Disruption of the Mammalian Ccr4–Not Complex Contributes to Transcription-Mediated Genome Instability
Abstract
1. Introduction
2. Methods and Materials
2.1. Cell Culture
2.2. Cell Line Generation
Doxycycline Inducible CNOT1 Knockdown Cell Lines
2.3. Doxycycline Inducible RNase H1-DEAD U2OS Cell Lines
2.4. HEK293T/CNOT7-KO and HEK293T/CNOT8-KO Cell Lines
2.5. siRNA Transfection of Cell Lines
2.6. Transient Transfection of DNA
2.7. Colony Survival Assays
2.8. Natural Comet Assay
2.9. Nocodazole Mitotic Shake Off
2.10. FACS Analysis: Cell Cycle Analysis via Quantitation of DNA Content (Using Propidium Iodide Staining)
2.11. FACS Analysis: G1/S Cell Cycle Sub-Population Analysis Using EdU and BrdU Dual Pulse Labeling
2.12. Metaphase Spread Analysis
2.13. Co-Immunoprecipitation
2.14. DNA Fibre Analysis
2.15. Immunofluorescence Microscopy
2.16. EU Incorporation Assay
2.17. Western Blotting
2.18. Slot-Blot Experiments
2.19. Quantitative Real-Time PCR
2.20. Total RNA-Sequencing
3. Results
3.1. CNOT1-Depleted Cells Show Increased Global Transcriptional Activity
3.2. Depletion of CNOT1 Leads to Increased R-Loop Formation
3.3. Depletion of CNOT1 Induces Replication Stress through Ongoing Transcription
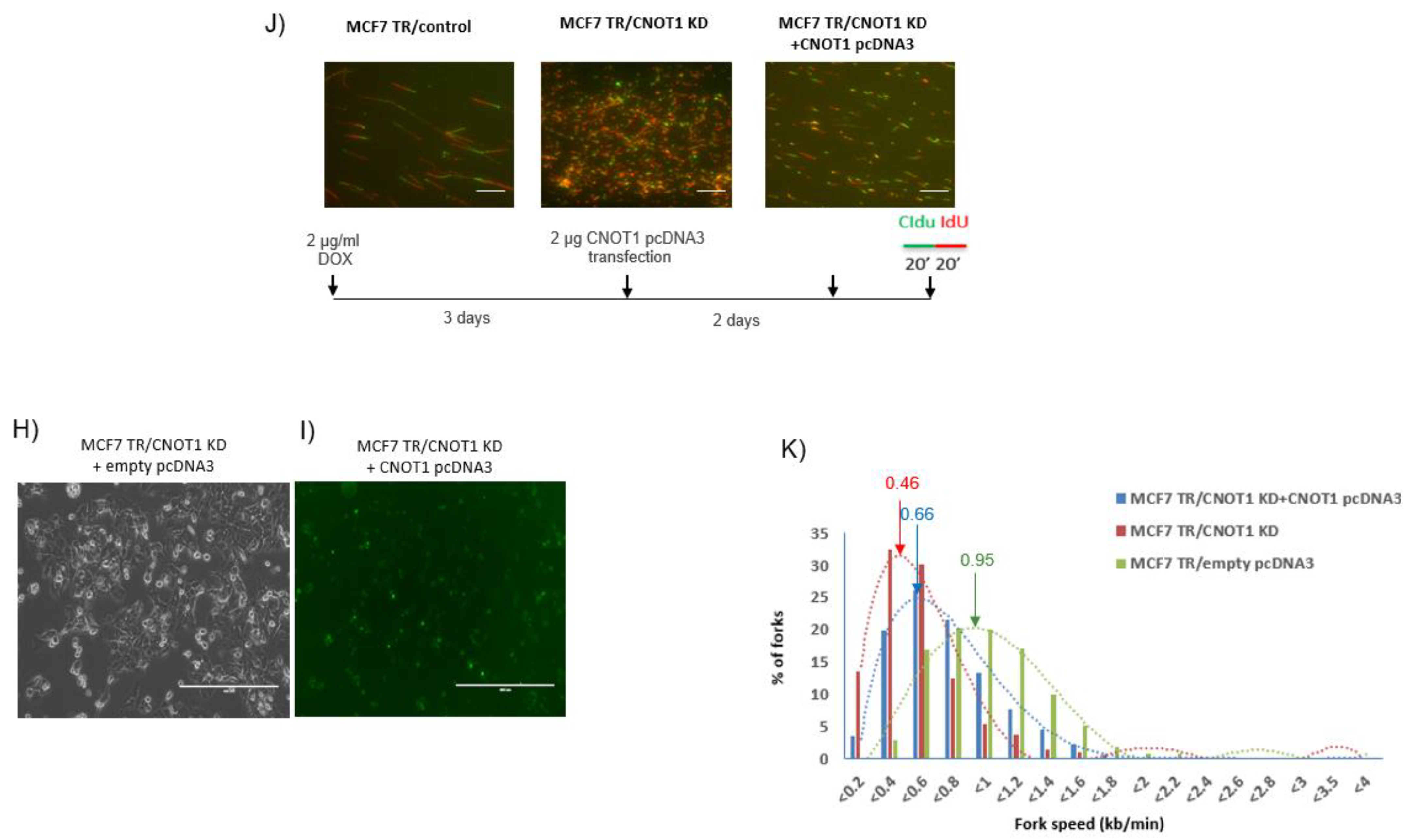
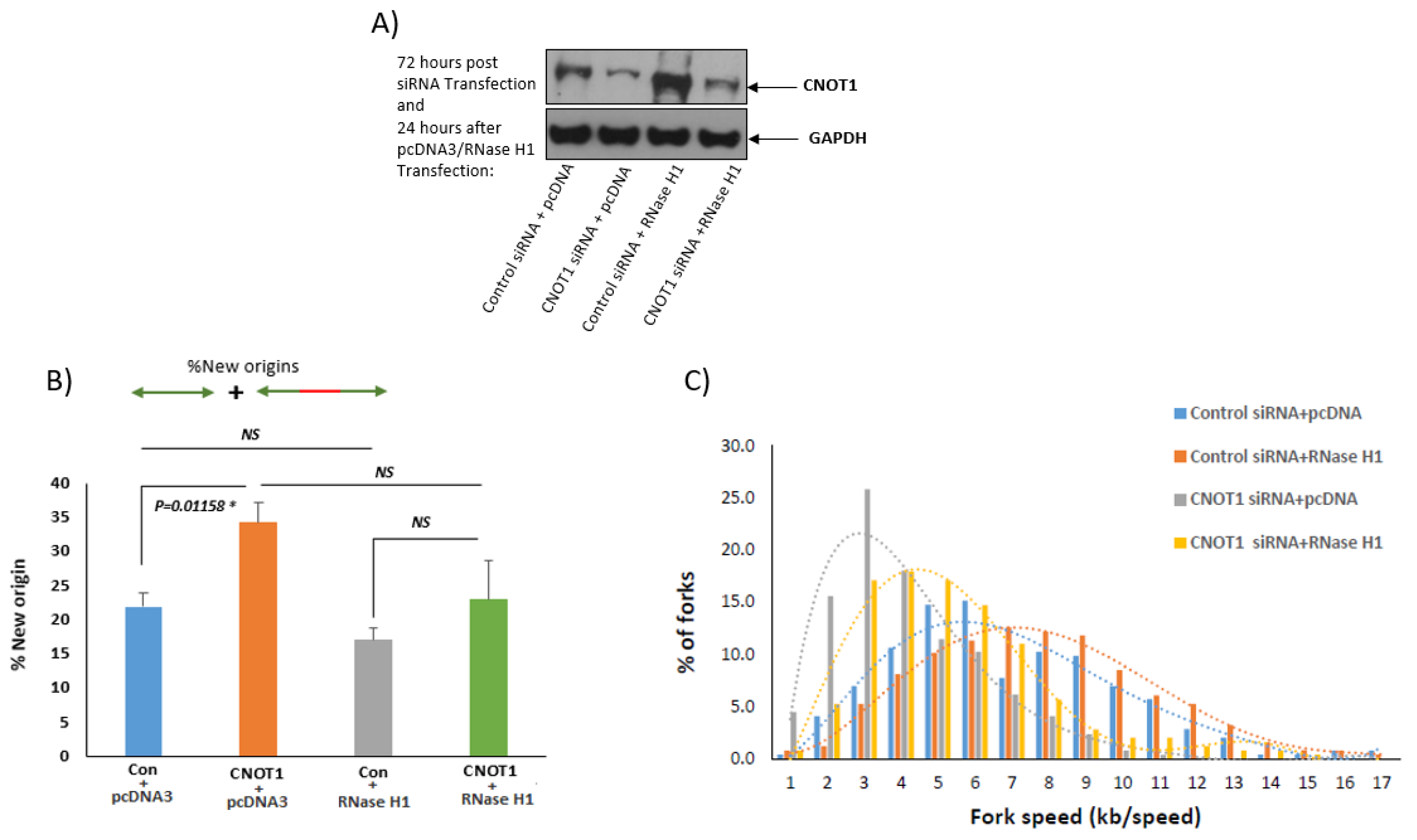
3.4. Depletion of CNOT1 Induces Replication Stress through R-Loop Formation
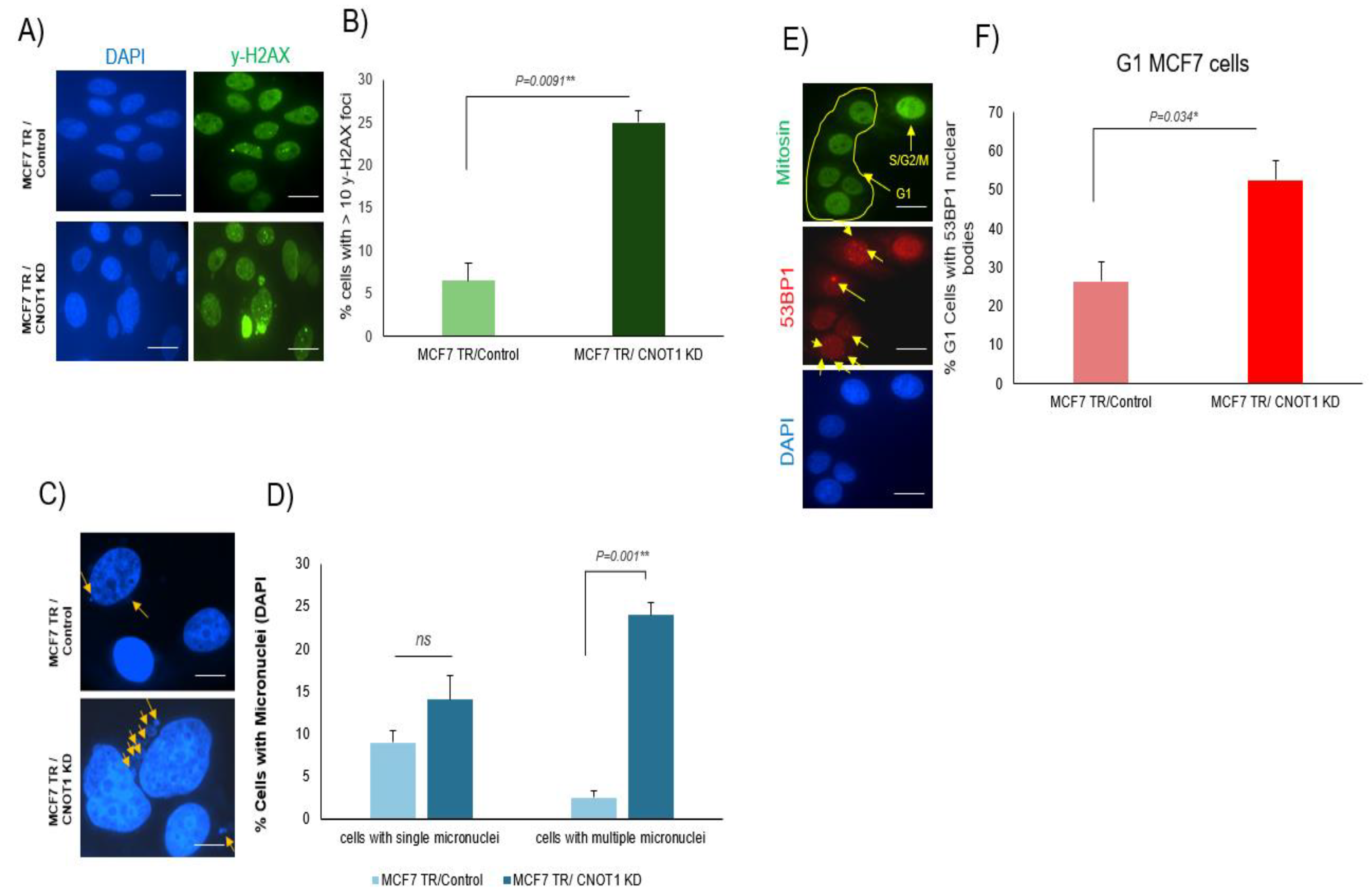
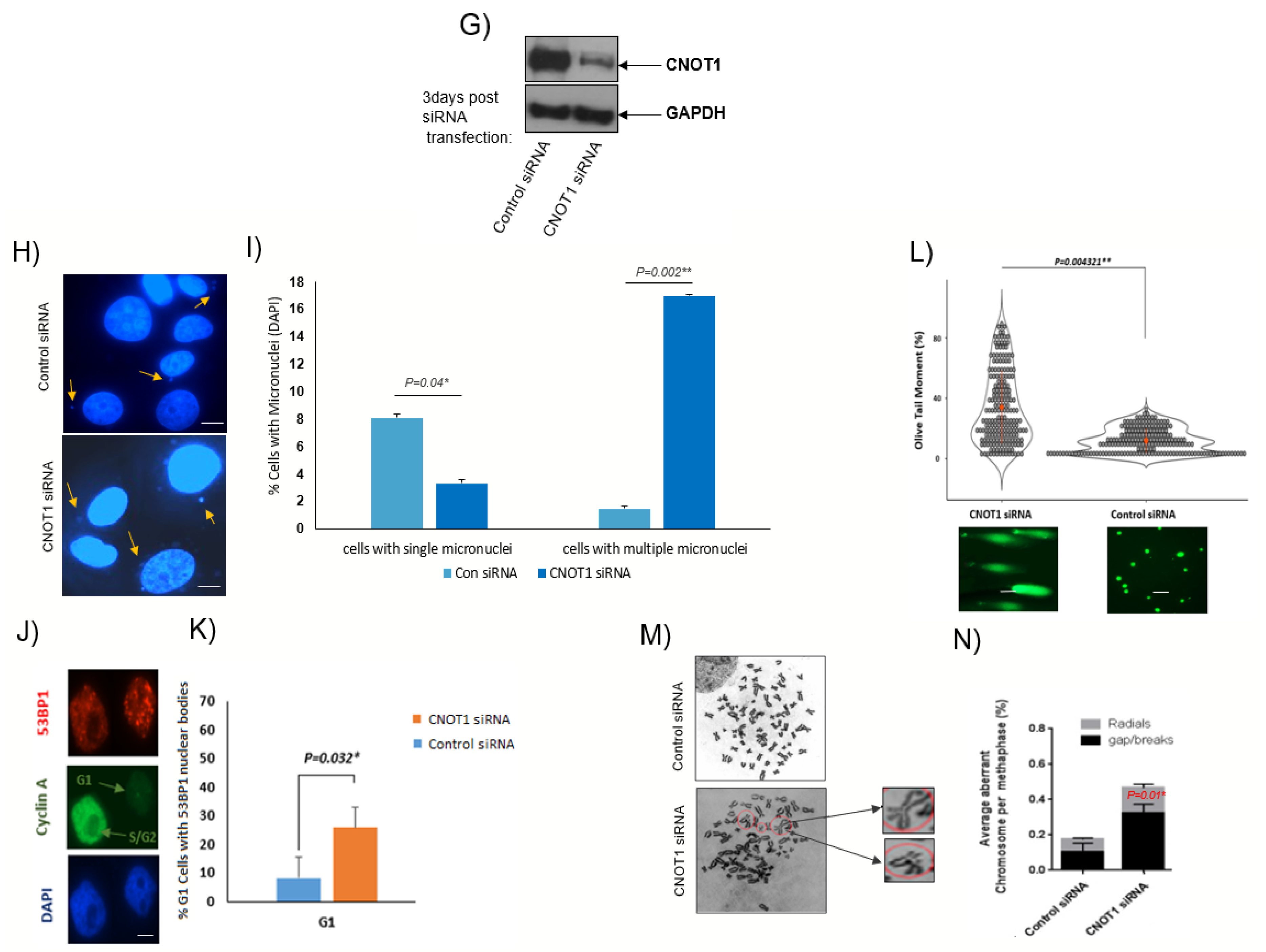
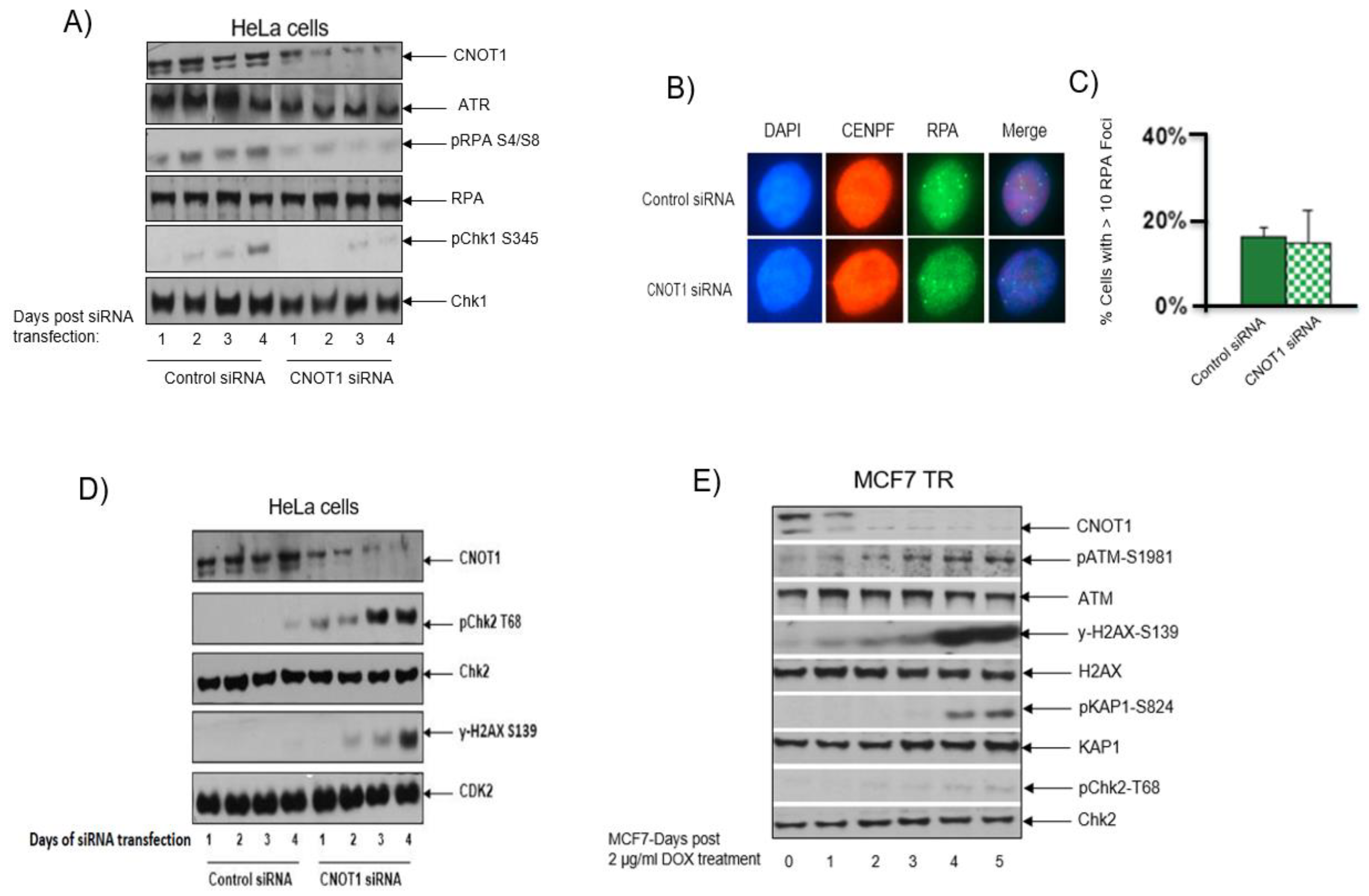
3.5. Increased Level of Double-Strand Breaks in Cells Lacking CNOT1
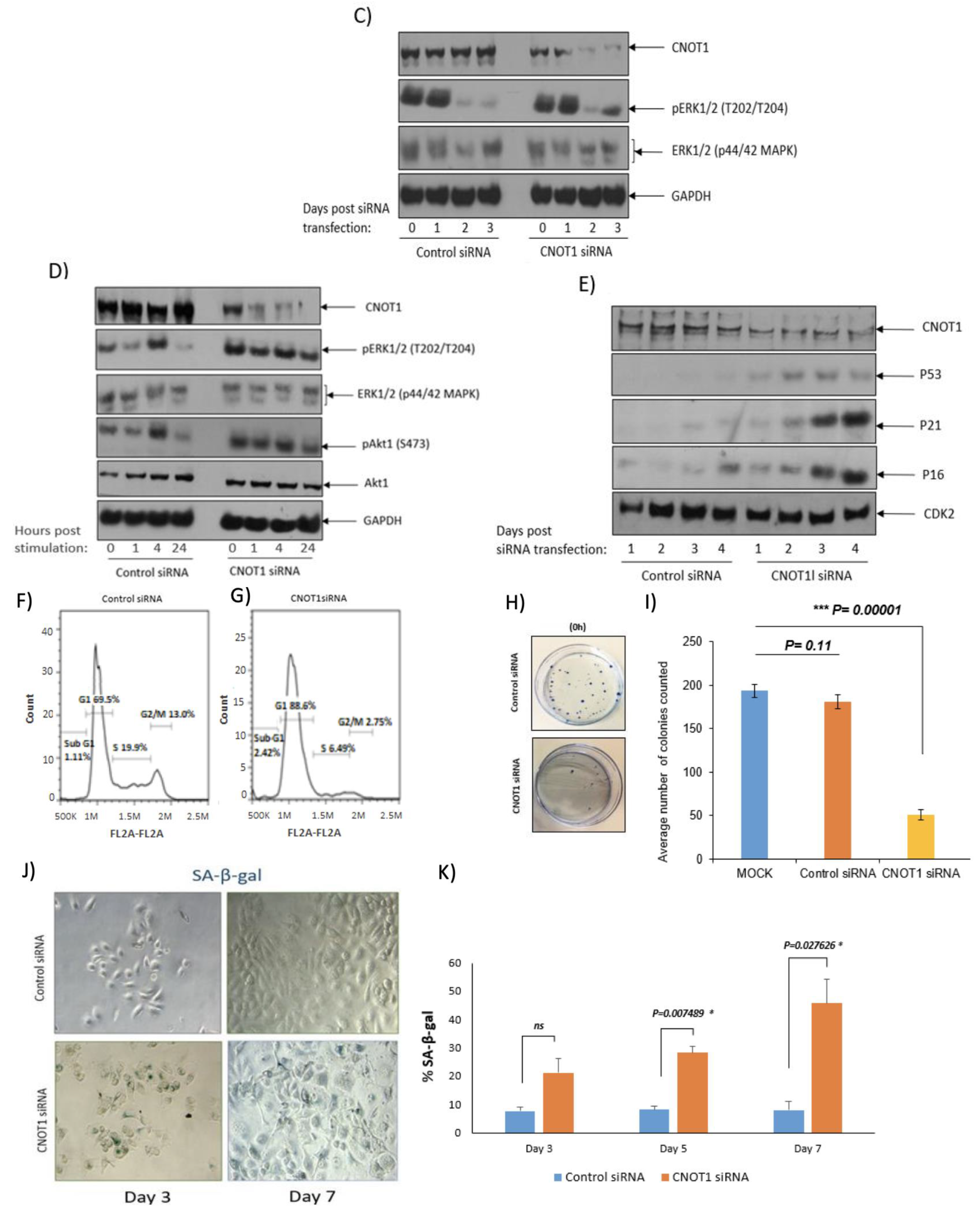
3.6. CNOT1 Depletion Leads to Activation of the ATM/Chk2 and Downregulation of the ATR/Chk1 Repair Pathways
3.7. DNA Damage-Induced Cell Cycle Arrest and Senescence in CNOT1-Depleted Cells
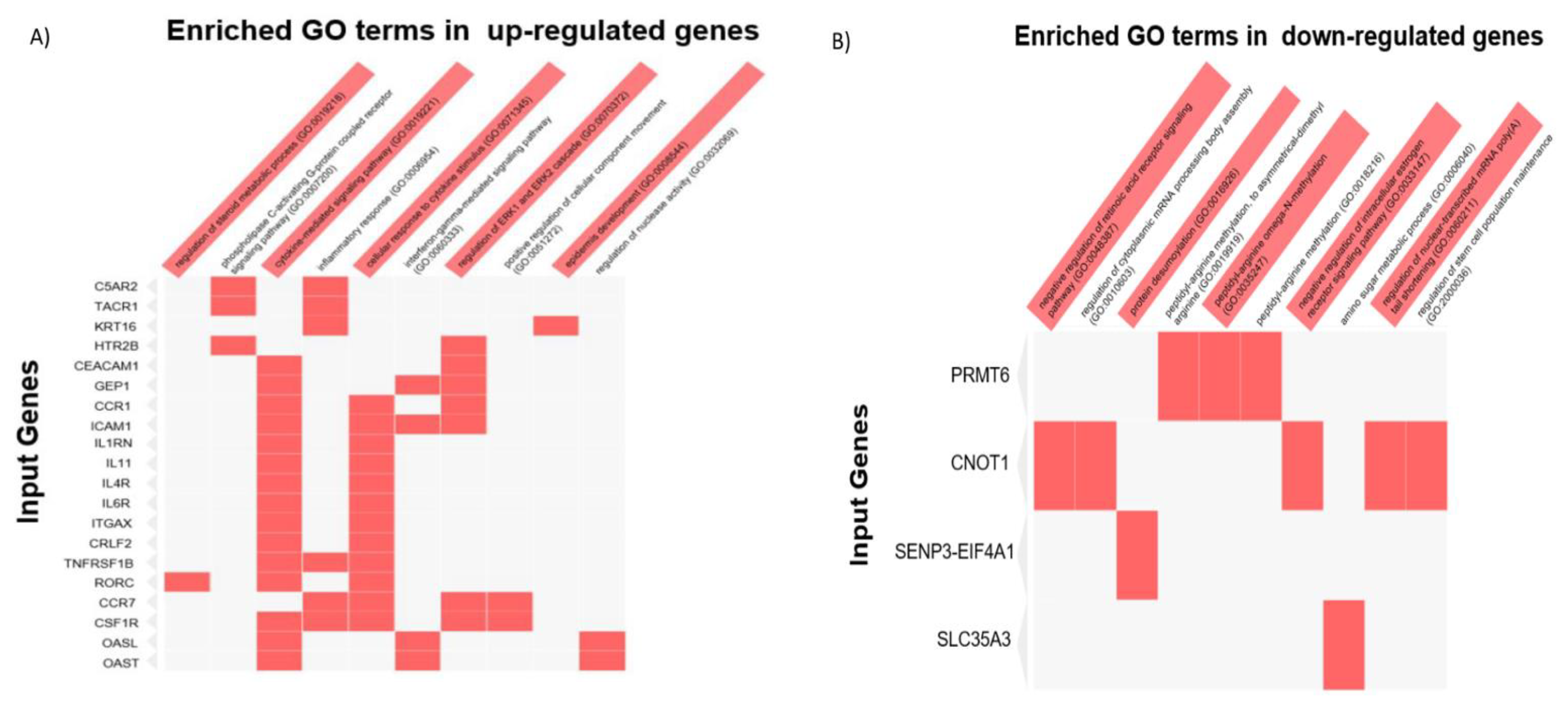
3.8. RNA-Seq Analysis of CNOT1-Depleted HeLa Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, J.; Rappsilber, J.; Chiang, Y.-C.; Russell, P.; Mann, M.; Denis, C.L. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J. Mol. Biol. 2001, 314, 683–694. [Google Scholar] [CrossRef]
- Denis, C.L.; Chen, J. The CCR4–NOT Complex Plays Diverse Roles in mRNA Metabolism. Prog. Nucleic Acid Res. Mol. Biol. 2003, 73, 221–250. [Google Scholar] [CrossRef]
- Kruk, J.A.; Dutta, A.; Fu, J.; Gilmour, D.S.; Reese, J.C. The multifunctional Ccr4–Not complex directly promotes transcription elongation. Genes Dev. 2011, 25, 581–593. [Google Scholar] [CrossRef]
- Collart, M.A.; Panasenko, O.O. The Ccr4–not complex. Gene 2012, 492, 42–53. [Google Scholar] [CrossRef]
- Inada, T.; Makino, S. Novel roles of the multi-functional CCR4-NOT complex in post-transcriptional regulation. Front. Genet. 2014, 5, 135. [Google Scholar] [CrossRef]
- Hagkarim, N.C.; Grand, R.J. The Regulatory Properties of the Ccr4–Not Complex. Cells 2020, 9, 2379. [Google Scholar] [CrossRef]
- Collart, M. Global control of gene expression in yeast by the Ccr4-Not complex. Gene 2003, 313, 1–16. [Google Scholar] [CrossRef]
- Collart, M.A. The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip. Rev. RNA 2016, 7, 438–454. [Google Scholar] [CrossRef]
- Stowell, J.A.; Webster, M.W.; Kögel, A.; Wolf, J.; Shelley, K.L.; Passmore, L.A. Reconstitution of Targeted Deadenylation by the Ccr4-Not Complex and the YTH Domain Protein Mmi1. Cell Rep. 2016, 17, 1978–1989. [Google Scholar] [CrossRef]
- Tucker, M.; Valencia-Sanchez, M.A.; Staples, R.R.; Chen, J.; Denis, C.L.; Parker, R. The Transcription Factor Associated Ccr4 and Caf1 Proteins Are Components of the Major Cytoplasmic mRNA Deadenylase in Saccharomyces cerevisiae. Cell 2001, 104, 377–386. [Google Scholar] [CrossRef]
- Tucker, M.; Staples, R.R.; Valencia-Sanchez, M.A.; Muhlrad, D.; Parker, R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002, 21, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Temme, C.; Zhang, L.; Kremmer, E.; Ihling, C.; Chartier, A.; Sinz, A.; Simonelig, M.; Wahle, E. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA 2010, 16, 1356–1370. [Google Scholar] [CrossRef] [PubMed]
- Basquin, J.; Roudko, V.V.; Rode, M.; Basquin, C.; Séraphin, B.; Conti, E. Architecture of the Nuclease Module of the Yeast Ccr4-Not Complex: The Not1-Caf1-Ccr4 Interaction. Mol. Cell 2012, 48, 207–218. [Google Scholar] [CrossRef]
- Petit, A.-P.; Wohlbold, L.; Bawankar, P.; Huntzinger, E.; Schmidt, S.; Izaurralde, E.; Weichenrieder, O. The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4–NOT deadenylase complex. Nucleic Acids Res. 2012, 40, 11058–11072. [Google Scholar] [CrossRef] [PubMed]
- Collart, M.A. The Not4 RING E3 Ligase: A Relevant Player in Cotranslational Quality Control. ISRN Mol. Biol. 2013, 2013, 548359. [Google Scholar] [CrossRef]
- Bhaskar, V.; Roudko, V.; Basquin, J.; Sharma, K.; Urlaub, H.; Séraphin, B.; Conti, E. Structure and RNA-binding properties of the Not1–Not2–Not5 module of the yeast Ccr4–Not complex. Nat. Struct. Mol. Biol. 2013, 20, 1281–1288. [Google Scholar] [CrossRef] [PubMed]
- Buschauer, R.; Matsuo, Y.; Sugiyama, T.; Chen, Y.-H.; Alhusaini, N.; Sweet, T.; Ikeuchi, K.; Cheng, J.; Matsuki, Y.; Nobuta, R.; et al. The Ccr4-Not complex monitors the translating ribosome for codon optimality. Science 2020, 368. [Google Scholar] [CrossRef]
- Collart, M.A.; Struhl, K. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 1994, 8, 525–537. [Google Scholar] [CrossRef]
- Sun, M.; Schwalb, B.; Schulz, D.; Pirkl, N.; Etzold, S.; Larivière, L.; Maier, K.C.; Seizl, M.; Tresch, A.; Cramer, P. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res. 2012, 22, 1350–1359. [Google Scholar] [CrossRef]
- Rambout, X.; Detiffe, C.; Bruyr, J.; Mariavelle, E.; Cherkaoui, M.; Brohée, S.; Demoitié, P.; Lebrun, M.; Soin, R.; Lesage, B.; et al. The transcription factor ERG recruits CCR4–NOT to control mRNA decay and mitotic progression. Nat. Struct. Mol. Biol. 2016, 23, 663–672. [Google Scholar] [CrossRef]
- Timmers, H.M.; Tora, L. Transcript Buffering: A Balancing Act between mRNA Synthesis and mRNA Degradation. Mol. Cell 2018, 72, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Begley, V.; Corzo, D.; Jordán-Pla, A.; Cuevas-Bermúdez, A.; De Miguel-Jiménez, L.; Pérez-Aguado, D.; Machuca-Ostos, M.; Navarro, F.; Chávez, M.J.; Pérez-Ortín, J.E.; et al. The mRNA degradation factor Xrn1 regulates transcription elongation in parallel to Ccr4. Nucleic Acids Res. 2019, 47, 9524–9541. [Google Scholar] [CrossRef]
- Slobodin, B.; Bahat, A.; Sehrawat, U.; Becker-Herman, S.; Zuckerman, B.; Weiss, A.; Han, R.; Elkon, R.; Agami, R.; Ulitsky, I.; et al. Transcription Dynamics Regulate Poly(A) Tails and Expression of the RNA Degradation Machinery to Balance mRNA Levels. Mol. Cell 2020, 78, 434–444.e5. [Google Scholar] [CrossRef] [PubMed]
- Reese, J.C. The control of elongation by the yeast Ccr4–Not complex. Biochim. et Biophys. Acta (BBA)-Gene Regul. Mech. 2013, 1829, 127–133. [Google Scholar] [CrossRef]
- Laribee, R.N.; Hosni-Ahmed, A.; Workman, J.J.; Chen, H. Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis. PLoS Genet. 2015, 11, e1005113. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wolgast, M.; Beebe, L.; Reese, J. Ccr4–Not maintains genomic integrity by controlling the ubiquitylation and degradation of arrested RNAPII. Genes Dev. 2019, 33, 705–717. [Google Scholar] [CrossRef]
- Benson, J.D.; Benson, M.; Howley, P.; Struhl, K. Association of distinct yeast Not2 functional domains with components of Gcn5 histone acetylase and Ccr4 transcriptional regulatory complexes. EMBO J. 1998, 17, 6714–6722. [Google Scholar] [CrossRef]
- Chang, M.; French-Cornay, D.; Fan, H.-Y.; Klein, H.; Denis, C.L.; Jaehning, J.A. A Complex Containing RNA Polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p Plays a Role in Protein Kinase C Signaling. Mol. Cell. Biol. 1999, 19, 1056–1067. [Google Scholar] [CrossRef]
- Badarinarayana, V.; Chiang, Y.-C.; Denis, C.L. Functional Interaction of CCR4-NOT Proteins with TATAA-Binding Protein (TBP) and Its Associated Factors in Yeast. Genetics 2000, 155, 1045–1054. [Google Scholar] [CrossRef]
- Lemaire, M.; Collart, M.A. The TATA-binding Protein-associated Factor yTafII19p Functionally Interacts with Components of the Global Transcriptional Regulator Ccr4-Not Complex and Physically Interacts with the Not5 Subunit. J. Biol. Chem. 2000, 275, 26925–26934. [Google Scholar] [CrossRef]
- Deluen, C.; James, N.; Maillet, L.; Molinete, M.; Theiler, G.; Lemaire, M.; Collart, M.A. The Ccr4-Not complex and yTAF1 (yTafII130p/yTafII145p) show physical and functional interactions. Mol. Cell. Biol. 2002, 22, 6735–6749. [Google Scholar] [CrossRef] [PubMed]
- Sanders, S.L.; Jennings, J.; Canutescu, A.; Link, A.J.; Weil, P.A. Proteomics of the Eukaryotic Transcription Machinery: Identification of Proteins Associated with Components of Yeast TFIID by Multidimensional Mass Spectrometry. Mol. Cell. Biol. 2002, 22, 4723–4738. [Google Scholar] [CrossRef]
- Swanson, M.J.; Qiu, H.; Sumibcay, L.; Krueger, A.; Kim, S.-J.; Natarajan, K.; Yoon, S.; Hinnebusch, A.G. A Multiplicity of Coactivators Is Required by Gcn4p at Individual Promoters In Vivo. Mol. Cell. Biol. 2003, 23, 2800–2820. [Google Scholar] [CrossRef] [PubMed]
- Oberholzer, U.; Collart, M.A. In Vitro Transcription of a TATA-Less Promoter: Negative Regulation by the Not1 Protein. Biol. Chem. 1999, 380, 1365–1370. [Google Scholar] [CrossRef]
- Maillet, L.; Tu, C.; Hong, Y.K.; Shuster, E.O.; Collart, M.A. The essential function of not1 lies within the Ccr4-not complex. J. Mol. Biol. 2000, 303, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Huisinga, K.L.; Pugh, B.F. A Genome-Wide Housekeeping Role for TFIID and a Highly Regulated Stress-Related Role for SAGA in Saccharomyces cerevisiae. Mol. Cell 2004, 13, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Lenssen, E.; James, N.; Pedruzzi, I.; Dubouloz, F.; Cameroni, E.; Bisig, R.; De Virgilio, C. The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation—Via a newly identified Glc7/Bud14 type I protein phosphatase module—And TFIID promoter distribution. Mol. Cell. Biol. 2005, 25, 488–498. [Google Scholar] [CrossRef]
- James, N.; Landrieux, E.; Collart, M.A. A SAGA-Independent Function of SPT3 Mediates Transcriptional Deregulation in a Mutant of the Ccr4-Not Complex in Saccharomyces cerevisiae. Genetics 2007, 177, 123–135. [Google Scholar] [CrossRef]
- Saponaro, M.; Kantidakis, T.; Mitter, R.; Kelly, G.; Heron, M.; Williams, H.; Söding, J.; Stewart, A.; Svejstrup, J.Q. RECQL5 Controls Transcript Elongation and Suppresses Genome Instability Associated with Transcription Stress. Cell 2014, 157, 1037–1049. [Google Scholar] [CrossRef]
- Kotsantis, P.; Silva, L.M.; Irmscher, S.; Jones, R.M.; Folkes, L.; Gromak, N.; Petermann, E. Increased global transcription activity as a mechanism of replication stress in cancer. Nat. Commun. 2016, 7, 13087. [Google Scholar] [CrossRef]
- Reines, D.; Chamberlin, M.J.; Kane, C.M. Transcription elongation factor SII (TFIIS) enables RNA polymerase II to elongate through a block to tran-scription in a human gene in vitro. J. Biol. Chem. 1989, 264, 10799–10809. [Google Scholar] [CrossRef]
- Cipres-Palacin, G.; Kane, C.M. Cleavage of the nascent transcript induced by TFIIS is insufficient to promote read-through of intrinsic blocks to elongation by RNA polymerase II. Proc. Natl. Acad. Sci. USA 1994, 91, 8087–8091. [Google Scholar] [CrossRef] [PubMed]
- Hausner, W.; Lange, U.; Musfeldt, M. Transcription Factor S, a Cleavage Induction Factor of the Archaeal RNA Polymerase. J. Biol. Chem. 2000, 275, 12393–12399. [Google Scholar] [CrossRef]
- Nudler, E. RNA Polymerase Backtracking in Gene Regulation and Genome Instability. Cell 2012, 149, 1438–1445. [Google Scholar] [CrossRef]
- Dutta, A.; Babbarwal, V.; Fu, J.; Brunke-Reese, D.; Libert, D.M.; Willis, J.; Reese, J.C. Ccr4-Not and TFIIS Function Cooperatively To Rescue Arrested RNA Polymerase II. Mol. Cell. Biol. 2015, 35, 1915–1925. [Google Scholar] [CrossRef] [PubMed]
- Kerr, S.C.; Azzouz, N.; Fuchs, S.M.; Collart, M.A.; Strahl, B.D.; Corbett, A.H.; Laribee, R.N. The Ccr4-Not Complex Interacts with the mRNA Export Machinery. PLoS ONE 2011, 6, e18302. [Google Scholar] [CrossRef] [PubMed]
- Kordyukova, M.; Sokolova, O.; Morgunova, V.; Ryazansky, S.; Akulenko, N.; Glukhov, S.; Kalmykova, A. Nuclear Ccr4-Not mediates the degradation of telomeric and transposon transcripts at chromatin in the Drosophila germline. Nucleic Acids Res. 2019, 48, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Sollier, J.; Stork, C.T.; Garcia-Rubio, M.L.; Paulsen, R.D.; Aguilera, A.; Cimprich, K.A. Transcription-Coupled Nucleotide Excision Repair Factors Promote R-Loop-Induced Genome Instability. Mol. Cell 2014, 56, 777–785. [Google Scholar] [CrossRef]
- Gaillard, H.; Tous, C.; Botet, J.; González-Aguilera, C.; Quintero, M.J.; Viladevall, L.; García-Rubio, M.L.; Rodríguez-Gil, A.; Marín, A.; Ariño, J.; et al. Genome-Wide Analysis of Factors Affecting Transcription Elongation and DNA Repair: A New Role for PAF and Ccr4-Not in Transcription-Coupled Repair. PLoS Genet. 2009, 5, e1000364. [Google Scholar] [CrossRef]
- Deshpande, G.P.; Hayles, J.; Hoe, K.-L.; Kim, D.-U.; Park, H.-O.; Hartsuiker, E. Screening a genome-wide S. pombe deletion library identifies novel genes and pathways involved in genome stability maintenance. DNA Repair 2009, 8, 672–679. [Google Scholar] [CrossRef]
- Westmoreland, T.J.; Marks, J.R.; Olson, J.A.; Thompson, E.M.; Resnick, M.A.; Bennett, C.B. Cell Cycle Progression in G 1 and S Phases Is CCR4 Dependent following Ionizing Radiation or Replication Stress in Saccharomyces cerevisiae. Eukaryot. Cell 2004, 3, 430–446. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Perez, I.; Manguan-Garcia, C.; Menacho-Marquez, M.; Murguía, J.; Perona, R. hCCR4/cNOT6 targets DNA-damage response proteins. Cancer Lett. 2009, 273, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.-H.; Shang, Z.-F.; Tan, W.; Liu, X.-D.; Xu, Q.-Z.; Song, M.; Wang, Y.; Guan, H.; Zhang, S.-M.; Yu, L.; et al. TNKS1BP1 functions in DNA double-strand break repair though facilitating DNA-PKcs autophosphorylation dependent on PARP-1. Oncotarget 2015, 6, 7011–7022. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R., III; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR Substrate Analysis Reveals Extensive Protein Networks Responsive to DNA Damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef]
- Ito, K.; Takahashi, A.; Morita, M.; Suzuki, T.; Yamamoto, T. The role of the CNOT1 subunit of the CCR4-NOT complex in mRNA deadenylation and cell viability. Protein Cell 2011, 2, 755–763. [Google Scholar] [CrossRef]
- Van De Wetering, M.; Oving, I.; Muncan, V.; Pon Fong, M.T.; Brantjes, H.; van Leenen, D.; Clevers, H. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003, 4, 609–615. [Google Scholar] [CrossRef]
- Lappin, K.M.; Barros, E.M.; Jhujh, S.S.; Irwin, G.W.; McMillan, H.; Liberante, F.G.; Latimer, C.; La Bonte, M.J.; Mills, K.I.; Harkin, D.P.; et al. Cancer-Associated SF3B1 Mutations Confer a BRCA-Like Cellular Phenotype and Synthetic Lethality to PARP Inhibitors. Cancer Res 2022, 82, 819–830. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Takahashi, A.; Suzuki, T.; Soeda, S.; Takaoka, S.; Kobori, S.; Yamaguchi, T.; Mohamed, H.M.A.; Yanagiya, A.; Abe, T.; Shigeta, M.; et al. The CCR4–NOT complex maintains liver homeostasis through mRNA deadenylation. Life Sci. Alliance 2020, 3, e201900494. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.-C.; Kolkman, A.; van Schaik, F.M.A.; Mulder, K.W.; Pijnappel, W.W.M.P.; Heck, A.J.R.; Timmers, H.T.M. Human Ccr4–Not complexes contain variable deadenylase subunits. Biochem. J. 2009, 422, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Albert, T.K.; Hanzawa, H.; Legtenberg, Y.I.; de Ruwe, M.J.; Heuvel, F.A.V.D.; Collart, M.A.; Boelens, R.; Timmers, H. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 2002, 21, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Keskeny, C.; Raisch, T.; Sgromo, A.; Igreja, C.; Bhandari, D.; Weichenrieder, O.; Izaurralde, E. A conserved CAF40-binding motif in metazoan NOT4 mediates association with the CCR4–NOT complex. Genes Dev. 2019, 33, 236–252. [Google Scholar] [CrossRef]
- Stathaki, K.; Proudfoot, N.J. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014, 28, 1384–1396. [Google Scholar] [CrossRef]
- Bou-Nader, C.; Bothra, A.; Garboczi, D.N.; Leppla, S.H.; Zhang, J. Structural basis of R-loop recognition by the S9.6 monoclonal antibody. Nat. Commun. 2022, 13, 1641. [Google Scholar] [CrossRef]
- Aslam, A.; Mittal, S.; Koch, F.; Andrau, J.-C.; Winkler, G.S. The Ccr4–Not Deadenylase Subunits CNOT7 and CNOT8 Have Overlapping Roles and Modulate Cell Proliferation. Mol. Biol. Cell 2009, 20, 3840–3850. [Google Scholar] [CrossRef]
- Jonkers, I.; Lis, J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015, 16, 167–177. [Google Scholar] [CrossRef]
- Chao, S.-H.; Price, D.H. Flavopiridol Inactivates P-TEFb and Blocks Most RNA Polymerase II Transcription in Vivo. J. Biol. Chem. 2001, 276, 31793–31799. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Oleinick, N.L.; Zhang, J. ATR/CHK1 inhibitors and cancer therapy. Radiother. Oncol. 2018, 126, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Wold, M.S. Replication protein A: Single-stranded DNA’s first responder: Dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays 2014, 36, 1156–1161. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, G.; Feng, X.; Shepherd, P.; Zhang, J.; Tang, M.; Chen, Z.; Srivastava, M.; McLaughlin, M.E.; Navone, N.M.; et al. Genome-wide CRISPR screens reveal synthetic lethality of RNASEH2 deficiency and ATR inhibition. Oncogene 2018, 38, 2451–2463. [Google Scholar] [CrossRef]
- Li, B.; Zhang, S.; Zhang, H.; Nu, W.; Cai, L.; Hertz, L.; Peng, L. Fluoxetine-mediated 5-HT2B receptor stimulation in astrocytes causes EGF receptor transactivation and ERK phosphorylation. Psychopharmacology 2008, 201, 443–458. [Google Scholar] [CrossRef]
- Hertz, L.; Rothman, D.L.; Li, B.; Peng, L. Chronic SSRI stimulation of astrocytic 5-HT2B receptors change multiple gene expressions/editings and me-tabolism of glutamate, glucose and glycogen: A potential paradigm shift. Front. Behav. Neurosci. 2015, 9, 25. [Google Scholar]
- Winkler, G.S.; Mulder, K.W.; Bardwell, V.J.; Kalkhoven, E.; Timmers, H.T.M. Human Ccr4-Not complex is a ligand-dependent repressor of nuclear receptor-mediated transcription. EMBO J. 2006, 25, 3089–3099. [Google Scholar] [CrossRef]
- Whitmarsh, A.J.; Davis, R.J. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J. Mol. Med. 1996, 74, 589–607. [Google Scholar] [CrossRef]
- Meloche, S.; Pouysségur, J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1-to S-phase transition. Oncogene 2007, 26, 3227–3239. [Google Scholar] [CrossRef]
- Pumiglia, K.M.; Decker, S.J. Cell cycle arrest mediated by the MEK/mitogen-activated protein kinase pathway. Proc. Natl. Acad. Sci. USA 1997, 94, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Sewing, A.; Wiseman, B.; Lloyd, A.C.; Land, H. High-Intensity Raf Signal Causes Cell Cycle Arrest Mediated by p21Cip1. Mol. Cell. Biol. 1997, 17, 5588–5597. [Google Scholar] [CrossRef] [PubMed]
- Woods, D.; Parry, D.; Cherwinski, H.; Bosch, E.; Lees, E.; McMahon, M. Raf-Induced Proliferation or Cell Cycle Arrest Is Determined by the Level of Raf Activity with Arrest Mediated by p21Cip1. Mol. Cell. Biol. 1997, 17, 5598–5611. [Google Scholar] [CrossRef] [PubMed]
- Kerkhoff, E.; Rapp, U.R. High-intensity Raf signals convert mitotic cell cycling into cellular growth. Cancer Res. 1998, 58. [Google Scholar]
- Tombes, M.R.; Auer, L.K.; Mikkelsen, R.; Valerie, K.; Wymann, M.; Marshall, J.C.; McMahon, M.; Dent, P. The mitogen-activated protein (MAP) kinase cascade can either stimulate or inhibit DNA synthesis in primary cultures of rat hepatocytes depending upon whether its activation is acute/phasic or chronic. Biochem. J. 1998, 330, 1451–1460. [Google Scholar] [CrossRef]
- Felsher, D.W.; Zetterberg, A.; Zhu, J.; Tlsty, T.; Bishop, J.M. Overexpression of MYC causes p53-dependent G2 arrest of normal fibroblasts. Proc. Natl. Acad. Sci. USA 2000, 97, 10544–10548. [Google Scholar] [CrossRef]
- Kadyrova, L.Y.; Habara, Y.; Lee, T.H.; Wharton, R.P. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 2007, 134, 1519–1527. [Google Scholar] [CrossRef]
- Dronamraju, R.; Hepperla, A.J.; Shibata, Y.; Adams, A.T.; Magnuson, T.; Davis, I.J.; Strahl, B.D. Spt6 Association with RNA Polymerase II Directs mRNA Turnover during Transcription. Mol. Cell 2018, 70, 1054–1066.e4. [Google Scholar] [CrossRef]
- Liu, H.; Toyn, J.H.; Chiang, Y.; Draper, M.P.; Johnston, L.H.; Denis, C.L. DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J. 1997, 16, 5289–5298. [Google Scholar] [CrossRef]
- Mittal, S.; Aslam, A.; Doidge, R.; Medica, R.; Winkler, G.S. The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4–Not complex contribute to the prevention of cell death and senescence. Mol. Biol. Cell 2011, 22, 748–758. [Google Scholar] [CrossRef]
- Robin-Lespinasse, Y.; Sentis, S.; Kolytcheff, C.; Rostan, M.-C.; Corbo, L.; Le Romancer, M. hCAF1, a new regulator of PRMT1-dependent arginine methylation. J. Cell Sci. 2007, 120, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Chapat, C.; Chettab, K.; Simonet, P.; Wang, P.; De La Grange, P.; Le Romancer, M.; Corbo, L. Alternative splicing of CNOT7 diversifies CCR4–NOT functions. Nucleic Acids Res. 2017, 45, 8508–8523. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Suzuki, T.; Sato, T.; Takahashi, A.; Watanabe, H.; Kadowaki, A.; Natsui, M.; Inagaki, H.; Arakawa, S.; Nakaoka, S.; et al. The CCR4-NOT deadenylase complex controls Atg7-dependent cell death and heart function. Sci. Signal. 2018, 11, eaan3638. [Google Scholar] [CrossRef]
- Jeong, K.; Kwon, H.Y.; Jeong, M.S.; Sohn, E.J.; Kim, S.H. CNOT2 promotes degradation of p62/SQSTM1 as a negative regulator in ATG5 dependent autophagy. Oncotarget 2017, 8, 46034. [Google Scholar] [CrossRef]
- Venturini, G.; Rose, A.M.; Shah, A.Z.; Bhattacharya, S.S.; Rivolta, C. CNOT3 Is a Modifier of PRPF31 Mutations in Retinitis Pigmentosa with Incomplete Penetrance. PLoS Genet. 2012, 8, e1003040. [Google Scholar] [CrossRef]
- Traven, A.; Hammet, A.; Tenis, N.; Denis, C.L.; Heierhorst, J. Ccr4-Not Complex mRNA Deadenylase Activity Contributes to DNA Damage Responses in Saccharomyces cerevisiae. Genetics 2005, 169, 65–75. [Google Scholar] [CrossRef]
- Song, P.; Liu, S.; Liu, D.; Keijzers, G.; Bakula, D.; Duan, S.; de Wind, N.; Ye, Z.; Vakhrushev, S.Y.; Scheibye-Knudsen, M.; et al. CNOT6: A Novel Regulator of DNA Mismatch Repair. Cells 2022, 11, 521. [Google Scholar] [CrossRef]
- Morel, A.-P.; Sentis, S.; Bianchin, C.; Le Romancer, M.; Jonard, L.; Rostan, M.-C.; Rimokh, R.; Corbo, L. BTG2 antiproliferative protein interacts with the human CCR4 complex existing in vivo in three cell-cycle-regulated forms. J. Cell Sci. 2003, 116, 2929–2936. [Google Scholar] [CrossRef] [PubMed]
- Shirai, Y.T.; Suzuki, T.; Morita, M.; Takahashi, A.; Yamamoto, T. Multifunctional roles of the mammalian CCR4–NOT complex in physiological phenomena. Front. Genet. 2014, 5, 286. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Camino, A.; Lopez-Lopez, E.; Martin-Guerrero, I.; Piñan, M.A.; Garcia-Miguel, P.; Sanchez-Toledo, J.; Bañeres, A.C.; Uriz, J.; Navajas, A.; Garcia Orad, A. Noncoding RNA–related polymorphisms in pediatric acute lymphoblastic leukemia susceptibility. Pediatr. Res. 2014, 75, 767–773. [Google Scholar] [CrossRef]
- Cheng, D.-D.; Li, J.; Li, S.-J.; Yang, Q.-C.; Fan, C.-Y. CNOT1 cooperates with LMNA to aggravate osteosarcoma tumorigenesis through the Hedgehog signaling pathway. Mol. Oncol. 2017, 11, 388–404. [Google Scholar] [CrossRef] [PubMed]
- Oberhuber, M.; Pecoraro, M.; Rusz, M.; Oberhuber, G.; Wieselberg, M.; Haslinger, P.; Kenner, L. STAT 3-dependent analysis reveals PDK 4 as independent predictor of recurrence in prostate cancer. Mol. Syst. Biol. 2020, 16, e9247. [Google Scholar] [CrossRef] [PubMed]

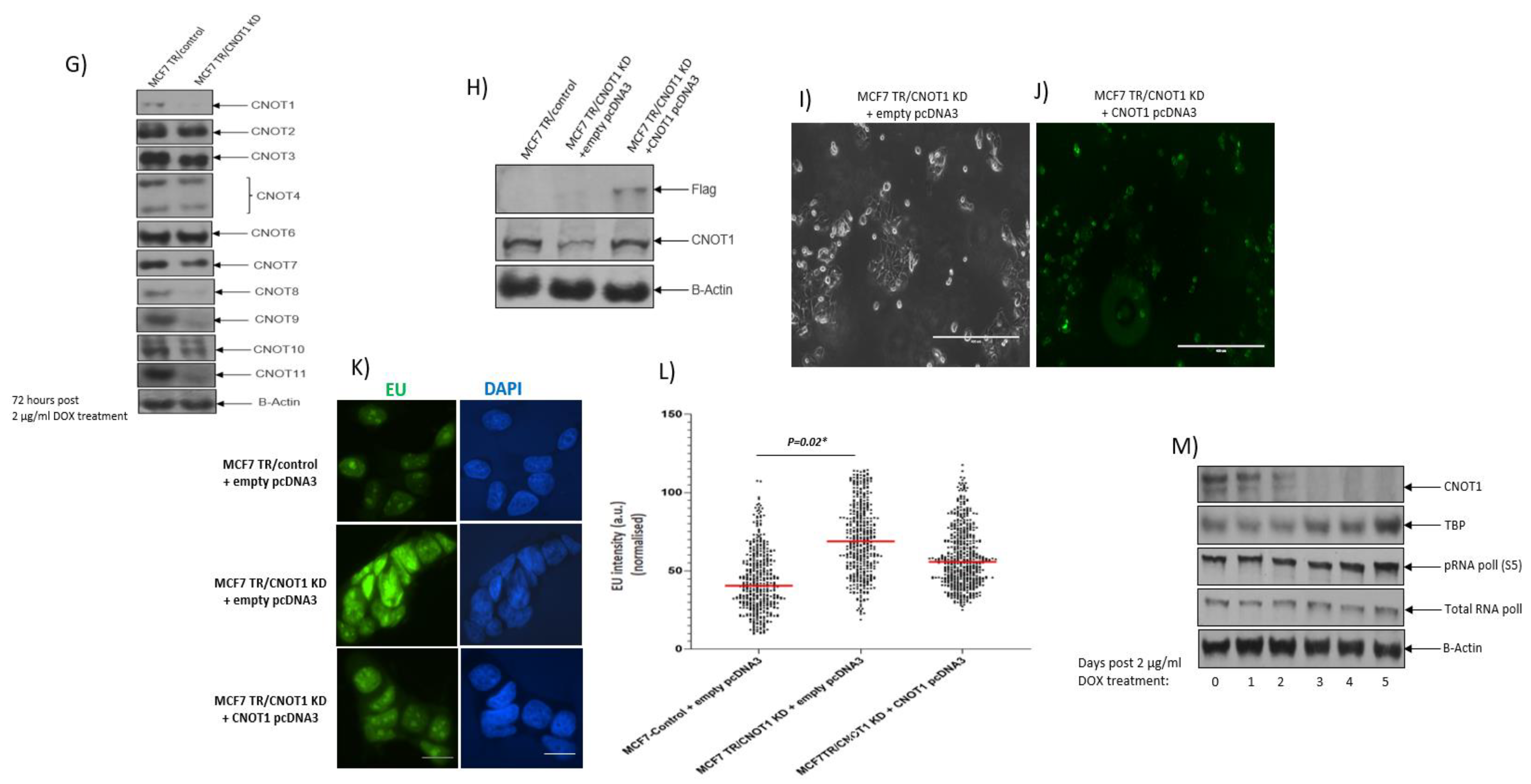
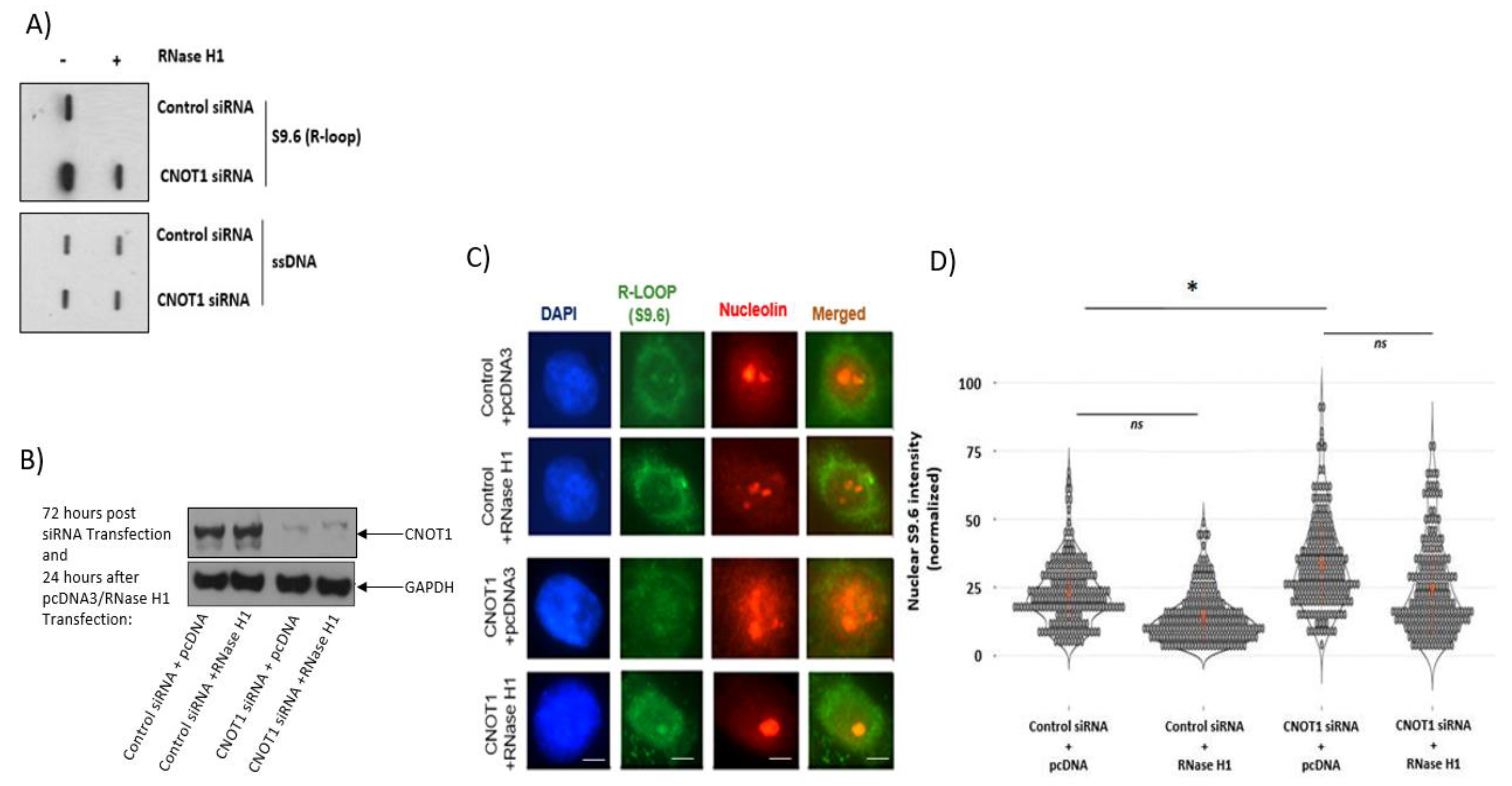
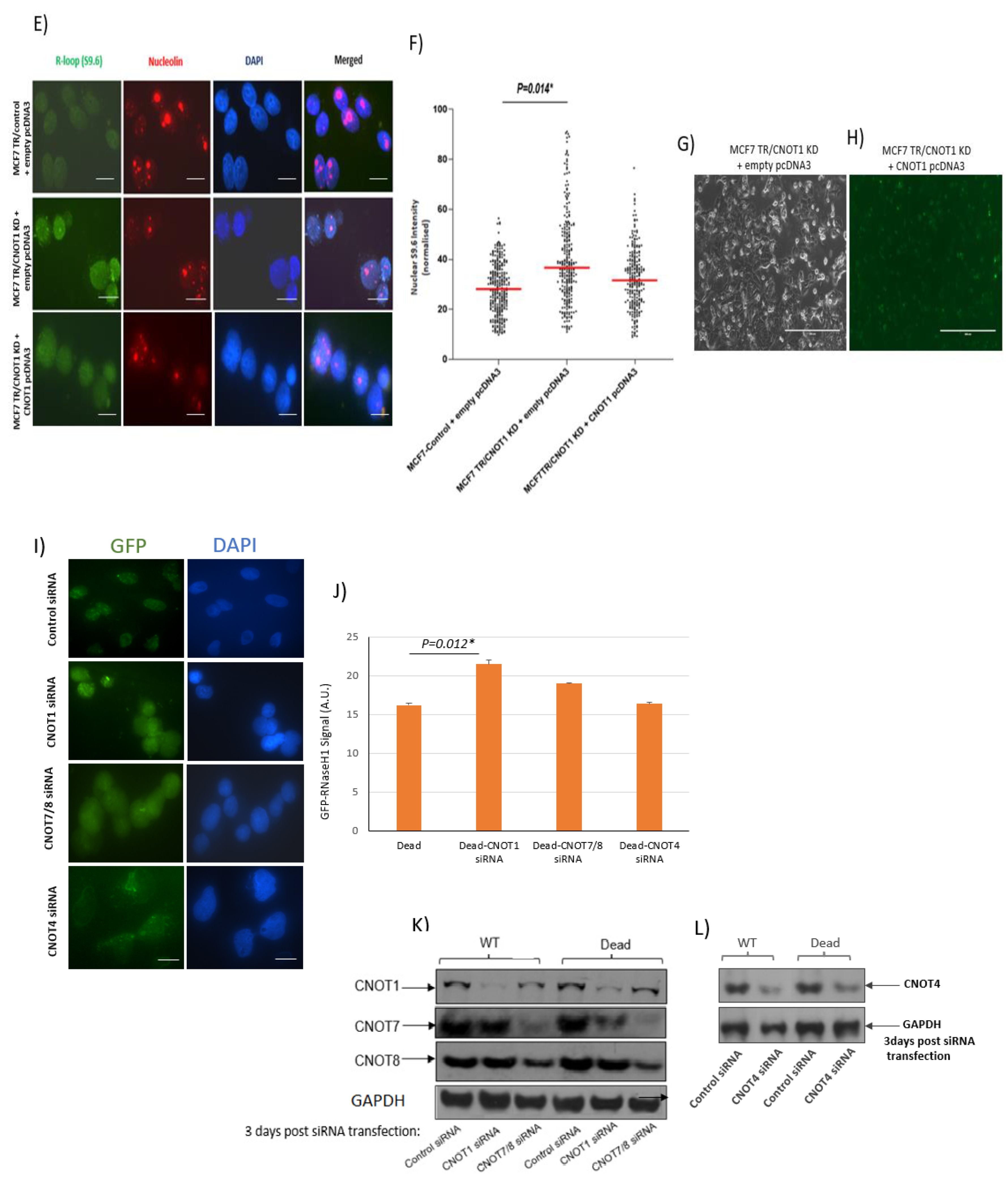
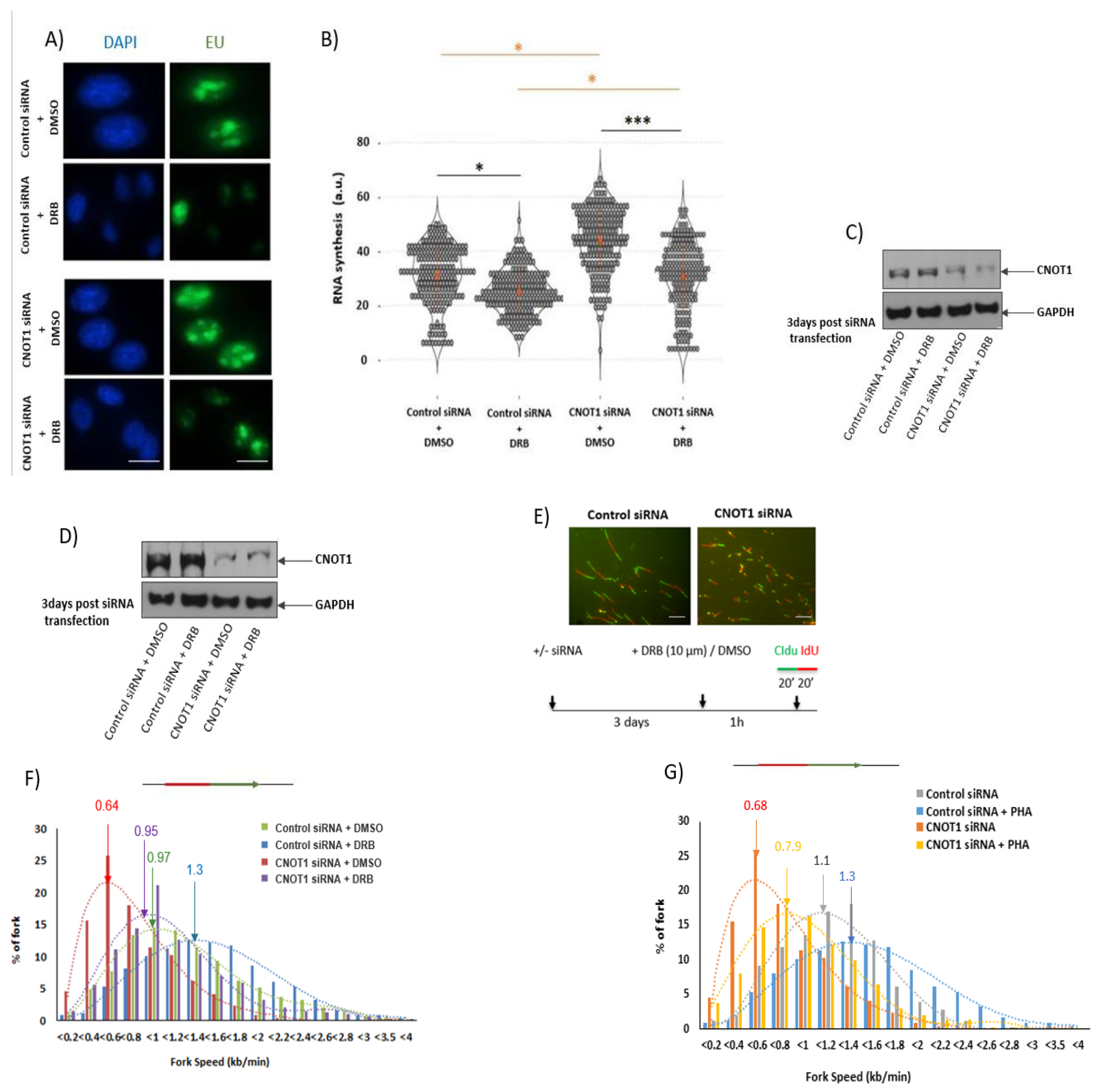

| Target | siRNA | Sense Sequence | Antisense Sequence | Supplier |
|---|---|---|---|---|
| CNOT1 | SMARTpool | CUAUAAAGAGGGAAGAGA CCAGAAACUUUGGCGACAA GGCCAAAUUGUCUCGAAUA CAAGUUAGCACUAUGGUAA | UCUCGUUCCCUCUUUAUAG UUGUCGCCAA AGUUUCUGG UAUUCGAGAC AAUUUGGCC UUACCAUAGU GCUAACUUG | Dharmacon |
| CNOT7 | SMARTpool | CAGCUAGGACUGACAUUUA GGAGAAUUCAGGAGCAAUG UCAUAGCGGUUACGACUUU GUUAGAGCUGGAACGGAUA | UCUCGUUCCCUCUUUAUAG UUGUCGCCAA AGUUUCUGG UAUUCGAGAC AAUUUGGCC UUACCAUAGU GCUAACUUG | Dharmacon |
| CNOT8 | SMARTpool | UUUCGUAGUUCCAUAGAU GAGAAUAGCCAGGUUAUCU AAUAUCAGCUUCUGCGGUG CCAUAGAUCUCCUUGCUAA | AUCUAUGGAACUACGAAA AGAUAACCUGGCUAUUCUC CACCGCAGAAGCUGAUAUU UUAGCAAGGAGAUCUAUGG | Dharmacon |
| lacZ 198-non silencing control | Custom | CGUACGCGGAAUAC UCGAdTdT | AhAhUCGAAG UAUUCCGCGUACG | Dharmacon |
| Primary Antibody | Species | Application | Dilution | Supplier |
|---|---|---|---|---|
| CNOT1 | Rabbit | WB/IP | 1:500 | Proteintech |
| CNOT2 | Rabbit | WB | 1:500 | Proteintech |
| CNOT 3 | Rabbit | WB | 1:500 | Proteintech |
| CNOT4 | Rabbit | WB | 1:500 | SantaCruz |
| CNOT4 | Rabbit | WB | 1:500 | Proteintech |
| CNOT6 | Rabbit | WB | 1:500 | SantaCruz |
| CNOT7 | Rabbit | WB | 1:500 | SantaCruz |
| CNOT8 | Rabbit | WB | 1:500 | Proteintech |
| CNOT9 | Rabbit | WB | 1:500 | Proteintech |
| CNOT10 | Rabbit | WB | 1:500 | SantaCruz |
| CNOT11 | Rabbit | WB | 1:500 | Proteintech |
| Pan3 | Rabbit | WB | 1:500 | SantaCruz |
| 53BP1 | Rabbit | IF/WB | 1:1000 | Novus |
| p53 (DO1) | Mouse | WB | 1:2000 | Gift from D. Lane |
| H2AX | Rabbit | WB | 1:1000 | Millipore |
| γH2AX (S139) | Mouse | WB/IF | 1:1000 | Millipore |
| Chk2 | Rabbit | WB | 1:1000 | Gift from S. Elledge |
| pChk2 (T68) | Rabbit | WB | 1:1000 | Cell Signalling |
| IdU | Mouse | DNA fibres | 1:500 | Becton Dickinson |
| CldU | Rat | DNA fibres | 1:750 | AbD SeroTec |
| ERK1/2 | Rabbit | WB/IF | 1:500 | Cell Signalling |
| pERK1/2 | Rabbit | WB/IF | 1:500 | Cell Signalling |
| p21Cip1 | Rabbit | WB | 1:500 | Abcam |
| TBP | Rabbit | WB | 1:1000 | SantaCruz |
| RNA Po II C-terminal domain (CTD) | Rabbit | WB | 1:1000 | SantaCruz |
| β-Actin | Mouse | WB | 1:1000 | Sigma-Aldrich |
| GAPDH | Mouse | WB | 1:500 | SantaCruz |
| R-loop (S9.6) | Mouse | WB | 1:1000 | Gift from E. Peterman |
| ssDNA | Mouse | WB | 1:5000 | Millipore |
| Cyclin A | Rabbit | WB/IF | 1:500 | SantaCruz |
| Cyclin B | Rabbit | WB | 1:500 | SantaCruz |
| Cyclin E | Rabbit | WB | 1:500 | Abcam |
| Cyclin D1 | Mouse | WB | 1:500 | SantaCruz |
| pHistone H3 (Ser- 10) | Rabbit | WB | 1:100 | Cell signalling |
| Mitosin | Mouse | WB/IF | 1:500 | BDBiosciences |
| ATM | Mouse | WB | 1:1000 | Gift from P. Byrd |
| pATM (S1981) | Rabbit | WB | 1:500 | R & D Systems |
| KAP1 | Rabbit | WB | 1:1000 | Bethyl Laboratories |
| pKAP1 (S824) | Rabbit | WB | 1:1000 | Bethyl Laboratories |
| MCM2 | Rabbit | WB/IP | 1:500 | SantaCruz |
| MCM7 | Rabbit | WB/IP | 1:500 | SantaCruz |
| Tab182 | Rabbit | WB/IP | 1:3000 | In house |
| TOPBP1 | Rabbit | WB | 1:1000 | Bethyl |
| ATRIP | Rabbit | WB, IP | 1:500 | Abcam |
| PARP1 | Mouse | WB | 1:1000 | SantaCruz |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hagkarim, N.C.; Hajkarim, M.C.; Suzuki, T.; Fujiwara, T.; Winkler, G.S.; Stewart, G.S.; Grand, R.J. Disruption of the Mammalian Ccr4–Not Complex Contributes to Transcription-Mediated Genome Instability. Cells 2023, 12, 1868. https://doi.org/10.3390/cells12141868
Hagkarim NC, Hajkarim MC, Suzuki T, Fujiwara T, Winkler GS, Stewart GS, Grand RJ. Disruption of the Mammalian Ccr4–Not Complex Contributes to Transcription-Mediated Genome Instability. Cells. 2023; 12(14):1868. https://doi.org/10.3390/cells12141868
Chicago/Turabian StyleHagkarim, Nafiseh Chalabi, Morteza Chalabi Hajkarim, Toru Suzuki, Toshinobu Fujiwara, G. Sebastiaan Winkler, Grant S. Stewart, and Roger J. Grand. 2023. "Disruption of the Mammalian Ccr4–Not Complex Contributes to Transcription-Mediated Genome Instability" Cells 12, no. 14: 1868. https://doi.org/10.3390/cells12141868
APA StyleHagkarim, N. C., Hajkarim, M. C., Suzuki, T., Fujiwara, T., Winkler, G. S., Stewart, G. S., & Grand, R. J. (2023). Disruption of the Mammalian Ccr4–Not Complex Contributes to Transcription-Mediated Genome Instability. Cells, 12(14), 1868. https://doi.org/10.3390/cells12141868








