A Shiga Toxin B-Subunit-Based Lectibody Boosts T Cell Cytotoxicity towards Gb3-Positive Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Antibodies and Chemical Reagents
2.2. Cell Lines
2.3. Isolation of PBMCs
2.4. Homology Modeling of the Lectibody Structure
2.5. Protein–Protein Docking for Monomeric Lectibody Design
2.6. Structure Optimization
2.7. Transformation of the StxB-scFv UCHT1 Lectibody Sequence
2.8. Protein Expression and Purification
2.9. Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Immunoblotting
2.10. Flow Cytometry
2.11. Cytotoxicity Assay
2.12. Cell Proliferation (MTT) Assay
2.13. Affinity Measurement Using Surface Plasmon Resonance (SPR)
2.14. Statistical Analysis
3. Results
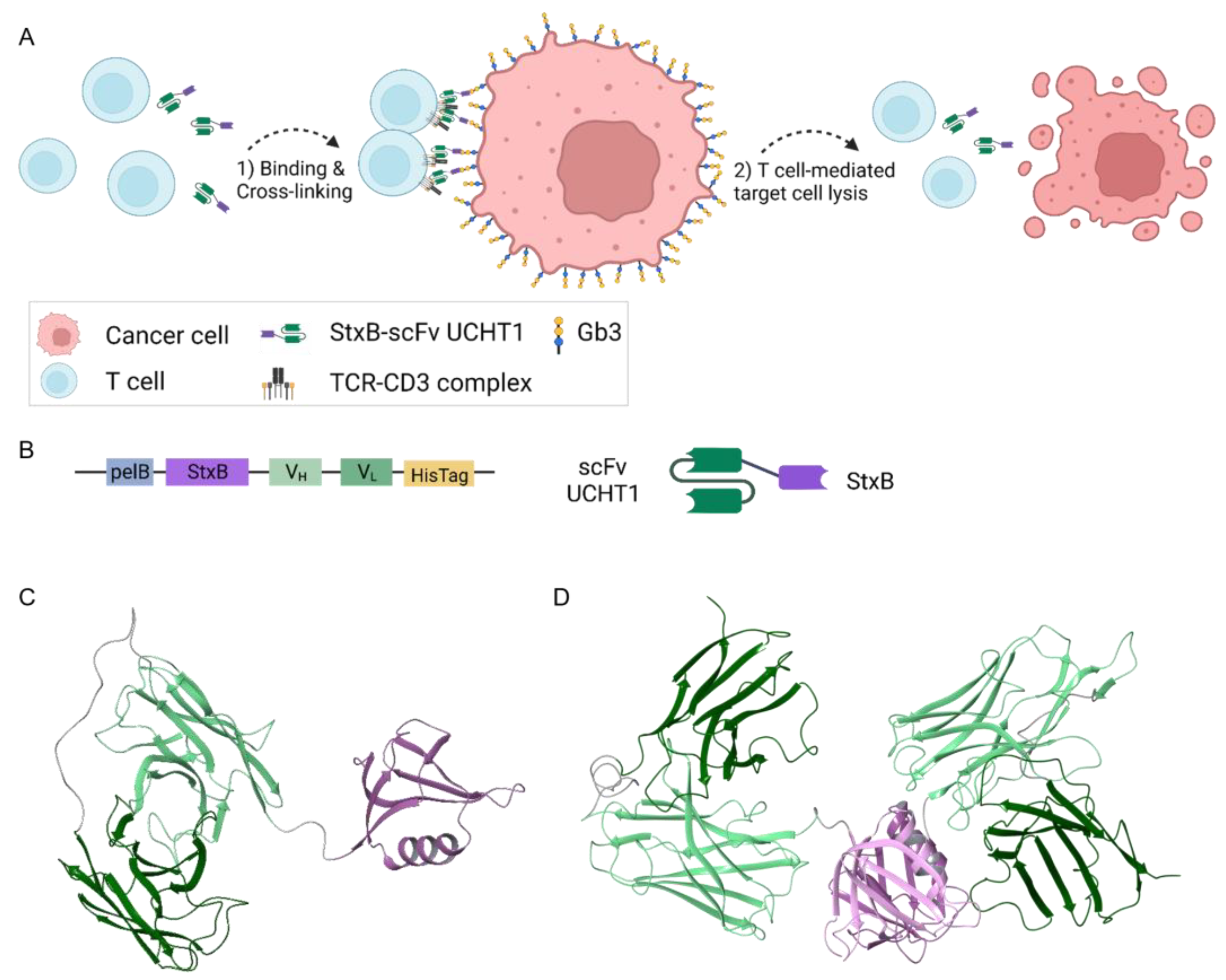
3.1. Design and Structural Prediction of the StxB-scFv UCHT1 Lectibody
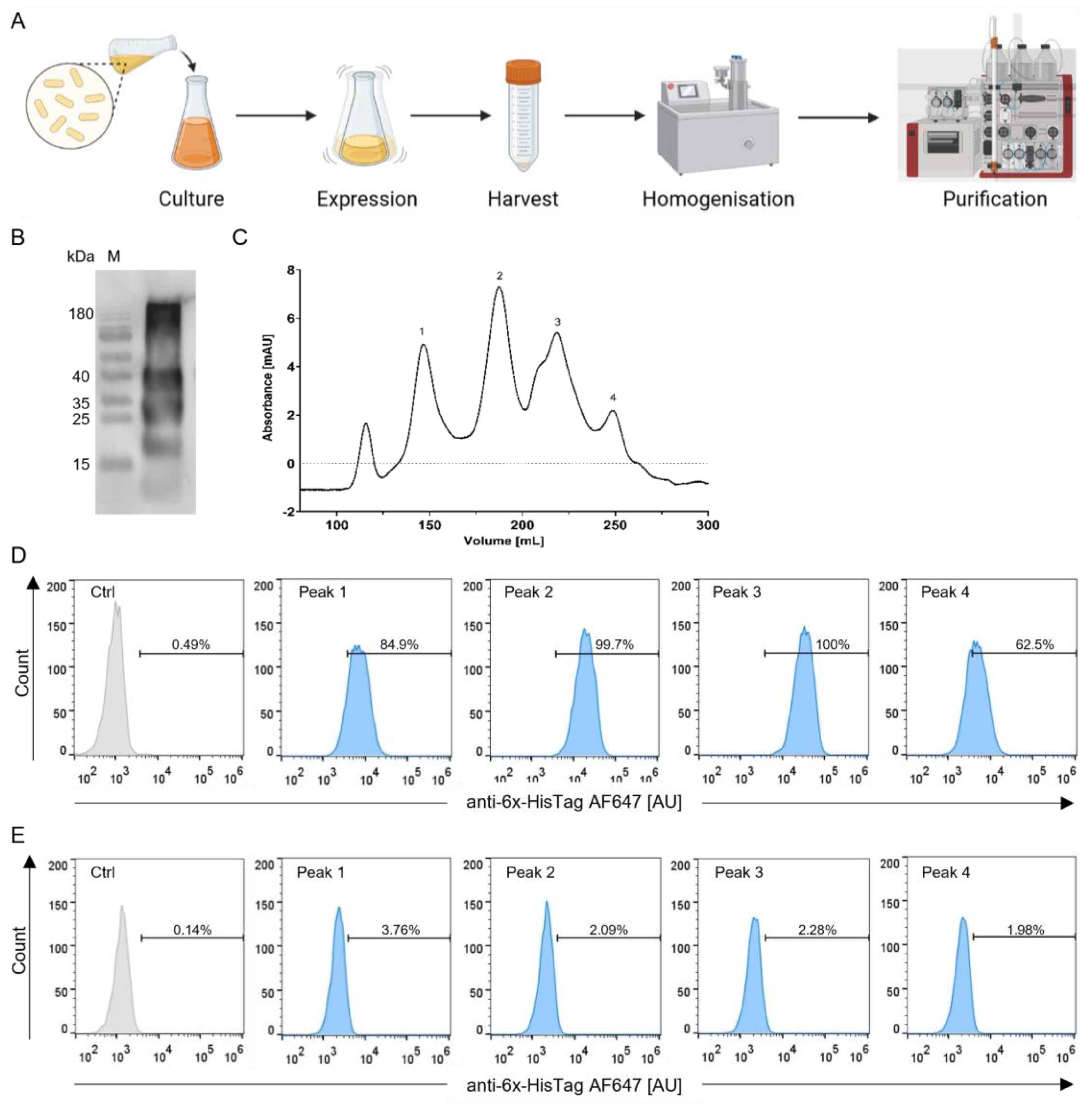
3.2. Production and Selection According to Size of the StxB-scFv UCHT1 Lectibody
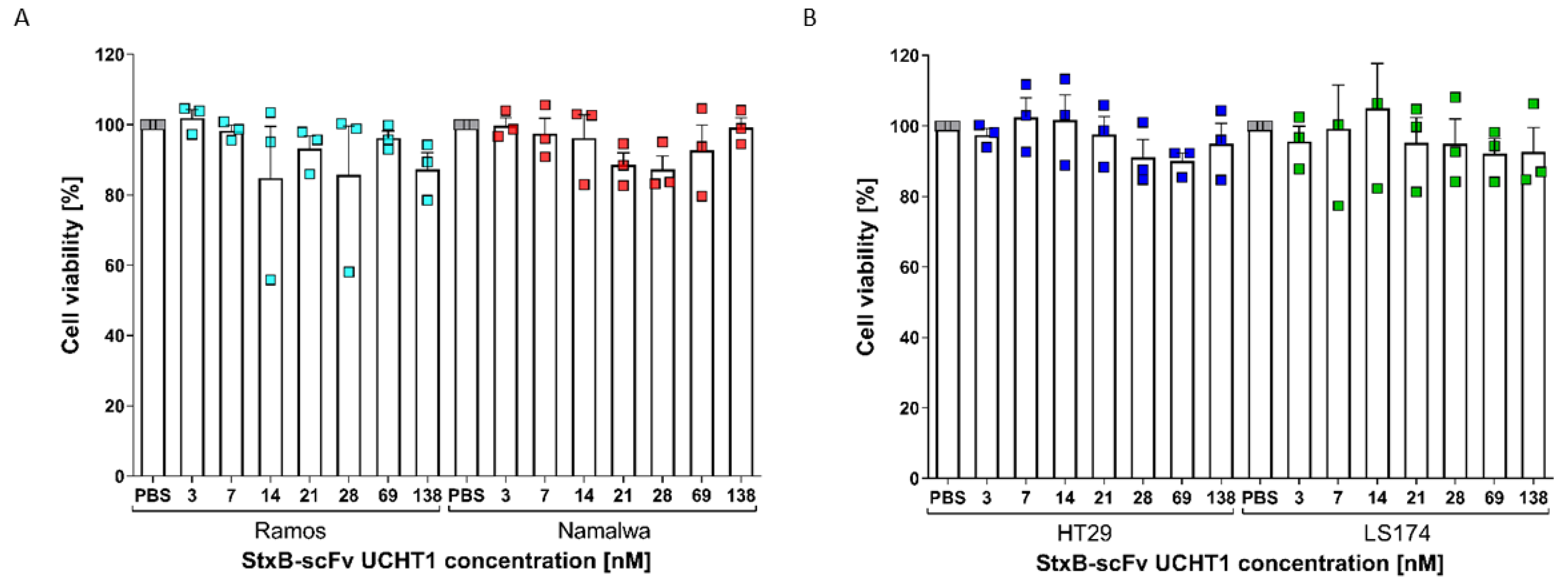
3.3. Ability of the StxB-scFv UCHT1 Lectibody to Induce T-Cell-Mediated Cancer Cell Killing
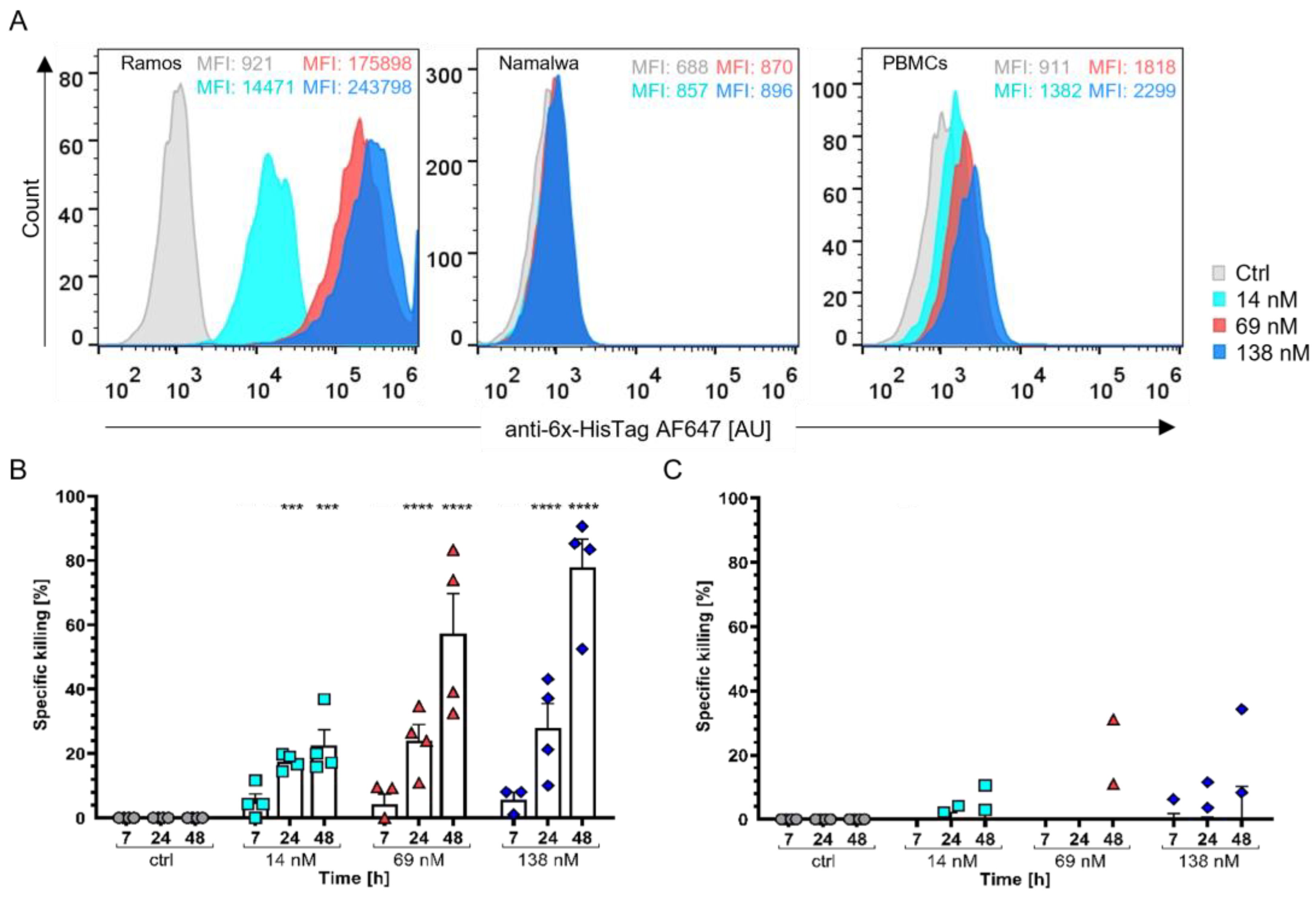
3.3.1. Burkitt’s Lymphoma Cells as a Model for Haematological Cancers
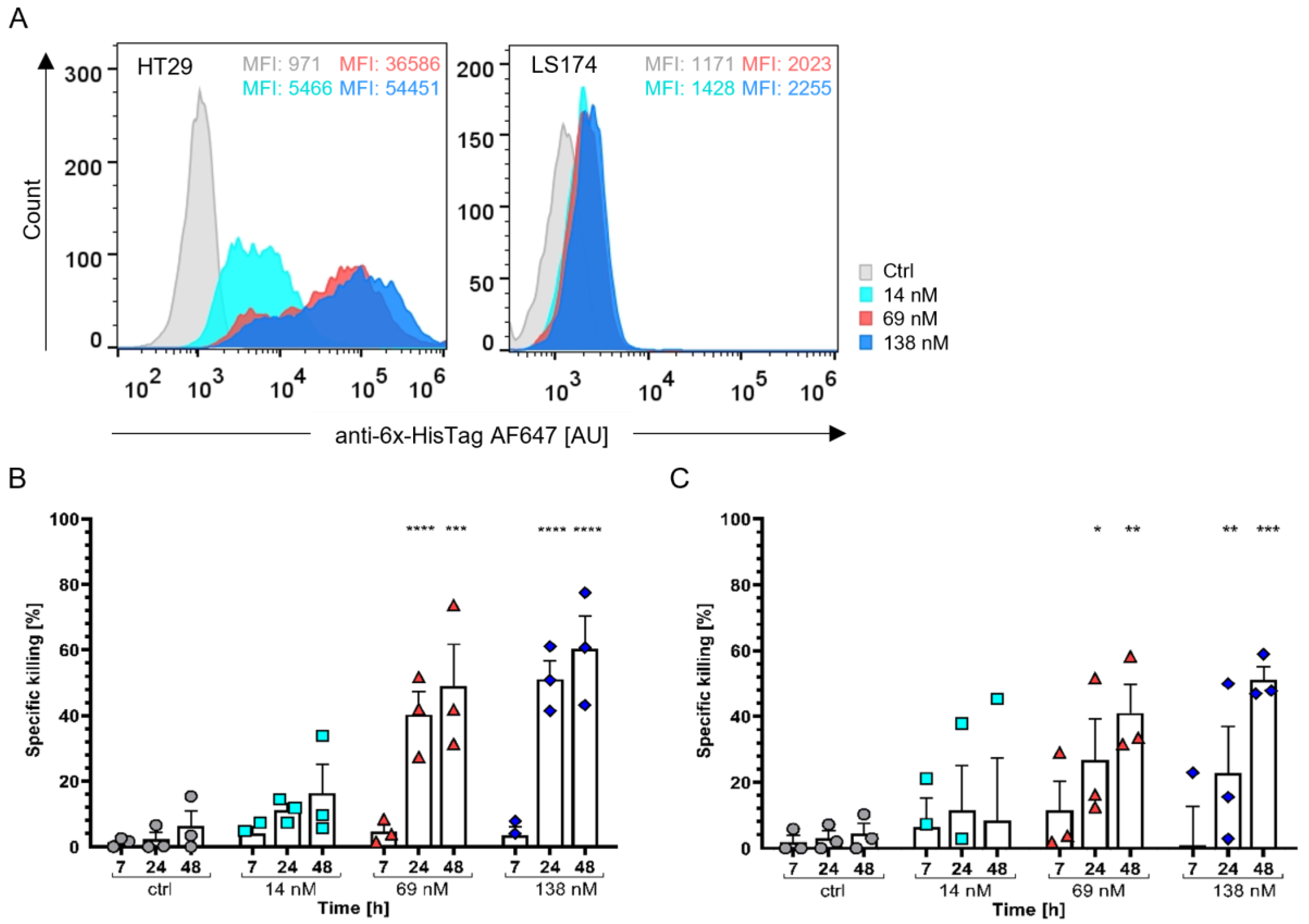
3.3.2. Colon Adenocarcinoma Cells as a Model for Solid Tumour Treatment
3.4. StxB-scFv UCHT1 Lectibody Cytotoxicity Is Strictly T-Cell-Mediated
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Wildgaard, K.; Ravn, J.; Nikolajsen, L.; Jakobsen, E.; Jensen, T.S.; Kehlet, H. Consequences of persistent pain after lung cancer surgery: A nationwide questionnaire study. Acta Anaesthesiol. Scand. 2011, 55, 60–68. [Google Scholar] [CrossRef]
- Tohme, S.; Simmons, R.L.; Tsung, A. Surgery for Cancer: A Trigger for Metastases. Cancer Res. 2017, 77, 1548–1552. [Google Scholar] [CrossRef]
- Carter, J.; Stabile, C.; Gunn, A.; Sonoda, Y. The Physical Consequences of Gynecologic Cancer Surgery and Their Impact on Sexual, Emotional, and Quality of Life Issues. J. Sex. Med. 2013, 10, 21–34. [Google Scholar] [CrossRef]
- Lindley, C.M.; Hirsch, J.D.; O’Neill, C.V.; Transau, M.C.; Gilbert, C.S.; Osterhaus, J.T. Quality of life consequences of chemotherapy-induced emesis. Qual. Life Res. 1992, 1, 331–340. [Google Scholar] [CrossRef]
- Baskar, R.; Lee, K.A.; Yeo, R.; Yeoh, K.W. Cancer and Radiation Therapy: Current Advances and Future Directions. Int. J. Med. Sci. 2012, 9, 193–199. [Google Scholar] [CrossRef]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef]
- Mellman, I.; Coukos, G.; Dranoff, G. Cancer immunotherapy comes of age. Nature 2011, 480, 480–489. [Google Scholar] [CrossRef]
- Till, S.J.; Francis, J.N.; Nouri-Aria, K.; Durham, S.R. Mechanisms of immunotherapy. J. Allergy Clin. Immunol. 2004, 113, 1025–1034. [Google Scholar] [CrossRef]
- Köhler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256, 495–497. [Google Scholar] [CrossRef]
- Kontermann, R.E.; Brinkmann, U. Bispecific antibodies. Drug Discov. Today 2015, 20, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Bird, R.E.; Walker, B.W. Single chain antibody variable regions. Trends Biotechnol. 1991, 9, 132–137. [Google Scholar] [CrossRef]
- Ahamadi-Fesharaki, R.; Fateh, A.; Vaziri, F.; Solgi, G.; Siadat, S.D.; Mahboudi, F.; Rahimi-Jamnani, F. Single-Chain Variable Fragment-Based Bispecific Antibodies: Hitting Two Targets with One Sophisticated Arrow. Mol. Ther.-Oncolytics 2019, 14, 38–56. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.P.; Lee, F.T.; Tahtis, K.; Power, B.E.; Smyth, F.E.; Brechbiel, M.W.; Hudson, P.J.; Scott, A.M. Tumor Targeting by a Multivalent Single-Chain Fv (scFv) Anti-Lewis Y Antibody Construct. Cancer Biother. Radiopharm. 2008, 23, 411–424. [Google Scholar] [CrossRef]
- Chowdhury, P.S.; Viner, J.L.; Beers, R.; Pastan, I. Isolation of a high-affinity stable single-chain Fv specific for mesothelin from DNA-immunized mice by phage display and construction of a recombinant immunotoxin with anti-tumor activity. Proc. Natl. Acad. Sci. USA 1998, 95, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Huehls, A.M.; Coupet, T.A.; Sentman, C.L. Bispecific T-cell engagers for cancer immunotherapy. Immunol. Cell Biol. 2015, 93, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Baeuerle, P.A.; Reinhardt, C. Bispecific T-Cell Engaging Antibodies for Cancer Therapy. Cancer Res. 2009, 69, 4941–4944. [Google Scholar] [CrossRef]
- Wolf, E.; Hofmeister, R.; Kufer, P.; Schlereth, B.; Baeuerle, P.A. BiTEs: Bispecific antibody constructs with unique anti-tumor activity. Drug Discov. Today 2005, 10, 1237–1244. [Google Scholar] [CrossRef]
- Zhou, S.; Liu, M.; Ren, F.; Meng, X.; Yu, J. The landscape of bispecific T cell engager in cancer treatment. Biomark. Res. 2021, 9, 38. [Google Scholar] [CrossRef]
- Baeuerle, P.A.; Kufer, P.; Bargou, R. BiTE: Teaching antibodies to engage T-cells for cancer therapy. Curr. Opin. Mol. Ther. 2009, 11, 22–30. [Google Scholar]
- Slaney, C.Y.; Wang, P.; Darcy, P.K.; Kershaw, M.H. CARs versus BiTEs: A Comparison between T Cell–Redirection Strategies for Cancer Treatment. Cancer Discov. 2018, 8, 924–934. [Google Scholar] [CrossRef] [PubMed]
- Trapani, J.A.; Smyth, M.J. Functional significance of the perforin/granzyme cell death pathway. Nat. Rev. Immunol. 2002, 2, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Haas, C.; Krinner, E.; Brischwein, K.; Hoffmann, P.; Lutterbüse, R.; Schlereth, B.; Kufer, B.; Baeurele, P.A. Mode of cytotoxic action of T cell-engaging BiTE antibody MT110. Immunobiology 2009, 214, 441–453. [Google Scholar] [CrossRef]
- Osińska, I.; Popko, K.; Demkow, U. Perforin: An important player in immune response. Cent. Eur. J. Immunol. 2014, 1, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Carrington, E.M.; Zhang, Y.; Heinzel, S.; Lew, A.M. Life and Death of Activated T Cells: How Are They Different from Naïve T Cells? Front. Immunol. 2017, 8, 1809. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.M.; An, J. Cytokines, Inflammation, and Pain. Int. Anesthesiol. Clin. 2007, 45, 27–37. [Google Scholar] [CrossRef]
- Przepiorka, D.; Ko, C.W.; Deisseroth, A.; Yancey, C.L.; Candau-Chacon, R.; Chiu, H.-J.; Gehrke, B.J.; Gomez-Broughton, C.; Kirshner, S. FDA Approval: Blinatumomab. Clin. Cancer Res. 2015, 21, 4035–4039. [Google Scholar] [CrossRef]
- Nagorsen, D.; Kufer, P.; Baeuerle, P.A.; Bargou, R. Blinatumomab: A historical perspective. Pharmacol. Ther. 2012, 136, 334–342. [Google Scholar] [CrossRef]
- Nair-Gupta, P.; Diem, M.; Reeves, D.; Wang, W.; Schulingkamp, R.; Sproesser, K.; Mattson, B.; Heidrich, B.; Mendonca, M.; Joseph, J.; et al. A novel C2 domain binding CD33xCD3 bispecific antibody with potent T-cell redirection activity against acute myeloid leukemia. Blood Adv. 2020, 4, 906–919. [Google Scholar] [CrossRef]
- Subklewe, M.; Stein, A.; Walter, R.B.; Bhatia, R.; Wei, A.H.; Ritchie, D.; Bücklein, V.; Vachhani, P.; Dai, T.; Hindoyan, A.; et al. Preliminary Results from a Phase 1 First-in-Human Study of AMG 673, a Novel Half-Life Extended (HLE) Anti-CD33/CD3 BiTE® (Bispecific T-Cell Engager) in Patients with Relapsed/Refractory (R/R) Acute Myeloid Leukemia (AML). Blood 2019, 134 (Suppl. S1), 833. [Google Scholar] [CrossRef]
- Hummel, H.D.; Kufer, P.; Grüllich, C.; Seggewiss-Bernhardt, R.; Deschler-Baier, B.; Chatterjee, M.; Goebeler, M.-E.; Miller, K.; de Santis, M.; Loidl, W.; et al. Pasotuxizumab, a BiTE ® immune therapy for castration-resistant prostate cancer: Phase I, dose-escalation study findings. Immunotherapy 2021, 13, 125–141. [Google Scholar] [CrossRef] [PubMed]
- Sternjak, A.; Lee, F.; Thomas, O.; Balazs, M.; Wahl, J.; Lorenczewski, G.; Ullrich, I.; Muenz, M.; Rattel, B.; Bailis, J.M.; et al. Preclinical Assessment of AMG 596, a Bispecific T-cell Engager (BiTE) Immunotherapy Targeting the Tumor-specific Antigen EGFRvIII. Mol. Cancer Ther. 2021, 20, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Hakomori, S.I. Possible functions of tumor-associated carbohydrate antigens. Curr. Opin. Immunol. 1991, 3, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Dube, D.H.; Bertozzi, C.R. Glycans in cancer and inflammation—Potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 2005, 4, 477–488. [Google Scholar] [CrossRef]
- Zhuo, D.; Li, X.; Guan, F. Biological Roles of Aberrantly Expressed Glycosphingolipids and Related Enzymes in Human Cancer Development and Progression. Front. Physiol. 2018, 9, 466. [Google Scholar] [CrossRef]
- Hakomori S itiroh Kannagi, R. Glycosphingolipids as Tumor-Associated and Differentiation Markers56. JNCI J. Natl. Cancer Inst. 1983, 71, 231–251. [Google Scholar] [CrossRef]
- Pellizzari, A.; Pang, H.; Lingwood, C.A. Binding of verocytotoxin 1 to its receptor is influenced by differences in receptor fatty acid content. Biochemistry 1992, 31, 1363–1370. [Google Scholar] [CrossRef]
- Merrill, A.H. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem. Rev. 2011, 111, 6387–6422. [Google Scholar] [CrossRef]
- Siukstaite, L.; Imberty, A.; Römer, W. Structural Diversities of Lectins Binding to the Glycosphingolipid Gb3. Front. Mol. Biosci. 2021, 8, 704685. [Google Scholar] [CrossRef]
- Okuda, T.; Tokuda, N.; Numata, S.I.; Ito, M.; Ohta, M.; Kawamura, K.; Wiels, J.; Urano, T.; Tajima, O. Targeted Disruption of Gb3/CD77 Synthase Gene Resulted in the Complete Deletion of Globo-series Glycosphingolipids and Loss of Sensitivity to Verotoxins*. J. Biol. Chem. 2006, 281, 10230–10235. [Google Scholar] [CrossRef]
- Okuda, T.; Nakayama, K.I. Identification and characterization of the human Gb3/CD77 synthase gene promoter. Glycobiology 2008, 18, 1028–1035. [Google Scholar] [CrossRef] [PubMed]
- Kannagi, R.; Fukuda, M.N.; Hakomori, S. A new glycolipid antigen isolated from human erythrocyte membranes reacting with antibodies directed to globo-N-tetraosylceramide (globoside). J. Biol. Chem. 1982, 257, 4438–4442. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.L.; Johnson, G.D.; Khan, M.; MacLennan, I.C. Quantitative analysis of molecules which distinguish functional compartments within germinal centers. Eur. J. Immunol. 1993, 23, 997–1004. [Google Scholar] [CrossRef]
- Mangeney, M.; Lingwood, C.A.; Taga, S.; Caillou, B.; Tursz, T.; Wiels, J. Apoptosis Induced in Burkitt’s Lymphoma Cells via Gb3/CD77, a Glycolipid Antigen1. Cancer Res. 1993, 53, 5314–5319. [Google Scholar] [PubMed]
- Kovbasnjuk, O.; Mourtazina, R.; Baibakov, B.; Wang, T.; Elowsky, C.; Choti, M.A.; Kane, A.; Donowitz, M. The glycosphingolipid globotriaosylceramide in the metastatic transformation of colon cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 19087–19092. [Google Scholar] [CrossRef] [PubMed]
- Geyer, P.E.; Maak, M.; Nitsche, U.; Perl, M.; Novotny, A.; Slotta-Huspenina, J.; Dransart, E.; Holtorf, A.; Johannes, L.; Janssen, K.-P. Gastric Adenocarcinomas Express the Glycosphingolipid Gb3/CD77: Targeting of Gastric Cancer Cells with Shiga Toxin B-Subunit. Mol. Cancer Ther. 2016, 15, 1008–1017. [Google Scholar] [CrossRef]
- Johansson, D.; Kosovac, E.; Moharer, J.; Ljuslinder, I.; Brännström, T.; Johansson, A.; Behnam-Motlagh, P. Expression of verotoxin-1 receptor Gb3 in breast cancer tissue and verotoxin-1 signal transduction to apoptosis. BMC Cancer 2009, 9, 67. [Google Scholar] [CrossRef]
- Maak, M.; Nitsche, U.; Keller, L.; Wolf, P.; Sarr, M.; Thiebaud, M.; Rosenberg, R.; Langer, R.; Kleef, J.; Friess, H.; et al. Tumor-Specific Targeting of Pancreatic Cancer with Shiga Toxin B-Subunit. Mol. Cancer Ther. 2011, 10, 1918–1928. [Google Scholar] [CrossRef]
- Distler, U.; Souady, J.; Hülsewig, M.; Drmić-Hofman, I.; Haier, J.; Friedrich, A.W.; Karch, H.; Senninger, N.; Dreisewerd, K.; Berenkamp, S.; et al. Shiga Toxin Receptor Gb3Cer/CD77: Tumor-Association and Promising Therapeutic Target in Pancreas and Colon Cancer. PLoS ONE 2009, 4, e6813. [Google Scholar] [CrossRef]
- Behnam-Motlagh, P.; Tyler, A.; Grankvist, K.; Johansson, A. Verotoxin-1 Treatment or Manipulation of its Receptor Globotriaosylceramide (Gb3) for Reversal of Multidrug Resistance to Cancer Chemotherapy. Toxins 2010, 2, 2467–2477. [Google Scholar] [CrossRef] [PubMed]
- Arab, S.; Russel, E.; Chapman, W.B.; Rosen, B.; Lingwood, C.A. Expression of the verotoxin receptor glycolipid, globotriaosylceramide, in ovarian hyperplasias. Oncol. Res. 1997, 9, 553–563. [Google Scholar] [PubMed]
- Nativi, C.; Papi, F.; Roelens, S. Tn antigen analogues: The synthetic way to “upgrade” an attracting tumour associated carbohydrate antigen (TACA). Chem. Commun. 2019, 55, 7729–7736. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, Q. Recent development in carbohydrate-based cancer vaccines. Curr. Opin. Chem. Biol. 2009, 13, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Beatty, G.L.; Gladney, W.L. Immune escape mechanisms as a guide for cancer immunotherapy. Clin. Cancer Res. 2015, 21, 687–692. [Google Scholar] [CrossRef]
- Bates, J.P.; Derakhshandeh, R.; Jones, L.; Webb, T.J. Mechanisms of immune evasion in breast cancer. BMC Cancer 2018, 18, 556. [Google Scholar] [CrossRef] [PubMed]
- Trofa, A.F.; Ueno-Olsen, H.; Oiwa, R.; Yoshikawa, M., Dr. Kiyoshi Shiga: Discoverer of the dysentery bacillus. Clin. Infect. Dis. 1999, 29, 1303–1306. [Google Scholar] [CrossRef]
- Gyles, C.L. Shiga toxin-producing Escherichia coli: An overview1. J. Anim. Sci. 2007, 85 (Suppl. S13), E45–E62. [Google Scholar] [CrossRef]
- Hasan, I.; Sugawara, S.; Fujii, Y.; Koide, Y.; Terada, D.; Iimura, N.; Fujiwara, T.; Takahashi, K.G.; Kojima, N.; Rajia, S.; et al. MytiLec, a Mussel R-Type Lectin, Interacts with Surface Glycan Gb3 on Burkitt’s Lymphoma Cells to Trigger Apoptosis through Multiple Pathways. Mar. Drugs 2015, 13, 7377–7389. [Google Scholar] [CrossRef]
- Terada, D.; Voet, A.R.D.; Noguchi, H.; Kamata, K.; Ohki, M.; Addy, C.; Fujii, Y.; Yamamoto, D.; Ozeki, Y.; Tame, J.R.H.; et al. Computational design of a symmetrical β-trefoil lectin with cancer cell binding activity. Sci. Rep. 2017, 7, 5943. [Google Scholar] [CrossRef]
- Johannes, L.; Römer, W. Shiga toxins—From cell biology to biomedical applications. Nat. Rev. Microbiol. 2010, 8, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Morabito, S. (Ed.) Pathogenic Escherichia Coli: Molecular and Cellular Microbiology; Caister Academic Press: Poole, UK, 2014. [Google Scholar]
- Römer, W.; Berland, L.; Chambon, V.; Gaus, K.; Windschiegl, B.; Tenza, D.; Aly, M.R.E.; Fraiser, V.; Florent, J.-C.; Perrais, D.; et al. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature 2007, 450, 670–675. [Google Scholar] [CrossRef]
- Römer, W.; Pontani, L.L.; Sorre, B.; Rentero, C.; Berland, L.; Chambon, V.; Lamaze, C.; Bassereau, P.; Sykes, C.; Gaus, K.; et al. Actin Dynamics Drive Membrane Reorganization and Scission in Clathrin-Independent Endocytosis. Cell 2010, 140, 540–553. [Google Scholar] [CrossRef] [PubMed]
- Kociurzynski, R.; Makshakova, O.N.; Knecht, V.; Römer, W. Multiscale Molecular Dynamics Studies Reveal Different Modes of Receptor Clustering by Gb3-Binding Lectins. J. Chem. Theory Comput. 2021, 17, 2488–2501. [Google Scholar] [CrossRef] [PubMed]
- Donohue-Rolfe, A.; Jacewicz, M.; Keusch, G.T. Isolation and characterization of functional Shiga toxin subunits and renatured holotoxin. Mol. Microbiol. 1989, 3, 1231–1236. [Google Scholar] [CrossRef]
- Batisse, C.; Dransart, E.; Ait Sarkouh, R.; Brulle, L.; Bai, S.-K.; Godefroy, S.; Johannes, L.; Schmidt, F. A new delivery system for auristatin in STxB-drug conjugate therapy. Eur. J. Med. Chem. 2015, 95, 483–491. [Google Scholar] [CrossRef]
- Rosato, F.; Pasupuleti, R.; Tomisch, J.; Meléndez, A.V.; Kolanovic, D.; Makshakova, O.N.; Wiltschi, B.; Römer, W. A bispecific, crosslinking lectibody activates cytotoxic T cells and induces cancer cell death. J. Transl. Med. 2022, 20, 578. [Google Scholar] [CrossRef]
- Meléndez, A.V.; Velasco Cárdenas, R.M.H.; Lagies, S.; Strietz, J.; Siukstaite, L.; Thomas, O.S.; Tomisch, J.; Weber, W.; Kammerer, B.; Römer, W.; et al. Novel lectin-based chimeric antigen receptors target Gb3-positive tumour cells. Cell Mol. Life Sci. 2022, 79, 513. [Google Scholar] [CrossRef]
- Danielewicz, N.; Rosato, F.; Tomisch, J.; Gräber, J.; Wiltschi, B.; Striedner, G.; Römer, W.; Mairhofer, J. Clickable Shiga Toxin B Subunit for Drug Delivery in Cancer Therapy. ACS Omega 2023, 8, 15406–15421. [Google Scholar] [CrossRef]
- Mohseni, Z.; Sedighian, H.; Halabian, R.; Amani, J.; Behzadi, E.; Imani Fooladi, A.A. Potent in vitro antitumor activity of B-subunit of Shiga toxin conjugated to the diphtheria toxin against breast cancer. Eur. J. Pharmacol. 2021, 899, 174057. [Google Scholar] [CrossRef]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 2020, 28, 1071–1081.e3. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gilson, M.K. AMBER 18; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Brischwein, K.; Parr, L.; Pflanz, S.; Volkland, J.; Lumsden, J.; Klinger, M.; Locher, M.; Hammond, S.A.; Kiener, P.; Kufer, P.; et al. Strictly Target Cell-dependent Activation of T Cells by Bispecific Single-chain Antibody Constructs of the BiTE Class. J. Immunother. 2007, 30, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Minguet, S.; Swamy, M.; Alarcón, B.; Luescher, I.F.; Schamel, W.W.A. Full Activation of the T Cell Receptor Requires Both Clustering and Conformational Changes at CD3. Immunity 2007, 26, 43–54. [Google Scholar] [CrossRef]
- Pina, D.G.; Gómez, J.; Villar, E.; Johannes, L.; Shnyrov, V.L. Thermodynamic Analysis of the Structural Stability of the Shiga Toxin B-Subunit. Biochemistry 2003, 42, 9498–9506. [Google Scholar] [CrossRef]
- Ling, H.; Boodhoo, A.; Hazes, B.; Cummings, M.D.; Armstrong, G.D.; Brunton, J.L.; Read, R.J. Structure of the Shiga-like Toxin I B-Pentamer Complexed with an Analogue of Its Receptor Gb3. Biochemistry 1998, 37, 1777–1788. [Google Scholar] [CrossRef]
- Batlevi, C.L.; Matsuki, E.; Brentjens, R.J.; Younes, A. Novel immunotherapies in lymphoid malignancies. Nat. Rev. Clin. Oncol. 2016, 13, 25–40. [Google Scholar] [CrossRef]
- Sinicrope, F.A. Increasing Incidence of Early-Onset Colorectal Cancer. N. Engl. J. Med. 2022, 386, 1547–1558. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Jakubowski, C.D.; Fedewa, S.A.; Davis, A.; Azad, N.S. Colorectal Cancer in the Young: Epidemiology, Prevention, Management. Am. Soc. Clin. Oncol. Educ. Book 2020, 40, e75–e88. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Peterson, D.A.; Kimura, H.; Schubert, D. Mechanism of Cellular 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) Reduction. J. Neurochem. 2002, 69, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Gupta, V.; Patwardhan, G.A.; Bhinge, K.; Zhao, Y.; Mehendale, H.; Cabot, M.C.; Li, Y.-T.; Jazwinski, S.M. Glycosylceramide Synthase Upregulates MDR1 Expression in the Regulation of Cancer Drug Resistance through cSrc and β-Catenin Signaling. Mol. Cancer 2010, 9, 145. [Google Scholar] [CrossRef]
- Padler-Karavani, V. Aiming at the sweet side of cancer: Aberrant glycosylation as possible target for personalized-medicine. Cancer Lett. 2014, 352, 102–112. [Google Scholar] [CrossRef]
- Feucht, J.; Kayser, S.; Gorodezki, D.; Hamieh, M.; Döring, M.; Blaescke, F.; Schlegel, P.; Bösmüller, H.; Quintanilla-Fend, L.; Ebinger, M.; et al. T-cell responses against CD19+ pediatric acute lymphoblastic leukemia mediated by bispecific T-cell engager (BiTE) are regulated contrarily by PD-L1 and CD80/CD86 on leukemic blasts. Oncotarget 2016, 7, 76902–76919. [Google Scholar] [CrossRef]
- Zugmaier, G.; Klinger, M.; Schmidt, M.; Subklewe, M. Clinical overview of anti-CD19 BiTE® and ex vivo data from anti-CD33 BiTE® as examples for retargeting T cells in hematologic malignancies. Mol. Immunol. 2015, 67, 58–66. [Google Scholar] [CrossRef]
- Löffler, A.; Gruen, M.; Wuchter, C.; Schriever, F.; Kufer, P.; Dreier, T.; Baeuerle, P.A.; Bommert, K.; Karaawajew, L.; Dörken, B.; et al. Efficient elimination of chronic lymphocytic leukaemia B cells by autologous T cells with a bispecific anti-CD19/anti-CD3 single-chain antibody construct. Leukemia 2003, 17, 900–909. [Google Scholar] [CrossRef]
- Pochechueva, T.; Jacob, F.; Fedier, A.; Heinzelmann-Schwarz, V. Tumor-Associated Glycans and Their Role in Gynecological Cancers: Accelerating Translational Research by Novel High-Throughput Approaches. Metabolites 2012, 2, 913–939. [Google Scholar] [CrossRef]
- Hakomori, S.I. Tumor-Associated Carbohydrate Antigens Defining Tumor Malignancy: Basis for Development of Anti-Cancer Vaccines. In The Molecular Immunology of Complex Carbohydrates—2; Wu, A.M., Ed.; Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2001; Volume 491, pp. 369–402. [Google Scholar] [CrossRef]
- Engedal, N.; Skotland, T.; Torgersen, M.L.; Sandvig, K. Shiga toxin and its use in targeted cancer therapy and imaging: Shiga toxin in cancer therapy and imaging. Microb. Biotechnol. 2011, 4, 32–46. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, Y.; Park, J.; Liu, X.; Hu, Y.; Wang, T.; McFArlad, K.; Betenbaugh, M.J. Design and Production of Bispecific Antibodies. Antibodies 2019, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, U.; Kontermann, R.E. The making of bispecific antibodies. mAbs 2017, 9, 182–212. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef]
- Kim, S.H.; Ryu, S.H.; Lee, S.H.; Lee, Y.-H.; Lee, S.-R.; Huh, J.-W.; Kim, S.-U.; Kim, E.; Kim, S.; Jon, S.; et al. Instability of toxin A subunit of AB5 toxins in the bacterial periplasm caused by deficiency of their cognate B subunits. Biochim. Biophys. Acta BBA-Biomembr. 2011, 1808, 2359–2365. [Google Scholar] [CrossRef]
- Le Gall, F.; Bové, J.M.; Garnier, M. Engineering of a Single-Chain Variable-Fragment (scFv) Antibody Specific for the Stolbur Phytoplasma (Mollicute) and Its Expression in Escherichia coli and Tobacco Plants. Appl. Env. Microbiol. 1998, 64, 4566–4572. [Google Scholar] [CrossRef] [PubMed]
- Dewi, K.S. Construction and Periplasmic Expression of the Anti-EGFRvIII ScFv Antibody Gene in Escherichia coli. Sci. Pharm. 2016, 84, 141–152. [Google Scholar] [CrossRef]
- Miller, K.D.; Weaver-Feldhaus, J.; Gray, S.A.; Siegel, R.W.; Feldhaus, M.J. Production, purification, and characterization of human scFv antibodies expressed in Saccharomyces cerevisiae, Pichia pastoris, and Escherichia coli. Protein Expr. Purif. 2005, 42, 255–267. [Google Scholar] [CrossRef]
- Denoncin, K.; Collet, J.F. Disulfide Bond Formation in the Bacterial Periplasm: Major Achievements and Challenges Ahead. Antioxid. Redox Signal. 2013, 19, 63–71. [Google Scholar] [CrossRef]
- Rodriguez, C.; Nam, D.H.; Kruchowy, E.; Ge, X. Efficient Antibody Assembly in E. coli Periplasm by Disulfide Bond Folding Factor Co-expression and Culture Optimization. Appl. Biochem. Biotechnol. 2017, 183, 520–529. [Google Scholar] [CrossRef]
- de Marco, A. Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microb. Cell Fact. 2009, 8, 26. [Google Scholar] [CrossRef]
- Seo, M.J.; Jeong, K.J.; Leysath, C.E.; Ellington, A.D.; Iverson, B.L.; Georgiou, G. Engineering antibody fragments to fold in the absence of disulfide bonds. Protein Sci. 2009, 18, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Malherbe, G.; Humphreys, D.P.; Davé, E. A robust fractionation method for protein subcellular localization studies in Escherichia coli. BioTechniques 2019, 66, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.H.; Hecht, M.H. Recombinant Proteins Can Be Isolated from E. coli Cells by Repeated Cycles of Freezing and Thawing. Nat. Biotechnol. 1994, 12, 1357–1360. [Google Scholar] [CrossRef] [PubMed]
- Ghamghami, E.; Abri Aghdam, M.; Tohidkia, M.R.; Ahmadikhah, A.; Khanmohammadi, M.; Mehdipour, T.; Mokhtarzadeh, A.; Bradaran, B. Optimization of Tris/EDTA/Sucrose (TES) periplasmic extraction for the recovery of functional scFv antibodies. AMB Expr. 2020, 10, 129. [Google Scholar] [CrossRef]
- Gallegos, K.M.; Conrady, D.G.; Karve, S.S.; Gunasekera, T.S.; Herr, A.B.; Weiss, A.A. Shiga Toxin Binding to Glycolipids and Glycans. PLoS ONE 2012, 7, e30368. [Google Scholar] [CrossRef]
- St Hilaire, P.M.; Boyd, M.K.; Toone, E.J. Interaction of the Shiga-like Toxin Type 1 B-Subunit with Its Carbohydrate Receptor. Biochemistry 1994, 33, 14452–14463. [Google Scholar] [CrossRef]
- Hexham, J.M.; Dudas, D.; Hugo, R.; Thompson, J.; King, V.; Dowling, C.; Neville, D.M.; Digan, M.E.; Lake, P. Influence of relative binding affinity on efficacy in a panel of anti-CD3 scFv immunotoxins. Mol. Immunol. 2001, 38, 397–408. [Google Scholar] [CrossRef]
- Adair, J.R.; Athwal, D.S.; Bodmer, M.W.; Bright, S.M.; Collins, A.M.; Pulito, V.L.; Rao, P.E.; Reedman, R.; Rothermel, A.L.; Xu, D. Humanization of the murine anti-human CD3 monoclonal antibody OKT3. Hum. Antibodies Hybrid. 1994, 5, 41–47. [Google Scholar] [CrossRef]
- Philipp, N.; Kazerani, M.; Nicholls, A.; Vick, B.; Wulf, J.; Straub, T.; Scheurer, M.; Muth, A.; Hänel, G.; Nixdorf, D.; et al. T-cell exhaustion induced by continuous bispecific molecule exposure is ameliorated by treatment-free intervals. Blood 2022, 140, 1104–1118. [Google Scholar] [CrossRef]
- Dopfer, E.P.; Hartl, F.A.; Oberg, H.H.; Siegers, G.M.; Yousefi, O.S.; Kock, S.; Fiala, G.J.; Garcillàn, B.; Sandstrom, A.; Alarcón, B.; et al. The CD3 Conformational Change in the γδ T Cell Receptor Is Not Triggered by Antigens but Can Be Enforced to Enhance Tumor Killing. Cell Rep. 2014, 7, 1704–1715. [Google Scholar] [CrossRef]
- Guha, P.; Heatherton, K.R.; O’Connell, K.P.; Alexander, I.S.; Katz, S.C. Assessing the Future of Solid Tumor Immunotherapy. Biomedicines 2022, 10, 655. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.M.; Duffy, M.R.; Freedman, J.D.; Fisher, K.D.; Seymour, L.W. Solid Tumor Immunotherapy with T Cell Engager-Armed Oncolytic Viruses. Macromol. Biosci. 2018, 18, 1700187. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Hou, S.; Li, W.; Li, K.; Xue, L.; Hu, Q.; Zhu, L.; Chen, Y.; Sun, H.; Ju, C.; et al. Combination of metabolic intervention and T cell therapy enhances solid tumor immunotherapy. Sci. Transl. Med. 2020, 12, eaaz6667. [Google Scholar] [CrossRef] [PubMed]
- Vierboom, M.P.M.; Bos, G.M.J.; Ooms, M.; Offringa, R.; Melief, C.J.M. Cyclophosphamide enhances anti-tumor effect of wild-type p53-specific CTL. Int. J. Cancer 2000, 87, 253–260. [Google Scholar] [CrossRef]
- Ebrahimnejad, P.; Dinarvand, R.; Sajadi, A.; Jaafari, M.R.; Nomani, A.R.; Azizi, E.; Rad-Malekshahi, M.; Atyabi, F. Preparation and in vitro evaluation of actively targetable nanoparticles for SN-38 delivery against HT-29 cell lines. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 478–485. [Google Scholar] [CrossRef]
- Johannes, L.; Goud, B. Surfing on a retrograde wave: How does Shiga toxin reach the endoplasmic reticulum? Trends Cell Biol. 1998, 8, 5. [Google Scholar] [CrossRef]
- Kim, J.H.; Lingwood, C.A.; Williams, D.B.; Furuya, W.; Manolson, M.F.; Grinstein, S. Dynamic measurement of the pH of the Golgi complex in living cells using retrograde transport of the verotoxin receptor. J. Cell Biol. 1996, 134, 1387–1399. [Google Scholar] [CrossRef]
- Sandvig, K.; Garred, Ø.; Prydz, K.; Kozlov, J.V.; Hansen, S.H.; van Deurs, B. Retrograde transport of endocytosed Shiga toxin to the endoplasmic reticulum. Nature 1992, 358, 510–512. [Google Scholar] [CrossRef]
- Sandvig, K.; Bergan, J.; Dyve, A.B.; Skotland, T.; Torgersen, M.L. Endocytosis and retrograde transport of Shiga toxin. Toxicon 2010, 56, 1181–1185. [Google Scholar] [CrossRef]
- Lingwood, C.A. Verotoxin/Globotriaosyl Ceramide Recognition: Angiopathy, Angiogenesis and Antineoplasia. Biosci. Rep. 1999, 19, 345–354. [Google Scholar] [CrossRef]
- Lingwood, C.A. Verotoxin-Binding in Human Renal Sections. Nephron 1994, 66, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, K.; Yokoyama, K.; Sato, T.; Wiels, J.; Hirayama, Y.; Ohta, M.; Furukawa, K. Expression of the Gb3/CD77 Synthase Gene in Megakaryoblastic Leukemia Cells: Implication in the Sensitivity to Verotoxins. J. Biol. Chem. 2002, 277, 11247–11254. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.A.; Vieira, F.L. Negative potential level in the outer layer of the toad skin. J. Membr. Biol. 1975, 24, 161–181. [Google Scholar] [CrossRef]
- Kontermann, R.E. Recombinant bispecific antibodies for cancer therapy. Acta Pharmacol. Sin. 2005, 26, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Huhalov, A.; Chester, K.A. Engineered single chain antibody fragments for radioimmunotherapy. Q. J. Nucl. Med. Mol. Imaging 2004, 48, 279–288. [Google Scholar]
- Kipriyanov, S.M.; Moldenhauer, G.; Schuhmacher, J.; Cochlovius, B.; Von Der Lieth, C.-W.; Matys, E.R.; Little, M. Bispecific tandem diabody for tumor therapy with improved antigen binding and pharmacokinetics. J. Mol. Biol. 1999, 293, 41–56. [Google Scholar] [CrossRef]
- Klinger, M.; Brandl, C.; Zugmaier, G.; Hijazi, Y.; Bargou, R.C.; Topp, M.S.; Gökbuget, N.; Neumann, S.; Goebeler, M.; Viardot, A.; et al. Immunopharmacologic response of patients with B-lineage acute lymphoblastic leukemia to continuous infusion of T cell–engaging CD19/CD3-bispecific BiTE antibody blinatumomab. Blood 2012, 119, 6226–6233. [Google Scholar] [CrossRef]
- Binnington, B.; Lingwood, D.; Nutikka, A.; Lingwood, C.A. Effect of Globotriaosyl Ceramide Fatty Acid-alpha-Hydroxylation on the Binding by Verotoxin 1 and Verotoxin 2. Neurochem. Res. 2002, 27, 807–813. [Google Scholar] [CrossRef]
- Kiarash, A.; Boyd, B.; Lingwood, C.A. Glycosphingolipid receptor function is modified by fatty acid content. Verotoxin 1 and verotoxin 2c preferentially recognize different globotriaosyl ceramide fatty acid homologues. J. Biol. Chem. 1994, 269, 11138–11146. [Google Scholar] [CrossRef]
- Wong, M.; Xu, G.; Park, D.; Barboza, M.; Lebrilla, C.B. Intact glycosphingolipidomic analysis of the cell membrane during differentiation yields extensive glycan and lipid changes. Sci. Rep. 2018, 8, 10993. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomisch, J.; Busse, V.; Rosato, F.; Makshakova, O.N.; Salavei, P.; Kittel, A.-S.; Gillon, E.; Lataster, L.; Imberty, A.; Meléndez, A.V.; et al. A Shiga Toxin B-Subunit-Based Lectibody Boosts T Cell Cytotoxicity towards Gb3-Positive Cancer Cells. Cells 2023, 12, 1896. https://doi.org/10.3390/cells12141896
Tomisch J, Busse V, Rosato F, Makshakova ON, Salavei P, Kittel A-S, Gillon E, Lataster L, Imberty A, Meléndez AV, et al. A Shiga Toxin B-Subunit-Based Lectibody Boosts T Cell Cytotoxicity towards Gb3-Positive Cancer Cells. Cells. 2023; 12(14):1896. https://doi.org/10.3390/cells12141896
Chicago/Turabian StyleTomisch, Jana, Vincent Busse, Francesca Rosato, Olga N. Makshakova, Pavel Salavei, Anna-Sophia Kittel, Emilie Gillon, Levin Lataster, Anne Imberty, Ana Valeria Meléndez, and et al. 2023. "A Shiga Toxin B-Subunit-Based Lectibody Boosts T Cell Cytotoxicity towards Gb3-Positive Cancer Cells" Cells 12, no. 14: 1896. https://doi.org/10.3390/cells12141896
APA StyleTomisch, J., Busse, V., Rosato, F., Makshakova, O. N., Salavei, P., Kittel, A.-S., Gillon, E., Lataster, L., Imberty, A., Meléndez, A. V., & Römer, W. (2023). A Shiga Toxin B-Subunit-Based Lectibody Boosts T Cell Cytotoxicity towards Gb3-Positive Cancer Cells. Cells, 12(14), 1896. https://doi.org/10.3390/cells12141896





