Origin, Diversity, and Multiple Roles of Enzymes with Metallo-β-Lactamase Fold from Different Organisms
Abstract
1. Introduction
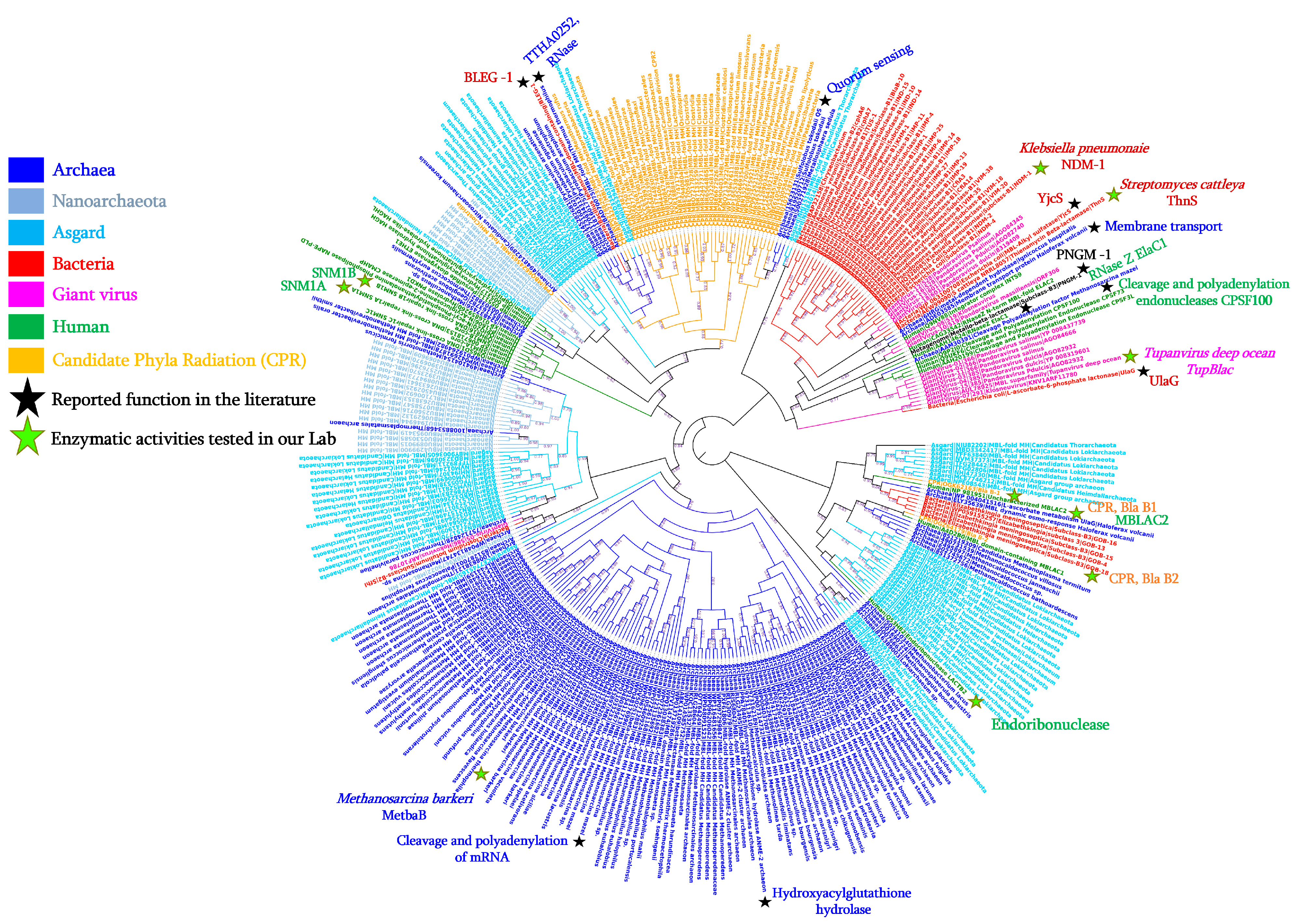
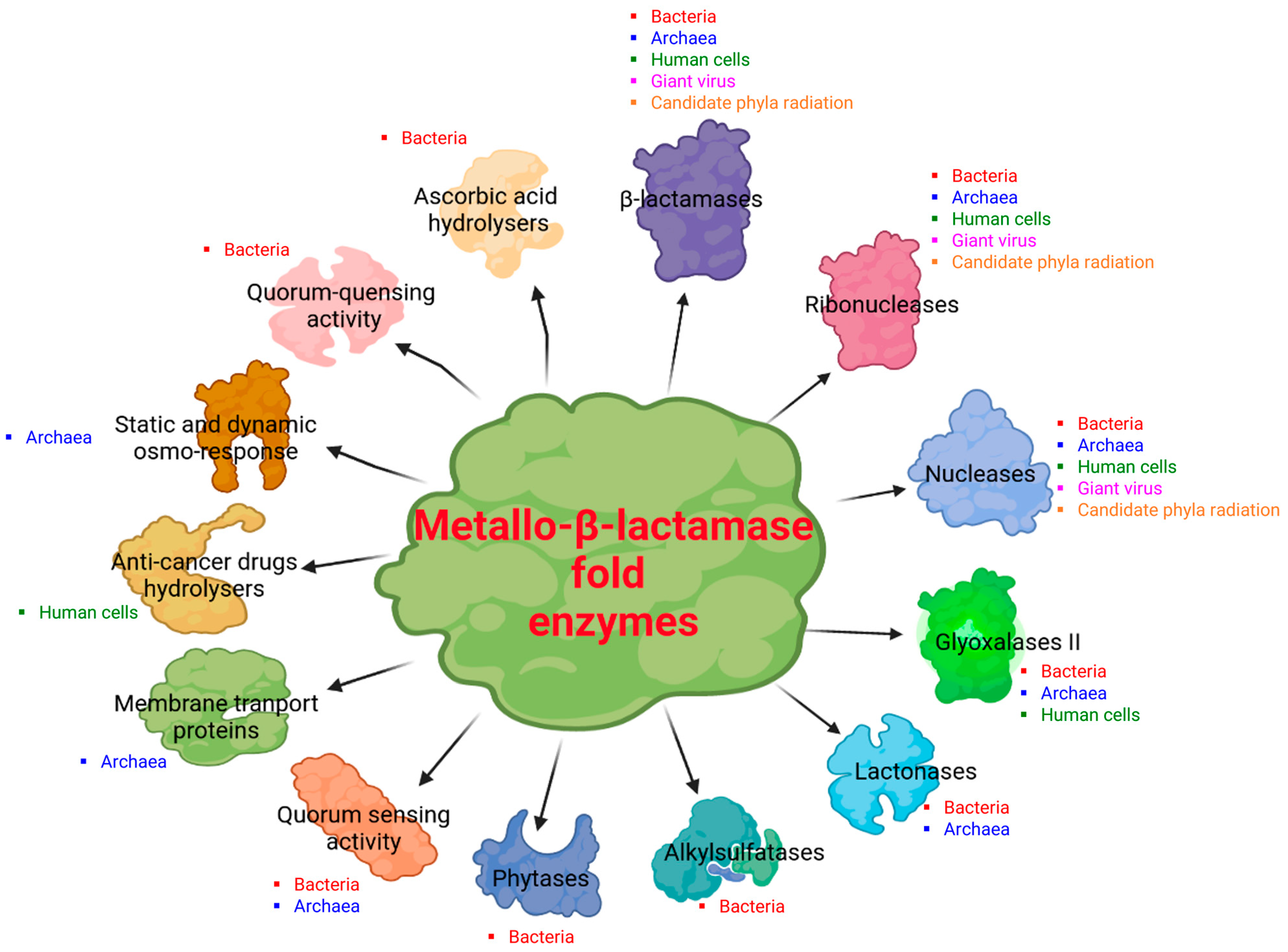
2. Diversity of the Superfamily of Metallo-β-Lactamase (MβL) Fold Enzymes
3. MβL Fold Enzymes in Bacteria: Class B β-Lactamases
3.1. Distribution and Diversity of MβL Fold Enzymes in Bacteria
3.2. Reported Activities of Bacterial MβL Enzymes Other Than β-Lactams Hydrolysis
4. MβL Fold Enzymes in Archaea
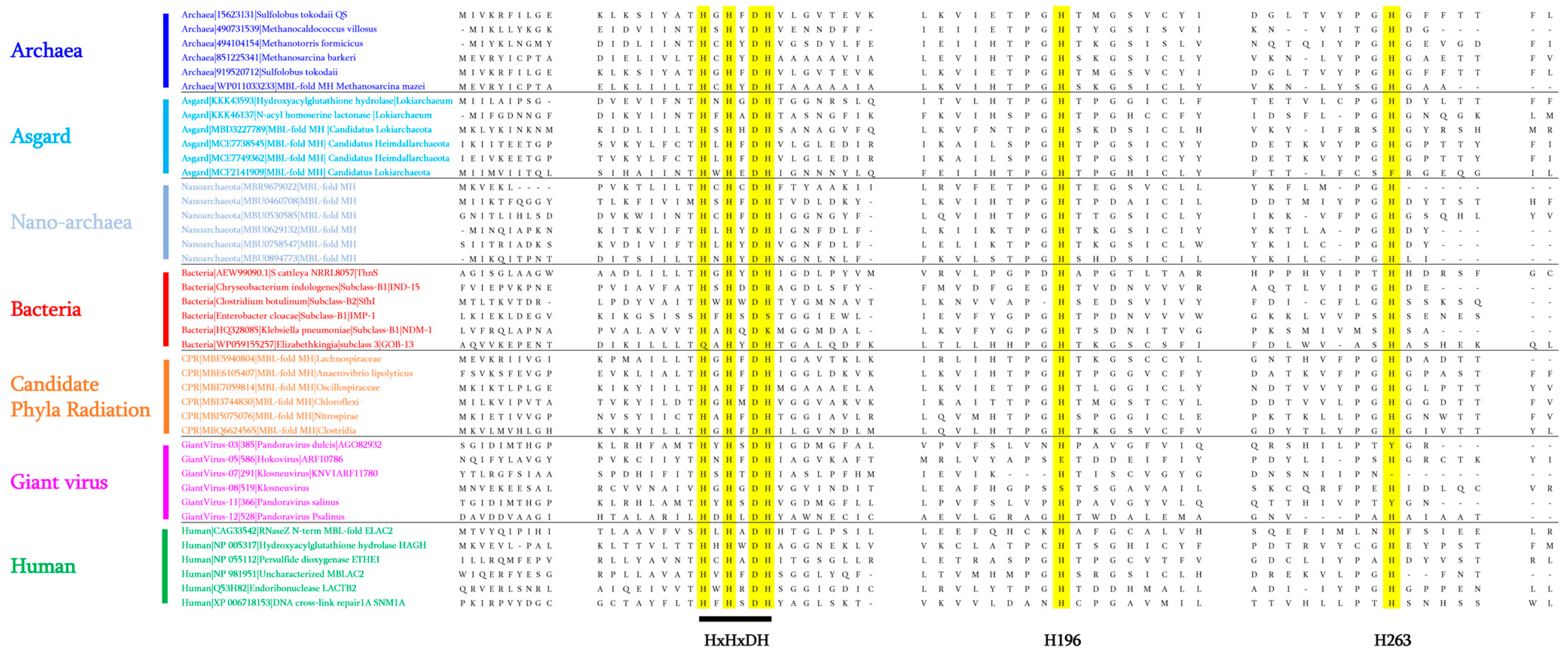
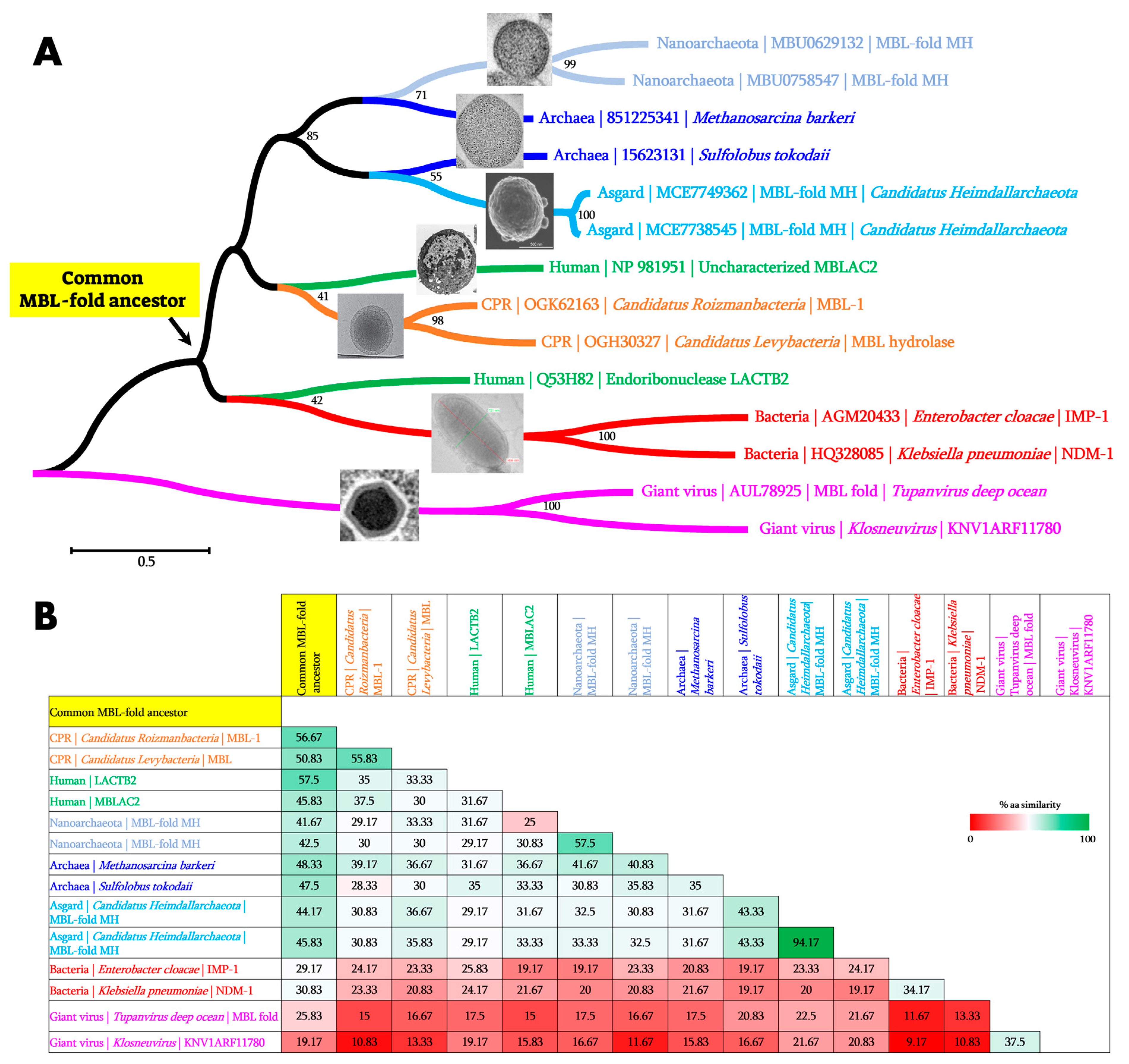
4.1. Reconstruction of Common Ancestral Sequences and Blast Analyses
4.2. The Presence of MβL Fold Enzymes in Subgroups of Archaea: Nanoarchaeota and Asgard Groups
4.3. Reported Activities of Archaeal MβL Fold Enzymes
5. MβL Fold Enzymes in Eukaryotes
5.1. MβL Fold Enzymes Described in the Eukaryote Domain of Life
5.2. Reported Enzymatic Activities of Human MβL Fold Enzymes
6. MβL Fold Enzymes in Giant Viruses
6.1. Discovery of MβL Fold Enzymes in Giant Viruses
6.2. Description of Dual Enzymatic Activity of a Giant Virus MβL Fold Enzyme
7. MβL Fold Enzymes in Candidate Phyla Radiation (CPR)
7.1. Wide Diversity of MβL Fold Enzymes in the CPR Domain
7.2. Reported Enzymatic Activity of CPR MβL Fold Enzymes on β-Lactam Antibiotics
8. Discussion
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jacoby George, A. β-Lactamase Nomenclature. Antimicrob. Agents Chemother. 2006, 50, 1123–1129. [Google Scholar] [CrossRef]
- Garau, J. Beta-lactamases: Current situation and clinical importance. Intensive Care Med. 1994, 20, S5–S9. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, D.; Kim, K.M.; Mittenthal, J.E.; Caetano-Anollés, G. Proteome evolution and the metabolic origins of translation and cellular life. J. Mol. Evol. 2011, 72, 14–33. [Google Scholar] [CrossRef] [PubMed]
- Risso, V.A.; Gavira, J.A.; Mejia-Carmona, D.F.; Gaucher, E.A.; Sanchez-Ruiz, J.M. Hyperstability and substrate promiscuity in laboratory resurrections of precambrian β-lactamases. J. Am. Chem. Soc. 2013, 135, 2899–2902. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.W. Resurrecting ancient genes: Experimental analysis of extinct molecules. Nat. Rev. Genet. 2004, 5, 366–375. [Google Scholar] [CrossRef]
- Diene, S.M.; Pinault, L.; Armstrong, N.; Azza, S.; Keshri, V.; Khelaifia, S.; Chabrière, E.; Caetano-Anolles, G.; Rolain, J.M.; Pontarotti, P.; et al. Dual rnase and β-lactamase activity of a single enzyme encoded in archaea. Life 2020, 10, 280. [Google Scholar] [CrossRef]
- Bush, K. The ABCD’s of β-lactamase nomenclature. J. Infect. Chemother. 2013, 19, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. Past and Present Perspectives on beta-lactamases. Antimicrob. Agents Chemother. 2018, 62, e01076-18. [Google Scholar] [CrossRef]
- Bebrone, C. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 2007, 74, 1686–1701. [Google Scholar] [CrossRef]
- Fröhlich, C.; Chen, J.Z.; Gholipour, S.; Erdogan, A.N.; Tokuriki, N. Evolution of β-lactamases and enzyme promiscuity. Protein Eng. Des. Sel. 2021, 34, gzab013. [Google Scholar] [CrossRef]
- Marasinghe, G.P.K.; Sander, I.M.; Bennett, B.; Periyannan, G.; Yang, K.W.; Makaroff, C.A.; Crowder, M.W. Structural studies on a mitochondrial glyoxalase II. J. Biol. Chem. 2005, 280, 40668–40675. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L. An evolutionary classification of the metallo-beta-lactamase fold proteins. Silico Biol. 1999, 1, 69–91. [Google Scholar]
- Baier, F.; Tokuriki, N. Connectivity between catalytic landscapes of the metallo-β-lactamase superfamily. J. Mol. Biol. 2014, 426, 2442–2456. [Google Scholar] [CrossRef] [PubMed]
- Bergonzi, C.; Schwab, M.; Naik, T.; Daudé, D.; Chabrière, E.; Elias, M. Structural and Biochemical Characterization of AaL, a Quorum Quenching Lactonase with Unusual Kinetic Properties. Sci. Rep. 2018, 8, 19–21. [Google Scholar] [CrossRef]
- Perez-Garcia, P.; Kobus, S.; Gertzen, C.G.W.; Hoeppner, A.; Holzscheck, N.; Strunk, C.H.; Huber, H.; Jaeger, K.E.; Gohlke, H.; Kovacic, F.; et al. A promiscuous ancestral enzyme’s structure unveils protein variable regions of the highly diverse metallo-β-lactamase family. Commun. Biol. 2021, 4, 132. [Google Scholar] [CrossRef]
- Palzkill, T. Metallo-β-lactamase structure and function. Ann. N. Y. Acad. Sci. 2013, 1277, 91–104. [Google Scholar] [CrossRef]
- Laham, S.M.; Gonsalkorale, K.; Von Hippel, W. Darwinian grandparenting: Preferential investment in more certain kin. Personal. Soc. Psychol. Bull. 2005, 31, 63–72. [Google Scholar] [CrossRef]
- Keshri, V.; Panda, A.; Levasseur, A.; Rolain, J.-M.; Pontarotti, P.; Raoult, D. Phylogenomic Analysis of β-Lactamase in Archaea and Bacteria Enables the Identification of Putative New Members. Genome Biol. Evol. 2018, 10, 1106–1114. [Google Scholar] [CrossRef]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef]
- Atkinson, H.J.; Morris, J.H.; Ferrin, T.E.; Babbitt, P.C. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE 2009, 4, e4345. [Google Scholar] [CrossRef]
- Somboro, A.M.; Sekyere, J.O.; Amoako, D.G.; Essack, S.Y.; Bester, L.A. Diversity and proliferation of metallo-β-lactamases: A clarion call for clinically effective metallo-β-lactamase inhibitors. Appl. Environ. Microbiol. 2018, 84, e00698-18. [Google Scholar] [CrossRef]
- Salahuddin, P.; Kumar, A.; Khan, A.U. Structure, Function of Serine and Metallo-β-lactamases and their Inhibitors. Curr. Protein Pept. Sci. 2018, 19, 130–144. [Google Scholar] [CrossRef]
- Heinz, U.; Adolph, H.W. Metallo-β-lactamases: Two binding sites for one catalytic metal ion? Cell. Mol. Life Sci. 2004, 61, 2827–2839. [Google Scholar] [CrossRef]
- Sabath, L.D.; Abraham, E.P. Zinc as a cofactor for cephalosporinase from Bacillus cereus 569. Biochem. J. 1966, 98, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Carfi, A.; Pares, S.; Duée, E.; Galleni, M.; Duez, C.; Frère, J.M.; Dideberg, O. The 3-D structure of a zinc metallo-β-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995, 14, 4914–4921. [Google Scholar] [CrossRef] [PubMed]
- Daiyasu, H.; Osaka, K.; Ishino, Y.; Toh, H. Expansion of the zinc metallo-hydrolase family of the β-lactamase fold. FEBS Lett. 2001, 503, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.Y.; Lin, H.J.; Li, B.R.; Li, Y.K. A Novel metallo-β-lactamase involved in the ampicillin resistance of Streptococcus pneumoniae ATCC 49136 strain. PLoS ONE 2016, 11, e0155905. [Google Scholar] [CrossRef]
- Garces, F.; Fernández, F.J.; Montellà, C.; Penya-Soler, E.; Prohens, R.; Aguilar, J.; Baldomà, L.; Coll, M.; Badia, J.; Vega, M.C. Molecular architecture of the Mn2+-dependent lactonase UlaG reveals an RNase-like metallo-β-lactamase fold and a novel quaternary structure. J. Mol. Biol. 2010, 398, 715–729. [Google Scholar] [CrossRef]
- Kato, Y.; Takahashi, M.; Seki, M.; Nashimoto, M.; Shimizu-Ibuka, A. RNA-hydrolyzing activity of metallo-βlactamase IMP-1. PLoS ONE 2020, 15, e0241557. [Google Scholar] [CrossRef]
- Diene, S.M.; Pinault, L.; Baron, S.A.; Azza, S.; Armstrong, N.; Hadjadj, L.; Chabrière, E.; Rolain, J.-M.; Pontarotti, P.; Raoult, D. A metallo-β-lactamase enzyme for internal detoxification of the antibiotic thienamycin. Sci. Rep. 2021, 11, 10062. [Google Scholar] [CrossRef]
- Au, S.X.; Dzulkifly, N.S.; Noor, N.D.M.; Matsumura, H.; Rahman, R.N.Z.R.A.; Normi, Y.M. Dual activity bleg-1 from bacillus lehensis g1 revealed structural resemblance to b3 metallo-β-lactamase and glyoxalase ii: An insight into its enzyme promiscuity and evolutionary divergence. Int. J. Mol. Sci. 2021, 22, 9377. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.H.; Normi, Y.M.; Leow, A.T.C.; Salleh, A.B.; Murad, A.M.A.; Mahadi, N.M.; Abdul Rahman, M.B. Danger lurking in the “unknowns”: Structure-to-Function Studies of Hypothetical Protein Bleg1_2437 from Bacillus lehensis G1 Alkaliphile Revealed an Evolutionary Divergent B3 Metallo-beta-lactamase. J. Biochem. Adv. 2016, 161, 167–186. [Google Scholar] [CrossRef]
- Liu, D.; Thomas, P.W.; Momb, J.; Hoang, Q.Q.; Petsko, G.A.; Ringe, D.; Fast, W. Structure and Specificity of a Quorum-Quenching Lactonase ( AiiB ) from. Biochemistry 2007, 46, 11789–11799. [Google Scholar] [CrossRef] [PubMed]
- Castillo Villamizar, G.A.; Funkner, K.; Nacke, H.; Foerster, K.; Daniel, R. Functional Metagenomics Reveals a New Catalytic Domain, the Metallo-β-Lactamase Superfamily Domain, Associated with Phytase Activity. mSphere 2019, 4. [Google Scholar] [CrossRef]
- Lee, J.H.; Takahashi, M.; Jeon, J.H.; Kang, L.-W.; Seki, M.; Park, K.S.; Hong, M.-K.; Park, Y.S.; Kim, T.Y.; Karim, A.M.; et al. Dual activity of PNGM-1, a metallo-β-lactamase and tRNase Z, pinpoints the evolutionary origin of subclass B3 metallo-β-lactamases. bioRxiv 2019, bioRxiv:575373. [Google Scholar] [CrossRef]
- Liang, Y.; Gao, Z.; Dong, Y.; Liu, Q. Structural and functional analysis show that the Escherichia coli uncharacterized protein YjcS is likely an alkylsulfatase. Protein Sci. 2014, 23, 1442–1450. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; John von Freyend, S.; Sabag-Daigle, A.; Daniels, C.J.; Allers, T.; Marchfelder, A. Assigning a function to a conserved archaeal metallo-β-lactamase from Haloferax volcanii. Extremophiles 2012, 16, 333–343. [Google Scholar] [CrossRef]
- Shimada, A.; Ishikawa, H.; Nakagawa, N.; Kuramitsu, S.; Masui, R. The first crystal structure of an archaeal metallo-β-lactamase superfamily protein; ST1585 from Sulfolobus tokodaii. Proteins Struct. Funct. Bioinform. 2010, 78, 2399–2402. [Google Scholar] [CrossRef] [PubMed]
- Mir-Montazeri, B.; Ammelburg, M.; Forouzan, D.; Lupas, A.N.; Hartmann, M.D. Crystal structure of a dimeric archaeal Cleavage and Polyadenylation Specificity Factor. J. Struct. Biol. 2011, 173, 191–195. [Google Scholar] [CrossRef]
- Ishikawa, H.; Nakagawa, N.; Kuramitsu, S.; Masui, R. Crystal structure of TTHA0252 from Thermus thermophilus HB8, a RNA degradation protein of the metallo-??-lactamase superfamily. J. Biochem. 2006, 140, 535–542. [Google Scholar] [CrossRef]
- Levy, S.; Portnoy, V.; Admon, J.; Schuster, G. Distinct activities of several RNase J proteins in methanogenic archaea. RNA Biol. 2011, 8, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Buschow, S.I.; van Balkom, B.W.M.; Aalberts, M.; Heck, A.J.R.; Wauben, M.; Stoorvogel, W. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunol. Cell Biol. 2010, 88, 851–856. [Google Scholar] [CrossRef]
- Diene, S.M.; Pinault, L.; Keshri, V.; Armstrong, N.; Khelaifia, S.; Chabrière, E.; Caetano-Anolles, G.; Colson, P.; La Scola, B.; Rolain, J.-M.; et al. Human metallo-β-lactamase enzymes degrade penicillin. Sci. Rep. 2019, 9, 12173. [Google Scholar] [CrossRef]
- Pettinati, I.; Brem, J.; Lee, S.Y.; McHugh, P.J.; Schofield, C.J. The Chemical Biology of Human Metallo-β-Lactamase Fold Proteins. Trends Biochem. Sci. 2016, 41, 338–355. [Google Scholar] [CrossRef]
- Colson, P.; Pinault, L.; Azza, S.; Armstrong, N.; Chabriere, E.; La Scola, B.; Pontarotti, P.; Raoult, D. A protein of the metallo-hydrolase/oxidoreductase superfamily with both beta-lactamase and ribonuclease activity is linked with translation in giant viruses. Sci. Rep. 2020, 10, 21685. [Google Scholar] [CrossRef]
- Maatouk, M.; Ibrahim, A.; Pinault, L.; Armstrong, N.; Azza, S.; Rolain, J.M.; Bittar, F.; Raoult, D. New Beta-lactamases in Candidate Phyla Radiation: Owning Pleiotropic Enzymes Is a Smart Paradigm for Microorganisms with a Reduced Genome. Int. J. Mol. Sci. 2022, 23, 5446. [Google Scholar] [CrossRef]
- Li, Z.; Huang, H.; Zhao, H.; Meng, K.; Zhao, J.; Shi, P.; Yang, P.; Luo, H.; Wang, Y.; Yao, B. Genetic diversity and expression profiles of cysteine phytases in the sheep rumen during a feeding cycle. Lett. Appl. Microbiol. 2014, 59, 615–620. [Google Scholar] [CrossRef]
- Dersjant-Li, Y.; Awati, A.; Schulze, H.; Partridge, G. Phytase in non-ruminant animal nutrition: A critical review on phytase activities in the gastrointestinal tract and influencing factors. J. Sci. Food Agric. 2015, 95, 878–896. [Google Scholar] [CrossRef] [PubMed]
- Kostelecky, B.; Pohl, E.; Vogel, A.; Schilling, O.; Meyer-Klaucke, W. The crystal structure of the zinc phosphodiesterase from Escherichia coli provides insight into function and cooperativity of tRNase Z-family proteins. J. Bacteriol. 2006, 188, 1607–1614. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S.; Kim, T.Y.; Kim, J.H.; Lee, J.H.; Jeon, J.H.; Karim, A.M.; Malik, S.K.; Lee, S.H. PNGM-1, a novel subclass B3 metallo-β-lactamase from a deep-sea sediment metagenome. J. Glob. Antimicrob. Resist. 2018, 14, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Bergonzi, C.; Schwab, M.; Chabriere, E.; Elias, M. The quorum-quenching lactonase from Alicyclobacter acidoterrestris: Purification, kinetic characterization, crystallization and crystallographic analysis. Acta Cryst. F Struct. Biol. Commun. 2017, 73, 476–480. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Lepore, B.W.; Petsko, G.A.; Thomas, P.W.; Stone, E.M.; Fast, W.; Ringe, D. Three-dimensional structure of the quorum-quenching N-acyl homoserine lactone hydrolase from Bacillus thuringiensis. Proc. Natl. Acad. Sci. USA 2005, 102, 11882–11887. [Google Scholar] [CrossRef] [PubMed]
- Mion, S.; Carriot, N.; Lopez, J.; Plener, L.; Ortalo-Magné, A.; Chabrière, E.; Culioli, G.; Daudé, D. Disrupting quorum sensing alters social interactions in Chromobacterium violaceum. npj Biofilms Microbiomes 2021, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- Bergonzi, C.; Schwab, M.; Elias, M. The quorum-quenching lactonase from Geobacillus caldoxylosilyticus: Purification, characterization, crystallization and crystallographic analysis. Acta Crystallogr. Sect. Struct. Biol. Commun. 2016, 72, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Yu, M.; Shan, H.; Tian, X.; Zheng, Y.; Xue, C.; Zhang, X.H. Characterization of a novel N-acylhomoserine lactonase RmmL from Ruegeria mobilis YJ3. Mar. Drugs 2018, 16, 370. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Lee, S.J.; Oh, T.K.; Oh, J.W.; Koo, B.T.; Yum, D.Y.; Lee, J.K. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology 2003, 149, 1541–1550. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Su, Y.; Brackman, G.; Cui, F.; Zhang, Y.; Shi, X.; Coenye, T.; Zhang, X.H. MomL, a novel marine-derived N-Acyl homoserine lactonase from Muricauda olearia. Appl. Environ. Microbiol. 2015, 81, 774–782. [Google Scholar] [CrossRef]
- Wang, W.Z.; Morohoshi, T.; Ikenoya, M.; Someya, N.; Ikeda, T. AiiM, a Novel Class ot N-Acylhomosenne Lactonase from the Leaf-Associated Bacterium Microbacterium testaceum. Appl. Environ. Microbiol. 2010, 76, 2524–2530. [Google Scholar] [CrossRef]
- Wang, W.Z.; Morohoshi, T.; Someya, N.; Ikeda, T. Aidc, a Novel N-Acylhomoserine Lactonase from the Potato Root-Associated Cytophaga-Flavobacteria-Bacteroides (CFB) Group Bacterium Chryseobacterium sp. Strain strb126. Appl. Environ. Microbiol. 2012, 78, 7985–7992. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Yu, S.; Jensen, V.; Seeliger, J.; Feldmann, I.; Weber, S.; Schleicher, E.; Häussler, S.; Blankenfeldt, W. Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein. Biochemistry 2009, 48, 10298–10307. [Google Scholar] [CrossRef] [PubMed]
- Condon, C. What is the role of RNase J in mRNA turnover? RNA Biol. 2010, 7, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Garau, G.; Marie, A.; Guilmi, D.; Hall, B.G. Structure-Based Phylogeny of the Metallo-β-Lactamases. Antimicrob. Agents Chemother. 2005, 49, 2778–2784. [Google Scholar] [CrossRef] [PubMed]
- Dronkert, M.L.G.; de Wit, J.; Boeve, M.; Vasconcelos, M.L.; van Steeg, H.; Tan, T.L.R.; Hoeijmakers, J.H.J.; Kanaar, R. Disruption of Mouse SNM1 Causes Increased Sensitivity to the DNA Interstrand Cross-Linking Agent Mitomycin C. Mol. Cell. Biol. 2000, 20, 4553–4561. [Google Scholar] [CrossRef]
- Ishiai, M.; Kimura, M.; Namikoshi, K.; Yamazoe, M.; Yamamoto, K.; Arakawa, H.; Agematsu, K.; Matsushita, N.; Takeda, S.; Buerstedde, J.-M.; et al. DNA Cross-Link Repair Protein SNM1A Interacts with PIAS1 in Nuclear Focus Formation. Mol. Cell. Biol. 2004, 24, 10733–10741. [Google Scholar] [CrossRef]
- Levy, S.; Allerston, C.K.; Liveanu, V.; Habib, M.R.; Gileadi, O.; Schuster, G. Identification of LACTB2, a metallo-β-lactamase protein, as a human mitochondrial endoribonuclease. Nucleic Acids Res. 2016, 44, 1813–1832. [Google Scholar] [CrossRef]
- Allerston, C.K.; Lee, S.Y.; Newman, J.A.; Schofield, C.J.; Mchugh, P.J.; Gileadi, O. The structures of the SNM1A and SNM1B/Apollo nuclease domains reveal a potential basis for their distinct DNA processing activities. Nucleic Acids Res. 2015, 43, 11047–11060. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Zheng, C.; Chitsaz, F.; Derbyshire, M.K.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Lanczycki, C.J.; et al. CDD: Conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013, 41, 348–352. [Google Scholar] [CrossRef]
- Brown, C.T.; Hug, L.A.; Thomas, B.C.; Sharon, I.; Castelle, C.J.; Singh, A.; Wilkins, M.J.; Wrighton, K.C.; Williams, K.H.; Banfield, J.F. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 2015, 523, 208–211. [Google Scholar] [CrossRef]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A new view of the tree of life. Nat. Microbiol. 2016, 1, 16048. [Google Scholar] [CrossRef]
- Maatouk, M.; Ibrahim, A.; Rolain, J.-M.; Merhej, V.; Bittar, F. Small and Equipped: The Rich Repertoire of Antibiotic. mSystems 2021, 6, e00898-21. [Google Scholar] [CrossRef] [PubMed]
- Dominski, Z. Nucleases of the Metallo-β-lactamase Family and Their Role in DNA and RNA Metabolism. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, F.J.; Lopez-Estepa, M.; Vega, M.C. Nucleases of Metallo-β-Lactamase and Protein Phosphatase Families in DNA Repair. In DNA Repair: On the Pathways to Fixing DNA Damage and Errors; Intech: Acton, MA, USA, 2011; Available online: www.intechopen.com (accessed on 5 November 2022).
- Visweswaran, G.R.R.; Dijkstra, B.W.; Kok, J. Murein and pseudomurein cell wall binding domains of bacteria and archaea-a comparative view. Appl. Microbiol. Biotechnol. 2011, 92, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Dridi, B.; Fardeau, M.L.; Ollivier, B.; Raoult, D.; Drancourt, M. The antimicrobial resistance pattern of cultured human methanogens reflects the unique phylogenetic position of archaea. J. Antimicrob. Chemother. 2011, 66, 2038–2044. [Google Scholar] [CrossRef]
- Crofts, T.S.; Wang, B.; Spivak, A.; Gianoulis, T.A.; Forsberg, K.J.; Gibson, M.K.; Johnsky, L.A.; Broomall, S.M.; Rosenzweig, C.N.; Skowronski, E.W.; et al. Shared strategies for β-lactam catabolism in the soil microbiome. Nat. Chem. Biol. 2018, 14, 556–564. [Google Scholar] [CrossRef]
- Woappi, Y.; Gabani, P.; Singh, A.; Singh, O.V. Antibiotrophs: The complexity of antibiotic-subsisting and antibiotic-resistant microorganisms. Crit. Rev. Microbiol. 2016, 42, 17–30. [Google Scholar] [CrossRef]
- Robertson, C.E. Electron Microscopy of Archaea. Methods Cell Biol. 2007, 79, 169–191. [Google Scholar] [CrossRef]
- Imachi, H.; Nobu, M.K.; Nakahara, N.; Morono, Y.; Ogawara, M.; Takaki, Y.; Takano, Y.; Uematsu, K.; Ikuta, T.; Ito, M.; et al. Isolation of an archaeon at the prokaryote–eukaryote interface. Nature 2020, 577, 519–525. [Google Scholar] [CrossRef]
- Luef, B.; Frischkorn, K.R.; Wrighton, K.C.; Holman, H.Y.N.; Birarda, G.; Thomas, B.C.; Singh, A.; Williams, K.H.; Siegerist, C.E.; Tringe, S.G.; et al. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat. Commun. 2015, 6, 6372. [Google Scholar] [CrossRef]
- Saier, M.H. Bacterial and archaeal cell membranes. Encycl. Microbiol. 2019, 333–347. [Google Scholar] [CrossRef]
- Boyer, M.; Yutin, N.; Pagnier, I.; Barrassi, L.; Fournous, G.; Espinosa, L.; Robert, C.; Azza, S.; Sun, S.; Rossmann, M.G.; et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. USA 2009, 106, 21848–21853. [Google Scholar] [CrossRef] [PubMed]
- Diene, S.M.; Merhej, V.; Henry, M.; El Filali, A.; Roux, V.; Robert, C.; Azza, S.; Gavory, F.; Barbe, V.; La Scola, B.; et al. The rhizome of the multidrug-resistant Enterobacter aerogenes genome reveals how new “killer bugs” are created because of a sympatric lifestyle. Mol. Biol. Evol. 2013, 30, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Colson, P.; Giorgi, R.; Pontarotti, P.; Raoult, D. DNA-dependent RNA polymerase detects hidden giant viruses in published databanks. Genome Biol. Evol. 2014, 6, 1603–1610. [Google Scholar] [CrossRef] [PubMed]
- Schnoes, A.M.; Brown, S.D.; Dodevski, I.; Babbitt, P.C. Annotation error in public databases: Misannotation of molecular function in enzyme superfamilies. PLoS Comput. Biol. 2009, 5, e1000605. [Google Scholar] [CrossRef]
| Kingdom | Acc. Number | Species | Gene Name | Size (aa) | Default Annotation | Reported Activities | References |
|---|---|---|---|---|---|---|---|
| Bacteria | AGM20433 | Enterobacter cloacae | IMP-1 | 247 | Imipenem hydrolysing β-lactamase | β-lactamase; Ribonuclease | [29] |
| AEW99090 | Streptomyces cattleya | ThnS | 330 | Putative β-lactamase | Imipenemase; Ascorbic acid degradation; Nuclease; Ribonuclease | [30] | |
| HQ328085 | Klebsiella pneumoniae | NDM-1 | 270 | New Delhi metallo-β-lactamase | β-lactamase; Ribonuclease | In this study | |
| NA | Streptococcus pneumoniae | - | 363 | L-ascorbate 6-phosphate lactonase | β-lactamase | [27] | |
| P39300.2 | Escherichia coli | UlaG | 354 | L-ascorbate-6-phosphate lactonase | β-lactamase; Ribonuclease | [28] | |
| WP_010974862 | Agrobacterium tumefaciens | AiiB | 276 | Zn-dependent hydrolases | Quorum-quenching lactonase | [33] | |
| PDB: 7EV5_A | Bacillus lehensis | BLEG-1 | 210 | β-lactamase domain containing protein | β-lactamase; Glyoxalase II | [31] | |
| NA | Soil metagenome | MβLp01 | 312 | Metallo-β-lactamase fold protein | β-lactamase; Phytase | [34] | |
| NA | Soil metagenome | MβLP02 | 355 | Metallo-β-lactamase fold protein | β-lactamase; Phytase | [34] | |
| PDB: 7BZ4_B | Deep-seep sediments metagenome | PNGM-1 | 372 | Metallo-β-lactamase | β-lactamase; Ribonuclease | [35] | |
| WP_063100708 | Escherichia coli | YjcS | 661 | Uncharacterised protein | Alkyl sulfatase | [36] | |
| Archaea | ELY35639 | Haloferax volcanii | HVO_2763 | 278 | Zn-dependent hydrolases of the β-lactamase fold | Membrane transport; Static and dynamic osmo-response | [37] |
| 851225341 | Methanosarcina barkeri | MetbaB | 214 | MβL fold metallo-hydrolase | β-lactamase; Ribonuclease; and D-lactate hydrolase | [6] | |
| 15623131 | Sulfolobus tokodaii | - | 200 | Putative hydrolase | Quorum sensing activity | [38] | |
| AAM30391 | Methanosarcina mazei | - | 638 | Metal-dependent RNase, contains metallo-β-lactamase and KH domains | Cleavage and Polyadenylation of mRNA | [39] | |
| BAD70075 | Thermus thermophilus | TTHA0252 | Metallo-β-lactamase superfamily | Ribonuclease (RNase) | [40] | ||
| NA | Methanocaldococcus jannaschii | MjRNase J1 | Ribonuclease Rnase J | Ribonuclease (RNase) | [41] | ||
| NA | Methanocaldococcus jannaschii | MjRNase J2 | Ribonuclease Rnase J | Ribonuclease (RNase); Nuclease | |||
| NA | Methanocaldococcus jannaschii | MjRNase J3 | Ribonuclease Rnase J | Ribonuclease (RNase); Nuclease | |||
| Human | NP_981951 | Homo sapiens | MβLAC2 | 280 | Metallo-β-lactamase domain | Exosome biogenesis enzyme; β-lactamase | [42,43] |
| Q6PJP8 | Homo sapiens | SNM1A | 365 | DNA cross-link repair 1A | Cisplatin or Mitomycin hydrolase; β-lactamase | [43,44] | |
| Q9H816 | Homo sapiens | SNM1B | 335 | 5′ exonuclease Apollo isoform X1 | β-lactamase | [43] | |
| Giant viruses | AUL78925 | Tupanvirus deep ocean | TupBlac | 322 | β-lactamase superfamily domain | β-lactamase; Ribonuclease | [45] |
| Candidate Phyla Radiation (CPR) | KKR15801 | Candidatus Levybacteria | - | 287 | β-lactamase class A-like protein | β-lactamase; Ribonuclease | [46] |
| KKR17584 | Candidatus Levybacteria | - | 303 | β-lactamase class A-like protein | β-lactamase; Ribonuclease | ||
| OGK62163 | Candidatus Roizmanbacteria | - | 263 | Hypothetical protein | β-lactamase; Ribonuclease | ||
| OGZ64179 | Candidatus Staskawiczbacteria | - | 251 | Hypothetical protein | β-lactamase; Ribonuclease | ||
| QHU90009 | Candidatus Saccharibacteria | - | 722 | RNase J family beta-CASP ribonuclease | β-lactamase; Ribonuclease |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diene, S.M.; Pontarotti, P.; Azza, S.; Armstrong, N.; Pinault, L.; Chabrière, E.; Colson, P.; Rolain, J.-M.; Raoult, D. Origin, Diversity, and Multiple Roles of Enzymes with Metallo-β-Lactamase Fold from Different Organisms. Cells 2023, 12, 1752. https://doi.org/10.3390/cells12131752
Diene SM, Pontarotti P, Azza S, Armstrong N, Pinault L, Chabrière E, Colson P, Rolain J-M, Raoult D. Origin, Diversity, and Multiple Roles of Enzymes with Metallo-β-Lactamase Fold from Different Organisms. Cells. 2023; 12(13):1752. https://doi.org/10.3390/cells12131752
Chicago/Turabian StyleDiene, Seydina M., Pierre Pontarotti, Saïd Azza, Nicholas Armstrong, Lucile Pinault, Eric Chabrière, Philippe Colson, Jean-Marc Rolain, and Didier Raoult. 2023. "Origin, Diversity, and Multiple Roles of Enzymes with Metallo-β-Lactamase Fold from Different Organisms" Cells 12, no. 13: 1752. https://doi.org/10.3390/cells12131752
APA StyleDiene, S. M., Pontarotti, P., Azza, S., Armstrong, N., Pinault, L., Chabrière, E., Colson, P., Rolain, J.-M., & Raoult, D. (2023). Origin, Diversity, and Multiple Roles of Enzymes with Metallo-β-Lactamase Fold from Different Organisms. Cells, 12(13), 1752. https://doi.org/10.3390/cells12131752









