Synaptonemal Complex in Human Biology and Disease
Abstract
1. Introduction
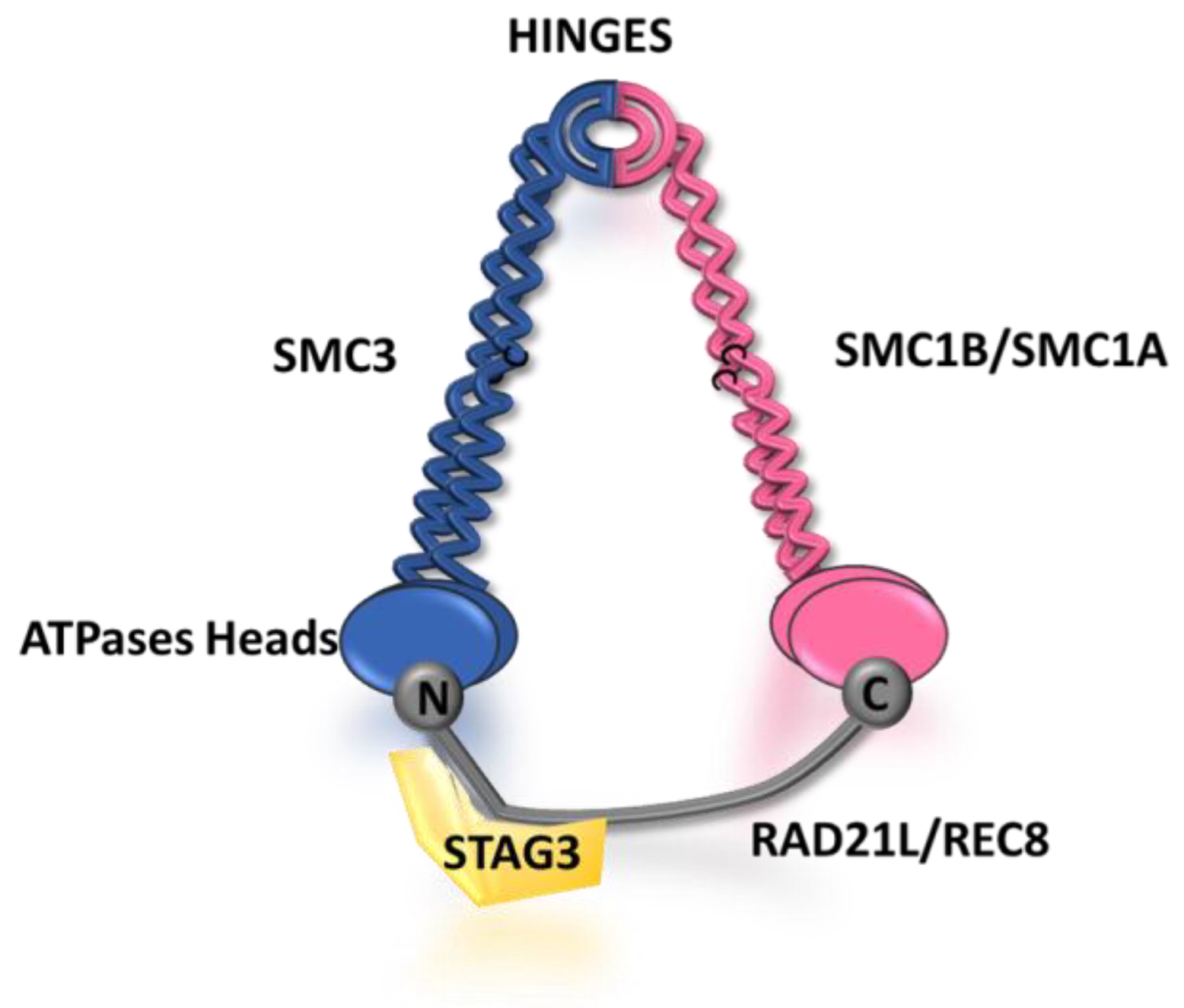
2. Structure and Function of the Synaptonemal Complex
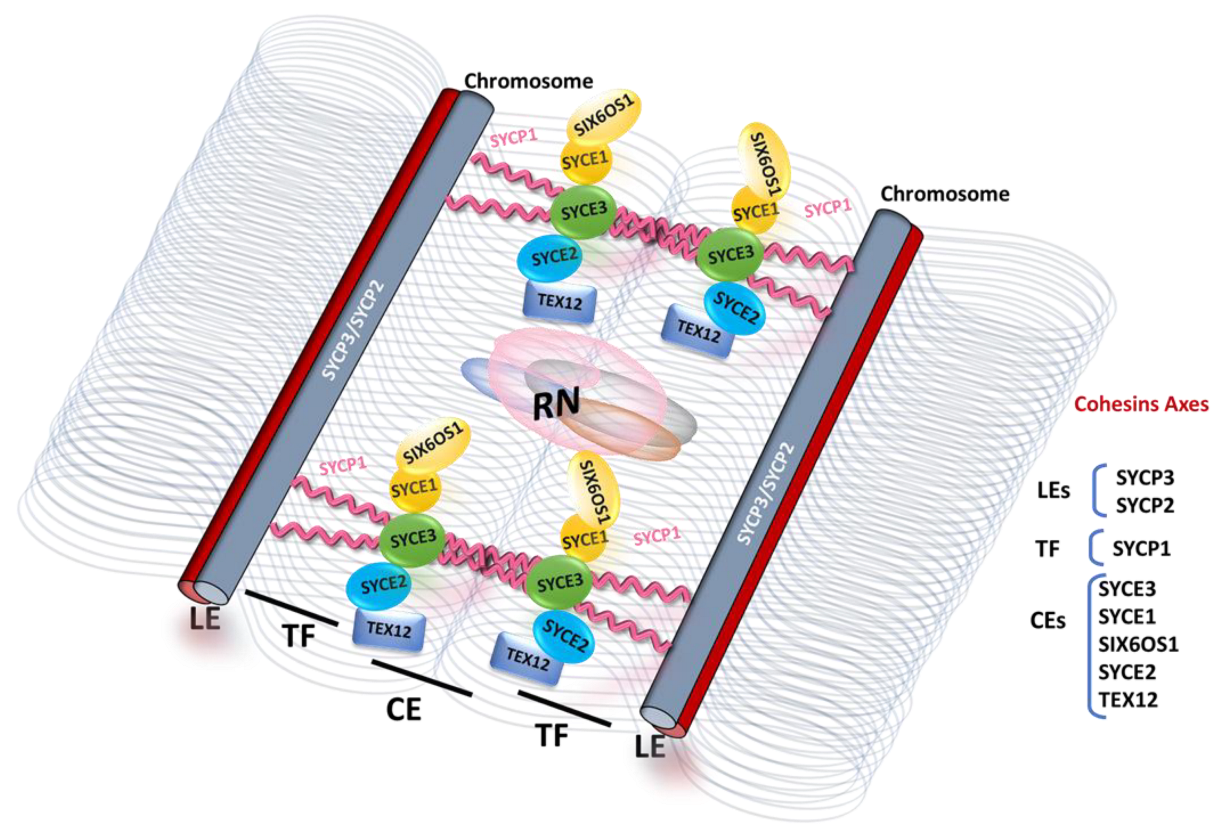
3. Assembly of the SC in Mammals
4. The Role of the SC in Sustaining Meiotic Recombination
5. Infertility
6. Cancer
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Handel, M.A.; Schimenti, J.C. Genetics of Mammalian Meiosis: Regulation, Dynamics and Impact on Fertility. Nat. Rev. Genet. 2010, 11, 124–136. [Google Scholar] [CrossRef]
- Veller, C.; Kleckner, N.; Nowak, M.A. A Rigorous Measure of Genome-Wide Genetic Shuffling That Takes into Account Crossover Positions and Mendel’s Second Law. Proc. Natl. Acad. Sci. USA 2019, 116, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Hann, M.C.; Lau, P.E.; Tempest, H.G. Meiotic Recombination and Male Infertility: From Basic Science to Clinical Reality? Asian J. Androl. 2011, 13, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Hassold, T.; Hall, H.; Hunt, P. The Origin of Human Aneuploidy: Where We Have Been, Where We Are Going. Hum. Mol. Genet. 2007, 16, R203–R208. [Google Scholar] [CrossRef] [PubMed]
- Moses, M.J. Chromosomal Structures in Crayfish Spermatocytes. J. Biophys. Biochem. Cytol. 1956, 2, 215–218. [Google Scholar] [CrossRef]
- Fawcett, D.W. The Fine Structure of Chromosomes in the Meiotic Prophase of Vertebrate Spermatocytes. J. Biophys. Biochem. Cytol. 1956, 2, 403–406. [Google Scholar] [CrossRef]
- Gillies, C.B. Synaptonemal Complex and Chromosome Structure. Annu. Rev. Genet. 1975, 9, 91–109. [Google Scholar] [CrossRef]
- Fraune, J.; Brochier-Armanet, C.; Alsheimer, M.; Benavente, R. Phylogenies of Central Element Proteins Reveal the Dynamic Evolutionary History of the Mammalian Synaptonemal Complex: Ancient and Recent Components. Genetics 2013, 195, 781–793. [Google Scholar] [CrossRef]
- Gao, J.; Colaiácovo, M.P. Zipping and Unzipping: Protein Modifications Regulating Synaptonemal Complex Dynamics. Trends Genet. TIG 2018, 34, 232–245. [Google Scholar] [CrossRef]
- Zhang, F.-G.; Zhang, R.-R.; Gao, J.-M. The Organization, Regulation, and Biological Functions of the Synaptonemal Complex. Asian J. Androl. 2021, 23, 580–589. [Google Scholar] [CrossRef]
- Cahoon, C.K.; Hawley, R.S. Regulating the Construction and Demolition of the Synaptonemal Complex. Nat. Struct. Mol. Biol. 2016, 23, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, K.-I. The Cohesin Complex in Mammalian Meiosis. Genes Cells Devoted Mol. Cell. Mech. 2019, 24, 6–30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Miley, N.; Zastrow, M.S.; MacQueen, A.J.; Sato, A.; Nabeshima, K.; Martinez-Perez, E.; Mlynarczyk-Evans, S.; Carlton, P.M.; Villeneuve, A.M. HAL-2 Promotes Homologous Pairing during Caenorhabditis Elegans Meiosis by Antagonizing Inhibitory Effects of Synaptonemal Complex Precursors. PLOS Genet. 2012, 8, e1002880. [Google Scholar] [CrossRef] [PubMed]
- Page, S.L.; Hawley, R.S. Chromosome Choreography: The Meiotic Ballet. Science 2003, 301, 785–789. [Google Scholar] [CrossRef]
- Zhang, F.; Liu, M.; Gao, J. Alterations in Synaptonemal Complex Coding Genes and Human Infertility. Int. J. Biol. Sci. 2022, 18, 1933–1943. [Google Scholar] [CrossRef]
- Turner, J.M.A. Meiotic Sex Chromosome Inactivation. Dev. Camb. Engl. 2007, 134, 1823–1831. [Google Scholar] [CrossRef]
- Meuwissen, R.L.; Offenberg, H.H.; Dietrich, A.J.; Riesewijk, A.; van Iersel, M.; Heyting, C. A Coiled-Coil Related Protein Specific for Synapsed Regions of Meiotic Prophase Chromosomes. EMBO J. 1992, 11, 5091–5100. [Google Scholar] [CrossRef]
- Lammers, J.H.; Offenberg, H.H.; van Aalderen, M.; Vink, A.C.; Dietrich, A.J.; Heyting, C. The Gene Encoding a Major Component of the Lateral Elements of Synaptonemal Complexes of the Rat Is Related to X-Linked Lymphocyte-Regulated Genes. Mol. Cell. Biol. 1994, 14, 1137–1146. [Google Scholar] [CrossRef]
- Offenberg, H.H.; Schalk, J.A.; Meuwissen, R.L.; van Aalderen, M.; Kester, H.A.; Dietrich, A.J.; Heyting, C. SCP2: A Major Protein Component of the Axial Elements of Synaptonemal Complexes of the Rat. Nucleic Acids Res. 1998, 26, 2572–2579. [Google Scholar] [CrossRef]
- Costa, Y.; Speed, R.; Ollinger, R.; Alsheimer, M.; Semple, C.A.; Gautier, P.; Maratou, K.; Novak, I.; Höög, C.; Benavente, R.; et al. Two Novel Proteins Recruited by Synaptonemal Complex Protein 1 (SYCP1) Are at the Centre of Meiosis. J. Cell Sci. 2005, 118, 2755–2762. [Google Scholar] [CrossRef]
- Hamer, G.; Gell, K.; Kouznetsova, A.; Novak, I.; Benavente, R.; Höög, C. Characterization of a Novel Meiosis-Specific Protein within the Central Element of the Synaptonemal Complex. J. Cell Sci. 2006, 119, 4025–4032. [Google Scholar] [CrossRef] [PubMed]
- Schramm, S.; Fraune, J.; Naumann, R.; Hernandez-Hernandez, A.; Höög, C.; Cooke, H.J.; Alsheimer, M.; Benavente, R. A Novel Mouse Synaptonemal Complex Protein Is Essential for Loading of Central Element Proteins, Recombination, and Fertility. PLoS Genet. 2011, 7, e1002088. [Google Scholar] [CrossRef] [PubMed]
- Gómez-H, L.; Felipe-Medina, N.; Sánchez-Martín, M.; Davies, O.R.; Ramos, I.; García-Tuñón, I.; de Rooij, D.G.; Dereli, I.; Tóth, A.; Barbero, J.L.; et al. C14ORF39/SIX6OS1 Is a Constituent of the Synaptonemal Complex and Is Essential for Mouse Fertility. Nat. Commun. 2016, 7, 13298. [Google Scholar] [CrossRef]
- West, A.M.; Rosenberg, S.C.; Ur, S.N.; Lehmer, M.K.; Ye, Q.; Hagemann, G.; Caballero, I.; Usón, I.; MacQueen, A.J.; Herzog, F.; et al. A Conserved Filamentous Assembly Underlies the Structure of the Meiotic Chromosome Axis. eLife 2019, 8, e40372. [Google Scholar] [CrossRef] [PubMed]
- Crichton, J.H.; Dunce, J.M.; Dunne, O.M.; Salmon, L.J.; Devenney, P.S.; Lawson, J.; Adams, I.R.; Davies, O.R. Structural Maturation of SYCP1-Mediated Meiotic Chromosome Synapsis by SYCE3. Nat. Struct. Mol. Biol. 2023, 30, 188–199. [Google Scholar] [CrossRef]
- Dunce, J.M.; Dunne, O.M.; Ratcliff, M.; Millán, C.; Madgwick, S.; Usón, I.; Davies, O.R. Structural Basis of Meiotic Chromosome Synapsis through SYCP1 Self-Assembly. Nat. Struct. Mol. Biol. 2018, 25, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Biswas, U.; Hempel, K.; Llano, E.; Pendas, A.; Jessberger, R. Distinct Roles of Meiosis-Specific Cohesin Complexes in Mammalian Spermatogenesis. PLoS Genet. 2016, 12, e1006389. [Google Scholar] [CrossRef]
- Llano, E.; Gomez-H, L.; García-Tuñón, I.; Sánchez-Martín, M.; Caburet, S.; Barbero, J.L.; Schimenti, J.C.; Veitia, R.A.; Pendas, A.M. STAG3 Is a Strong Candidate Gene for Male Infertility. Hum. Mol. Genet. 2014, 23, 3421–3431. [Google Scholar] [CrossRef]
- Mehta, G.D.; Kumar, R.; Srivastava, S.; Ghosh, S.K. Cohesin: Functions beyond Sister Chromatid Cohesion. FEBS Lett. 2013, 587, 2299–2312. [Google Scholar] [CrossRef]
- Michaelis, C.; Ciosk, R.; Nasmyth, K. Cohesins: Chromosomal Proteins That Prevent Premature Separation of Sister Chromatids. Cell 1997, 91, 35–45. [Google Scholar] [CrossRef]
- Severson, A.F.; Ling, L.; van Zuylen, V.; Meyer, B.J. The Axial Element Protein HTP-3 Promotes Cohesin Loading and Meiotic Axis Assembly in C. Elegans to Implement the Meiotic Program of Chromosome Segregation. Genes Dev. 2009, 23, 1763–1778. [Google Scholar] [CrossRef] [PubMed]
- Llano, E.; Herrán, Y.; García-Tuñón, I.; Gutiérrez-Caballero, C.; de Álava, E.; Barbero, J.L.; Schimenti, J.; de Rooij, D.G.; Sánchez-Martín, M.; Pendás, A.M. Meiotic Cohesin Complexes Are Essential for the Formation of the Axial Element in Mice. J. Cell Biol. 2012, 197, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Klein, F.; Mahr, P.; Galova, M.; Buonomo, S.B.; Michaelis, C.; Nairz, K.; Nasmyth, K. A Central Role for Cohesins in Sister Chromatid Cohesion, Formation of Axial Elements, and Recombination during Yeast Meiosis. Cell 1999, 98, 91–103. [Google Scholar] [CrossRef]
- Ward, A.; Hopkins, J.; Mckay, M.; Murray, S.; Jordan, P.W. Genetic Interactions Between the Meiosis-Specific Cohesin Components, STAG3, REC8, and RAD21L. G3 Bethesda Md 2016, 6, 1713–1724. [Google Scholar] [CrossRef] [PubMed]
- Llano, E.; Gómez, R.; Gutiérrez-Caballero, C.; Herrán, Y.; Sánchez-Martín, M.; Vázquez-Quiñones, L.; Hernández, T.; de Alava, E.; Cuadrado, A.; Barbero, J.L.; et al. Shugoshin-2 Is Essential for the Completion of Meiosis but Not for Mitotic Cell Division in Mice. Genes Dev. 2008, 22, 2400–2413. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Iwai, T.; Yokota, T.; Yamashita, M. Temporally and Spatially Selective Loss of Rec8 Protein from Meiotic Chromosomes during Mammalian Meiosis. J. Cell Sci. 2003, 116, 2781–2790. [Google Scholar] [CrossRef]
- Eijpe, M.; Offenberg, H.; Jessberger, R.; Revenkova, E.; Heyting, C. Meiotic Cohesin REC8 Marks the Axial Elements of Rat Synaptonemal Complexes before Cohesins SMC1beta and SMC3. J. Cell Biol. 2003, 160, 657–670. [Google Scholar] [CrossRef]
- Clift, D.; McEwan, W.A.; Labzin, L.I.; Konieczny, V.; Mogessie, B.; James, L.C.; Schuh, M. A Method for the Acute and Rapid Degradation of Endogenous Proteins. Cell 2017, 171, 1692–1706.e18. [Google Scholar] [CrossRef]
- Herrán, Y.; Gutiérrez-Caballero, C.; Sánchez-Martín, M.; Hernández, T.; Viera, A.; Barbero, J.L.; de Álava, E.; de Rooij, D.G.; Suja, J.Á.; Llano, E.; et al. The Cohesin Subunit RAD21L Functions in Meiotic Synapsis and Exhibits Sexual Dimorphism in Fertility. EMBO J. 2011, 30, 3091–3105. [Google Scholar] [CrossRef]
- Bannister, L.A.; Reinholdt, L.G.; Munroe, R.J.; Schimenti, J.C. Positional Cloning and Characterization of Mouse Mei8, a Disrupted Allelle of the Meiotic Cohesin Rec8. Genesis 2004, 40, 184–194. [Google Scholar] [CrossRef]
- Ishiguro, K.-I.; Kim, J.; Shibuya, H.; Hernández-Hernández, A.; Suzuki, A.; Fukagawa, T.; Shioi, G.; Kiyonari, H.; Li, X.C.; Schimenti, J.; et al. Meiosis-Specific Cohesin Mediates Homolog Recognition in Mouse Spermatocytes. Genes Dev. 2014, 28, 594–607. [Google Scholar] [CrossRef] [PubMed]
- Prince, J.P.; Martinez-Perez, E. Functions and Regulation of Meiotic HORMA-Domain Proteins. Genes 2022, 13, 777. [Google Scholar] [CrossRef] [PubMed]
- Woglar, A.; Yamaya, K.; Roelens, B.; Boettiger, A.; Köhler, S.; Villeneuve, A.M. Quantitative Cytogenetics Reveals Molecular Stoichiometry and Longitudinal Organization of Meiotic Chromosome Axes and Loops. PLoS Biol. 2020, 18, e3000817. [Google Scholar] [CrossRef]
- Wojtasz, L.; Cloutier, J.M.; Baumann, M.; Daniel, K.; Varga, J.; Fu, J.; Anastassiadis, K.; Stewart, A.F.; Reményi, A.; Turner, J.M.A.; et al. Meiotic DNA Double-Strand Breaks and Chromosome Asynapsis in Mice Are Monitored by Distinct HORMAD2-Independent and -Dependent Mechanisms. Genes Dev. 2012, 26, 958–973. [Google Scholar] [CrossRef]
- Syrjänen, J.L.; Heller, I.; Candelli, A.; Davies, O.R.; Peterman, E.J.G.; Wuite, G.J.L.; Pellegrini, L. Single-Molecule Observation of DNA Compaction by Meiotic Protein SYCP3. eLife 2017, 6, e22582. [Google Scholar] [CrossRef] [PubMed]
- Pelttari, J.; Hoja, M.-R.; Yuan, L.; Liu, J.-G.; Brundell, E.; Moens, P.; Santucci-Darmanin, S.; Jessberger, R.; Barbero, J.L.; Heyting, C.; et al. A Meiotic Chromosomal Core Consisting of Cohesin Complex Proteins Recruits DNA Recombination Proteins and Promotes Synapsis in the Absence of an Axial Element in Mammalian Meiotic Cells. Mol. Cell. Biol. 2001, 21, 5667–5677. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Tong, Z.; Ye, Q.; Sun, T.; Hong, Z.; Zhang, L.; Bortnick, A.; Cho, S.; Beuzer, P.; Axelrod, J.; et al. Molecular Organization of Mammalian Meiotic Chromosome Axis Revealed by Expansion STORM Microscopy. Proc. Natl. Acad. Sci. USA 2019, 116, 18423–18428. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Horisawa-Takada, Y.; Inoue, E.; Tani, N.; Shibuya, H.; Fujimura, S.; Kariyazono, R.; Sakata, T.; Ohta, K.; Araki, K.; et al. Meiotic Cohesins Mediate Initial Loading of HORMAD1 to the Chromosomes and Coordinate SC Formation during Meiotic Prophase. PLoS Genet. 2020, 16, e1009048. [Google Scholar] [CrossRef]
- Liu, J.G.; Yuan, L.; Brundell, E.; Björkroth, B.; Daneholt, B.; Höög, C. Localization of the N-Terminus of SCP1 to the Central Element of the Synaptonemal Complex and Evidence for Direct Interactions between the N-Termini of SCP1 Molecules Organized Head-to-Head. Exp. Cell Res. 1996, 226, 11–19. [Google Scholar] [CrossRef]
- Fraune, J.; Brochier-Armanet, C.; Alsheimer, M.; Volff, J.-N.; Schücker, K.; Benavente, R. Evolutionary History of the Mammalian Synaptonemal Complex. Chromosoma 2016, 125, 355–360. [Google Scholar] [CrossRef]
- Bolcun-Filas, E.; Hall, E.; Speed, R.; Taggart, M.; Grey, C.; de Massy, B.; Benavente, R.; Cooke, H.J. Mutation of the Mouse Syce1 Gene Disrupts Synapsis and Suggests a Link between Synaptonemal Complex Structural Components and DNA Repair. PLoS Genet. 2009, 5, e1000393. [Google Scholar] [CrossRef]
- Bolcun-Filas, E.; Costa, Y.; Speed, R.; Taggart, M.; Benavente, R.; De Rooij, D.G.; Cooke, H.J. SYCE2 Is Required for Synaptonemal Complex Assembly, Double Strand Break Repair, and Homologous Recombination. J. Cell Biol. 2007, 176, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Grey, C.; de Massy, B. Coupling Crossover and Synaptonemal Complex in Meiosis. Genes Dev. 2022, 36, 4–6. [Google Scholar] [CrossRef]
- Brick, K.; Smagulova, F.; Khil, P.; Camerini-Otero, R.D.; Petukhova, G.V. Genetic Recombination Is Directed Away from Functional Genomic Elements in Mice. Nature 2012, 485, 642–645. [Google Scholar] [CrossRef]
- Smagulova, F.; Gregoretti, I.V.; Brick, K.; Khil, P.; Camerini-Otero, R.D.; Petukhova, G.V. Genome-Wide Analysis Reveals Novel Molecular Features of Mouse Recombination Hotspots. Nature 2011, 472, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Bourbon, H.-M.; de Massy, B. Functional Conservation of Mei4 for Meiotic DNA Double-Strand Break Formation from Yeasts to Mice. Genes Dev. 2010, 24, 1266–1280. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Ghyselinck, N.; Ishiguro, K.; Watanabe, Y.; Kouznetsova, A.; Höög, C.; Strong, E.; Schimenti, J.; Daniel, K.; Toth, A.; et al. MEI4—A Central Player in the Regulation of Meiotic DNA Double-Strand Break Formation in the Mouse. J. Cell Sci. 2015, 128, 1800–1811. [Google Scholar] [CrossRef]
- Stanzione, M.; Baumann, M.; Papanikos, F.; Dereli, I.; Lange, J.; Ramlal, A.; Tränkner, D.; Shibuya, H.; de Massy, B.; Watanabe, Y.; et al. Meiotic DNA Break Formation Requires the Unsynapsed Chromosome Axis-Binding Protein IHO1 (CCDC36) in Mice. Nat. Cell Biol. 2016, 18, 1208–1220. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lisby, M.; Symington, L.S. RPA Coordinates DNA End Resection and Prevents Formation of DNA Hairpins. Mol. Cell 2013, 50, 589–600. [Google Scholar] [CrossRef]
- Felipe-Medina, N.; Caburet, S.; Sánchez-Sáez, F.; Condezo, Y.B.; de Rooij, D.G.; Gómez-H, L.; Garcia-Valiente, R.; Todeschini, A.L.; Duque, P.; Sánchez-Martin, M.A.; et al. A Missense in HSF2BP Causing Primary Ovarian Insufficiency Affects Meiotic Recombination by Its Novel Interactor C19ORF57/BRME1. eLife 2020, 9, e56996. [Google Scholar] [CrossRef]
- Huang, T.; Wu, X.; Wang, S.; Bao, Z.; Wan, Y.; Wang, Z.; Li, M.; Yu, X.; Lv, Y.; Liu, Z.; et al. SPIDR Is Required for Homologous Recombination during Mammalian Meiosis. Nucleic Acids Res. 2023, 51, gkad154. [Google Scholar] [CrossRef] [PubMed]
- Tsubouchi, H.; Argunhan, B.; Ito, K.; Takahashi, M.; Iwasaki, H. Two Auxiliary Factors Promote Dmc1-Driven DNA Strand Exchange via Stepwise Mechanisms. Proc. Natl. Acad. Sci. USA 2020, 117, 12062–12070. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-S.; Higashide, M.T.; Choi, H.; Li, K.; Hong, S.; Lee, K.; Shinohara, A.; Shinohara, M.; Kim, K.P. The Synaptonemal Complex Central Region Modulates Crossover Pathways and Feedback Control of Meiotic Double-Strand Break Formation. Nucleic Acids Res. 2021, 49, 7537–7553. [Google Scholar] [CrossRef]
- Baudat, F.; de Massy, B. Regulating Double-Stranded DNA Break Repair towards Crossover or Non-Crossover during Mammalian Meiosis. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2007, 15, 565–577. [Google Scholar] [CrossRef]
- Hunter, N. Meiotic Recombination: The Essence of Heredity. Cold Spring Harb. Perspect. Biol. 2015, 7, a016618. [Google Scholar] [CrossRef] [PubMed]
- Guiraldelli, M.F.; Eyster, C.; Wilkerson, J.L.; Dresser, M.E.; Pezza, R.J. Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis. PLOS Genet. 2013, 9, e1003383. [Google Scholar] [CrossRef] [PubMed]
- Baudat, F.; Imai, Y.; de Massy, B. Meiotic Recombination in Mammals: Localization and Regulation. Nat. Rev. Genet. 2013, 14, 794–806. [Google Scholar] [CrossRef]
- Zickler, D.; Kleckner, N. Recombination, Pairing, and Synapsis of Homologs during Meiosis. Cold Spring Harb. Perspect. Biol. 2015, 7, a016626. [Google Scholar] [CrossRef]
- Baker, S.M.; Plug, A.W.; Prolla, T.A.; Bronner, C.E.; Harris, A.C.; Yao, X.; Christie, D.M.; Monell, C.; Arnheim, N.; Bradley, A.; et al. Involvement of Mouse Mlh1 in DNA Mismatch Repair and Meiotic Crossing Over. Nat. Genet. 1996, 13, 336–342. [Google Scholar] [CrossRef]
- Wei, K.; Clark, A.B.; Wong, E.; Kane, M.F.; Mazur, D.J.; Parris, T.; Kolas, N.K.; Russell, R.; Hou, H.; Kneitz, B.; et al. Inactivation of Exonuclease 1 in Mice Results in DNA Mismatch Repair Defects, Increased Cancer Susceptibility, and Male and Female Sterility. Genes Dev. 2003, 17, 603–614. [Google Scholar] [CrossRef]
- Rog, O.; Köhler, S.; Dernburg, A.F. The Synaptonemal Complex Has Liquid Crystalline Properties and Spatially Regulates Meiotic Recombination Factors. eLife 2017, 6, e21455. [Google Scholar] [CrossRef]
- Di Giacomo, M.; Barchi, M.; Baudat, F.; Edelmann, W.; Keeney, S.; Jasin, M. Distinct DNA-Damage-Dependent and -Independent Responses Drive the Loss of Oocytes in Recombination-Defective Mouse Mutants. Proc. Natl. Acad. Sci. USA 2005, 102, 737–742. [Google Scholar] [CrossRef] [PubMed]
- Pittman, D.L.; Cobb, J.; Schimenti, K.J.; Wilson, L.A.; Cooper, D.M.; Brignull, E.; Handel, M.A.; Schimenti, J.C. Meiotic Prophase Arrest with Failure of Chromosome Synapsis in Mice Deficient for Dmc1, a Germline-Specific RecA Homolog. Mol. Cell 1998, 1, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Caburet, S.; Arboleda, V.A.; Llano, E.; Overbeek, P.A.; Barbero, J.L.; Oka, K.; Harrison, W.; Vaiman, D.; Ben-Neriah, Z.; García-Tuñón, I.; et al. Mutant Cohesin in Premature Ovarian Failure. N. Engl. J. Med. 2014, 370, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Riera-Escamilla, A.; Enguita-Marruedo, A.; Moreno-Mendoza, D.; Chianese, C.; Sleddens-Linkels, E.; Contini, E.; Benelli, M.; Natali, A.; Colpi, G.M.; Ruiz-Castañé, E.; et al. Sequencing of a “mouse Azoospermia” Gene Panel in Azoospermic Men: Identification of RNF212 and STAG3 Mutations as Novel Genetic Causes of Meiotic Arrest. Hum. Reprod. Oxf. Engl. 2019, 34, 978–988. [Google Scholar] [CrossRef]
- Krausz, C.; Riera-Escamilla, A.; Moreno-Mendoza, D.; Holleman, K.; Cioppi, F.; Algaba, F.; Pybus, M.; Friedrich, C.; Wyrwoll, M.J.; Casamonti, E.; et al. Genetic Dissection of Spermatogenic Arrest through Exome Analysis: Clinical Implications for the Management of Azoospermic Men. Genet. Med. Off. J. Am. Coll. Med. Genet. 2020, 22, 1956–1966. [Google Scholar] [CrossRef]
- van der Bijl, N.; Röpke, A.; Biswas, U.; Wöste, M.; Jessberger, R.; Kliesch, S.; Friedrich, C.; Tüttelmann, F. Mutations in the Stromal Antigen 3 (STAG3) Gene Cause Male Infertility Due to Meiotic Arrest. Hum. Reprod. Oxf. Engl. 2019, 34, 2112–2119. [Google Scholar] [CrossRef]
- Akbari, A.; Zoha Tabatabaei, S.; Salehi, N.; Padidar, K.; Almadani, N.; Ali Sadighi Gilani, M.; Mashayekhi, M.; Motevaseli, E.; Totonchi, M. Novel STAG3 Variant Associated with Primary Ovarian Insufficiency and Non-Obstructive Azoospermia in an Iranian Consanguineous Family. Gene 2022, 821, 146281. [Google Scholar] [CrossRef]
- Bouilly, J.; Beau, I.; Barraud, S.; Bernard, V.; Azibi, K.; Fagart, J.; Fèvre, A.; Todeschini, A.L.; Veitia, R.A.; Beldjord, C.; et al. Identification of Multiple Gene Mutations Accounts for a New Genetic Architecture of Primary Ovarian Insufficiency. J. Clin. Endocrinol. Metab. 2016, 101, 4541–4550. [Google Scholar] [CrossRef]
- Minase, G.; Miyamoto, T.; Miyagawa, Y.; Iijima, M.; Ueda, H.; Saijo, Y.; Namiki, M.; Sengoku, K. Single-Nucleotide Polymorphisms in the Human RAD21L Gene May Be a Genetic Risk Factor for Japanese Patients with Azoospermia Caused by Meiotic Arrest and Sertoli Cell-Only Syndrome. Hum. Fertil. Camb. Engl. 2017, 20, 217–220. [Google Scholar] [CrossRef]
- Kherraf, Z.-E.; Cazin, C.; Bouker, A.; Fourati Ben Mustapha, S.; Hennebicq, S.; Septier, A.; Coutton, C.; Raymond, L.; Nouchy, M.; Thierry-Mieg, N.; et al. Whole-Exome Sequencing Improves the Diagnosis and Care of Men with Non-Obstructive Azoospermia. Am. J. Hum. Genet. 2022, 109, 508–517. [Google Scholar] [CrossRef]
- Tucker, E.J.; Bell, K.M.; Robevska, G.; van den Bergen, J.; Ayers, K.L.; Listyasari, N.; Faradz, S.M.; Dulon, J.; Bakhshalizadeh, S.; Sreenivasan, R.; et al. Meiotic Genes in Premature Ovarian Insufficiency: Variants in HROB and REC8 as Likely Genetic Causes. Eur. J. Hum. Genet. EJHG 2022, 30, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Chen, Q.; Nguyen, C.; Meerzaman, D.; Singer, D.S. Cohesin Regulates Alternative Splicing. Sci. Adv. 2023, 9, eade3876. [Google Scholar] [CrossRef] [PubMed]
- Okutman, O.; Boivin, M.; Muller, J.; Charlet-Berguerand, N.; Viville, S. A Biallelic Loss of Function Variant in HORMAD1 within a Large Consanguineous Turkish Family Is Associated with Spermatogenic Arrest. Hum. Reprod. Oxf. Engl. 2023, 38, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, B.; Fu, J.; Li, R.; Diao, F.; Li, C.; Chen, B.; Du, J.; Zhou, Z.; Mu, J.; et al. Bi-Allelic Missense Pathogenic Variants in TRIP13 Cause Female Infertility Characterized by Oocyte Maturation Arrest. Am. J. Hum. Genet. 2020, 107, 15–23. [Google Scholar] [CrossRef]
- Ali, H.; Unar, A.; Zubair, M.; Dil, S.; Ullah, F.; Khan, I.; Hussain, A.; Shi, Q. In Silico Analysis of a Novel Pathogenic Variant c.7G > A in C14orf39 Gene Identified by WES in a Pakistani Family with Azoospermia. Mol. Genet. Genom. 2022, 297, 719–730. [Google Scholar] [CrossRef]
- Sánchez-Sáez, F.; Gómez-H, L.; Dunne, O.M.; Gallego-Páramo, C.; Felipe-Medina, N.; Sánchez-Martín, M.; Llano, E.; Pendas, A.M.; Davies, O.R. Meiotic Chromosome Synapsis Depends on Multivalent SYCE1-SIX6OS1 Interactions That Are Disrupted in Cases of Human Infertility. Sci. Adv. 2020, 6, eabb1660. [Google Scholar] [CrossRef]
- Qian, M.-X.; Pang, Y.; Liu, C.H.; Haratake, K.; Du, B.-Y.; Ji, D.-Y.; Wang, G.-F.; Zhu, Q.-Q.; Song, W.; Yu, Y.; et al. Acetylation-Mediated Proteasomal Degradation of Core Histones during DNA Repair and Spermatogenesis. Cell 2013, 153, 1012–1024. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ji, S.-Y.; Busayavalasa, K.; Shao, J.; Yu, C. Meiosis I Progression in Spermatogenesis Requires a Type of Testis-Specific 20S Core Proteasome. Nat. Commun. 2019, 10, 3387. [Google Scholar] [CrossRef] [PubMed]
- Keeney, S.; Lange, J.; Mohibullah, N. Self-Organization of Meiotic Recombination Initiation: General Principles and Molecular Pathways. Annu. Rev. Genet. 2014, 48, 187–214. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Y.-F.; Jiang, H.-W.; Khan, R.; Han, Q.-Q.; Iqbal, F.; Jiang, X.-H.; Shi, Q.-H. The Molecular Control of Meiotic Double-Strand Break (DSB) Formation and Its Significance in Human Infertility. Asian J. Androl. 2021, 23, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Dong, J.; Chen, B.; Du, J.; Kuang, Y.; Sun, X.; Fu, J.; Li, B.; Mu, J.; Zhang, Z.; et al. Homozygous Mutations in REC114 Cause Female Infertility Characterised by Multiple Pronuclei Formation and Early Embryonic Arrest. J. Med. Genet. 2020, 57, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Fan, S.; Jabeen, N.; Zhang, H.; Khan, R.; Murtaza, G.; Jiang, H.; Ali, A.; Li, Y.; Bao, J.; et al. A TOP6BL Mutation Abolishes Meiotic DNA Double-Strand Break Formation and Causes Human Infertility. Sci. Bull. 2020, 65, 2120–2129. [Google Scholar] [CrossRef]
- Fakhro, K.A.; Elbardisi, H.; Arafa, M.; Robay, A.; Rodriguez-Flores, J.L.; Al-Shakaki, A.; Syed, N.; Mezey, J.G.; Abi Khalil, C.; Malek, J.A.; et al. Point-of-Care Whole-Exome Sequencing of Idiopathic Male Infertility. Genet. Med. Off. J. Am. Coll. Med. Genet. 2018, 20, 1365–1373. [Google Scholar] [CrossRef]
- Irie, S.; Tsujimura, A.; Miyagawa, Y.; Ueda, T.; Matsuoka, Y.; Matsui, Y.; Okuyama, A.; Nishimune, Y.; Tanaka, H. Single-Nucleotide Polymorphisms of the PRDM9 (MEISETZ) Gene in Patients with Nonobstructive Azoospermia. J. Androl. 2009, 30, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, N.; Ji, Z.; Bai, H.; Ou, N.; Tian, R.; Li, P.; Zhi, E.; Huang, Y.; Zhao, J.; et al. Bi-Allelic MEI1 Variants Cause Meiosis Arrest and Non-Obstructive Azoospermia. J. Hum. Genet. 2023, 68, 383–392. [Google Scholar] [CrossRef]
- Boekhout, M.; Karasu, M.E.; Wang, J.; Acquaviva, L.; Pratto, F.; Brick, K.; Eng, D.Y.; Xu, J.; Camerini-Otero, R.D.; Patel, D.J.; et al. REC114 Partner ANKRD31 Controls Number, Timing, and Location of Meiotic DNA Breaks. Mol. Cell 2019, 74, 1053–1068.e8. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, T.; Ke, H.; Zhang, Q.; Li, S.; Luo, W.; Qin, Y. Pathogenic Variants of Meiotic Double Strand Break (DSB) Formation Genes PRDM9 and ANKRD31 in Premature Ovarian Insufficiency. Genet. Med. Off. J. Am. Coll. Med. Genet. 2021, 23, 2309–2315. [Google Scholar] [CrossRef]
- Wyrwoll, M.J.; van Walree, E.S.; Hamer, G.; Rotte, N.; Motazacker, M.M.; Meijers-Heijboer, H.; Alders, M.; Meißner, A.; Kaminsky, E.; Wöste, M.; et al. Bi-Allelic Variants in DNA Mismatch Repair Proteins MutS Homolog MSH4 and MSH5 Cause Infertility in Both Sexes. Hum. Reprod. Oxf. Engl. 2021, 37, 178–189. [Google Scholar] [CrossRef]
- Li, P.; Ji, Z.; Zhi, E.; Zhang, Y.; Han, S.; Zhao, L.; Tian, R.; Chen, H.; Huang, Y.; Zhang, J.; et al. Novel Bi-Allelic MSH4 Variants Causes Meiotic Arrest and Non-Obstructive Azoospermia. Reprod. Biol. Endocrinol. 2022, 20, 21. [Google Scholar] [CrossRef]
- Xie, X.; Murtaza, G.; Li, Y.; Zhou, J.; Ye, J.; Khan, R.; Jiang, L.; Khan, I.; Zubair, M.; Yin, H.; et al. Biallelic HFM1 Variants Cause Non-Obstructive Azoospermia with Meiotic Arrest in Humans by Impairing Crossover Formation to Varying Degrees. Hum. Reprod. 2022, 37, 1664–1677. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Lv, M.; Gao, Y.; Cheng, H.; Li, K.; Xu, C.; Geng, H.; Li, G.; Shen, Q.; Wang, C.; et al. Novel Variants in Helicase for Meiosis 1 Lead to Male Infertility Due to Non-Obstructive Azoospermia. Reprod. Biol. Endocrinol. RBE 2021, 19, 129. [Google Scholar] [CrossRef]
- Yu, X.-C.; Li, M.-J.; Cai, F.-F.; Yang, S.-J.; Liu, H.-B.; Zhang, H.-B. A New TEX11 Mutation Causes Azoospermia and Testicular Meiotic Arrest. Asian J. Androl. 2021, 23, 510–515. [Google Scholar] [CrossRef] [PubMed]
- He, W.-B.; Tu, C.-F.; Liu, Q.; Meng, L.-L.; Yuan, S.-M.; Luo, A.-X.; He, F.-S.; Shen, J.; Li, W.; Du, J.; et al. DMC1 Mutation That Causes Human Non-Obstructive Azoospermia and Premature Ovarian Insufficiency Identified by Whole-Exome Sequencing. J. Med. Genet. 2018, 55, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Zangen, D.; Kaufman, Y.; Zeligson, S.; Perlberg, S.; Fridman, H.; Kanaan, M.; Abdulhadi-Atwan, M.; Abu Libdeh, A.; Gussow, A.; Kisslov, I.; et al. XX Ovarian Dysgenesis Is Caused by a PSMC3IP/HOP2 Mutation That Abolishes Coactivation of Estrogen-Driven Transcription. Am. J. Hum. Genet. 2011, 89, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Smirin-Yosef, P.; Zuckerman-Levin, N.; Tzur, S.; Granot, Y.; Cohen, L.; Sachsenweger, J.; Borck, G.; Lagovsky, I.; Salmon-Divon, M.; Wiesmüller, L.; et al. A Biallelic Mutation in the Homologous Recombination Repair Gene SPIDR Is Associated With Human Gonadal Dysgenesis. J. Clin. Endocrinol. Metab. 2017, 102, 681–688. [Google Scholar] [CrossRef]
- Heddar, A.; Ogur, C.; Da Costa, S.; Braham, I.; Billaud-Rist, L.; Findikli, N.; Beneteau, C.; Reynaud, R.; Mahmoud, K.; Legrand, S.; et al. Genetic Landscape of a Large Cohort of Primary Ovarian Insufficiency: New Genes and Pathways and Implications for Personalized Medicine. EBioMedicine 2022, 84, 104246. [Google Scholar] [CrossRef]
- Nawaz, S.; Ullah, M.I.; Hamid, B.S.; Nargis, J.; Nawaz, M.; Hussain, S.; Ahmad, W. A Loss-of-Function Variant in DNA Mismatch Repair Gene MLH3 Underlies Severe Oligozoospermia. J. Hum. Genet. 2021, 66, 725–730. [Google Scholar] [CrossRef]
- Nishimura, K.; Ishiai, M.; Horikawa, K.; Fukagawa, T.; Takata, M.; Takisawa, H.; Kanemaki, M.T. Mcm8 and Mcm9 Form a Complex That Functions in Homologous Recombination Repair Induced by DNA Interstrand Crosslinks. Mol. Cell 2012, 47, 511–522. [Google Scholar] [CrossRef]
- Takata, K.; Reh, S.; Tomida, J.; Person, M.D.; Wood, R.D. Human DNA Helicase HELQ Participates in DNA Interstrand Crosslink Tolerance with ATR and RAD51 Paralogs. Nat. Commun. 2013, 4, 2338. [Google Scholar] [CrossRef]
- Griffin, W.C.; Trakselis, M.A. The MCM8/9 Complex: A Recent Recruit to the Roster of Helicases Involved in Genome Maintenance. DNA Repair 2019, 76, 1–10. [Google Scholar] [CrossRef]
- Hustedt, N.; Saito, Y.; Zimmermann, M.; Álvarez-Quilón, A.; Setiaputra, D.; Adam, S.; McEwan, A.; Yuan, J.Y.; Olivieri, M.; Zhao, Y.; et al. Control of Homologous Recombination by the HROB-MCM8-MCM9 Pathway. Genes Dev. 2019, 33, 1397–1415. [Google Scholar] [CrossRef]
- Franca, M.M.; Condezo, Y.B.; Elzaiat, M.; Felipe-Medina, N.; Sánchez-Sáez, F.; Muñoz, S.; Sainz-Urruela, R.; Martín-Hervás, M.R.; García-Valiente, R.; Sánchez-Martín, M.A.; et al. A Truncating Variant of RAD51B Associated with Primary Ovarian Insufficiency Provides Insights into Its Meiotic and Somatic Functions. Cell Death Differ. 2022, 29, 2347–2361. [Google Scholar] [CrossRef] [PubMed]
- Daum, H.; Zlotogora, J. Fanconi Anemia Gene Variants in Patients with Gonadal Dysfunction. Reprod. Sci. 2022, 29, 1408–1413. [Google Scholar] [CrossRef] [PubMed]
- Tsui, V.; Crismani, W. The Fanconi Anemia Pathway and Fertility. Trends Genet. 2019, 35, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Caburet, S.; Heddar, A.; Dardillac, E.; Creux, H.; Lambert, M.; Messiaen, S.; Tourpin, S.; Livera, G.; Lopez, B.S.; Misrahi, M. Homozygous Hypomorphic BRCA2 Variant in Primary Ovarian Insufficiency without Cancer or Fanconi Anaemia Trait. J. Med. Genet. 2020, 58, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Lutzmann, M.; Grey, C.; Traver, S.; Ganier, O.; Maya-Mendoza, A.; Ranisavljevic, N.; Bernex, F.; Nishiyama, A.; Montel, N.; Gavois, E.; et al. MCM8- and MCM9-Deficient Mice Reveal Gametogenesis Defects and Genome Instability Due to Impaired Homologous Recombination. Mol. Cell 2012, 47, 523–534. [Google Scholar] [CrossRef]
- Adelman, C.A.; Lolo, R.L.; Birkbak, N.J.; Murina, O.; Matsuzaki, K.; Horejsi, Z.; Parmar, K.; Borel, V.; Skehel, J.M.; Stamp, G.; et al. HELQ Promotes RAD51 Paralogue-Dependent Repair to Avert Germ Cell Loss and Tumorigenesis. Nature 2013, 502, 381–384. [Google Scholar] [CrossRef]
- Lipkin, S.M.; Moens, P.B.; Wang, V.; Lenzi, M.; Shanmugarajah, D.; Gilgeous, A.; Thomas, J.; Cheng, J.; Touchman, J.W.; Green, E.D.; et al. Meiotic Arrest and Aneuploidy in MLH3-Deficient Mice. Nat. Genet. 2002, 31, 385–390. [Google Scholar] [CrossRef]
- Singh, P.; Fragoza, R.; Blengini, C.S.; Tran, T.N.; Pannafino, G.; Al-Sweel, N.; Schimenti, K.J.; Schindler, K.; Alani, E.A.; Yu, H.; et al. Human MLH1/3 Variants Causing Aneuploidy, Pregnancy Loss, and Premature Reproductive Aging. Nat. Commun. 2021, 12, 5005. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, R.J.; Feichtinger, J.; Larcombe, L. Cancer Germline Gene Activation. Cell Cycle 2014, 13, 2151–2152. [Google Scholar] [CrossRef] [PubMed]
- Lingg, L.; Rottenberg, S.; Francica, P. Meiotic Genes and DNA Double Strand Break Repair in Cancer. Front. Genet. 2022, 13, 831620. [Google Scholar] [CrossRef]
- Fratta, E.; Coral, S.; Covre, A.; Parisi, G.; Colizzi, F.; Danielli, R.; Marie Nicolay, H.J.; Sigalotti, L.; Maio, M. The Biology of Cancer Testis Antigens: Putative Function, Regulation and Therapeutic Potential. Mol. Oncol. 2011, 5, 164–182. [Google Scholar] [CrossRef]
- Simpson, A.J.G.; Caballero, O.L.; Jungbluth, A.; Chen, Y.-T.; Old, L.J. Cancer/Testis Antigens, Gametogenesis and Cancer. Nat. Rev. Cancer 2005, 5, 615–625. [Google Scholar] [CrossRef]
- Hosoya, N.; Miyagawa, K. Synaptonemal Complex Proteins Modulate the Level of Genome Integrity in Cancers. Cancer Sci. 2021, 112, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Masterson, L.; Sorgeloos, F.; Winder, D.; Lechner, M.; Marker, A.; Malhotra, S.; Sudhoff, H.; Jani, P.; Goon, P.; Sterling, J. Deregulation of SYCP2 Predicts Early Stage Human Papillomavirus-Positive Oropharyngeal Carcinoma: A Prospective Whole Transcriptome Analysis. Cancer Sci. 2015, 106, 1568–1575. [Google Scholar] [CrossRef] [PubMed]
- Mobasheri, M.B.; Shirkoohi, R.; Modarressi, M.H. Synaptonemal Complex Protein 3 Transcript Analysis in Breast Cancer. Iran. J. Public Health 2016, 45, 1618–1624. [Google Scholar]
- Neumann, F.; Wagner, C.; Preuss, K.-D.; Kubuschok, B.; Schormann, C.; Stevanovic, S.; Pfreundschuh, M. Identification of an Epitope Derived from the Cancer Testis Antigen HOM-TES-14/SCP1 and Presented by Dendritic Cells to Circulating CD4+ T Cells. Blood 2005, 106, 3105–3113. [Google Scholar] [CrossRef]
- Sandhu, S.; Sou, I.F.; Hunter, J.E.; Salmon, L.; Wilson, C.L.; Perkins, N.D.; Hunter, N.; Davies, O.R.; McClurg, U.L. Centrosome Dysfunction Associated with Somatic Expression of the Synaptonemal Complex Protein TEX12. Commun. Biol. 2021, 4, 1371. [Google Scholar] [CrossRef] [PubMed]
- Hosoya, N.; Okajima, M.; Kinomura, A.; Fujii, Y.; Hiyama, T.; Sun, J.; Tashiro, S.; Miyagawa, K. Synaptonemal Complex Protein SYCP3 Impairs Mitotic Recombination by Interfering with BRCA2. EMBO Rep. 2012, 13, 44–51. [Google Scholar] [CrossRef]
- Watkins, J.; Weekes, D.; Shah, V.; Gazinska, P.; Joshi, S.; Sidhu, B.; Gillett, C.; Pinder, S.; Vanoli, F.; Jasin, M.; et al. Genomic Complexity Profiling Reveals That HORMAD1 Overexpression Contributes to Homologous Recombination Deficiency in Triple-Negative Breast Cancers. Cancer Discov. 2015, 5, 488–505. [Google Scholar] [CrossRef]
- Gao, Y.; Kardos, J.; Yang, Y.; Tamir, T.Y.; Mutter-Rottmayer, E.; Weissman, B.; Major, M.B.; Kim, W.Y.; Vaziri, C. The Cancer/Testes (CT) Antigen HORMAD1 Promotes Homologous Recombinational DNA Repair and Radioresistance in Lung Adenocarcinoma Cells. Sci. Rep. 2018, 8, 15304. [Google Scholar] [CrossRef]
- Nichols, B.A.; Oswald, N.W.; McMillan, E.A.; McGlynn, K.; Yan, J.; Kim, M.S.; Saha, J.; Mallipeddi, P.L.; LaDuke, S.A.; Villalobos, P.A.; et al. HORMAD1 Is a Negative Prognostic Indicator in Lung Adenocarcinoma and Specifies Resistance to Oxidative and Genotoxic Stress. Cancer Res. 2018, 78, 6196–6208. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tan, Y.; Cao, X.; Kim, J.A.; Chen, T.; Hu, Y.; Wexler, M.; Wang, X. Epigenetic Activation of HORMAD1 in Basal-like Breast Cancer: Role in Rucaparib Sensitivity. Oncotarget 2018, 9, 30115–30127. [Google Scholar] [CrossRef]
- Feichtinger, J.; McFarlane, R.J. Meiotic Gene Activation in Somatic and Germ Cell Tumours. Andrology 2019, 7, 415–427. [Google Scholar] [CrossRef]
- Boukaba, A.; Liu, J.; Ward, C.; Wu, Q.; Arnaoutov, A.; Liang, J.; Pugacheva, E.M.; Dasso, M.; Lobanenkov, V.; Esteban, M.; et al. Ectopic Expression of Meiotic Cohesin Generates Chromosome Instability in Cancer Cell Line. Proc. Natl. Acad. Sci. USA 2022, 119, e2204071119. [Google Scholar] [CrossRef]
- Houle, A.A.; Gibling, H.; Lamaze, F.C.; Edgington, H.A.; Soave, D.; Fave, M.-J.; Agbessi, M.; Bruat, V.; Stein, L.D.; Awadalla, P. Aberrant PRDM9 Expression Impacts the Pan-Cancer Genomic Landscape. Genome Res. 2018, 28, 1611–1620. [Google Scholar] [CrossRef] [PubMed]
- Jay, A.; Reitz, D.; Namekawa, S.H.; Heyer, W.-D. Cancer Testis Antigens and Genomic Instability: More than Immunology. DNA Repair 2021, 108, 103214. [Google Scholar] [CrossRef] [PubMed]
- Cho, N.W.; Dilley, R.L.; Lampson, M.A.; Greenberg, R.A. Interchromosomal Homology Searches Drive Directional ALT Telomere Movement and Synapsis. Cell 2014, 159, 108–121. [Google Scholar] [CrossRef]
- Ianzini, F.; Kosmacek, E.A.; Nelson, E.S.; Napoli, E.; Erenpreisa, J.; Kalejs, M.; Mackey, M.A. Activation of Meiosis-Specific Genes Is Associated with Depolyploidization of Human Tumor Cells Following Radiation-Induced Mitotic Catastrophe. Cancer Res. 2009, 69, 2296–2304. [Google Scholar] [CrossRef]
- Rivera, M.; Wu, Q.; Hamerlik, P.; Hjelmeland, A.B.; Bao, S.; Rich, J.N. Acquisition of Meiotic DNA Repair Regulators Maintain Genome Stability in Glioblastoma. Cell Death Dis. 2015, 6, e1732. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, P.; Bandlamudi, C.; Cheng, M.L.; Srinivasan, P.; Chavan, S.S.; Friedman, N.D.; Rosen, E.Y.; Richards, A.L.; Bouvier, N.; Selcuklu, S.D.; et al. Tumour Lineage Shapes BRCA-Mediated Phenotypes. Nature 2019, 571, 576–579. [Google Scholar] [CrossRef] [PubMed]
- Peake, J.D.; Noguchi, E. Fanconi Anemia: Current Insights Regarding Epidemiology, Cancer, and DNA Repair. Hum. Genet. 2022, 141, 1811–1836. [Google Scholar] [CrossRef]
- Pećina-Šlaus, N.; Kafka, A.; Salamon, I.; Bukovac, A. Mismatch Repair Pathway, Genome Stability and Cancer. Front. Mol. Biosci. 2020, 7, 122. [Google Scholar] [CrossRef] [PubMed]
- Gausachs, M.; Mur, P.; Corral, J.; Pineda, M.; González, S.; Benito, L.; Menéndez, M.; Espinàs, J.A.; Brunet, J.; Iniesta, M.D.; et al. MLH1 Promoter Hypermethylation in the Analytical Algorithm of Lynch Syndrome: A Cost-Effectiveness Study. Eur. J. Hum. Genet. 2012, 20, 762–768. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llano, E.; Pendás, A.M. Synaptonemal Complex in Human Biology and Disease. Cells 2023, 12, 1718. https://doi.org/10.3390/cells12131718
Llano E, Pendás AM. Synaptonemal Complex in Human Biology and Disease. Cells. 2023; 12(13):1718. https://doi.org/10.3390/cells12131718
Chicago/Turabian StyleLlano, Elena, and Alberto M. Pendás. 2023. "Synaptonemal Complex in Human Biology and Disease" Cells 12, no. 13: 1718. https://doi.org/10.3390/cells12131718
APA StyleLlano, E., & Pendás, A. M. (2023). Synaptonemal Complex in Human Biology and Disease. Cells, 12(13), 1718. https://doi.org/10.3390/cells12131718




