biol. 2014, 35, 10333–10340. [Google Scholar] [CrossRef]
- El-shaarawy, F.; Abo ElAzm, M.M.; Mohamed, R.H.; Radwan, M.I.; Abo-Elmatty, D.M.; Mehanna, E.T. Relation of the Methylation State of RUNX3 and P16 Gene Promoters with Hepatocellular Carcinoma in Egyptian Patients. Egypt. J. Med. Hum. Genet. 2022, 23, 48. [Google Scholar] [CrossRef]
- Sun, G.; Zhang, C.; Feng, M.; Liu, W.; Xie, H.; Qin, Q.; Zhao, E.; Wan, L. Methylation Analysis of P16, SLIT2, SCARA5, and Runx3 Genes in Hepatocellular Carcinoma. Medicine 2017, 96, e8279. [Google Scholar] [CrossRef]
- El-Bendary, M.; Nour, D.; Arafa, M.; Neamatallah, M. Methylation of Tumour Suppressor Genes RUNX3, RASSF1A and E-Cadherin in HCV-Related Liver Cirrhosis and Hepatocellular Carcinoma. Br. J. Biomed. Sci. 2020, 77, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Liu, X.; Bai, J.; Pei, D.; Zheng, J. The Emerging Role of RUNX3 in Cancer Metastasis (Review). Oncol. Rep. 2016, 35, 1227–1236. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Chen, Y.; Wu, K.-C.; Liu, J.; Zhao, Y.-Q.; Pan, Y.-L.; Du, R.; Zheng, G.-R.; Xiong, Y.-M.; Xu, H.-L.; et al. RUNX3 Directly Interacts with Intracellular Domain of Notch1 and Suppresses Notch Signaling in Hepatocellular Carcinoma Cells. Exp. Cell Res. 2010, 316, 149–157. [Google Scholar] [CrossRef]
- Chi, X.-Z.; Lee, J.-W.; Lee, Y.-S.; Park, I.Y.; Ito, Y.; Bae, S.-C. Runx3 Plays a Critical Role in Restriction-Point and Defense against Cellular Transformation. Oncogene 2017, 36, 6884–6894. [Google Scholar] [CrossRef] [PubMed]
- Shiraha, H.; Nishina, S.; Yamamoto, K. Loss of Runt-Related Transcription Factor 3 Causes Development and Progression of Hepatocellular Carcinoma. J. Cell. Biochem. 2011, 112, 745–749. [Google Scholar] [CrossRef]
- Tanaka, S.; Shiraha, H.; Nakanishi, Y.; Nishina, S.-I.; Matsubara, M.; Horiguchi, S.; Takaoka, N.; Iwamuro, M.; Kataoka, J.; Kuwaki, K.; et al. Runt-Related Transcription Factor 3 Reverses Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma. Int. J. Cancer 2012, 131, 2537–2546. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, J.; Shiraha, H.; Horiguchi, S.; Sawahara, H.; Uchida, D.; Nagahara, T.; Iwamuro, M.; Morimoto, H.; Takeuchi, Y.; Kuwaki, K.; et al. Loss of Runt-Related Transcription Factor 3 Induces Resistance to 5-Fluorouracil and Cisplatin in Hepatocellular Carcinoma. Oncol. Rep. 2016, 35, 2576–2582. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, N.; Shen, C.; Luo, Y.; Xia, L.; Xue, F.; Xia, Q.; Zhang, J. Upregulated MiR-130a Increases Drug Resistance by Regulating RUNX3 and Wnt Signaling in Cisplatin-Treated HCC Cell. Biochem. Biophys. Res. Commun. 2012, 425, 468–472. [Google Scholar] [CrossRef]
- Chen, W.; Gao, N.; Shen, Y.; Cen, J. Hypermethylation Downregulates Runx3 Gene Expression and Its Restoration Suppresses Gastric Epithelial Cell Growth by Inducing P27 and Caspase3 in Human Gastric Cancer. J. Gastroenterol. Hepatol. 2010, 25, 823–831. [Google Scholar] [CrossRef]
- Wei, D.; Gong, W.; Oh, S.C.; Li, Q.; Kim, W.D.; Wang, L.; Le, X.; Yao, J.; Wu, T.T.; Huang, S.; et al. Loss of RUNX3 Expression Significantly Affects the Clinical Outcome of Gastric Cancer Patients and Its Restoration Causes Drastic Suppression of Tumor Growth and Metastasis. Cancer Res. 2005, 65, 4809–4816. [Google Scholar] [CrossRef]
- Chi, X.-Z.; Yang, J.-O.; Lee, K.-Y.; Ito, K.; Sakakura, C.; Li, Q.-L.; Kim, H.-R.; Cha, E.-J.; Lee, Y.-H.; Kaneda, A.; et al. RUNX3 Suppresses Gastric Epithelial Cell Growth by Inducing P21WAF1/Cip1 Expression in Cooperation with Transforming Growth Factor β-Activated SMAD. Mol. Cell. Biol. 2005, 25, 8097–8107. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Miyazono, K. RUNX Transcription Factors as Key Targets of TGF-β Superfamily Signaling. Curr. Opin. Genet. Dev. 2003, 13, 43–47. [Google Scholar] [CrossRef]
- Yano, T.; Ito, K.; Fukamachi, H.; Chi, X.-Z.; Wee, H.-J.; Inoue, K.; Ida, H.; Bouillet, P.; Strasser, A.; Bae, S.-C.; et al. The RUNX3 Tumor Suppressor Upregulates Bim in Gastric Epithelial Cells Undergoing Transforming Growth Factorβ-Induced Apoptosis. Mol. Cell. Biol. 2006, 26, 4474–4488. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.-C.; Choi, J.-K. Tumor Suppressor Activity of RUNX3. Oncogene 2004, 23, 4336–4340. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.K.; Sullivan, A.J.; Van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. Integration of Runx and Smad Regulatory Signals at Transcriptionally Active Subnuclear Sites. Proc. Natl. Acad. Sci. USA 2002, 99, 8048–8053. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Shen, W.; Liu, L. Senescence and Cancer. Cancer Transl. Med. 2018, 4, 70. [Google Scholar] [CrossRef]
- Sweeney, K.; Cameron, E.R.; Blyth, K. Complex Interplay between the RUNX Transcription Factors and Wnt/β-Catenin Pathway in Cancer: A Tango in the Night. Mol. Cells 2020, 43, 188–197. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, W.; Cheng, N.; Wang, K.; Li, B.; Jiang, X.; Sun, S. Hepatitis C Virus-Induced up-Regulation of MicroRNA-155 Promotes Hepatocarcinogenesis by Activating Wnt Signaling. Hepatology 2012, 56, 1631–1640. [Google Scholar] [CrossRef]
- Giovannini, C.; Fornari, F.; Piscaglia, F.; Gramantieri, L. Notch Signaling Regulation in HCC: From Hepatitis Virus to Non-Coding RNAs. Cells 2021, 10, 521. [Google Scholar] [CrossRef]
- Ahn, S.; Hyeon, J.; Park, C.-K. Notchl and Notch4 Are Markers for Poor Prognosis of Hepatocellular Carcinoma. Hepatobiliary Pancreat. Dis. Int. 2013, 12, 286–294. [Google Scholar] [CrossRef]
- Nishina, S.-I. Restored Expression of the Tumor Suppressor Gene RUNX3 Reduces Cancer Stem Cells in Hepatocellular Carcinoma by Suppressing Jagged1-Notch Signaling. Oncol. Rep. 2011, 26, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Chai, M.Y.; Kou, B.X.; Fu, Z.; Wei, F.L.; Dou, S.S.; Chen, D.X.; Liu, X.N. Sorafenib regulates vascular endothelial growth factor by runt-related transcription factor-3 to inhibit angiogenesis in hepatocellular carcinoma. Zhonghua Gan Zang Bing Za Zhi 2022, 30, 770–776. [Google Scholar] [PubMed]
- Lee, S.H.; Bae, S.C.; Kim, K.W.; Lee, Y.M. RUNX3 Inhibits Hypoxia-Inducible Factor-1α Protein Stability by Interacting with Prolyl Hydroxylases in Gastric Cancer Cells. Oncogene 2014, 33, 1458–1467. [Google Scholar] [CrossRef] [PubMed]
- Galle, P.R.; Forner, A.; Llovet, J.M.; Mazzaferro, V.; Piscaglia, F.; Raoul, J.-L.; Schirmacher, P.; Vilgrain, V. EASL Clinical Practice Guidelines: Management of Hepatocellular Carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular Mechanisms of Epithelial–Mesenchymal Transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Voon, D.C.-C.; Wang, H.; Koo, J.K.W.; Nguyen, T.A.P.; Hor, Y.T.; Chu, Y.-S.; Ito, K.; Fukamachi, H.; Chan, S.L.; Thiery, J.P.; et al. Runx3 Protects Gastric Epithelial Cells Against Epithelial-Mesenchymal Transition-Induced Cellular Plasticity and Tumorigenicity. Stem Cells 2012, 30, 2088–2099. [Google Scholar] [CrossRef]
- Loh, C.-Y.; Chai, J.; Tang, T.; Wong, W.; Sethi, G.; Shanmugam, M.; Chong, P.; Looi, C. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef]
- Lee, S.H.; Jung, Y.D.; Choi, Y.S.; Lee, Y.M. Targeting of RUNX3 by MiR-130a and MiR-495 Cooperatively Increases Cell Proliferation and Tumor Angiogenesis in Gastric Cancer Cells. Oncotarget 2015, 6, 33269–33278. [Google Scholar] [CrossRef]
- Wang, M.; Wang, X.; Liu, W. MicroRNA-130a-3p Promotes the Proliferation and Inhibits the Apoptosis of Cervical Cancer Cells via Negative Regulation of RUNX3. Mol. Med. Rep. 2020, 22, 2990–3000. [Google Scholar] [CrossRef]
- Tang, L.; Chen, R.; Xu, X. Synthetic Lethality: A Promising Therapeutic Strategy for Hepatocellular Carcinoma. Cancer Lett. 2020, 476, 120–128. [Google Scholar] [CrossRef]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.-F.; De Oliveira, A.C.; Santoro, A.; Raoul, J.-L.; Forner, A.; et al. Sorafenib in Advanced Hepatocellular Carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Boyault, S.; Rickman, D.S.; De Reyniès, A.; Balabaud, C.; Rebouissou, S.; Jeannot, E.; Hérault, A.; Saric, J.; Belghiti, J.; Franco, D.; et al. Transcriptome Classification of HCC Is Related to Gene Alterations and to New Therapeutic Targets. Hepatology 2007, 45, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Schulze, K.; Nault, J.-C.; Villanueva, A. Genetic Profiling of Hepatocellular Carcinoma Using Next-Generation Sequencing. J. Hepatol. 2016, 65, 1031–1042. [Google Scholar] [CrossRef] [PubMed]
- Jhunjhunwala, S.; Jiang, Z.; Stawiski, E.W.; Gnad, F.; Liu, J.; Mayba, O.; Du, P.; Diao, J.; Johnson, S.; Wong, K.-F.; et al. Diverse Modes of Genomic Alteration in Hepatocellular Carcinoma. Genome Biol. 2014, 15, 436. [Google Scholar] [CrossRef] [PubMed]
- Ally, A.; Balasundaram, M.; Carlsen, R.; Chuah, E.; Clarke, A.; Dhalla, N.; Holt, R.A.; Jones, S.J.M.; Lee, D.; Ma, Y.; et al. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341. [Google Scholar] [CrossRef]
- Yang, C.; Guo, Y.; Qian, R.; Huang, Y.; Zhang, L.; Wang, J.; Huang, X.; Liu, Z.; Qin, W.; Wang, C.; et al. Mapping the Landscape of Synthetic Lethal Interactions in Liver Cancer. Theranostics 2021, 11, 9038–9053. [Google Scholar] [CrossRef]
- Bae, S.-C.; Kolinjivadi, A.M.; Ito, Y. Functional Relationship between P53 and RUNX Proteins. J. Mol. Cell Biol. 2019, 11, 224–230. [Google Scholar] [CrossRef]
- Krishnan, V. The RUNX Family of Proteins, DNA Repair, and Cancer. Cells 2023, 12, 1106. [Google Scholar] [CrossRef]
- Pharos. Available online: https://pharos.nih.gov/targets/RUNX2 (accessed on 25 August 2023).
- Lu, X.X.; Zhu, L.Q.; Pang, F.; Sun, W.; Ou, C.; Li, Y.; Cao, J.; Hu, Y.L. Relationship between RUNX3 Methylation and Hepatocellular Carcinoma in Asian Populations: A Systematic Review. Genet. Mol. Res. 2014, 13, 5182–5189. [Google Scholar] [CrossRef]
- Jovanovic-Cupic, S.; Bozovic, A.; Krajnovic, M.; Petrovic, N. Hepatitis C: Host and Viral Factors Associated with Response to Therapy and Progression of Liver Fibrosis. In Hepatitis C—From Infection to Cure; Shahid, I., Ed.; InTech: München, Germany, 2018; ISBN 978-1-78984-207-4. [Google Scholar]
| Gene | Regulation Mechanisms | Disease | References |
|---|---|---|---|
| RUNX1 | Point mutations | AML, FPD/AML | [57] |
| Frameshift mutations | AML FPD | [27,58] [57] | |
| Translocations | AML CML ALL | [29] [30] [26] | |
| Decreased expression of RUNX1 and increased of VEGF | HCC | [32] | |
| Increased expression | Colorectal cancer Glioblastoma Epithelial ovarian cancer | [33] [34] [35] [31] | |
| Loss of RUNX1 | Breast cancer cell lines | [36] | |
| RUNX2 | Missense, nonsense, nonstop, deletions, and frameshift | Different types of cancers | [37] |
| Gene amplification | Osteosarcoma Melanoma | [38] [39] | |
| Increased expression | Clear cell renal cell carcinoma Colorectal cancer Bladder cancer Lung adenocarcinoma | [59] [60] [61] [62] | |
| RUNX3 | Promoter hypermethylation | Gastric cancer Sporadic colon cancer Prostate cancer Breast cancer Lung adenocarcinoma Melanoma Bladder cancer HCC | [41,63] [42,53] [43] [44] [40] [46] [48] [51,64] |
| Histone modification | Gastric cancer cells | [55] | |
| LOH | HCC | [51] | |
| Protein mislocalisation | Breast cancer | [44] | |
| R122C point mutation | Gastric cancer | [41] | |
| L89P, P102T, A119D, and M128V | Bladder cancer | [48] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krajnović, M.; Kožik, B.; Božović, A.; Jovanović-Ćupić, S. Multiple Roles of the RUNX Gene Family in Hepatocellular Carcinoma and Their Potential Clinical Implications. Cells 2023, 12, 2303. https://doi.org/10.3390/cells12182303
Krajnović M, Kožik B, Božović A, Jovanović-Ćupić S. Multiple Roles of the RUNX Gene Family in Hepatocellular Carcinoma and Their Potential Clinical Implications. Cells. 2023; 12(18):2303. https://doi.org/10.3390/cells12182303
Chicago/Turabian StyleKrajnović, Milena, Bojana Kožik, Ana Božović, and Snežana Jovanović-Ćupić. 2023. "Multiple Roles of the RUNX Gene Family in Hepatocellular Carcinoma and Their Potential Clinical Implications" Cells 12, no. 18: 2303. https://doi.org/10.3390/cells12182303
APA StyleKrajnović, M., Kožik, B., Božović, A., & Jovanović-Ćupić, S. (2023). Multiple Roles of the RUNX Gene Family in Hepatocellular Carcinoma and Their Potential Clinical Implications. Cells, 12(18), 2303. https://doi.org/10.3390/cells12182303







