Breast Cancer Cell Type and Biomechanical Properties of Decellularized Mouse Organs Drives Tumor Cell Colonization
Abstract
1. Introduction
2. Materials and Methods
2.1. Laboratory Animals and Organ Harvesting
2.2. Decellularization of the Organ Samples
2.3. Total DNA Content Analysis
2.4. Cell Culture
2.5. Recellularization of the Matrices
2.6. 3D Tumor Growth Models in Collagen Hydrogel
2.7. Histomorphological Analysis
2.8. Scanning Electron Microscopy
2.9. Macroindentation
2.10. Atomic Force Microscopy (Nanoindentation)
2.11. Rheometry
3. Results
3.1. Obtaining DCL Matrices of Murine Organs
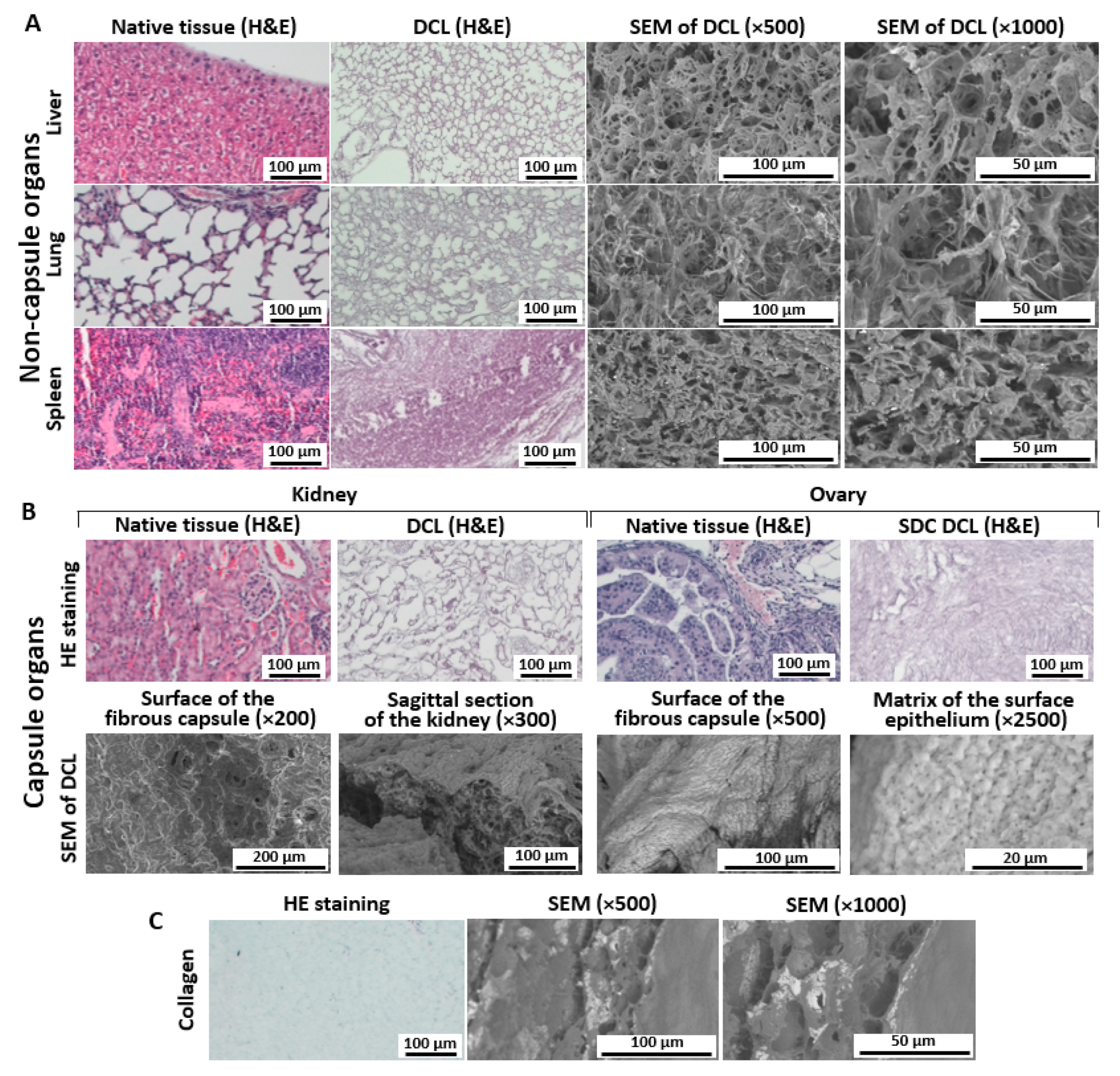
3.2. Histological Analysis of Decellularized Matrices
3.2.1. Liver
3.2.2. Lung
3.2.3. Spleen
3.2.4. Kidney
3.2.5. Ovary
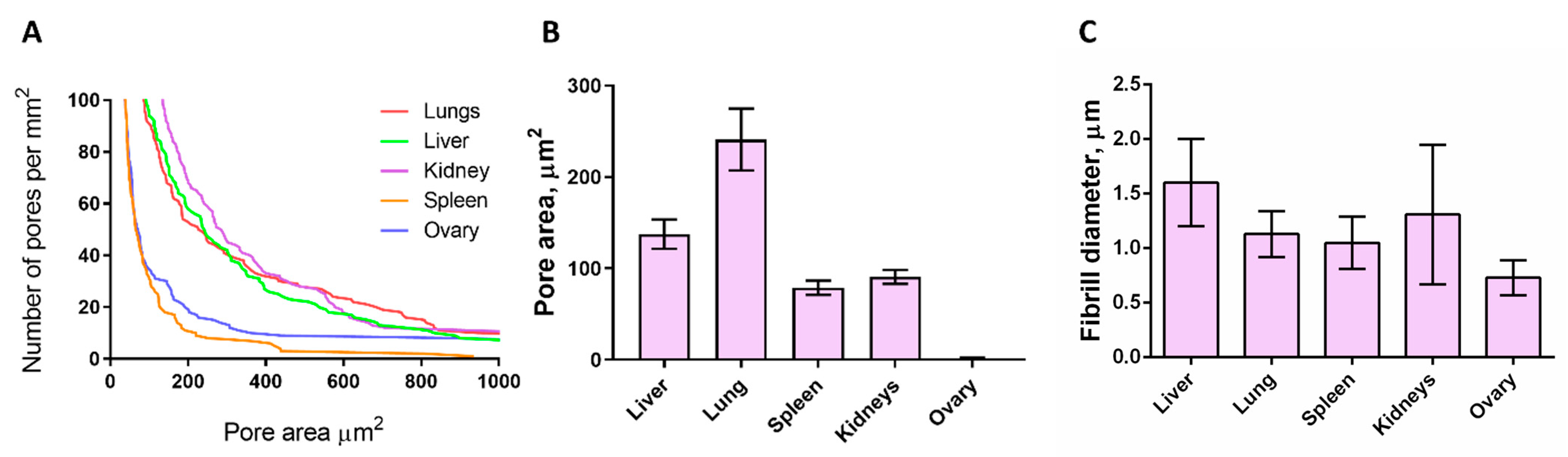
3.3. SEM Analysis of Matrices
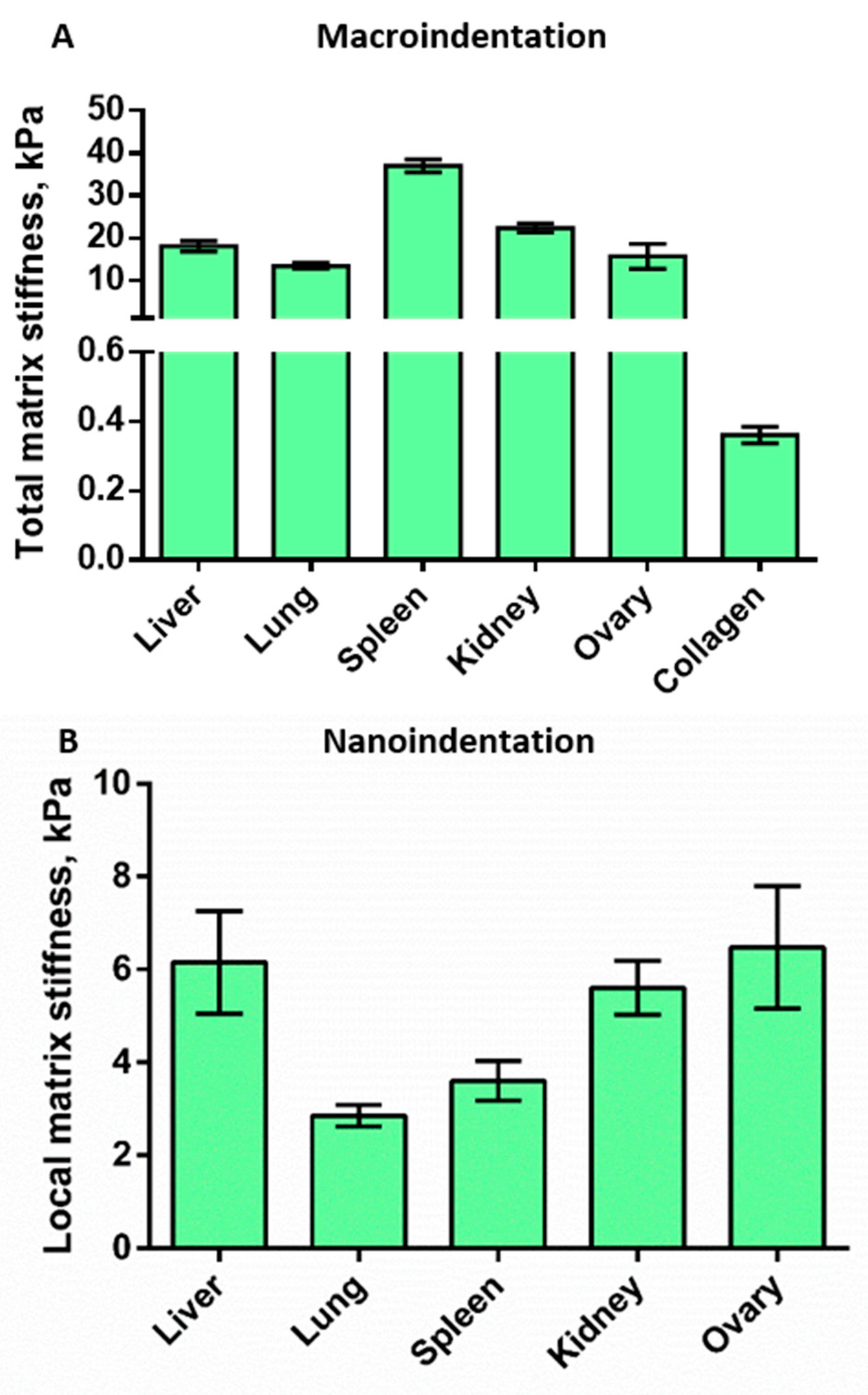
3.4. Mechanical Properties of the Decellularized Matrices
3.4.1. Macroindentation
3.4.2. Nanoindentation
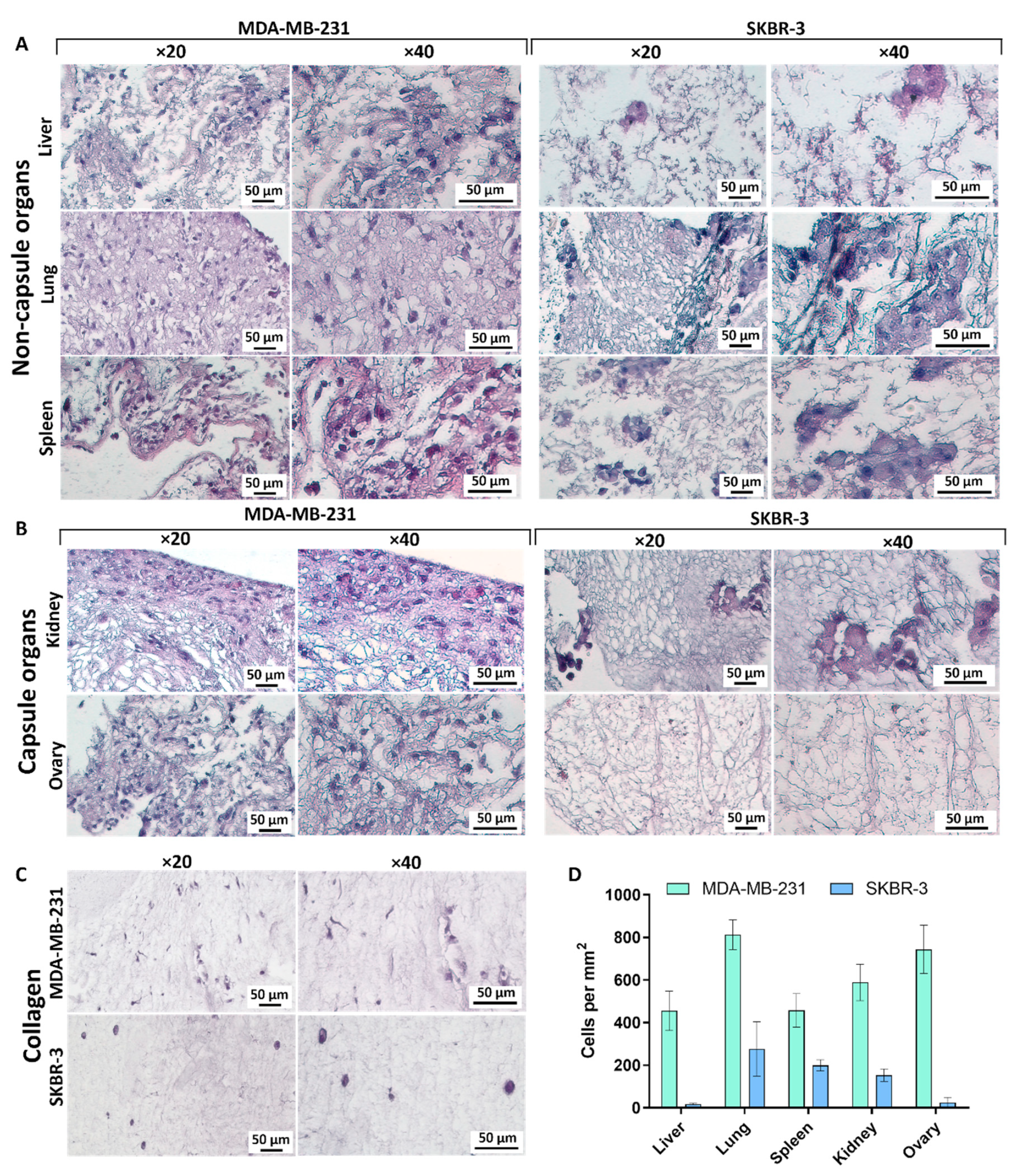
3.5. Repopulation of the Decellularized Matrices
3.5.1. Liver
3.5.2. Lungs
3.5.3. Spleen
3.5.4. Kidney
3.5.5. Ovary
3.5.6. Collagen
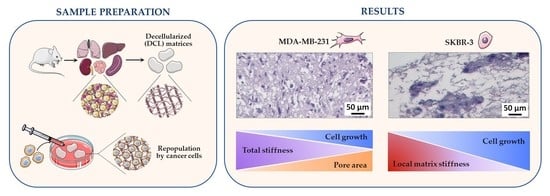
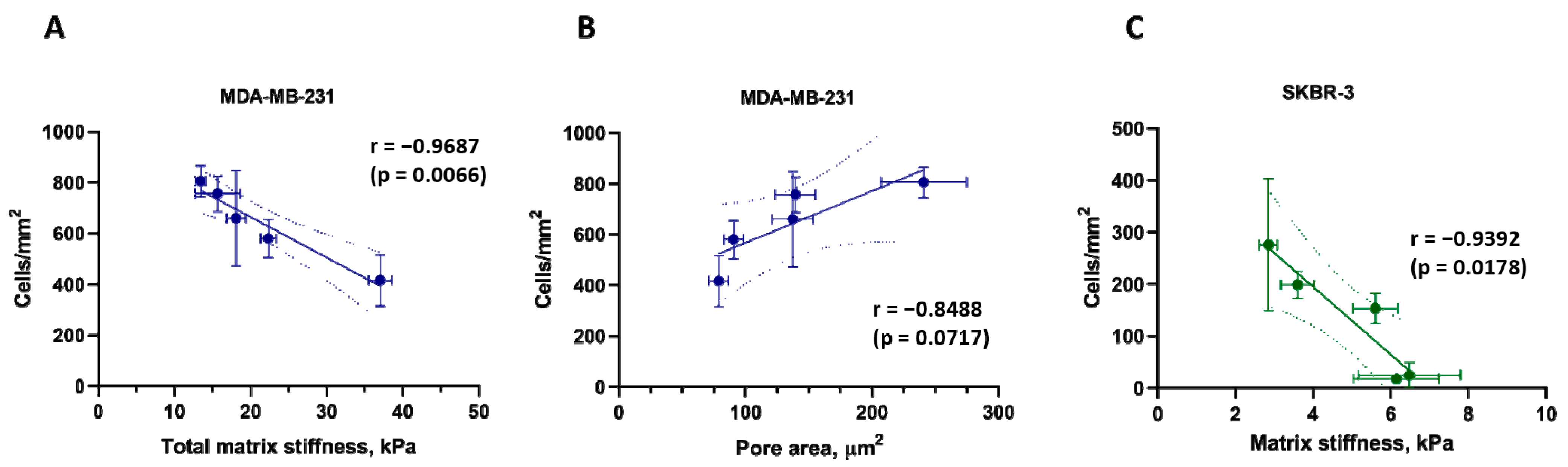
3.6. Correlations between Matrix Properties and Cancer Cell Growth
4. Discussion
4.1. Decellularization Protocol for Murine Organs
4.2. Biomechanical and Structural Properties of DCL Matrices of Murine Organs
4.3. The Current Ideas about Stiffness-Dependent Behavior of Cells on 2D and 3D Substrates
4.4. Behavior of Breast Cancer Cells in DCL Matrices of Murine Organs with Different Biomechanical Characteristics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shimpi, A.A.; Fischbach, C. Engineered ECM models: Opportunities to advance understanding of tumor heterogeneity. Curr. Opin. Cell Biol. 2021, 72, 1–9. [Google Scholar] [CrossRef]
- Kutova, O.M.; Pospelov, A.D.; Balalaeva, I.V. The Multifaceted Role of Connexins in Tumor Microenvironment Initiation and Maintenance. Biology 2023, 12, 204. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular matrix structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef]
- Nicolas-Boluda, A.; Vaquero, J.; Vimeux, L.; Guilbert, T.; Barrin, S.; Kantari-Mimoun, C.; Ponzo, M.; Renault, G.; Deptula, P.; Pogoda, K.; et al. Tumor stiffening reversion through collagen crosslinking inhibition improves T cell migration and anti-PD-1 treatment. eLife 2021, 10, 58688. [Google Scholar] [CrossRef] [PubMed]
- Cox, T.R. The matrix in cancer. Nat. Rev. Cancer 2021, 21, 217–238. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Lee, C.; Park, J. Extracellular matrix remodeling facilitates obesity-associated cancer progression. Trends Cell Biol. 2022, 32, 825–834. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Manou, D.; Karamanos, N.K. The extracellular matrix as a multitasking player in disease. FEBS J. 2019, 286, 2830–2869. [Google Scholar] [CrossRef]
- Lepucki, A.; Orlińska, K.; Mielczarek-Palacz, A.; Kabut, J.; Olczyk, P.; Komosińska-Vassev, K. The Role of Extracellular Matrix Proteins in Breast Cancer. J. Clin. Med. 2022, 11, 1250. [Google Scholar] [CrossRef]
- Northcott, J.M.; Dean, I.S.; Mouw, J.K.; Weaver, V.M. Feeling Stress: The Mechanics of Cancer Progression and Aggression. Front. Cell Dev. Biol. 2018, 6, 17. [Google Scholar] [CrossRef]
- Walker, C.; Mojares, E.; Del Río Hernández, A. Role of Extracellular Matrix in Development and Cancer Progression. Int. J. Mol. Sci. 2018, 19, 3028. [Google Scholar] [CrossRef]
- Mierke, C.T. The matrix environmental and cell mechanical properties regulate cell migration and contribute to the invasive phenotype of cancer cells. Reports on progress in physics. Phys. Soc. 2019, 82, 064602. [Google Scholar] [CrossRef]
- Espina, J.A.; Marchant, C.L.; Barriga, E.H. Durotaxis: The mechanical control of directed cell migration. FEBS J. 2022, 289, 2736–2754. [Google Scholar] [CrossRef] [PubMed]
- Reuten, R.; Mayorca-Guiliani, A.E.; Erler, J.T. Matritecture: Mapping the extracellular matrix architecture during health and disease. Matrix Biol. Plus 2022, 14, 100102. [Google Scholar] [CrossRef] [PubMed]
- Zanotelli, M.R.; Zhang, J.; Reinhart-King, C.A. Mechanoresponsive metabolism in cancer cell migration and metastasis. Cell Metab. 2021, 33, 1307–1321. [Google Scholar] [CrossRef]
- Choudhury, D.; Yee, M.; Sheng, Z.L.J.; Amirul, A.; Naing, M.W. Decellularization systems and devices: State-of-the-art. Acta Biomater. 2020, 115, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Pospelov, A.D.; Timofeeva, L.B.; Cherkasova, E.I.; Balalaeva, I.V. Comparative Analysis of Two Protocols of Mouse Tissues Decellularization for Application in Experimental Oncology. Opera Medica Physiol. 2020, 7, 13–21. [Google Scholar] [CrossRef]
- Lv, Y.; Wang, H.; Li, G.; Zhao, B. Three-dimensional decellularized tumor extracellular matrices with different stiffness as bioengineered tumor scaffolds. Bioact. Mater. 2021, 6, 2767–2782. [Google Scholar] [CrossRef]
- van Tienderen, G.S.; Rosmark, O.; Lieshout, R.; Willemse, J.; de Weijer, F.; Elowsson Rendin, L.; Westergren-Thorsson, G.; Doukas, M.; Groot Koerkamp, B.; van Royen, M.E.; et al. Extracellular matrix drives tumor organoids toward desmoplastic matrix deposition and mesenchymal transition. Acta Biomater. 2023, 158, 115–131. [Google Scholar] [CrossRef]
- Wilcox, H.E.; Korossis, S.A.; Booth, C.; Watterson, K.G.; Kearney, J.N.; Fisher, J.; Ingham, E. Biocompatibility and recellularization potential of an acellular porcine heart valve matrix. J. Heart Valve Dis. 2005, 14, 228–236, discussion 236-227. [Google Scholar]
- Vukadinovic-Nikolic, Z.; Andrée, B.; Dorfman, S.E.; Pflaum, M.; Horvath, T.; Lux, M.; Venturini, L.; Bär, A.; Kensah, G.; Lara, A.R.; et al. Generation of bioartificial heart tissue by combining a three-dimensional gel-based cardiac construct with decellularized small intestinal submucosa. Tissue Eng. Part A 2014, 20, 799–809. [Google Scholar] [CrossRef]
- Kutova, O.M.; Sencha, L.M.; Pospelov, A.D.; Dobrynina, O.E.; Brilkina, A.A.; Cherkasova, E.I.; Balalaeva, I.V. Comparative Analysis of Cell-Cell Contact Abundance in Ovarian Carcinoma Cells Cultured in Two- and Three-Dimensional In Vitro Models. Biology 2020, 9, 446. [Google Scholar] [CrossRef]
- Efremov, Y.M.; Shpichka, A.I.; Kotova, S.L.; Timashev, P.S. Viscoelastic mapping of cells based on fast force volume and PeakForce Tapping. Soft Matter 2019, 15, 5455–5463. [Google Scholar] [CrossRef] [PubMed]
- Bikmulina, P.; Kosheleva, N.; Efremov, Y.; Antoshin, A.; Heydari, Z.; Kapustina, V.; Royuk, V.; Mikhaylov, V.; Fomin, V.; Vosough, M.; et al. 3D or not 3D: A guide to assess cell viability in 3D cell systems. Soft Matter 2022, 18, 2222–2233. [Google Scholar] [CrossRef] [PubMed]
- Mayorca-Guiliani, A.E.; Rafaeva, M.; Willacy, O.; Madsen, C.D.; Reuten, R.; Erler, J.T. Decellularization of the Murine Cardiopulmonary Complex. J. Vis. Exp. JoVE 2021, 171, e61854. [Google Scholar] [CrossRef]
- Miranda, C.; Leonel, L.; Cañada, R.R.; Maria, D.A.; Miglino, M.A.; Del Sol, M.; Lobo, S.E. Effects of chemical and physical methods on decellularization of murine skeletal muscles. An. Acad. Bras. Cienc. 2021, 93, e20190942. [Google Scholar] [CrossRef] [PubMed]
- Crapo, P.M.; Gilbert, T.W.; Badylak, S.F. An overview of tissue and whole organ decellularization processes. Biomaterials 2011, 32, 3233–3243. [Google Scholar] [CrossRef] [PubMed]
- Somuncu, Ö.S. Decellularization Concept in Regenerative Medicine. Adv. Exp. Med. Biol. 2020, 1212, 71–85. [Google Scholar] [CrossRef]
- Edgar, L.; Pu, T.; Porter, B.; Aziz, J.M.; La Pointe, C.; Asthana, A.; Orlando, G. Regenerative medicine, organ bioengineering and transplantation. Br. J. Surg. 2020, 107, 793–800. [Google Scholar] [CrossRef]
- Akram, M.; Iqbal, M.; Daniyal, M.; Khan, A.U. Awareness and current knowledge of breast cancer. Biol. Res. 2017, 50, 33. [Google Scholar] [CrossRef]
- Boughton, O.R.; Ma, S.; Zhao, S.; Arnold, M.; Lewis, A.; Hansen, U.; Cobb, J.P.; Giuliani, F.; Abel, R.L. Measuring bone stiffness using spherical indentation. PLoS ONE 2018, 13, e0200475. [Google Scholar] [CrossRef]
- Levental, I.; Levental, K.R.; Klein, E.A.; Assoian, R.; Miller, R.T.; Wells, R.G.; Janmey, P.A. A simple indentation device for measuring micrometer-scale tissue stiffness. J. Phys. Condens. Matter Inst. Phys. J. 2010, 22, 194120. [Google Scholar] [CrossRef]
- Sim, S.; Chevrier, A.; Garon, M.; Quenneville, E.; Lavigne, P.; Yaroshinsky, A.; Hoemann, C.D.; Buschmann, M.D. Electromechanical probe and automated indentation maps are sensitive techniques in assessing early degenerated human articular cartilage. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2017, 35, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Griffin, M.; Naik, A.; Szarko, M.; Butler, P.E.M. Optimising the decellularization of human elastic cartilage with trypsin for future use in ear reconstruction. Sci. Rep. 2018, 8, 3097. [Google Scholar] [CrossRef] [PubMed]
- Tse, J.R.; Long, J.L. Microstructure characterization of a decellularized vocal fold scaffold for laryngeal tissue engineering. Laryngoscope 2014, 124, E326–E331. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, C.F.; Gasperini, L.; Marques, A.P.; Reis, R.L. The stiffness of living tissues and its implications for tissue engineering. Nat. Rev. Mater. 2020, 5, 351–370. [Google Scholar] [CrossRef]
- Jorba, I.; Uriarte, J.J.; Campillo, N.; Farré, R.; Navajas, D. Probing Micromechanical Properties of the Extracellular Matrix of Soft Tissues by Atomic Force Microscopy. J. Cell. Physiol. 2017, 232, 19–26. [Google Scholar] [CrossRef]
- Luque, T.; Melo, E.; Garreta, E.; Cortiella, J.; Nichols, J.; Farré, R.; Navajas, D. Local micromechanical properties of decellularized lung scaffolds measured with atomic force microscopy. Acta Biomater. 2013, 9, 6852–6859. [Google Scholar] [CrossRef]
- Cailleau, R.; Olivé, M.; Cruciger, Q.V. Long-term human breast carcinoma cell lines of metastatic origin: Preliminary characterization. In Vitro 1978, 14, 911–915. [Google Scholar] [CrossRef]
- Bean, M.A.; Hajdu, S.I. Cytological Characterization of Human Tumor Cells from Monolayer Cultures. In Human Tumor Cells In Vitro; Fogh, J., Ed.; Springer: Boston, MA, USA, 1975; pp. 333–358. [Google Scholar]
- Fahad Ullah, M. Breast Cancer: Current Perspectives on the Disease Status. Adv. Exp. Med. Biol. 2019, 1152, 51–64. [Google Scholar] [CrossRef]
- Song, Y.H.; Maynes, M.A.; Hlavac, N.; Visosevic, D.; Daramola, K.O.; Porvasnik, S.L.; Schmidt, C.E. Development of novel apoptosis-assisted lung tissue decellularization methods. Biomater. Sci. 2021, 9, 3485–3498. [Google Scholar] [CrossRef]
- Atat, O.E.; Farzaneh, Z.; Pourhamzeh, M.; Taki, F.; Abi-Habib, R.; Vosough, M.; El-Sibai, M. 3D modeling in cancer studies. Hum. Cell 2022, 35, 23–36. [Google Scholar] [CrossRef]
- Kretzschmar, K. Cancer research using organoid technology. J. Mol. Med. 2021, 99, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Amirghasemi, F.; Adjei-Sowah, E.; Pockaj, B.A.; Nikkhah, M. Microengineered 3D Tumor Models for Anti-Cancer Drug Discovery in Female-Related Cancers. Ann. Biomed. Eng. 2021, 49, 1943–1972. [Google Scholar] [CrossRef] [PubMed]
- Caon, I.; Bartolini, B.; Parnigoni, A.; Caravà, E.; Moretto, P.; Viola, M.; Karousou, E.; Vigetti, D.; Passi, A. Revisiting the hallmarks of cancer: The role of hyaluronan. Semin. Cancer Biol. 2020, 62, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Kechagia, J.Z.; Ivaska, J.; Roca-Cusachs, P. Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Biol. 2019, 20, 457–473. [Google Scholar] [CrossRef] [PubMed]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Mhaidly, R.; Mechta-Grigoriou, F. Fibroblast heterogeneity in tumor micro-environment: Role in immunosuppression and new therapies. Semin. Immunol. 2020, 48, 101417. [Google Scholar] [CrossRef]
- Abdelgaied, A.; Stanley, M.; Galfe, M.; Berry, H.; Ingham, E.; Fisher, J. Comparison of the biomechanical tensile and compressive properties of decellularised and natural porcine meniscus. J. Biomech. 2015, 48, 1389–1396. [Google Scholar] [CrossRef] [PubMed]
- Khorramirouz, R.; Sabetkish, S.; Akbarzadeh, A.; Muhammadnejad, A.; Heidari, R.; Kajbafzadeh, A.M. Effect of three decellularisation protocols on the mechanical behaviour and structural properties of sheep aortic valve conduits. Adv. Med. Sci. 2014, 59, 299–307. [Google Scholar] [CrossRef]
- Hoshiba, T. Decellularized Extracellular Matrix for Cancer Research. Materials 2019, 12, 1311. [Google Scholar] [CrossRef]
- Fan, Y.; Sun, Q.; Li, X.; Feng, J.; Ao, Z.; Li, X.; Wang, J. Substrate Stiffness Modulates the Growth, Phenotype, and Chemoresistance of Ovarian Cancer Cells. Front. Cell Dev. Biol. 2021, 9, 718834. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, S.; Haga, H. Matrix Stiffness Contributes to Cancer Progression by Regulating Transcription Factors. Cancers 2022, 14, 1049. [Google Scholar] [CrossRef] [PubMed]
- Piersma, B.; Hayward, M.K.; Weaver, V.M. Fibrosis and cancer: A strained relationship. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188356. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wang, X.; Lu, J.; Salfenmoser, M.; Wirsik, N.M.; Schleussner, N.; Imle, A.; Freire Valls, A.; Radhakrishnan, P.; Liang, J.; et al. Reduction of Liver Metastasis Stiffness Improves Response to Bevacizumab in Metastatic Colorectal Cancer. Cancer Cell 2020, 37, 800–817.e807. [Google Scholar] [CrossRef]
- Wei, X.; Lou, H.; Zhou, D.; Jia, Y.; Li, H.; Huang, Q.; Ma, J.; Yang, Z.; Sun, C.; Meng, Y.; et al. TAGLN mediated stiffness-regulated ovarian cancer progression via RhoA/ROCK pathway. J. Exp. Clin. Cancer Res. 2021, 40, 292. [Google Scholar] [CrossRef]
- Nia, H.T.; Munn, L.L.; Jain, R.K. Physical traits of cancer. Science 2020, 370, eaaz0868. [Google Scholar] [CrossRef]
- Li, J.; Wu, Y.; Schimmel, N.; Al-Ameen, M.A.; Ghosh, G. Breast cancer cells mechanosensing in engineered matrices: Correlation with aggressive phenotype. J. Mech. Behav. Biomed. Mater. 2016, 61, 208–220. [Google Scholar] [CrossRef]
- Efremov, Y.M.; Zurina, I.M.; Presniakova, V.S.; Kosheleva, N.V.; Butnaru, D.V.; Svistunov, A.A.; Rochev, Y.A.; Timashev, P.S. Mechanical properties of cell sheets and spheroids: The link between single cells and complex tissues. Biophys. Rev. 2021, 13, 541–561. [Google Scholar] [CrossRef]
- Kosheleva, N.V.; Efremov, Y.M.; Koteneva, P.I.; Ilina, I.V.; Zurina, I.M.; Bikmulina, P.Y.; Shpichka, A.I.; Timashev, P.S. Building a tissue: Mesenchymal and epithelial cell spheroids mechanical properties at micro- and nanoscale. Acta Biomater. 2022, 165, 140–152. [Google Scholar] [CrossRef]
- Xu, D.; Harvey, T.; Begiristain, E.; Domínguez, C.; Sánchez-Abella, L.; Browne, M.; Cook, R.B. Measuring the elastic modulus of soft biomaterials using nanoindentation. J. Mech. Behav. Biomed. Mater. 2022, 133, 105329. [Google Scholar] [CrossRef]
- Mattei, G.; Di Patria, V.; Tirella, A.; Alaimo, A.; Elia, G.; Corti, A.; Paolicchi, A.; Ahluwalia, A. Mechanostructure and composition of highly reproducible decellularized liver matrices. Acta Biomater. 2014, 10, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.W.; Moran, E.C.; Baptista, P.M.; Soker, S.; Sparks, J.L. Scale-dependent mechanical properties of native and decellularized liver tissue. Biomech. Model. Mechanobiol. 2013, 12, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Liu, P.; Zheng, X.; Dong, D.; Fan, S.; Dong, J.; Zhang, X.; Liu, X.; Wang, B.; Lv, Y. The effect of riboflavin/UVA cross-linking on anti-degeneration and promoting angiogenic capability of decellularized liver matrix. J. Biomed. Mater. Res. Part. A 2017, 105, 2662–2669. [Google Scholar] [CrossRef]
- Grant, R.; Hallett, J.; Forbes, S.; Hay, D.; Callanan, A. Blended electrospinning with human liver extracellular matrix for engineering new hepatic microenvironments. Sci. Rep. 2019, 9, 6293. [Google Scholar] [CrossRef] [PubMed]
- Shirani, A.; Ganji, F.; Golmohammadi, M.; Hashemi, S.M.; Mozafari, M.; Amoabediny, G.; Karkuki Osguei, N.; Samadikuchaksaraei, A. Cross-linked acellular lung for application in tissue engineering: Effects on biocompatibility, mechanical properties and immunological responses. Mater. Sci. Eng. C Mater. Biol. Appl. 2021, 122, 111938. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Z.; Yu, Q.; Xi, H.; Weng, J.; Du, X.; Chen, D.; Ma, J.; Mei, J.; Chen, C. Comparative study of two perfusion routes with different flow in decellularization to harvest an optimal pulmonary scaffold for recellularization. J. Biomed. Mater. Res. Part A 2016, 104, 2567–2575. [Google Scholar] [CrossRef]
- Jorba, I.; Beltrán, G.; Falcones, B.; Suki, B.; Farré, R.; García-Aznar, J.M.; Navajas, D. Nonlinear elasticity of the lung extracellular microenvironment is regulated by macroscale tissue strain. Acta Biomater. 2019, 92, 265–276. [Google Scholar] [CrossRef]
- Poornejad, N.; Frost, T.S.; Scott, D.R.; Elton, B.B.; Reynolds, P.R.; Roeder, B.L.; Cook, A.D. Freezing/Thawing without Cryoprotectant Damages Native but not Decellularized Porcine Renal Tissue. Organogenesis 2015, 11, 30–45. [Google Scholar] [CrossRef]
- Taylor, D.A.; Kren, S.M.; Rhett, K.; Robertson, M.J.; Morrissey, J.; Rodriguez, O.E.; Virk, H.; Chacon-Alberty, L.; Curty da Costa, E.; Mesquita, F.C.P.; et al. Characterization of perfusion decellularized whole animal body, isolated organs, and multi-organ systems for tissue engineering applications. Physiol. Rep. 2021, 9, e14817. [Google Scholar] [CrossRef]
- Melica, M.E.; La Regina, G.; Parri, M.; Peired, A.J.; Romagnani, P.; Lasagni, L. Substrate Stiffness Modulates Renal Progenitor Cell Properties via a ROCK-Mediated Mechanotransduction Mechanism. Cells 2019, 8, 1561. [Google Scholar] [CrossRef]
- Liu, B.; Kilpatrick, J.I.; Lukasz, B.; Jarvis, S.P.; McDonnell, F.; Wallace, D.M.; Clark, A.F.; O’Brien, C.J. Increased Substrate Stiffness Elicits a Myofibroblastic Phenotype in Human Lamina Cribrosa Cells. Investig. Ophthalmol. Vis. Sci. 2018, 59, 803–814. [Google Scholar] [CrossRef]
- Pandamooz, S.; Jafari, A.; Salehi, M.S.; Jurek, B.; Ahmadiani, A.; Safari, A.; Hassanajili, S.; Borhani-Haghighi, A.; Dianatpour, M.; Niknejad, H.; et al. Substrate stiffness affects the morphology and gene expression of epidermal neural crest stem cells in a short term culture. Biotechnol. Bioeng. 2020, 117, 305–317. [Google Scholar] [CrossRef]
- Piao, J.; You, K.; Guo, Y.; Zhang, Y.; Li, Z.; Geng, L. Substrate stiffness affects epithelial-mesenchymal transition of cervical cancer cells through miR-106b and its target protein DAB2. Int. J. Oncol. 2017, 50, 2033–2042. [Google Scholar] [CrossRef]
- Li, S.; Bai, H.; Chen, X.; Gong, S.; Xiao, J.; Li, D.; Li, L.; Jiang, Y.; Li, T.; Qin, X.; et al. Soft Substrate Promotes Osteosarcoma Cell Self-Renewal, Differentiation, and Drug Resistance Through miR-29b and Its Target Protein Spin 1. ACS Biomater. Sci. Eng. 2020, 6, 5588–5598. [Google Scholar] [CrossRef]
- Zhao, D.; Li, Q.; Liu, M.; Ma, W.; Zhou, T.; Xue, C.; Cai, X. Substrate stiffness regulated migration and invasion ability of adenoid cystic carcinoma cells via RhoA/ROCK pathway. Cell Prolif. 2018, 51, e12442. [Google Scholar] [CrossRef]
- Sencha, L.M.; Dobrynina, O.E.; Pospelov, A.D.; Guryev, E.L.; Peskova, N.N.; Brilkina, A.A.; Cherkasova, E.I.; Balalaeva, I.V. Real-Time Fluorescence Visualization and Quantitation of Cell Growth and Death in Response to Treatment in 3D Collagen-Based Tumor Model. Int. J. Mol. Sci. 2022, 23, 8837. [Google Scholar] [CrossRef]
- Sencha, L.M.; Gorokhova, A.A.; Peskova, N.N.; Cherkasova, E.I.; Balalaeva, I.V. Dynamic Study of PDT-Induced Oxidative Stress in Cancer Cells Embedded in 3D Collagen Hydrogel Using Genetically Encoded H2O2-Sensor. J. Biomed. Photonics Eng. 2022, 2022, 8. [Google Scholar] [CrossRef]
- McKenzie, A.J.; Hicks, S.R.; Svec, K.V.; Naughton, H.; Edmunds, Z.L.; Howe, A.K. The mechanical microenvironment regulates ovarian cancer cell morphology, migration, and spheroid disaggregation. Sci. Rep. 2018, 8, 7228. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.C.; Fattet, L.; Tsai, J.H.; Guo, Y.; Pai, V.H.; Majeski, H.E.; Chen, A.C.; Sah, R.L.; Taylor, S.S.; Engler, A.J.; et al. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat. Cell Biol. 2015, 17, 678–688. [Google Scholar] [CrossRef]
- Prauzner-Bechcicki, S.; Raczkowska, J.; Madej, E.; Pabijan, J.; Lukes, J.; Sepitka, J.; Rysz, J.; Awsiuk, K.; Bernasik, A.; Budkowski, A.; et al. PDMS substrate stiffness affects the morphology and growth profiles of cancerous prostate and melanoma cells. J. Mech. Behav. Biomed. Mater. 2015, 41, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Chen, Z.; Chen, Y.; Li, S.; Jiang, Y.; Yang, H.; Wu, C.; You, F.; Zheng, C.; Zhu, J.; et al. ROCK isoforms differentially modulate cancer cell motility by mechanosensing the substrate stiffness. Acta Biomater. 2019, 88, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.W.; Grant, A.D.; Parker, S.S.; Hill, S.; Whalen, M.B.; Chakrabarti, J.; Harman, M.W.; Roman, M.R.; Forte, B.L.; Gowan, C.C.; et al. Breast tumor stiffness instructs bone metastasis via maintenance of mechanical conditioning. Cell Rep. 2021, 35, 109293. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Sinha, S.; Jiang, X.; Murphy, L.; Fitch, S.; Wilson, C.; Grant, G.; Yang, F. Matrix Stiffness Modulates Patient-Derived Glioblastoma Cell Fates in Three-Dimensional Hydrogels. Tissue Eng. Part A 2021, 27, 390–401. [Google Scholar] [CrossRef]
- van Tienderen, G.S.; Conboy, J.; Muntz, I.; Willemse, J.; Tieleman, J.; Monfils, K.; Schurink, I.J.; Demmers, J.A.A.; Doukas, M.; Koenderink, G.H.; et al. Tumor decellularization reveals proteomic and mechanical characteristics of the extracellular matrix of primary liver cancer. Biomater. Adv. 2023, 146, 213289. [Google Scholar] [CrossRef] [PubMed]
- Shah, L.; Latif, A.; Williams, K.J.; Mancuso, E.; Tirella, A. Invasion and Secondary Site Colonization as a Function of In Vitro Primary Tumor Matrix Stiffness: Breast to Bone Metastasis. Adv. Healthc. Mater. 2023, 12, 2201898. [Google Scholar] [CrossRef] [PubMed]
- Firatligil-Yildirir, B.; Bati-Ayaz, G.; Tahmaz, I.; Bilgen, M.; Pesen-Okvur, D.; Yalcin-Ozuysal, O. On-chip determination of tissue-specific metastatic potential of breast cancer cells. Biotechnol. Bioeng. 2021, 118, 3799–3810. [Google Scholar] [CrossRef]
- Amaro, A.; Angelini, G.; Mirisola, V.; Esposito, A.I.; Reverberi, D.; Matis, S.; Maffei, M.; Giaretti, W.; Viale, M.; Gangemi, R.; et al. A highly invasive subpopulation of MDA-MB-231 breast cancer cells shows accelerated growth, differential chemoresistance, features of apocrine tumors and reduced tumorigenicity in vivo. Oncotarget 2016, 7, 68803–68820. [Google Scholar] [CrossRef]
- Huang, Y.; Huang, H.; Wang, S.; Chen, F.; Zheng, G. Dehydrocorydaline inhibits the tumorigenesis of breast cancer MDA-MB-231 cells. Mol. Med. Rep. 2020, 22, 43–50. [Google Scholar] [CrossRef]
- Meng, Z.; Zhang, R.; Wang, Y.; Zhu, G.; Jin, T.; Li, C.; Zhang, S. miR-200c/PAI-2 promotes the progression of triple negative breast cancer via M1/M2 polarization induction of macrophage. Int. Immunopharmacol. 2020, 81, 106028. [Google Scholar] [CrossRef]
- Yang, Q.; Yang, D.; Zhu, H.B.; Li, P.; Wu, X.P.; Tan, Y.R. LncRNA MORT negatively regulates FGF1 to suppress malignant progression of breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 6179–6186. [Google Scholar] [CrossRef]
- Dong, H.; Hu, J.; Zou, K.; Ye, M.; Chen, Y.; Wu, C.; Chen, X.; Han, M. Activation of LncRNA TINCR by H3K27 acetylation promotes Trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast Cancer. Mol. Cancer 2019, 18, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.N.; Qian, X.; Granick, M.S.; Solomon, M.P.; Rothman, V.L.; Berger, D.H.; Tuszynski, G.P. Thrombospondin-1 (TSP-1) promotes the invasive properties of human breast cancer. J. Surg. Res. 1996, 63, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Cai, Y.; Long, J.; Zeng, H.; Xu, W.; Li, Y.; Liu, M.; Zhang, H.; He, Z.L.; Chen, W.G. Cathepsin L is involved in proliferation and invasion of breast cancer cells. Neoplasma 2016, 63, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, H.; Song, X.; Yang, Q. Metastatic heterogeneity of breast cancer: Molecular mechanism and potential therapeutic targets. Semin. Cancer Biol. 2020, 60, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, I.; Yogev, O.; Mattsson, J.; Schurich, A. The Metabolic Profile of Tumor and Virally Infected Cells Shapes Their Microenvironment Counteracting T Cell Immunity. Front. Immunol. 2019, 10, 2309. [Google Scholar] [CrossRef]
- Elia, I.; Haigis, M.C. Metabolites and the tumour microenvironment: From cellular mechanisms to systemic metabolism. Nat. Metab. 2021, 3, 21–32. [Google Scholar] [CrossRef]
- Kubeček, O.; Laco, J.; Špaček, J.; Petera, J.; Kopecký, J.; Kubečková, A.; Filip, S. The pathogenesis, diagnosis, and management of metastatic tumors to the ovary: A comprehensive review. Clin. Exp. Metastasis 2017, 34, 295–307. [Google Scholar] [CrossRef]
- Bogliolo, S.; Morotti, M.; Valenzano Menada, M.; Fulcheri, E.; Musizzano, Y.; Casabona, F. Breast cancer with synchronous massive metastasis in the uterine cervix: A case report and review of the literature. Arch. Gynecol. Obstet. 2010, 281, 769–773. [Google Scholar] [CrossRef]
- Karaosmanoglu, A.D.; Onur, M.R.; Salman, M.C.; Usubutun, A.; Karcaaltincaba, M.; Ozmen, M.N.; Akata, D. Imaging in secondary tumors of the ovary. Abdom. Radiol. 2019, 44, 1493–1505. [Google Scholar] [CrossRef]
- Fang, J.Y.; Tan, S.J.; Wu, Y.C.; Yang, Z.; Hoang, B.X.; Han, B. From competency to dormancy: A 3D model to study cancer cells and drug responsiveness. J. Transl. Med. 2016, 14, 38. [Google Scholar] [CrossRef]
- Sheykhzadeh, S.; Luo, M.; Peng, B.; White, J.; Abdalla, Y.; Tang, T.; Mäkilä, E.; Voelcker, N.H.; Tong, W.Y. Transferrin-targeted porous silicon nanoparticles reduce glioblastoma cell migration across tight extracellular space. Sci. Rep. 2020, 10, 2320. [Google Scholar] [CrossRef] [PubMed]
- Wolf, K.; Alexander, S.; Schacht, V.; Coussens, L.M.; von Andrian, U.H.; van Rheenen, J.; Deryugina, E.; Friedl, P. Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 2009, 20, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chen, L.; Wang, W.; Qin, D.; Jia, C.; Yuan, M.; Wang, H.; Guo, Y.; Zhu, J.; Zhou, Y.; et al. Emodin Suppresses the Migration and Invasion of Melanoma Cells. Biol. Pharm. Bull. 2021, 44, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Knowles, L.M.; Gurski, L.A.; Maranchie, J.K.; Pilch, J. Fibronectin Matrix Formation is a Prerequisite for Colonization of Kidney Tumor Cells in Fibrin. J. Cancer 2015, 6, 98–104. [Google Scholar] [CrossRef]
- Paget, S. The distribution of secondary growths in cancer of the breast. Cancer Metastasis Rev. 1989, 8, 98–101. [Google Scholar] [CrossRef]
- Chen, W.; Hoffmann, A.D.; Liu, H.; Liu, X. Organotropism: New insights into molecular mechanisms of breast cancer metastasis. NPJ Precis. Oncol. 2018, 2, 4. [Google Scholar] [CrossRef]
- Kaplan, R.N.; Riba, R.D.; Zacharoulis, S.; Bramley, A.H.; Vincent, L.; Costa, C.; MacDonald, D.D.; Jin, D.K.; Shido, K.; Kerns, S.A.; et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 2005, 438, 820–827. [Google Scholar] [CrossRef]
- Wakisaka, N.; Hasegawa, Y.; Yoshimoto, S.; Miura, K.; Shiotani, A.; Yokoyama, J.; Sugasawa, M.; Moriyama-Kita, M.; Endo, K.; Yoshizaki, T. Primary Tumor-Secreted Lymphangiogenic Factors Induce Pre-Metastatic Lymphvascular Niche Formation at Sentinel Lymph Nodes in Oral Squamous Cell Carcinoma. PLoS ONE 2015, 10, e0144056. [Google Scholar] [CrossRef]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef]
- Karaca, Z.; Tanriverdi, F.; Unluhizarci, K.; Ozturk, F.; Gokahmetoglu, S.; Elbuken, G.; Cakir, I.; Bayram, F.; Kelestimur, F. VEGFR1 expression is related to lymph node metastasis and serum VEGF may be a marker of progression in the follow-up of patients with differentiated thyroid carcinoma. Eur. J. Endocrinol. 2011, 164, 277–284. [Google Scholar] [CrossRef][Green Version]
- Qian, B.-Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Takahashi, H.; Lin, W.W.; Descargues, P.; Grivennikov, S.; Kim, Y.; Luo, J.L.; Karin, M. Carcinoma-produced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature 2009, 457, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.H.; Pickup, M.; Pang, Y.; Gorska, A.E.; Li, Z.; Chytil, A.; Geng, Y.; Gray, J.W.; Moses, H.L.; Yang, L. Gr-1+CD11b+ myeloid cells tip the balance of immune protection to tumor promotion in the premetastatic lung. Cancer Res. 2010, 70, 6139–6149. [Google Scholar] [CrossRef] [PubMed]
- Seubert, B.; Grünwald, B.; Kobuch, J.; Cui, H.; Schelter, F.; Schaten, S.; Siveke, J.T.; Lim, N.H.; Nagase, H.; Simonavicius, N.; et al. Tissue inhibitor of metalloproteinases (TIMP)-1 creates a premetastatic niche in the liver through SDF-1/CXCR4-dependent neutrophil recruitment in mice. Hepatology 2015, 61, 238–248. [Google Scholar] [CrossRef]
- Chaudhuri, O.; Koshy, S.T.; Branco da Cunha, C.; Shin, J.W.; Verbeke, C.S.; Allison, K.H.; Mooney, D.J. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat. Mater. 2014, 13, 970–978. [Google Scholar] [CrossRef]
- Kanchanawong, P.; Shtengel, G.; Pasapera, A.M.; Ramko, E.B.; Davidson, M.W.; Hess, H.F.; Waterman, C.M. Nanoscale architecture of integrin-based cell adhesions. Nature 2010, 468, 580–584. [Google Scholar] [CrossRef]
- Bae, Y.H.; Mui, K.L.; Hsu, B.Y.; Liu, S.L.; Cretu, A.; Razinia, Z.; Xu, T.; Puré, E.; Assoian, R.K. A FAK-Cas-Rac-lamellipodin signaling module transduces extracellular matrix stiffness into mechanosensitive cell cycling. Sci. Signal 2014, 7, ra57. [Google Scholar] [CrossRef]
- Moreno-Layseca, P.; Streuli, C.H. Signalling pathways linking integrins with cell cycle progression. Matrix Biol. 2014, 34, 144–153. [Google Scholar] [CrossRef]
- Aragona, M.; Panciera, T.; Manfrin, A.; Giulitti, S.; Michielin, F.; Elvassore, N.; Dupont, S.; Piccolo, S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 2013, 154, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, S.; Liu, J.; Tian, Y.; Ma, B.; Xu, S.; Fu, Y.; Luo, Y. Secreted Pyruvate Kinase M2 Promotes Lung Cancer Metastasis through Activating the Integrin Beta1/FAK Signaling Pathway. Cell Rep. 2020, 30, 1780–1797.e1786. [Google Scholar] [CrossRef]
- Borghi, N.; Lowndes, M.; Maruthamuthu, V.; Gardel, M.L.; Nelson, W.J. Regulation of cell motile behavior by crosstalk between cadherin- and integrin-mediated adhesions. Proc. Natl. Acad. Sci. USA 2010, 107, 13324–13329. [Google Scholar] [CrossRef]
- Chang, G.; Zhang, H.; Wang, J.; Zhang, Y.; Xu, H.; Wang, C.; Zhang, H.; Ma, L.; Li, Q.; Pang, T. CD44 targets Wnt/β-catenin pathway to mediate the proliferation of K562 cells. Cancer Cell Int. 2013, 13, 117. [Google Scholar] [CrossRef]
- Lai, C.J.; Lin, C.Y.; Liao, W.Y.; Hour, T.C.; Wang, H.D.; Chuu, C.P. CD44 Promotes Migration and Invasion of Docetaxel-Resistant Prostate Cancer Cells Likely via Induction of Hippo-Yap Signaling. Cells 2019, 8, 295. [Google Scholar] [CrossRef] [PubMed]
- SenGupta, S.; Parent, C.A.; Bear, J.E. The principles of directed cell migration. Nat. Rev. Mol. Cell Biol. 2021, 22, 529–547. [Google Scholar] [CrossRef]
- Franklin, O.; Billing, O.; Öhlund, D.; Berglund, A.; Herdenberg, C.; Wang, W.; Hellman, U.; Sund, M. Novel prognostic markers within the CD44-stromal ligand network in pancreatic cancer. J. Pathol. Clin. Res. 2019, 5, 130–141. [Google Scholar] [CrossRef]

| Total DNA Content | |||
|---|---|---|---|
| Native Tissue, µg/g Wet Tissue Weight | DCL Tissue, µg/g Wet Tissue Weight | DCL Tissue, % from Native | |
| Liver | 1032 ± 11 | 0.63 ± 0.28 | 0.06 |
| Lung | 1955 ± 29 | 0.90 ± 0.12 | 0.05 |
| Spleen | 2337 ± 30 | 2.12 ± 0.14 | 0.09 |
| Kidney | 1286 ± 45 | 1.03 ± 0.32 | 0.08 |
| Ovary | 1757 ± 63 | 1.22 ± 0.26 | 0.07 |
| Liver | Lung | Spleen | Kidney | Ovary | Collagen Gel | ||
|---|---|---|---|---|---|---|---|
| Matrix fiber density | Medium | Low | High | Medium | High | High | |
| Pore area, μm2 | 133 | 240 | 78 | 90 | 152,34 | N/A | |
| Local matrix stiffness (nanoindentation), kPa | 6.15 | 2.85 | 3.6 | 5.6 | 6.4 | 0.36 | |
| Total matrix stiffness (macroindentation), kPa | 18 | 13.4 | 37 | 25.4 | 15.7 | N/A | |
| Degree of repopulation | MDA-MB-231 | Medium | High | Medium | Medium | High | Low |
| SKBR-3 | Single | Low | Low | Low | Single | Low | |
| Cell morphology | MDA-MB-231 | Mes a/Ep b | Mes | Mes | Mes | Mes | Mes |
| SKBR-3 | Ep | Ep | Ep | Ep | Ep | Ep | |
| DCL Organ | Young’s Modulus | Method | Device and Measuring Conditions | Ref. |
|---|---|---|---|---|
| Porcine liver | 1.25 ± 0.07 kPa | Compression test | Zwick/Roell ProLine Z005 testing machine (Zwick/Roell, Ulm, Germany) equipped with a 10 N load cell. | [62] |
| Ferret liver | 1.18 kPa | Compression test | ElectroForce TestBench mechanical testing system equipped with a 1000-g load cell (Bose ElectroForce, Eden Prairie, MN, USA). | [63] |
| Rat liver | 145 ± 19.68 kPa | Tensile test | Uniaxial tensile testing machine (CMT8502, Shenzhen New Sans Test Technical Company, Nanshan, Shenzhen, China). The test was performed at loading speed of 1 mm/min until the final fracture of the specimen. | [64] |
| Human liver | 18,490 ± 1400 kPa | Tensile test | Instron 3367 dual column universal testing system (Instron, Glenview, IL, USA) fitted with Instron biopulse submersible pneumatic side action grips (Instron, USA) and a 50 N load cell. A gauge length of 20 mm and an extension rate of 20 mm/min were used. | [65] |
| Rat lung | ~80 kPa | Tensile test | Santam tensile testing machine STM-1 (SANTAM, Tehran, Iran). The uniaxial tensile test was carried out by a crosshead speed of 2 mm/min, elongation raised to 20–40 mm, and a load cell of 6 kg at a constant elongation speed until rupture of the specimen was observed. | [66] |
| Rat lung | 74.91 ± 5.78 kPa | Tensile test | Material testing machine BZ2.5/TN1S (Zwick/Roell, Ulm, Germany). Preloading of 0.015 N was imposed and the sample length was reported, then a preload of 0.003 N was set. | [67] |
| Rat lung | 0.38 ± 0.07 kPa | Tensile test | Displacement actuator 300C-LR (Aurora Scientific, Aurora, ON, Canada) with a force transducer 404A (Aurora Scientific, Canada). Uniaxial tensile measurements after preconditioning with triangular stretch at 0.1 mN force. | [68] |
| Porcine kidney (cortex) | 6.4 ± 2.7 kPa | Compression test | Instron 3342 Single Column Universal Testing System (Instron, USA) and Instron Model 1321 (Instron, Glenview, IL, USA) were used to perform compression tests at a rate of 0.07 mm/sec until a compression force of 45 N was reached. | [69] |
| Rat Kidney | 178.9 ± 50.2 kPa | Tensile test | Tensile mechanical testing machine (Instron, Glenview, IL, USA). Samples were preconditioned by cycles of loading and unloading. Strain rate was 0.01 s−1 throughout the test. | [70] |
| Rat liver | 4242.2 ± 891.6 kPa | |||
| Rat Lung | 233.6 ± 98.0 kPa |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pospelov, A.D.; Kutova, O.M.; Efremov, Y.M.; Nekrasova, A.A.; Trushina, D.B.; Gefter, S.D.; Cherkasova, E.I.; Timofeeva, L.B.; Timashev, P.S.; Zvyagin, A.V.; et al. Breast Cancer Cell Type and Biomechanical Properties of Decellularized Mouse Organs Drives Tumor Cell Colonization. Cells 2023, 12, 2030. https://doi.org/10.3390/cells12162030
Pospelov AD, Kutova OM, Efremov YM, Nekrasova AA, Trushina DB, Gefter SD, Cherkasova EI, Timofeeva LB, Timashev PS, Zvyagin AV, et al. Breast Cancer Cell Type and Biomechanical Properties of Decellularized Mouse Organs Drives Tumor Cell Colonization. Cells. 2023; 12(16):2030. https://doi.org/10.3390/cells12162030
Chicago/Turabian StylePospelov, Anton D., Olga M. Kutova, Yuri M. Efremov, Albina A. Nekrasova, Daria B. Trushina, Sofia D. Gefter, Elena I. Cherkasova, Lidia B. Timofeeva, Peter S. Timashev, Andrei V. Zvyagin, and et al. 2023. "Breast Cancer Cell Type and Biomechanical Properties of Decellularized Mouse Organs Drives Tumor Cell Colonization" Cells 12, no. 16: 2030. https://doi.org/10.3390/cells12162030
APA StylePospelov, A. D., Kutova, O. M., Efremov, Y. M., Nekrasova, A. A., Trushina, D. B., Gefter, S. D., Cherkasova, E. I., Timofeeva, L. B., Timashev, P. S., Zvyagin, A. V., & Balalaeva, I. V. (2023). Breast Cancer Cell Type and Biomechanical Properties of Decellularized Mouse Organs Drives Tumor Cell Colonization. Cells, 12(16), 2030. https://doi.org/10.3390/cells12162030