Dissecting the Molecular Mechanisms Driving Electropathology in Atrial Fibrillation: Deployment of RNA Sequencing and Transcriptomic Analyses
Abstract
1. Introduction
2. RNA Sequencing Studies in Human AF Cohorts
2.1. Large Cohort Studies
2.2. Differentially Expressed Protein Coding Genes
2.3. Non-Coding RNAs
2.4. Combining Genetic Variation and Differential Gene Expression Analysis
3. Transcriptomic Analyses in Experimental AF Model Systems
3.1. Scope of Cell, Rodent and Large Animal Model Research
3.2. Treatment and Mutation-Related Pathways
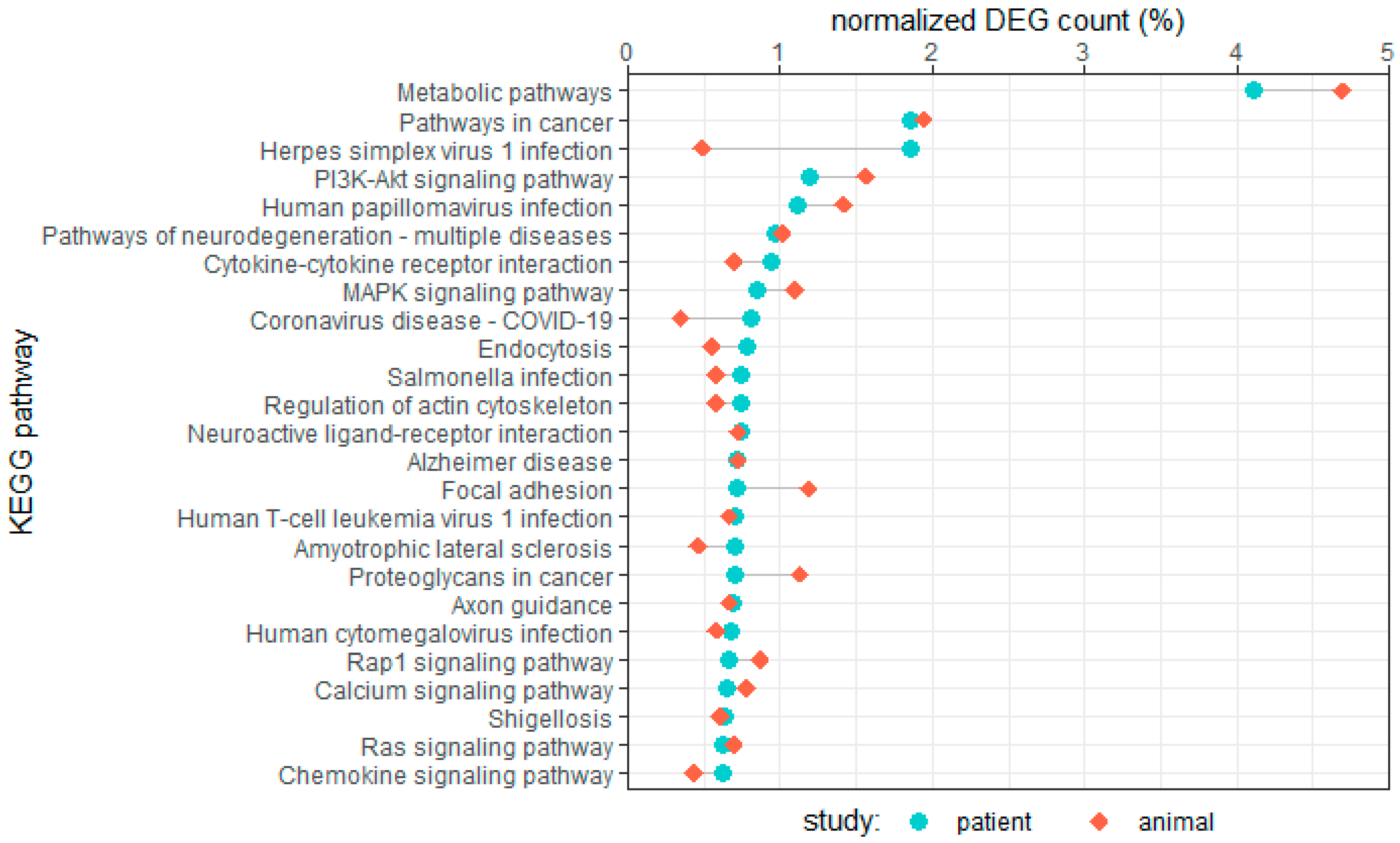
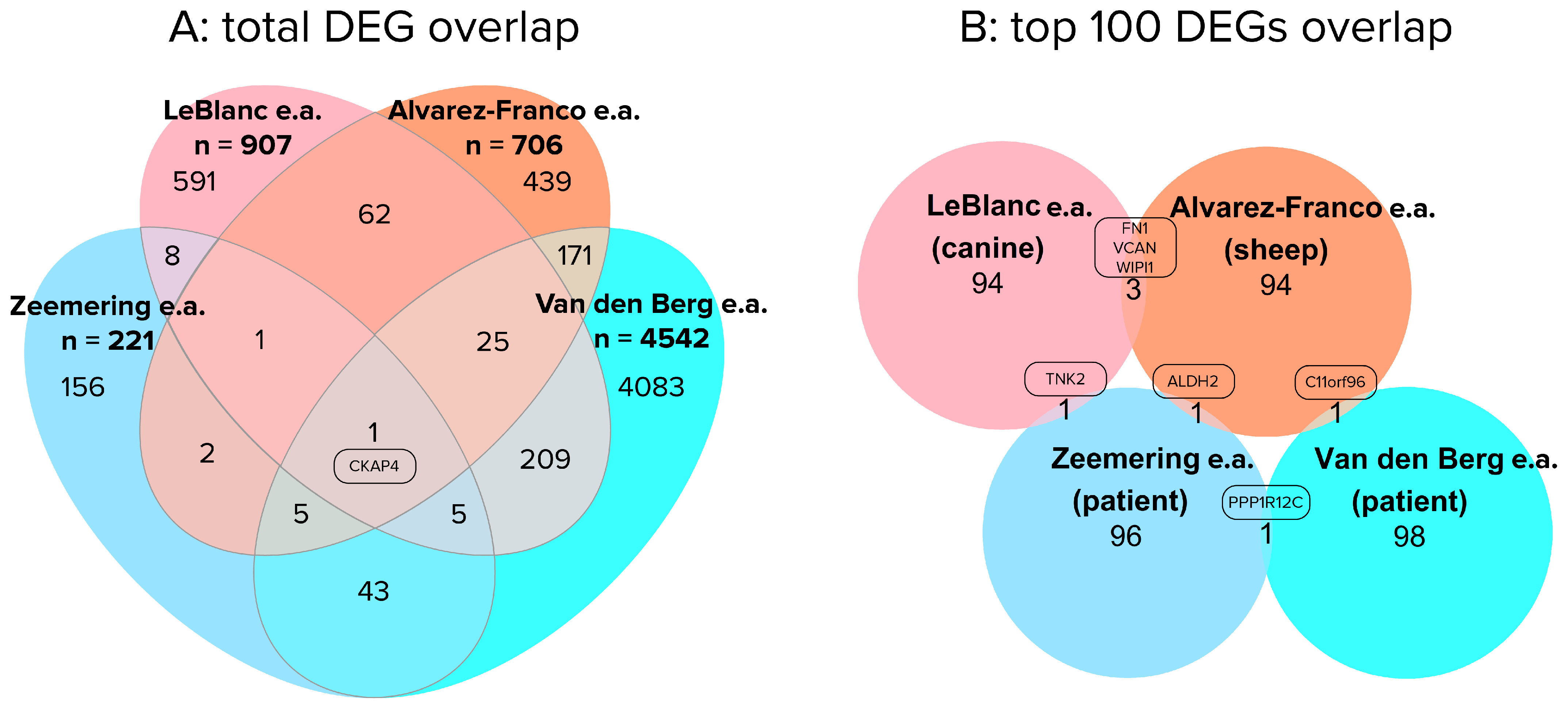
4. Comparing Results of Patient and Large Animal RNA-Seq Studies
5. Challenges and Perspectives in RNA Sequencing Technologies
6. Conclusions
Supplementary Materials
Funding
Conflicts of Interest
Abbreviations
| AF | atrial fibrillation |
| PAF | paroxysmal atrial fibrillation |
| PeAF | persistent atrial fibrillation |
| PoAF | post-operative atrial fibrillation |
| RNA-seq | RNA sequencing |
| HF | heart failure |
| SR | sinus rhythm |
| LA/RA | left/right atrium |
| LAA/RAA | left/right atrial appendage |
| LV/RV | left/right ventricle |
| mRNA | messenger RNA |
| ncRNA | non-coding RNA |
| lncRNA | long non-coding RNA |
| miRNA | micro RNA |
| circRNA | circular RNA |
| CABG | coronary artery bypass |
| DEG | differentially expressed gene |
| EMT | endothelial-to-mesenchymal transition |
| ECM | extracellular matrix |
| GWAS | genome-wide association studies |
| SNP | single-nucleotide polymorphism |
| eQTL | expression quantitative trait loci |
| iPSC | induced pluripotent stem cell |
| CM | cardiomyocyte |
| CF | cardiac fibroblast |
| RIN | RNA integrity number |
| scRNA-seq | single-cell RNA sequencing |
| snRNA-seq | single-nucleus RNA sequencing |
References
- Brundel, B.; Ai, X.; Hills, M.T.; Kuipers, M.F.; Lip, G.Y.H.; de Groot, N.M.S. Atrial fibrillation. Nat. Rev. Dis. Primers 2022, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Brundel, B. The Role of Proteostasis Derailment in Cardiac Diseases. Cells 2020, 9, 2317. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Brundel, B. Inflammasomes and Proteostasis Novel Molecular Mechanisms Associated with Atrial Fibrillation. Circ. Res. 2020, 127, 73–90. [Google Scholar] [CrossRef]
- Ai, X. SR calcium handling dysfunction, stress-response signaling pathways, and atrial fibrillation. Front. Physiol. 2015, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Steenman, M. Insight into atrial fibrillation through analysis of the coding transcriptome in humans. Biophys. Rev. 2020, 12, 817–826. [Google Scholar] [CrossRef]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- van den Hoogenhof, M.M.; Pinto, Y.M.; Creemers, E.E. RNA Splicing: Regulation and Dysregulation in the Heart. Circ. Res. 2016, 118, 454–468. [Google Scholar] [CrossRef]
- Strobel, E.J.; Yu, A.M.; Lucks, J.B. High-throughput determination of RNA structures. Nat. Rev. Genet. 2018, 19, 615–634. [Google Scholar] [CrossRef]
- Zeemering, S.; Isaacs, A.; Winters, J.; Maesen, B.; Bidar, E.; Dimopoulou, C.; Guasch, E.; Batlle, M.; Haase, D.; Hatem, S.N.; et al. Atrial fibrillation in the presence and absence of heart failure enhances expression of genes involved in cardiomyocyte structure, conduction properties, fibrosis, inflammation, and endothelial dysfunction. Heart Rhythm. 2022, 19, 2115–2124. [Google Scholar] [CrossRef]
- van den Berg, N.W.E.; Kawasaki, M.; Fabrizi, B.; Nariswari, F.A.; Verduijn, A.C.; Neefs, J.; Wesselink, R.; Al-Shama, R.F.M.; van der Wal, A.C.; de Boer, O.J.; et al. Epicardial and endothelial cell activation concurs with extracellular matrix remodeling in atrial fibrillation. Clin. Transl. Med. 2021, 11, e558. [Google Scholar] [CrossRef]
- Thomas, A.M.; Cabrera, C.P.; Finlay, M.; Lall, K.; Nobles, M.; Schilling, R.J.; Wood, K.; Mein, C.A.; Barnes, M.R.; Munroe, P.B.; et al. Differentially expressed genes for atrial fibrillation identified by RNA sequencing from paired human left and right atrial appendages. Physiol. Genom. 2019, 51, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.; Hanna, P.; Van Wagoner, D.R.; Barnard, J.; Serre, D.; Chung, M.K.; Smith, J.D. Whole genome expression differences in human left and right atria ascertained by RNA sequencing. Circ. Cardiovasc. Genet. 2012, 5, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Ke, Z.P.; Xu, Y.J.; Wang, Z.S.; Sun, J. RNA sequencing profiling reveals key mRNAs and long noncoding RNAs in atrial fibrillation. J. Cell Biochem. 2020, 121, 3752–3763. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Shao, Y. Transcriptome analysis reveals key pathways that vary in patients with paroxysmal and persistent atrial fibrillation. Exp. Ther. Med. 2021, 21, 571. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.X.; Li, M.Y.; Jiang, Y.Y.; Hou, H.T.; Wang, J.; Liu, X.C.; Yang, Q.; He, G.W. Role of the PPAR pathway in atrial fibrillation associated with heart valve disease: Transcriptomics and proteomics in human atrial tissue. Signal. Transduct. Target Ther. 2020, 5, 4. [Google Scholar] [CrossRef]
- Jiang, Y.Y.; Hou, H.T.; Yang, Q.; Liu, X.C.; He, G.W. Chloride Channels are Involved in the Development of Atrial Fibrillation-A Transcriptomic and proteomic Study. Sci. Rep. 2017, 7, 10215. [Google Scholar] [CrossRef]
- Zhang, Y.; Shen, H.; Wang, P.; Min, J.; Yu, Y.; Wang, Q.; Wang, S.; Xi, W.; Nguyen, Q.M.; Xiao, J.; et al. Identification and characterization of circular RNAs in atrial appendage of patients with atrial fibrillation. Exp. Cell Res. 2020, 389, 111821. [Google Scholar] [CrossRef]
- Zhang, Y.; Ke, X.; Liu, J.; Ma, X.; Liu, Y.; Liang, D.; Wang, L.; Guo, C.; Luo, Y. Characterization of circRNA-associated ceRNA networks in patients with nonvalvular persistent atrial fibrillation. Mol. Med. Rep. 2019, 19, 638–650. [Google Scholar] [CrossRef]
- Hu, M.; Wei, X.; Li, M.; Tao, L.; Wei, L.; Zhang, M.; Cheng, H.; Yuan, Y. Circular RNA expression profiles of persistent atrial fibrillation in patients with rheumatic heart disease. Anatol. J. Cardiol. 2019, 21, 2–10. [Google Scholar] [CrossRef]
- Costa, M.C.; Cortez-Dias, N.; Gabriel, A.; de Sousa, J.; Fiuza, M.; Gallego, J.; Nobre, A.; Pinto, F.J.; Enguita, F.J. circRNA-miRNA cross-talk in the transition from paroxysmal to permanent atrial fibrillation. Int. J. Cardiol. 2019, 290, 134–137. [Google Scholar] [CrossRef]
- Wang, R.; Bektik, E.; Sakon, P.; Wang, X.; Huang, S.; Meng, X.; Chen, M.; Han, W.; Chen, J.; Wang, Y.; et al. Integrated Analysis of the microRNA-mRNA Network Predicts Potential Regulators of Atrial Fibrillation in Humans. Cells 2022, 11, 2629. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Li, J.; Chen, X.; Xiang, Y.; Wu, L.; Li, C.; Zhang, H.; Tong, S.; Zhong, L.; Li, Y. Identification of Long Non-Coding RNA and Circular RNA Expression Profiles in Atrial Fibrillation. Heart Lung Circ. 2020, 29, e157–e167. [Google Scholar] [CrossRef] [PubMed]
- Lipovsky, C.E.; Jimenez, J.; Guo, Q.; Li, G.; Yin, T.; Hicks, S.C.; Bhatnagar, S.; Takahashi, K.; Zhang, D.M.; Brumback, B.D.; et al. Chamber-specific transcriptional responses in atrial fibrillation. JCI Insight 2020, 5, e135319. [Google Scholar] [CrossRef]
- Hsu, J.; Gore-Panter, S.; Tchou, G.; Castel, L.; Lovano, B.; Moravec, C.S.; Pettersson, G.B.; Roselli, E.E.; Gillinov, A.M.; McCurry, K.R.; et al. Genetic Control of Left Atrial Gene Expression Yields Insights into the Genetic Susceptibility for Atrial Fibrillation. Circ. Genom. Precis. Med. 2018, 11, e002107. [Google Scholar] [CrossRef] [PubMed]
- Sigurdsson, M.I.; Saddic, L.; Heydarpour, M.; Chang, T.W.; Shekar, P.; Aranki, S.; Couper, G.S.; Shernan, S.K.; Muehlschlegel, J.D.; Body, S.C. Post-operative atrial fibrillation examined using whole-genome RNA sequencing in human left atrial tissue. BMC Med. Genom. 2017, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Voigt, N.; Li, N.; Wang, Q.; Wang, W.; Trafford, A.W.; Abu-Taha, I.; Sun, Q.; Wieland, T.; Ravens, U.; Nattel, S.; et al. Enhanced sarcoplasmic reticulum Ca2+ leak and increased Na+-Ca2+ exchanger function underlie delayed afterdepolarizations in patients with chronic atrial fibrillation. Circulation 2012, 125, 2059–2070. [Google Scholar] [CrossRef] [PubMed]
- Goette, A.; Staack, T.; Rocken, C.; Arndt, M.; Geller, J.C.; Huth, C.; Ansorge, S.; Klein, H.U.; Lendeckel, U. Increased expression of extracellular signal-regulated kinase and angiotensin-converting enzyme in human atria during atrial fibrillation. J. Am. Coll. Cardiol. 2000, 35, 1669–1677. [Google Scholar] [CrossRef]
- Greiser, M.; Kerfant, B.G.; Williams, G.S.; Voigt, N.; Harks, E.; Dibb, K.M.; Giese, A.; Meszaros, J.; Verheule, S.; Ravens, U.; et al. Tachycardia-induced silencing of subcellular Ca2+ signaling in atrial myocytes. J. Clin. Investig. 2014, 124, 4759–4772. [Google Scholar] [CrossRef]
- Neef, S.; Dybkova, N.; Sossalla, S.; Ort, K.R.; Fluschnik, N.; Neumann, K.; Seipelt, R.; Schondube, F.A.; Hasenfuss, G.; Maier, L.S. CaMKII-dependent diastolic SR Ca2+ leak and elevated diastolic Ca2+ levels in right atrial myocardium of patients with atrial fibrillation. Circ. Res. 2010, 106, 1134–1144. [Google Scholar] [CrossRef]
- Yan, J.; Zhao, W.; Thomson, J.K.; Gao, X.; DeMarco, D.M.; Carrillo, E.; Chen, B.; Wu, X.; Ginsburg, K.S.; Bakhos, M.; et al. Stress Signaling JNK2 Crosstalk with CaMKII Underlies Enhanced Atrial Arrhythmogenesis. Circ. Res. 2018, 122, 821–835. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, W.; Wang, X.; Wang, X.; Song, Y.; Zhang, J.; Zhao, H. Focal adhesion kinase mediates atrial fibrosis via the AKT/S6K signaling pathway in chronic atrial fibrillation patients with rheumatic mitral valve disease. Int. J. Cardiol. 2013, 168, 3200–3207. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, F.; Beneke, K.; Pavlidou, N.G.; Conradi, L.; Reichenspurner, H.; Hove-Madsen, L.; Molina, C.E. Abnormal Calcium Handling in Atrial Fibrillation Is Linked to Changes in Cyclic AMP Dependent Signaling. Cells 2021, 10, 3042. [Google Scholar] [CrossRef]
- Ko, T.H.; Jeong, D.; Yu, B.; Song, J.E.; Le, Q.A.; Woo, S.H.; Choi, J.I. Inhibition of late sodium current via PI3K/Akt signaling prevents cellular remodeling in tachypacing-induced HL-1 atrial myocytes. Pflugers Arch. 2023, 475, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Lenaerts, I.; Bito, V.; Heinzel, F.R.; Driesen, R.B.; Holemans, P.; D’Hooge, J.; Heidbuchel, H.; Sipido, K.R.; Willems, R. Ultrastructural and functional remodeling of the coupling between Ca2+ influx and sarcoplasmic reticulum Ca2+ release in right atrial myocytes from experimental persistent atrial fibrillation. Circ. Res. 2009, 105, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Khandekar, A.; Springer, S.; Wang, W.; Hicks, S.; Weinheimer, C.; Diaz-Trelles, R.; Nerbonne, J.M.; Rentschler, S. Notch-Mediated Epigenetic Regulation of Voltage-Gated Potassium Currents. Circ. Res. 2016, 119, 1324–1338. [Google Scholar] [CrossRef] [PubMed]
- Niessen, K.; Karsan, A. Notch signaling in cardiac development. Circ. Res. 2008, 102, 1169–1181. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Roselli, C.; Chaffin, M.D.; Weng, L.C.; Aeschbacher, S.; Ahlberg, G.; Albert, C.M.; Almgren, P.; Alonso, A.; Anderson, C.D.; Aragam, K.G.; et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat. Genet. 2018, 50, 1225–1233. [Google Scholar] [CrossRef]
- Franco, D.; Aranega, A.; Dominguez, J.N. Non-coding RNAs and Atrial Fibrillation. In Non-Coding RNAs in Cardiovascular Diseases; Xiao, J., Ed.; Springer: Singapore, 2020; pp. 311–325. [Google Scholar] [CrossRef]
- Bennett, J.S. Structure and function of the platelet integrin αIIbβ3. J. Clin. Investig. 2005, 115, 3363–3369. [Google Scholar] [CrossRef]
- Jaeger, A.M.; Pemble, C.W.T.; Sistonen, L.; Thiele, D.J. Structures of HSF2 reveal mechanisms for differential regulation of human heat-shock factors. Nat. Struct. Mol. Biol. 2016, 23, 147–154. [Google Scholar] [CrossRef]
- Chen, C.; Li, R.; Ross, R.S.; Manso, A.M. Integrins and integrin-related proteins in cardiac fibrosis. J. Mol. Cell. Cardiol. 2016, 93, 162–174. [Google Scholar] [CrossRef] [PubMed]
- van Wijk, S.W.; Ramos, K.S.; Brundel, B. Cardioprotective Role of Heat Shock Proteins in Atrial Fibrillation: From Mechanism of Action to Therapeutic and Diagnostic Target. Int. J. Mol. Sci. 2021, 22, 442. [Google Scholar] [CrossRef] [PubMed]
- Henning, R.H.; Brundel, B. Proteostasis in cardiac health and disease. Nat. Rev. Cardiol. 2017, 14, 637–653. [Google Scholar] [CrossRef] [PubMed]
- Brundel, B.J.; Shiroshita-Takeshita, A.; Qi, X.; Yeh, Y.H.; Chartier, D.; van Gelder, I.C.; Henning, R.H.; Kampinga, H.H.; Nattel, S. Induction of heat shock response protects the heart against atrial fibrillation. Circ. Res. 2006, 99, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Aufiero, S.; Reckman, Y.J.; Pinto, Y.M.; Creemers, E.E. Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 2019, 16, 503–514. [Google Scholar] [CrossRef]
- Tao, H.; Zhang, M.; Yang, J.J.; Shi, K.H. MicroRNA-21 via Dysregulation of WW Domain-Containing Protein 1 Regulate Atrial Fibrosis in Atrial Fibrillation. Heart Lung Circ. 2018, 27, 104–113. [Google Scholar] [CrossRef]
- Canon, S.; Caballero, R.; Herraiz-Martinez, A.; Perez-Hernandez, M.; Lopez, B.; Atienza, F.; Jalife, J.; Hove-Madsen, L.; Delpon, E.; Bernad, A. miR-208b upregulation interferes with calcium handling in HL-1 atrial myocytes: Implications in human chronic atrial fibrillation. J. Mol. Cell Cardiol. 2016, 99, 162–173. [Google Scholar] [CrossRef]
- Schellings, M.W.; Vanhoutte, D.; van Almen, G.C.; Swinnen, M.; Leenders, J.J.; Kubben, N.; van Leeuwen, R.E.; Hofstra, L.; Heymans, S.; Pinto, Y.M. Syndecan-1 amplifies angiotensin II-induced cardiac fibrosis. Hypertension 2010, 55, 249–256. [Google Scholar] [CrossRef]
- Frangogiannis, N.G. Syndecan-1. Hypertension 2010, 55, 233–235. [Google Scholar] [CrossRef]
- Wolke, C.; Antileo, E.; Lendeckel, U. WNT signaling in atrial fibrillation. Exp. Biol. Med. 2021, 246, 1112–1120. [Google Scholar] [CrossRef]
- Lin, R.; Wu, S.; Zhu, D.; Qin, M.; Liu, X. Osteopontin induces atrial fibrosis by activating Akt/GSK-3β/β-catenin pathway and suppressing autophagy. Life Sci. 2020, 245, 117328. [Google Scholar] [CrossRef]
- Chilukoti, R.K.; Giese, A.; Malenke, W.; Homuth, G.; Bukowska, A.; Goette, A.; Felix, S.B.; Kanaan, J.; Wollert, H.G.; Evert, K.; et al. Atrial fibrillation and rapid acute pacing regulate adipocyte/adipositas-related gene expression in the atria. Int. J. Cardiol. 2015, 187, 604–613. [Google Scholar] [CrossRef]
- Schuttler, D.; Bapat, A.; Kaab, S.; Lee, K.; Tomsits, P.; Clauss, S.; Hucker, W.J. Animal Models of Atrial Fibrillation. Circ. Res. 2020, 127, 91–110. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Zhang, M.; Ly, O.T.; Chen, H.; Sridhar, A.; Lambers, E.; Chalazan, B.; Youn, S.W.; Maienschein-Cline, M.; Feferman, L.; et al. Human induced pluripotent stem cell-derived atrial cardiomyocytes carrying an SCN5A mutation identify nitric oxide signaling as a mediator of atrial fibrillation. Stem Cell Rep. 2021, 16, 1542–1554. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Deng, Y.; Zhao, Y.; Li, Z.; Gao, J.; Zhang, Y.; Yang, X.; Liu, Y.; Xia, Y. Time series RNA-seq analysis identifies MAPK10 as a critical gene in diabetes mellitus-induced atrial fibrillation in mice. J. Mol. Cell Cardiol. 2022, 168, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Yang, S.; Liu, X.; Jana, S.; Izaddoustdar, F.; Gao, X.; Debi, R.; Kim, D.K.; Kim, K.H.; Yang, P.; et al. Transcriptomic Bioinformatic Analyses of Atria Uncover Involvement of Pathways Related to Strain and Post-translational Modification of Collagen in Increased Atrial Fibrillation Vulnerability in Intensely Exercised Mice. Front. Physiol. 2020, 11, 605671. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wang, W.; Liu, Y.; Guan, S.; Wang, M.; Song, F.; Shangguan, W.; Miao, S.; Zhang, X.; Liu, H.; et al. Establishment of a lncRNA-miRNA-mRNA network in a rat model of atrial fibrosis by whole transcriptome sequencing. J. Interv. Card. Electrophysiol. 2022, 63, 723–736. [Google Scholar] [CrossRef]
- Yue, H.; Zhao, X.; Liang, W.; Qin, X.; Bian, L.; He, K.; Wu, Z. Curcumin, novel application in reversing myocardial fibrosis in the treatment for atrial fibrillation from the perspective of transcriptomics in rat model. Biomed. Pharmacother. 2022, 146, 112522. [Google Scholar] [CrossRef]
- Yue, H.; Liang, W.; Gu, J.; Zhao, X.; Zhang, T.; Qin, X.; Zhu, G.; Wu, Z. Comparative transcriptome analysis to elucidate the therapeutic mechanism of colchicine against atrial fibrillation. Biomed. Pharmacother. 2019, 119, 109422. [Google Scholar] [CrossRef]
- Cheng, C.; Liu, H.; Tan, C.; Tong, D.; Zhao, Y.; Liu, X.; Si, W.; Wang, L.; Liang, L.; Li, J.; et al. Mutation in NPPA causes atrial fibrillation by activating inflammation and cardiac fibrosis in a knock-in rat model. FASEB J. 2019, 33, 8878–8891. [Google Scholar] [CrossRef]
- Du, J.; Li, Z.; Wang, X.; Li, J.; Liu, D.; Wang, X.; Wei, J.; Ma, S.; Zhang, Y.; Hou, Y. Long noncoding RNA TCONS-00106987 promotes atrial electrical remodelling during atrial fibrillation by sponging miR-26 to regulate KCNJ2. J. Cell Mol. Med. 2020, 24, 12777–12788. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, X.; Wang, W.; Du, J.; Wei, J.; Zhang, Y.; Wang, J.; Hou, Y. Altered long non-coding RNA expression profile in rabbit atria with atrial fibrillation: TCONS_00075467 modulates atrial electrical remodeling by sponging miR-328 to regulate CACNA1C. J. Mol. Cell Cardiol. 2017, 108, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Leblanc, F.J.A.; Hassani, F.V.; Liesinger, L.; Qi, X.; Naud, P.; Birner-Gruenberger, R.; Lettre, G.; Nattel, S. Transcriptomic Profiling of Canine Atrial Fibrillation Models After One Week of Sustained Arrhythmia. Circ. Arrhythm. Electrophysiol. 2021, 14, e009887. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Franco, A.; Rouco, R.; Ramirez, R.J.; Guerrero-Serna, G.; Tiana, M.; Cogliati, S.; Kaur, K.; Saeed, M.; Magni, R.; Enriquez, J.A.; et al. Transcriptome and proteome mapping in the sheep atria reveal molecular featurets of atrial fibrillation progression. Cardiovasc. Res. 2021, 117, 1760–1775. [Google Scholar] [CrossRef]
- Anto Michel, N.; Ljubojevic-Holzer, S.; Bugger, H.; Zirlik, A. Cellular Heterogeneity of the Heart. Front. Cardiovasc. Med. 2022, 9, 868466. [Google Scholar] [CrossRef] [PubMed]
- Litvinukova, M.; Talavera-Lopez, C.; Maatz, H.; Reichart, D.; Worth, C.L.; Lindberg, E.L.; Kanda, M.; Polanski, K.; Heinig, M.; Lee, M.; et al. Cells of the adult human heart. Nature 2020, 588, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Gaspo, R.; Bosch, R.F.; Talajic, M.; Nattel, S. Functional Mechanisms Underlying Tachycardia-Induced Sustained Atrial Fibrillation in a Chronic Dog Model. Circulation 1997, 96, 4027–4035. [Google Scholar] [CrossRef]
- Wijffels, M.C.E.F.; Kirchhof, C.J.H.J.; Dorland, R.; Allessie, M.A. Atrial Fibrillation Begets Atrial Fibrillation. Circulation 1995, 92, 1954–1968. [Google Scholar] [CrossRef]
- Luo, T.; Chang, C.-X.; Zhou, X.; Gu, S.-K.; Jiang, T.-M.; Li, Y.-M. Characterization of atrial histopathological and electrophysiological changes in a mouse model of aging. Int. J. Mol. Med. 2013, 31, 138–146. [Google Scholar] [CrossRef][Green Version]
- Jansen, H.J.; Moghtadaei, M.; Mackasey, M.; Rafferty, S.A.; Bogachev, O.; Sapp, J.L.; Howlett, S.E.; Rose, R.A. Atrial structure, function and arrhythmogenesis in aged and frail mice. Sci. Rep. 2017, 7, 44336. [Google Scholar] [CrossRef]
- Hayashi, H.; Wang, C.; Miyauchi, Y.; Omichi, C.; Pak, H.N.; Zhou, S.; Ohara, T.; Mandel, W.J.; Lin, S.F.; Fishbein, M.C.; et al. Aging-related increase to inducible atrial fibrillation in the rat model. J. Cardiovasc. Electrophysiol. 2002, 13, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Anyukhovsky, E.P.; Sosunov, E.A.; Plotnikov, A.; Gainullin, R.Z.; Jhang, J.S.; Marboe, C.C.; Rosen, M.R. Cellular electrophysiologic properties of old canine atria provide a substrate for arrhythmogenesis. Cardiovasc. Res. 2002, 54, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhang, Y.; Zhang, C.; Xie, Z. RNA-Binding Profiles of CKAP4 as an RNA-Binding Protein in Myocardial Tissues. Front. Cardiovasc. Med. 2021, 8, 773573. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Chen, Z.; Chen, F.; Xu, W.; Liu, X. CKAP4 participates in tryptase-induced phenotypic conversion in atrial fibroblasts through PAR2/p38/JNK pathway. Am. J. Transl. Res. 2021, 13, 2270–2282. [Google Scholar]
- Harada, T.; Sada, R.; Osugi, Y.; Matsumoto, S.; Matsuda, T.; Hayashi-Nishino, M.; Nagai, T.; Harada, A.; Kikuchi, A. Palmitoylated CKAP4 regulates mitochondrial functions through an interaction with VDAC2 at ER-mitochondria contact sites. J. Cell Sci. 2020, 133, jcs249045. [Google Scholar] [CrossRef]
- Osugi, Y.; Fumoto, K.; Kikuchi, A. CKAP4 Regulates Cell Migration via the Interaction with and Recycling of Integrin. Mol. Cell Biol. 2019, 39, e00073-19. [Google Scholar] [CrossRef] [PubMed]
- Ramos, K.S.; Li, J.; Wijdeveld, L.F.J.; van Schie, M.S.; Taverne, Y.; Boon, R.A.; de Groot, N.M.S.; Brundel, B. Long Noncoding RNA UCA1 Correlates with Electropathology in Patients with Atrial Fibrillation. JACC Clin. Electrophysiol. 2023, 9 Pt 2, 1097–1107. [Google Scholar] [CrossRef]
- Ramos, K.S.; Pool, L.; van Schie, M.S.; Wijdeveld, L.F.J.M.; van der Does, W.F.B.; Baks, L.; Sultan, H.M.D.; van Wijk, S.W.; Bogers, A.J.J.C.; Verheule, S.; et al. Degree of Fibrosis in Human Atrial Tissue Is Not the Hallmark Driving AF. Cells 2022, 11, 427. [Google Scholar] [CrossRef]
- Miranda, A.M.A.; Janbandhu, V.; Maatz, H.; Kanemaru, K.; Cranley, J.; Teichmann, S.A.; Hubner, N.; Schneider, M.D.; Harvey, R.P.; Noseda, M. Single-cell transcriptomics for the assessment of cardiac disease. Nat. Rev. Cardiol. 2023, 20, 289–308. [Google Scholar] [CrossRef]
- Simonson, B.; Chaffin, M.; Hill, M.C.; Atwa, O.; Guedira, Y.; Bhasin, H.; Hall, A.W.; Hayat, S.; Baumgart, S.; Bedi, K.C., Jr.; et al. Single-nucleus RNA sequencing in ischemic cardiomyopathy reveals common transcriptional profile underlying end-stage heart failure. Cell Rep. 2023, 42, 112086. [Google Scholar] [CrossRef]
- Chaffin, M.; Papangeli, I.; Simonson, B.; Akkad, A.D.; Hill, M.C.; Arduini, A.; Fleming, S.J.; Melanson, M.; Hayat, S.; Kost-Alimova, M.; et al. Single-nucleus profiling of human dilated and hypertrophic cardiomyopathy. Nature 2022, 608, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yin, K.; Chen, L.; Chen, W.; Li, W.; Zhang, T.; Sun, Y.; Yuan, M.; Wang, H.; Song, Y.; et al. Lineage-specific regulatory changes in hypertrophic cardiomyopathy unraveled by single-nucleus RNA-seq and spatial transcriptomics. Cell Discov. 2023, 9, 6. [Google Scholar] [CrossRef] [PubMed]
| Source | Tissue | RNA | SR Patients | PAF Patients | PeAF Patients | Implicated Pathways | |||
|---|---|---|---|---|---|---|---|---|---|
| Number | Surgical Indication | Number | Surgical Indication | Number | Surgical Indication | ||||
| [9] | LAA, RAA | mRNA, lncRNA | 91 | CABG, valve | 53 | CABG, valve | 51 | CABG, valve | inflammation, cellular respiration, fibrosis, cell division |
| [10] | LAA | mRNA | 22 | CABG, valve | 22 | thoracoscopic ablation | 20 | thoracoscopic ablation | EMT, ECM remodeling, angiogenesis |
| [11] | LAA, RAA | mRNA | 5 | CABG, valve | 0 | - | 5 | CABG, valve | wnt, circadian entrainment, ligand-gated ion channels, intracellular transport, ECM and tissue structure |
| [12] | LAA, RAA | mRNA, miRNA | 1 | healthy donor | 0 | - | 3 | Maze surgery | - |
| [13] * | LAA, RAA | mRNA, lncRNA | 5 | CABG, valve | 0 | - | 5 | CABG, valve | Adipose tissue accumulation, TGFB signaling, ion channel remodeling |
| [14] | LAA | mRNA, lncRNA | 3 | healthy donor | 3 | Maze surgery | 3 | Maze surgery | ECM interaction, Insulin resistance, Ras/MAPK signaling |
| [15] | LA, RA | mRNA | 2 | valve | 0 | - | 3 | valve | cell differentiation and development, cellular metabolism |
| [16] | LA, RA | mRNA | 2 | valve | 0 | - | 3 | valve | chloride ion channels, focal adhesion pathway |
| [17] | LAA, RAA | miRNA, circRNA | 6 | valve | 0 | - | 9 | valve | protein binding, nucleotide binding, nucleoside phosphate binding |
| [18] | LAA | circRNA | 4 | healthy donor | 0 | - | 4 | surgical ablation | protein modification, tissue development, calcium ion signaling |
| [19] | LAA | circRNA | 6 | healthy donor; | 0 | - | 9 | valve | DCM, HCM, regulating pluripotency of stem cells, Hippo signaling, TGFB signaling |
| [20] | LA | miRNA, circRNA | 6 | valve | 5 | valve | 3 | valve | wnt, ECM remodeling, Cell junction organization, cardiac conduction *** |
| [21] | RA | mRNA, miRNA | 2 | atrial septal repair, tricuspid vegetation excision | 0 | - | 3 | valve | cell–cell adhesion, TNF-α signaling, p53 signaling, EMT |
| Source | Tissue | RNA | SR patients | AF patients ** | Implicated pathways | ||||
| number | surgical indication | number | surgical indication | ||||||
| [22] | RA | mRNA, lncRNA, miRNA, circRNA | 7 | valve | 7 | valve | Rap1 signaling, MAPK signaling, oxidative phosphorylation, cardiac muscle contraction, tight junction | ||
| [23] | LA, RA | mRNA | 7; 3 | healthy donor; heart transplant | 5 | heart transplant | ion channels, transcriptional regulation, Notch signaling | ||
| [24] | LAA | mRNA, lncRNA | 14; 38 | healthy donor; CABG, valve | 213 | CABG, valve, surgical ablation | calcineurin signaling, sarcomere organization, wnt RhoGAP | ||
| Source | Tissue | RNA | No PoAF patients | PoAF patients | Implicated pathways | ||||
| number | surgical indication | number | surgical indication | ||||||
| [25] | LA | mRNA, lncRNA | 41 | valve | 21 | valve | wnt, cGMP metabolism | ||
| Source | Model | Tissue | Model Groups | Control n= | AF n= | AF Induction or Simulation | Implicated Pathways |
|---|---|---|---|---|---|---|---|
| [55] | Human iPSCs | atrial CMs | control, SCN5a mutant, gene correction control | 2 | 1 | - | nitric oxide signaling, cardiac ion channels |
| [56] | Mouse | LA | WT, db/db | 18 | 18 | intracardial pacing | MAPK10 signaling, IL-17 signaling, TNF signaling, cAMP receptor signaling |
| [57] | Mouse | LAA, LV | sedentary/exercised, WT/TNF-KO | Unclear | - | - | ECM remodeling, fibrosis, intercellular communication |
| [23] | Mouse | RA/LA- CM nuclei | control, iNICD | 6 | 6 | - | NOTCH signaling, cardiac conduction |
| [58] | Rat | RA | control, CIH | 15 | 15 | Hypoxia | cell cycle, p53 signaling, IL-17 signaling, NLR signaling, cell adhesion |
| [59] | Rat | LA, venous blood | control, curcumin treatment | 4 | 6 | Ach-CaCl2 | collagen synthesis, lipid metabolism, inflammation, angiogenesis |
| [60] | Rat | LA | control, AF control, AF colchicine treatment | 4 | 7 | Ach-CaCl2 | phagosome, IL-17 signaling |
| [61] | Rat | neonatal CFs | wNPA treatment, mNPA treatment | 2 | 2 | mutant human NPA treatment | NLR signaling, TNF-α, NF-κB |
| [62] | Rabbit | unclear | control, paced | 3 | 3 | intracardial pacing | KCNJ2 expression |
| [63] | Rabbit | RA | control, paced | 3 | 3 | tachypacing | calcium signaling, gap-junction, focal adhesion |
| [64] | Canine | LA | control, paced | 6 | 12 | tachypacing, tachypacing + AVB | ECM structure, muscle structure development, striated muscle cell differentiation, glutamate signaling |
| [65] | Sheep | LAA/RAA | control, paced | 3 | 6 | tachypacing | calcineurin signaling, chromatin structure, cell adhesion, ion channels, TGF-β signaling, SLIT/ROBO |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huiskes, F.G.; Creemers, E.E.; Brundel, B.J.J.M. Dissecting the Molecular Mechanisms Driving Electropathology in Atrial Fibrillation: Deployment of RNA Sequencing and Transcriptomic Analyses. Cells 2023, 12, 2242. https://doi.org/10.3390/cells12182242
Huiskes FG, Creemers EE, Brundel BJJM. Dissecting the Molecular Mechanisms Driving Electropathology in Atrial Fibrillation: Deployment of RNA Sequencing and Transcriptomic Analyses. Cells. 2023; 12(18):2242. https://doi.org/10.3390/cells12182242
Chicago/Turabian StyleHuiskes, Fabries G., Esther E. Creemers, and Bianca J. J. M. Brundel. 2023. "Dissecting the Molecular Mechanisms Driving Electropathology in Atrial Fibrillation: Deployment of RNA Sequencing and Transcriptomic Analyses" Cells 12, no. 18: 2242. https://doi.org/10.3390/cells12182242
APA StyleHuiskes, F. G., Creemers, E. E., & Brundel, B. J. J. M. (2023). Dissecting the Molecular Mechanisms Driving Electropathology in Atrial Fibrillation: Deployment of RNA Sequencing and Transcriptomic Analyses. Cells, 12(18), 2242. https://doi.org/10.3390/cells12182242







