The Transcription Factor HOXA5: Novel Insights into Metabolic Diseases and Adipose Tissue Dysfunction
Abstract
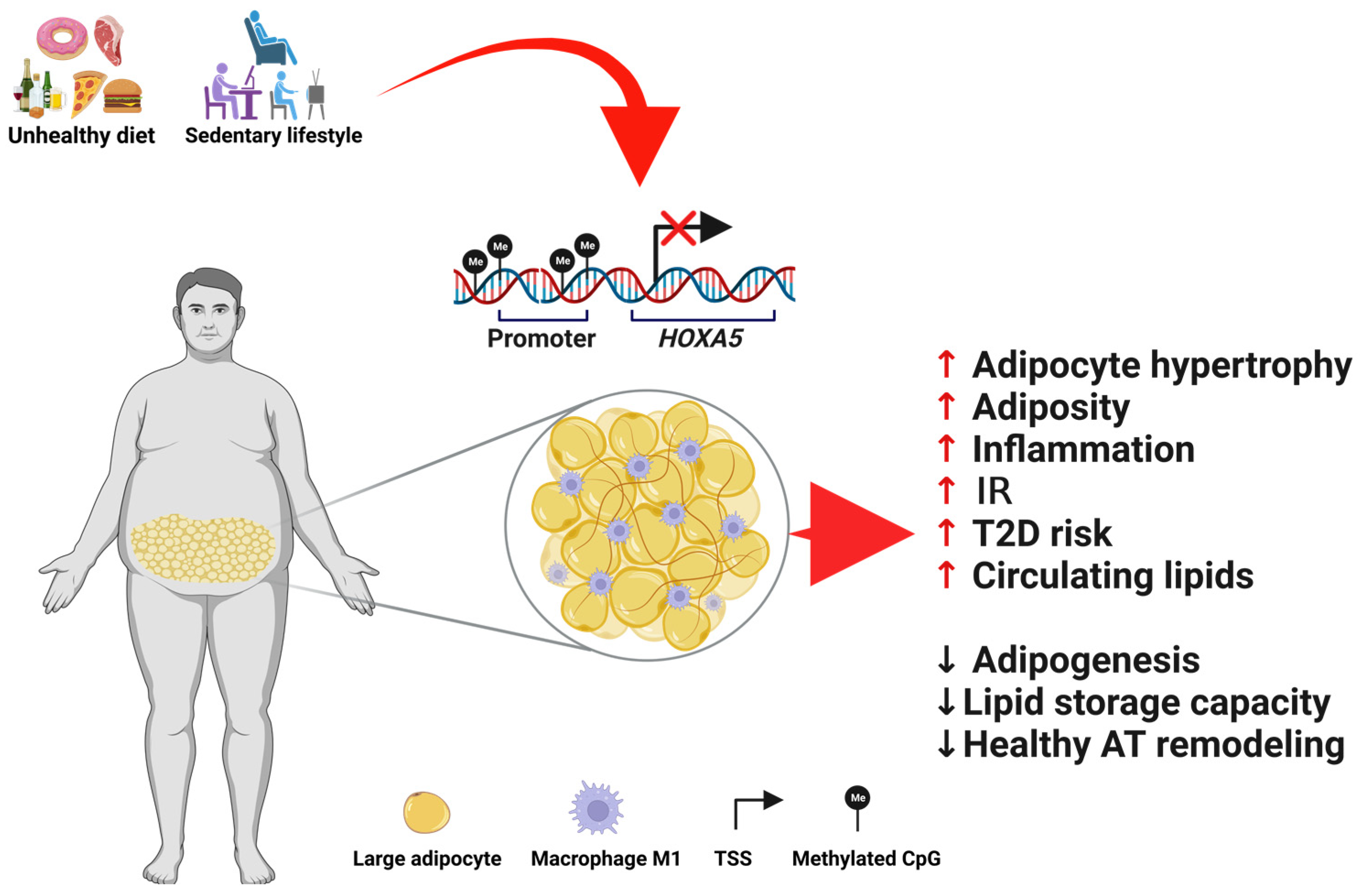
:1. Introduction
2. HOXA5: A Crucial Transcription Factor in Development and Disease
3. Role of HOXA5 in Metabolically Unhealthy States
3.1. HOXA5 and Hypertrophy of AT
3.2. HOXA5 in Obesity and Type 2 Diabetes
4. Changes in HOXA5 DNA Methylation Are Related to Metabolic Diseases
5. HOXA5 as a Potential Therapeutic Target
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shenoy, U.S.; Adiga, D.; Kabekkodu, S.P.; Hunter, K.D.; Radhakrishnan, R. Molecular implications of HOX genes targeting multiple signaling pathways in cancer. Cell Biol. Toxicol. 2022, 38, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Burglin, T.R.; Affolter, M. Homeodomain proteins: An update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef] [PubMed]
- Rezsohazy, R.; Saurin, A.J.; Maurel-Zaffran, C.; Graba, Y. Cellular and molecular insights into Hox protein action. Development 2015, 142, 1212–1227. [Google Scholar] [CrossRef] [PubMed]
- Quinonez, S.C.; Innis, J.W. Human HOX gene disorders. Mol. Genet. Metab. 2014, 111, 4–15. [Google Scholar] [CrossRef]
- Fan, F.; Mo, H.; Zhang, H.; Dai, Z.; Wang, Z.; Qu, C.; Liu, F.; Zhang, L.; Luo, P.; Zhang, J.; et al. HOXA5: A crucial transcriptional factor in cancer and a potential therapeutic target. Biomed. Pharmacother. 2022, 155, 113800. [Google Scholar] [CrossRef] [PubMed]
- Jeannotte, L.; Gotti, F.; Landry-Truchon, K. Hoxa5: A Key Player in Development and Disease. J. Dev. Biol. 2016, 4, 13. [Google Scholar] [CrossRef]
- Boucherat, O.; Guillou, F.; Aubin, J.; Jeannotte, L. Hoxa5: A master gene with multifaceted roles. Med. Sci. 2009, 25, 77–82. [Google Scholar] [CrossRef]
- Gesta, S.; Bluher, M.; Yamamoto, Y.; Norris, A.W.; Berndt, J.; Kralisch, S.; Boucher, J.; Lewis, C.; Kahn, C.R. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc. Natl. Acad. Sci. USA 2006, 103, 6676–6681. [Google Scholar] [CrossRef]
- Passaro, A.; Miselli, M.A.; Sanz, J.M.; Dalla Nora, E.; Morieri, M.L.; Colonna, R.; Pisot, R.; Zuliani, G. Gene expression regional differences in human subcutaneous adipose tissue. BMC Genom. 2017, 18, 202. [Google Scholar] [CrossRef]
- Dankel, S.N.; Fadnes, D.J.; Stavrum, A.K.; Stansberg, C.; Holdhus, R.; Hoang, T.; Veum, V.L.; Christensen, B.J.; Vage, V.; Sagen, J.V.; et al. Switch from stress response to homeobox transcription factors in adipose tissue after profound fat loss. PLoS ONE 2010, 5, e11033. [Google Scholar] [CrossRef]
- Parrillo, L.; Costa, V.; Raciti, G.A.; Longo, M.; Spinelli, R.; Esposito, R.; Nigro, C.; Vastolo, V.; Desiderio, A.; Zatterale, F.; et al. Hoxa5 undergoes dynamic DNA methylation and transcriptional repression in the adipose tissue of mice exposed to high-fat diet. Int. J. Obes. 2016, 40, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Ghaben, A.L.; Scherer, P.E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019, 20, 242–258. [Google Scholar] [CrossRef] [PubMed]
- Parrillo, L.; Spinelli, R.; Costanzo, M.; Florese, P.; Cabaro, S.; Desiderio, A.; Prevenzano, I.; Raciti, G.A.; Smith, U.; Miele, C.; et al. Epigenetic Dysregulation of the Homeobox A5 (HOXA5) Gene Associates with Subcutaneous Adipocyte Hypertrophy in Human Obesity. Cells 2022, 11, 728. [Google Scholar] [CrossRef] [PubMed]
- Jakab, J.; Miskic, B.; Miksic, S.; Juranic, B.; Cosic, V.; Schwarz, D.; Vcev, A. Adipogenesis as a Potential Anti-Obesity Target: A Review of Pharmacological Treatment and Natural Products. Diabetes Metab. Syndr. Obes. 2021, 14, 67–83. [Google Scholar] [CrossRef]
- Zatterale, F.; Raciti, G.A.; Prevenzano, I.; Leone, A.; Campitelli, M.; De Rosa, V.; Beguinot, F.; Parrillo, L. Epigenetic Reprogramming of the Inflammatory Response in Obesity and Type 2 Diabetes. Biomolecules 2022, 12, 982. [Google Scholar] [CrossRef]
- Ronn, T.; Perfilyev, A.; Jonsson, J.; Eriksson, K.F.; Jorgensen, S.W.; Brons, C.; Gillberg, L.; Vaag, A.; Stener-Victorin, E.; Ling, C. Circulating triglycerides are associated with human adipose tissue DNA methylation of genes linked to metabolic disease. Hum. Mol. Genet. 2023, 32, 1875–1887. [Google Scholar] [CrossRef]
- Parrillo, L.; Spinelli, R.; Nicolo, A.; Longo, M.; Mirra, P.; Raciti, G.A.; Miele, C.; Beguinot, F. Nutritional Factors, DNA Methylation, and Risk of Type 2 Diabetes and Obesity: Perspectives and Challenges. Int. J. Mol. Sci. 2019, 20, 2983. [Google Scholar] [CrossRef]
- Carless, M.A.; Kulkarni, H.; Kos, M.Z.; Charlesworth, J.; Peralta, J.M.; Goring, H.H.; Curran, J.E.; Almasy, L.; Dyer, T.D.; Comuzzie, A.G.; et al. Genetic effects on DNA methylation and its potential relevance for obesity in Mexican Americans. PLoS ONE 2013, 8, e73950. [Google Scholar] [CrossRef]
- Paul, R.; Peraldi, R.; Kmita, M. The pioneering function of the hox transcription factors. Semin. Cell Dev. Biol. 2022, 152–153, 85–92. [Google Scholar] [CrossRef]
- Jeannotte, L.; Lemieux, M.; Charron, J.; Poirier, F.; Robertson, E.J. Specification of axial identity in the mouse: Role of the Hoxa-5 (Hox1.3) gene. Genes. Dev. 1993, 7, 2085–2096. [Google Scholar] [CrossRef]
- Aubin, J.; Lemieux, M.; Tremblay, M.; Berard, J.; Jeannotte, L. Early postnatal lethality in Hoxa-5 mutant mice is attributable to respiratory tract defects. Dev. Biol. 1997, 192, 432–445. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, W.V. Transcription factors and pattern formation in the developing lung. Am. J. Physiol. 1995, 269, L429–L442. [Google Scholar] [CrossRef] [PubMed]
- Kinkead, R.; LeBlanc, M.; Gulemetova, R.; Lalancette-Hebert, M.; Lemieux, M.; Mandeville, I.; Jeannotte, L. Respiratory adaptations to lung morphological defects in adult mice lacking Hoxa5 gene function. Pediatr. Res. 2004, 56, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Golpon, H.A.; Geraci, M.W.; Moore, M.D.; Miller, H.L.; Miller, G.J.; Tuder, R.M.; Voelkel, N.F. HOX genes in human lung: Altered expression in primary pulmonary hypertension and emphysema. Am. J. Pathol. 2001, 158, 955–966. [Google Scholar] [CrossRef]
- Boucherat, O.; Franco-Montoya, M.L.; Thibault, C.; Incitti, R.; Chailley-Heu, B.; Delacourt, C.; Bourbon, J.R. Gene expression profiling in lung fibroblasts reveals new players in alveolarization. Physiol. Genom. 2007, 32, 128–141. [Google Scholar] [CrossRef]
- Liu, X.H.; Lu, K.H.; Wang, K.M.; Sun, M.; Zhang, E.B.; Yang, J.S.; Yin, D.D.; Liu, Z.L.; Zhou, J.; Liu, Z.J.; et al. MicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5. BMC Cancer 2012, 12, 348. [Google Scholar] [CrossRef]
- Shi, W.; Warburton, D. Is COPD in adulthood really so far removed from early development? Eur. Respir. J. 2010, 35, 12–13. [Google Scholar] [CrossRef]
- Breitfeld, J.; Kehr, S.; Muller, L.; Stadler, P.F.; Bottcher, Y.; Bluher, M.; Stumvoll, M.; Kovacs, P. Developmentally Driven Changes in Adipogenesis in Different Fat Depots Are Related to Obesity. Front. Endocrinol. 2020, 11, 138. [Google Scholar] [CrossRef]
- Aubin, J.; Dery, U.; Lemieux, M.; Chailler, P.; Jeannotte, L. Stomach regional specification requires Hoxa5-driven mesenchymal-epithelial signaling. Development 2002, 129, 4075–4087. [Google Scholar] [CrossRef] [PubMed]
- Meunier, D.; Aubin, J.; Jeannotte, L. Perturbed thyroid morphology and transient hypothyroidism symptoms in Hoxa5 mutant mice. Dev. Dyn. 2003, 227, 367–378. [Google Scholar] [CrossRef]
- Crooks, G.M.; Fuller, J.; Petersen, D.; Izadi, P.; Malik, P.; Pattengale, P.K.; Kohn, D.B.; Gasson, J.C. Constitutive HOXA5 expression inhibits erythropoiesis and increases myelopoiesis from human hematopoietic progenitors. Blood 1999, 94, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Fuller, J.F.; McAdara, J.; Yaron, Y.; Sakaguchi, M.; Fraser, J.K.; Gasson, J.C. Characterization of HOX gene expression during myelopoiesis: Role of HOX A5 in lineage commitment and maturation. Blood 1999, 93, 3391–3400. [Google Scholar] [CrossRef] [PubMed]
- Ptaschinski, C.; Hrycaj, S.M.; Schaller, M.A.; Wellik, D.M.; Lukacs, N.W. Hox5 Paralogous Genes Modulate Th2 Cell Function during Chronic Allergic Inflammation via Regulation of Gata3. J. Immunol. 2017, 199, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Zhang, T.; Feng, R.; Xia, T.; Huang, H.; Liu, C.; Sun, C. Hoxa5 alleviates obesity-induced chronic inflammation by reducing ER stress and promoting M2 macrophage polarization in mouse adipose tissue. J. Cell. Mol. Med. 2019, 23, 7029–7042. [Google Scholar] [CrossRef] [PubMed]
- Abate-Shen, C. Deregulated homeobox gene expression in cancer: Cause or consequence? Nat. Rev. Cancer 2002, 2, 777–785. [Google Scholar] [CrossRef]
- Henderson, G.S.; van Diest, P.J.; Burger, H.; Russo, J.; Raman, V. Expression pattern of a homeotic gene, HOXA5, in normal breast and in breast tumors. Cell. Oncol. 2006, 28, 305–313. [Google Scholar] [CrossRef]
- Raman, V.; Martensen, S.A.; Reisman, D.; Evron, E.; Odenwald, W.F.; Jaffee, E.; Marks, J.; Sukumar, S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature 2000, 405, 974–978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.L.; Nie, F.Q.; Sun, M.; Xia, R.; Xie, M.; Lu, K.H.; Li, W. HOXA5 indicates poor prognosis and suppresses cell proliferation by regulating p21 expression in non small cell lung cancer. Tumour Biol. 2015, 36, 3521–3531. [Google Scholar] [CrossRef]
- Wang, C.C.; Su, K.Y.; Chen, H.Y.; Chang, S.Y.; Shen, C.F.; Hsieh, C.H.; Hong, Q.S.; Chiang, C.C.; Chang, G.C.; Yu, S.L.; et al. HOXA5 inhibits metastasis via regulating cytoskeletal remodelling and associates with prolonged survival in non-small-cell lung carcinoma. PLoS ONE 2015, 10, e0124191. [Google Scholar] [CrossRef]
- Kim, D.S.; Kim, M.J.; Lee, J.Y.; Lee, S.M.; Choi, J.Y.; Yoon, G.S.; Na, Y.K.; Hong, H.S.; Kim, S.G.; Choi, J.E.; et al. Epigenetic inactivation of Homeobox A5 gene in nonsmall cell lung cancer and its relationship with clinicopathological features. Mol. Carcinog. 2009, 48, 1109–1115. [Google Scholar] [CrossRef]
- Loh, M.; Liem, N.; Vaithilingam, A.; Lim, P.L.; Sapari, N.S.; Elahi, E.; Mok, Z.Y.; Cheng, C.L.; Yan, B.; Pang, B.; et al. DNA methylation subgroups and the CpG island methylator phenotype in gastric cancer: A comprehensive profiling approach. BMC Gastroenterol. 2014, 14, 55. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Bai, Y.; Feng, Z.; Li, W.; Yang, C.; Guo, Y.; Lin, C.; Zhang, Y.; He, Q.; Hu, G.; et al. Study of Promoter Methylation Patterns of HOXA2, HOXA5, and HOXA6 and Its Clinicopathological Characteristics in Colorectal Cancer. Front. Oncol. 2019, 9, 394. [Google Scholar] [CrossRef] [PubMed]
- Ordonez-Moran, P.; Dafflon, C.; Imajo, M.; Nishida, E.; Huelsken, J. HOXA5 Counteracts Stem Cell Traits by Inhibiting Wnt Signaling in Colorectal Cancer. Cancer Cell 2015, 28, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Hwang, S.H.; Song, E.J.; Shin, H.J.; Jung, J.S.; Lee, E.Y. Level of HOXA5 hypermethylation in acute myeloid leukemia is associated with short-term outcome. Korean J. Lab. Med. 2010, 30, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Strathdee, G.; Holyoake, T.L.; Sim, A.; Parker, A.; Oscier, D.G.; Melo, J.V.; Meyer, S.; Eden, T.; Dickinson, A.M.; Mountford, J.C.; et al. Inactivation of HOXA genes by hypermethylation in myeloid and lymphoid malignancy is frequent and associated with poor prognosis. Clin. Cancer Res. 2007, 13, 5048–5055. [Google Scholar] [CrossRef] [PubMed]
- Cimino, P.J.; Kim, Y.; Wu, H.J.; Alexander, J.; Wirsching, H.G.; Szulzewsky, F.; Pitter, K.; Ozawa, T.; Wang, J.; Vazquez, J.; et al. Increased HOXA5 expression provides a selective advantage for gain of whole chromosome 7 in IDH wild-type glioblastoma. Genes. Dev. 2018, 32, 512–523. [Google Scholar] [CrossRef]
- Ding, F.; Chen, P.; Bie, P.; Piao, W.; Cheng, Q. HOXA5 Is Recognized as a Prognostic-Related Biomarker and Promotes Glioma Progression Through Affecting Cell Cycle. Front. Oncol. 2021, 11, 633430. [Google Scholar] [CrossRef]
- Scherer, P.E. Adipose tissue: From lipid storage compartment to endocrine organ. Diabetes 2006, 55, 1537–1545. [Google Scholar] [CrossRef]
- Spinelli, R.; Parrillo, L.; Longo, M.; Florese, P.; Desiderio, A.; Zatterale, F.; Miele, C.; Raciti, G.A.; Beguinot, F. Molecular basis of ageing in chronic metabolic diseases. J. Endocrinol. Investig. 2020, 43, 1373–1389. [Google Scholar] [CrossRef]
- Parrillo, L.; Spinelli, R.; Longo, M.; Desiderio, A.; Mirra, P.; Nigro, C.; Fiory, F.; Hedjazifar, S.; Mutarelli, M.; Carissimo, A.; et al. Altered PTPRD DNA methylation associates with restricted adipogenesis in healthy first-degree relatives of Type 2 diabetes subjects. Epigenomics 2020, 12, 873–888. [Google Scholar] [CrossRef]
- Longo, M.; Zatterale, F.; Naderi, J.; Parrillo, L.; Formisano, P.; Raciti, G.A.; Beguinot, F.; Miele, C. Adipose Tissue Dysfunction as Determinant of Obesity-Associated Metabolic Complications. Int. J. Mol. Sci. 2019, 20, 2358. [Google Scholar] [CrossRef] [PubMed]
- Siersbaek, R.; Mandrup, S. Transcriptional networks controlling adipocyte differentiation. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Karastergiou, K.; Fried, S.K.; Xie, H.; Lee, M.J.; Divoux, A.; Rosencrantz, M.A.; Chang, R.J.; Smith, S.R. Distinct developmental signatures of human abdominal and gluteal subcutaneous adipose tissue depots. J. Clin. Endocrinol. Metab. 2013, 98, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Xu, Y.; Luo, D.; Saeed, M.; Sun, C. Hoxa5 Promotes Adipose Differentiation via Increasing DNA Methylation Level and Inhibiting PKA/HSL Signal Pathway in Mice. Cell. Physiol. Biochem. 2018, 45, 1023–1033. [Google Scholar] [CrossRef]
- Koga, T.; Matsui, Y.; Asagiri, M.; Kodama, T.; de Crombrugghe, B.; Nakashima, K.; Takayanagi, H. NFAT and Osterix cooperatively regulate bone formation. Nat. Med. 2005, 11, 880–885. [Google Scholar] [CrossRef]
- Jia, B.; Wang, Z.; Sun, X.; Chen, J.; Zhao, J.; Qiu, X. Long noncoding RNA LINC00707 sponges miR-370-3p to promote osteogenesis of human bone marrow-derived mesenchymal stem cells through upregulating WNT2B. Stem Cell Res. Ther. 2019, 10, 67. [Google Scholar] [CrossRef]
- Christodoulides, C.; Lagathu, C.; Sethi, J.K.; Vidal-Puig, A. Adipogenesis and WNT signalling. Trends Endocrinol. Metab. 2009, 20, 16–24. [Google Scholar] [CrossRef]
- Osuga, J.; Ishibashi, S.; Oka, T.; Yagyu, H.; Tozawa, R.; Fujimoto, A.; Shionoiri, F.; Yahagi, N.; Kraemer, F.B.; Tsutsumi, O.; et al. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc. Natl. Acad. Sci. USA 2000, 97, 787–792. [Google Scholar] [CrossRef]
- Holzman, M.A.; Ryckman, A.; Finkelstein, T.M.; Landry-Truchon, K.; Schindler, K.A.; Bergmann, J.M.; Jeannotte, L.; Mansfield, J.H. HOXA5 Participates in Brown Adipose Tissue and Epaxial Skeletal Muscle Patterning and in Brown Adipocyte Differentiation. Front. Cell Dev. Biol. 2021, 9, 632303. [Google Scholar] [CrossRef]
- Osorio-Conles, O.; Ibarzabal, A.; Balibrea, J.M.; Vidal, J.; Ortega, E.; de Hollanda, A. FABP4 Expression in Subcutaneous Adipose Tissue Is Independently Associated with Circulating Triglycerides in Obesity. J. Clin. Med. 2023, 12, 1013. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.; Yu, H.; Liu, Y.; Song, H.; Tian, X.; Liu, D.; Yan, C.; Han, Y. HOXA5-miR-574-5p axis promotes adipogenesis and alleviates insulin resistance. Mol. Ther. Nucleic Acids 2022, 27, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Alkhouri, N.; Gornicka, A.; Berk, M.P.; Thapaliya, S.; Dixon, L.J.; Kashyap, S.; Schauer, P.R.; Feldstein, A.E. Adipocyte apoptosis, a link between obesity, insulin resistance, and hepatic steatosis. J. Biol. Chem. 2010, 285, 3428–3438. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Ren, Q.; Wu, S.; Saeed, M.; Sun, C. Hoxa5 increases mitochondrial apoptosis by inhibiting Akt/mTORC1/S6K1 pathway in mice white adipocytes. Oncotarget 2017, 8, 95332–95345. [Google Scholar] [CrossRef] [PubMed]
- Gesta, S.; Tseng, Y.H.; Kahn, C.R. Developmental origin of fat: Tracking obesity to its source. Cell 2007, 131, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Gesta, S.; Lee, K.Y.; Tran, T.T.; Saadatirad, P.; Kahn, C.R. Adipose depots possess unique developmental gene signatures. Obesity 2010, 18, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Arner, P.; Arner, E.; Hammarstedt, A.; Smith, U. Genetic predisposition for Type 2 diabetes, but not for overweight/obesity, is associated with a restricted adipogenesis. PLoS ONE 2011, 6, e18284. [Google Scholar] [CrossRef]
- Lee, Y.H.; Nair, S.; Rousseau, E.; Allison, D.B.; Page, G.P.; Tataranni, P.A.; Bogardus, C.; Permana, P.A. Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima Indians: Increased expression of inflammation-related genes. Diabetologia 2005, 48, 1776–1783. [Google Scholar] [CrossRef]
- Nono Nankam, P.A.; Bluher, M.; Kehr, S.; Kloting, N.; Krohn, K.; Adams, K.; Stadler, P.F.; Mendham, A.E.; Goedecke, J.H. Distinct abdominal and gluteal adipose tissue transcriptome signatures are altered by exercise training in African women with obesity. Sci. Rep. 2020, 10, 10240. [Google Scholar] [CrossRef]
- Wang, K.C.; Helms, J.A.; Chang, H.Y. Regeneration, repair and remembering identity: The three Rs of Hox gene expression. Trends Cell Biol. 2009, 19, 268–275. [Google Scholar] [CrossRef]
- Cao, W.; Huang, H.; Xia, T.; Liu, C.; Muhammad, S.; Sun, C. Homeobox a5 Promotes White Adipose Tissue Browning Through Inhibition of the Tenascin C/Toll-Like Receptor 4/Nuclear Factor Kappa B Inflammatory Signaling in Mice. Front. Immunol. 2018, 9, 647. [Google Scholar] [CrossRef]
- Jing, Y.; Gao, B.; Han, Z.; Xin, S. HOXA5 induces M2 macrophage polarization to attenuate carotid atherosclerosis by activating MED1. IUBMB Life 2021, 73, 1142–1152. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y. MED1 is a lipogenesis coactivator required for postnatal adipose expansion. FASEB J. 2021, 35, 713–728. [Google Scholar] [CrossRef]
- Mandeville, I.; Aubin, J.; LeBlanc, M.; Lalancette-Hebert, M.; Janelle, M.F.; Tremblay, G.M.; Jeannotte, L. Impact of the loss of Hoxa5 function on lung alveogenesis. Am. J. Pathol. 2006, 169, 1312–1327. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ren, Y.; Chang, K.; Wu, W.; Griffiths, H.R.; Lu, S.; Gao, D. Adipose tissue macrophages as potential targets for obesity and metabolic diseases. Front. Immunol. 2023, 14, 1153915. [Google Scholar] [CrossRef] [PubMed]
- Stone, T.W.; McPherson, M.; Gail Darlington, L. Obesity and Cancer: Existing and New Hypotheses for a Causal Connection. EBioMedicine 2018, 30, 14–28. [Google Scholar] [CrossRef]
- Pai, P.; Wang, G.; Teo, W.W.; Raez-Rodriguez, D.; Gabrielson, K.L.; Gyorffy, B.; Downs, B.M.; Aggarwal, A.; Sukumar, S. HOXA5-Mediated Stabilization of IkappaBalpha Inhibits the NF-kappaB Pathway and Suppresses Malignant Transformation of Breast Epithelial Cells. Cancer Res. 2022, 82, 3802–3814. [Google Scholar] [CrossRef]
- Ma, H.M.; Cui, N.; Zheng, P.S. HOXA5 inhibits the proliferation and neoplasia of cervical cancer cells via downregulating the activity of the Wnt/beta-catenin pathway and transactivating TP53. Cell Death Dis. 2020, 11, 420. [Google Scholar] [CrossRef]
- Henninger, A.M.; Eliasson, B.; Jenndahl, L.E.; Hammarstedt, A. Adipocyte hypertrophy, inflammation and fibrosis characterize subcutaneous adipose tissue of healthy, non-obese subjects predisposed to type 2 diabetes. PLoS ONE 2014, 9, e105262. [Google Scholar] [CrossRef]
- Szabo, M.; Mate, B.; Csep, K.; Benedek, T. Epigenetic Modifications Linked to T2D, the Heritability Gap, and Potential Therapeutic Targets. Biochem. Genet. 2018, 56, 553–574. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Formichi, C.; Nigi, L.; Grieco, G.E.; Maccora, C.; Fignani, D.; Brusco, N.; Licata, G.; Sebastiani, G.; Dotta, F. Non-Coding RNAs: Novel Players in Insulin Resistance and Related Diseases. Int. J. Mol. Sci. 2021, 22, 7716. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Lin, Z.J.; Li, C.C.; Lin, X.; Shan, S.K.; Guo, B.; Zheng, M.H.; Li, F.; Yuan, L.Q.; Li, Z.H. Epigenetic regulation in metabolic diseases: Mechanisms and advances in clinical study. Signal Transduct. Target. Ther. 2023, 8, 98. [Google Scholar] [CrossRef] [PubMed]
- Werge, M.P.; McCann, A.; Galsgaard, E.D.; Holst, D.; Bugge, A.; Albrechtsen, N.J.W.; Gluud, L.L. The Role of the Transsulfuration Pathway in Non-Alcoholic Fatty Liver Disease. J. Clin. Med. 2021, 10, 1081. [Google Scholar] [CrossRef]
- Hershko, A.Y.; Kafri, T.; Fainsod, A.; Razin, A. Methylation of HoxA5 and HoxB5 and its relevance to expression during mouse development. Gene 2003, 302, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.; Ronn, T. Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab. 2019, 29, 1028–1044. [Google Scholar] [CrossRef] [PubMed]
- Raciti, G.A.; Spinelli, R.; Desiderio, A.; Longo, M.; Parrillo, L.; Nigro, C.; D'Esposito, V.; Mirra, P.; Fiory, F.; Pilone, V.; et al. Specific CpG hyper-methylation leads to Ankrd26 gene down-regulation in white adipose tissue of a mouse model of diet-induced obesity. Sci. Rep. 2017, 7, 43526. [Google Scholar] [CrossRef]
- Willmer, T.; Johnson, R.; Louw, J.; Pheiffer, C. Blood-Based DNA Methylation Biomarkers for Type 2 Diabetes: Potential for Clinical Applications. Front. Endocrinol. 2018, 9, 744. [Google Scholar] [CrossRef]
- Gillberg, L.; Ling, C. The potential use of DNA methylation biomarkers to identify risk and progression of type 2 diabetes. Front. Endocrinol. 2015, 6, 43. [Google Scholar] [CrossRef] [PubMed]
- Weyer, C.; Foley, J.E.; Bogardus, C.; Tataranni, P.A.; Pratley, R.E. Enlarged subcutaneous abdominal adipocyte size, but not obesity itself, predicts type II diabetes independent of insulin resistance. Diabetologia 2000, 43, 1498–1506. [Google Scholar] [CrossRef]
- Cheng, Y.J.; Kanaya, A.M.; Araneta, M.R.G.; Saydah, S.H.; Kahn, H.S.; Gregg, E.W.; Fujimoto, W.Y.; Imperatore, G. Prevalence of Diabetes by Race and Ethnicity in the United States, 2011–2016. JAMA 2019, 322, 2389–2398. [Google Scholar] [CrossRef]
- Chilunga, F.P.; Meeks, K.A.C.; Henneman, P.; Agyemang, C.; Doumatey, A.P.; Rotimi, C.N.; Adeyemo, A.A. An epigenome-wide association study of insulin resistance in African Americans. Clin. Epigenetics 2022, 14, 88. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, H.; Kos, M.Z.; Neary, J.; Dyer, T.D.; Kent, J.W., Jr.; Goring, H.H.; Cole, S.A.; Comuzzie, A.G.; Almasy, L.; Mahaney, M.C.; et al. Novel epigenetic determinants of type 2 diabetes in Mexican-American families. Hum. Mol. Genet. 2015, 24, 5330–5344. [Google Scholar] [CrossRef] [PubMed]
- Nepali, K.; Liou, J.P. Recent developments in epigenetic cancer therapeutics: Clinical advancement and emerging trends. J. Biomed. Sci. 2021, 28, 27. [Google Scholar] [CrossRef]
- Bernard, H.; Teijeiro, A.; Chaves-Perez, A.; Perna, C.; Satish, B.; Novials, A.; Wang, J.P.; Djouder, N. Coxsackievirus B Type 4 Infection in beta Cells Downregulates the Chaperone Prefoldin URI to Induce a MODY4-like Diabetes via Pdx1 Silencing. Cell Rep. Med. 2020, 1, 100125. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Chen, X.; Nie, S.; Chang, Y.; Meng, F.; Zhou, J.; Mao, C.; Li, T.; Yan, X.; Huang, J.; et al. Decitabine: An effective and safe treatment for myelodysplastic syndrome and acute myeloid leukemia. J. Cancer Res. Ther. 2019, 15, 1471–1476. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Tang, D.; Du, Y.L.; Cao, C.Y.; Nie, Y.Q.; Cao, J.; Zhou, Y.J. Fatty liver mediated by peroxisome proliferator-activated receptor-alpha DNA methylation can be reversed by a methylation inhibitor and curcumin. J. Dig. Dis. 2018, 19, 421–430. [Google Scholar] [CrossRef]
- Gao, J.; Cheng, Y.; Hao, H.; Yin, Y.; Xue, J.; Zhang, Q.; Li, L.; Liu, J.; Xie, Z.; Yu, S.; et al. Decitabine assists umbilical cord-derived mesenchymal stem cells in improving glucose homeostasis by modulating macrophage polarization in type 2 diabetic mice. Stem Cell Res. Ther. 2019, 10, 259. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L.; Zhang, M.; Huang, K.; Yao, Z.; Rao, P.; Cai, X.; Xiao, J. Advanced glycation end products inhibit the osteogenic differentiation potential of adipose-derived stem cells by modulating Wnt/beta-catenin signalling pathway via DNA methylation. Cell Prolif. 2020, 53, e12834. [Google Scholar] [CrossRef]
| Position of DNA Methylation Change | Tissue | Disease and/or Clinical Manifestation | Reference |
|---|---|---|---|
| Promoter region | Peripheral blood leukocytes | Adipocyte hypertrophy; family history and risk of T2D | [13] |
| Promoter region | Peripheral blood leukocytes | Obesity; BMI | [13] |
| TSS 1500 | Peripheral blood | Obesity; BMI | [18] |
| cg14013695 | Whole blood | IR; HOMA-IR index | [91] |
| cg14013695 | Peripheral blood cells | T2D risk | [92] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parrillo, L.; Spinelli, R.; Longo, M.; Zatterale, F.; Santamaria, G.; Leone, A.; Campitelli, M.; Raciti, G.A.; Beguinot, F. The Transcription Factor HOXA5: Novel Insights into Metabolic Diseases and Adipose Tissue Dysfunction. Cells 2023, 12, 2090. https://doi.org/10.3390/cells12162090
Parrillo L, Spinelli R, Longo M, Zatterale F, Santamaria G, Leone A, Campitelli M, Raciti GA, Beguinot F. The Transcription Factor HOXA5: Novel Insights into Metabolic Diseases and Adipose Tissue Dysfunction. Cells. 2023; 12(16):2090. https://doi.org/10.3390/cells12162090
Chicago/Turabian StyleParrillo, Luca, Rosa Spinelli, Michele Longo, Federica Zatterale, Gianluca Santamaria, Alessia Leone, Michele Campitelli, Gregory Alexander Raciti, and Francesco Beguinot. 2023. "The Transcription Factor HOXA5: Novel Insights into Metabolic Diseases and Adipose Tissue Dysfunction" Cells 12, no. 16: 2090. https://doi.org/10.3390/cells12162090
APA StyleParrillo, L., Spinelli, R., Longo, M., Zatterale, F., Santamaria, G., Leone, A., Campitelli, M., Raciti, G. A., & Beguinot, F. (2023). The Transcription Factor HOXA5: Novel Insights into Metabolic Diseases and Adipose Tissue Dysfunction. Cells, 12(16), 2090. https://doi.org/10.3390/cells12162090









