Abstract
Intracellular trafficking plays a critical role in the functioning of highly polarized cells, such as neurons. Transport of mRNAs, proteins, and other molecules to synaptic terminals maintains contact between neurons and ensures the transmission of nerve impulses. Cytoplasmic polyadenylation element binding (CPEB) proteins play an essential role in long-term memory (LTM) formation by regulating local translation in synapses. Here, we show that the 3′UTR of the Drosophila CPEB gene orb2 is required for targeting the orb2 mRNA and protein to synapses and that this localization is important for LTM formation. When the orb2 3′UTR is deleted, the orb2 mRNAs and proteins fail to localize in synaptic fractions, and pronounced LTM deficits arise. We found that the phenotypic effects of the orb2 3′UTR deletion were rescued by introducing the 3′UTR from the orb, another Drosophila CPEB gene. In contrast, the phenotypic effects of the 3′UTR deletion were not rescued by the 3′UTR from one of the Drosophila α-tubulin genes. Our results show that the orb2 mRNAs must be targeted to the correct locations in neurons and that proper targeting depends upon sequences in the 3′UTR.
1. Introduction
Proper distribution of mRNAs and proteins within the cell plays a critical role in embryogenesis, cell differentiation, and the functioning of specialized cell types [1,2,3,4]. As is known, mRNA localization coupled with on-site translation is one of the common strategies used to localize proteins to appropriate cell compartments [5,6,7]. Neurons, which have long outgrowths, termed axons, and much shorter outgrowths, dendrites, are a context in which mRNA localization and on-site translation are particularly important. Specific mRNAs and proteins must be localized in the distal part of neuronal outgrowths (neuritis) [8,9] to ensure synaptic transduction of nerve impulses between neurons [10].
Cytoplasmic polyadenylation element (CPE) binding proteins (CPEBs) are a family of proteins that are known to play a key role in on-site translational regulation [11]. Depending on the context and the mRNA, CPEBs can either repress or activate translation through a polyA-dependent mechanism [12,13]. In addition to regulating translation, CPEBs can mediate the intracellular transport of mRNAs in neurons according to several studies [14,15,16].
Neuron-specific mRNAs typically have longer 3′UTRs compared with mRNAs of other cell types [17]. These long 3′UTRs have regulatory elements to target the mRNAs to specific compartments within the cell and to control their translation. Recent studies have shown that long 3′UTRs can additionally play noncanonical roles, mediating the protein–protein interactions [18], forming new intracellular domains [19], and acting independently of the mRNA-coding region [20,21].
The orb2 gene has multiple mRNA isoforms of different lengths, including a very long one, which is expressed in nerve cells [22]. The orb2 gene encodes one of the two Drosophila melanogaster CPEBs and plays an important role in the functioning of the nervous system [23,24]. There are two Orb2 protein isoforms, Orb2A and Orb2B. The shorter isoform Orb2A forms oligomers with amyloid-like properties and can induce a similar conversion of the larger Orb2B isoform [25]. Depending on its oligomer state, the Orb2 protein can control the local translation of mRNAs in synapses in response to external stimuli. As such, Orb2 plays a central role in the formation of long-term memory (LTM) [26]. Local translation in a stimulated synapse is thought to change the composition and physiological properties of receptors in the postsynaptic membrane. This results in long-term potentiation (or, much less frequently, long-term depression), that is, changes in the strength of nerve impulse transmission. At the next step, the activated synapse undergoes morphological alterations, which depend upon transcription. Together the changes form a stable memory engram [27,28,29].
The Orb2 protein has been identified as a critical participant in LTM formation in Drosophila [30,31]. A deletion of the Q-domain, which mediates Orb2 oligomerization, or even a single amino acid substitution in this part of the protein results in the inability to form LTM. The Orb2A isoform is expressed at very low levels in the fly brain, but its expression in dopaminergic neurons of the mushroom bodies (a center of memory acquisition and consolidation) is critical for LTM formation [26].
We have shown previously that a deletion of the 3′UTR sequences from the mRNA isoforms that encode the larger Orb2 protein disrupts LTM formation [32]. Our experiments suggest that LTM formation is disrupted because insufficient amounts of the Orb2 protein are expressed in the synaptic zone of neurons. To provide further evidence that LTM formation requires the localization of orb2 mRNAs to the synaptic zones of neurons, we generated heterologous 3′ UTR replacements. For the control replacement, we selected the 3′UTR of the Drosophila α-tubulin gene (αTub84B) as it is not expected to be localized in a pattern like orb2. For the experiment, we selected the 3′UTR from the orb gene, which encodes the other Drosophila CPEB protein. Like orb2, its mRNA has localization signals and it also has functions in LTM formation [33,34].
2. Materials and Methods
2.1. Generation of Plasmid Constructs for Drosophila Transgenesis: Fly Strains
Full-length 3′UTR sequences from the alpha-tubulin (αTub84B) and orb genes were amplified from genomic DNA with primers containing BamHI and BglII restriction sites. The sequences of primers and the 3′UTRs are given in the Supplementary Materials. Next, the amplified fragments were cloned in a pOT2-attB-containing vector. The resulting plasmids were checked by sequencing.
In the starting orb2 3′UTR deletion, orb2R, an attP site for recombination was substituted for the deleted 3′UTR sequences [35]. To generate the replacements, the orb2R fly strain was crossed with a phiC31-expressing strain (34772, DBSC, USA, genotype: y (1) w(*) P[y[+t7.7] = nos-phiC311]X; L (1)/CyO; TM2/TM6B, Tb (1)). Embryos obtained from the crosses were injected with a plasmid containing attB and the full-length sequences of the αTub84B and orb 3′UTRs. Flies with recombination events were selected by dsRed expression in the eyes; the dsRed marker was removed via Cre recombinase-mediated excision. Five independent flies with recombination events were used to generate homozygote stocks; the integrated fragments were sequenced. The resulting strains were designated as orb2tub and orb2orb.
The Drosophila fly strain 51324 (genotype: w (1118); PBacVK00027) from the Drosophila Bloomington Stock Center (DBSC, Bloomington, IN, USA) was additionally used in the study as a wild-type (WT) control to measure the mRNA and protein levels. It was shown previously that the 51,324 fly strain has the same ability to form memory as the standard CantonS strain, which is usually used in behavioral assays. The CantonS fly strain was used as a WT control in behavioral experiments.
2.2. Antibodies
The 4G8 (1:50 dilution) antibody against the Orb2 protein was used in Western blotting. The antibody was deposited in the Developmental Studies Hybridoma Bank (DSHB, Iowa, IA, USA) by P. Schedl. The DCSP-1 (ab49) antibody against the Cysteine-string protein (CSP) was deposited in the DSHB by E. Buchner and A. Hofbauer and was used in a 1:500 dilution. The 3C11 antibody against Synapsin was deposited in the DSHB by B.R.E. Klagge and colleagues and used at a 1:200 dilution. An anti-polychaetoid (PYD1) antibody was deposited in the DSHB by A.S. Fanning and used at a 1:500 dilution. An HRP-conjugated goat anti-mouse IgG antibody (Jackson ImmunoResearch, UK, catalog number 115-035-174) was used as a secondary antibody in Western blotting at 1:2000.
2.3. Behavioral Assay
A courtship-suppression paradigm was used to assess memory formation [32,36]. Flies were raised on standard Drosophila yeast–raisin medium at 25 °C and 60–70% humidity with a 12 h light–dark cycle. Males of a studied strain were collected without anesthesia 0–10 h after eclosion and kept individually in food vials until the tests. The training was performed using 5-day-old CS females fertilized 1 day before. Testing was performed at the age of 5 days. For training, naive males having no courtship experience were placed in a chamber with a fertilized female for 30 min for further assessment of either STM (short-term memory, immediately after training) or MTM (middle-term memory, 180 min. after training). For LTM formation, a male and a female were kept together for 5 h and assessed 2 days after training, using newly fertilized 5-day-old CS females. Naive males were used as a control. For each point, 20 males were tested. The time spent in courtship (orientation, following, wing vibration, licking, and attempted copulation) was recorded for 300 s. The courtship index (percentage of time spent in courtship) was calculated for each male. A courtship index (CI) was calculated as a ratio of courtship time to the entire observation time [36]. A learning index (LI) was calculated as LI = [(CIn − CIexp)/CIn] × 100%, where CIn is the average CI of two independent samples of males without courtship experience, and CIexp is the average CI of two independent samples of males after training. Statistical comparisons of behavioral data were conducted by a randomization test, by directly calculating the probability of rejection of the null hypothesis αR. A sampled randomization test with 10,000 permutations was used. The null hypothesis was rejected at αR < 0.05. A two-sided test was used to compare CIs, and a one-sided test for the comparison of LIs. In the latter case, no SEMs were required [36].
2.4. Fluorescent In Situ Hybridization (FISH)
To perform in situ hybridization, at least 20 brains were dissected from 4- to 5-day-old flies for each genotype used in the study. The brains were washed twice with PBS supplemented with 0.05% Tween-20 (PBSt) for 10 min and fixed with 3.5% paraformaldehyde for 20 min at room temperature. The brains were treated with 100% methanol at –20 °C for 5 min, and then methanol was replaced with PBS through a series of washes with descending vol/vol methanol/PBS solutions (70/30, 50/50, 30/70). Each rinse was combined with rotation and lasted 10 min (room temperature). After the final wash, the brains were washed with PBSt twice for 5 min and incubated in a prewarmed wash buffer (4× SSC, 35% formamide, 0.1% Tween-20) at 37 °C for 15 min. After incubation, the samples were placed in a hybridization buffer (Stellaris RNA FISH Hybridization Buffer, LGC Biosearch Technologies, Middlesex, UK, cat. No SMF-HB1-10) supplemented with FISH probes at a 1:10 dilution and incubated at 37 °C with continuous shaking. The next day, the samples were washed with a wash buffer at 37 °C for 4 h and with PBSt at room temperature for 2 h. The samples were mounted on a slide glass in the VectaShield mounting medium with DAPI (Vector Labs, Newark, CA, USA) and examined by microscopy. The FISH probes were manufactured by LGC Biosearch Technologies. A probe set consisted of twenty 20-nt probes complementary to part of the last common exon of the orb2 gene. Each 20-nt probe was conjugated with Quasar 670 at the 3′ end. The sequences of all probes are presented in the Supplementary Materials.
2.5. RNA Isolation, Reverse Transcription, qPCR
To isolate RNA, the Trizol reagent was used according to the manufacturer’s recommendations (Molecular Research Center, Cincinnati, OH, USA, catalog number TR118). Twenty fly heads of each genotype were taken; the final RNA concentration was measured with an RNA assay kit (Thermo Fisher Scientific, Waltham, MA, USA, catalog number Q32852). Equal amounts of RNAs obtained from different biological samples were used to generate cDNAs. Reverse transcription was carried out with a reverse transcriptase RNAscribe RT kit (Biolabmix, Novosibirsk, Russia, catalog number R04-10) according to the manufacturer’s recommendations.
When performing quantitative PCR, each sample was used in technical triplicate. At least five independent biological replicas were used in every experiment. To assess the relative amount of the orb2 mRNA in total fly head extracts, gapdh2 was used as a reference gene to calculate 2−ΔΔCt. The 28S RNA was used as a reference when 2−ΔΔCt was calculated to compare the distribution of the orb2 mRNA and its potential mRNA targets (act5C, csp, and pyd) between the cell body and synaptic fractions of neurons. Sequences of primers used in the study are presented in the Supplementary Materials.
2.6. Crude Synaptosome Preparation
To study the protein distribution in nerve cells, 50 fly heads of each genotype were homogenized in 50 µL of 0.32 M sucrose buffer (5 mM HEPES, pH 7.4, 145 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 5 mM glucose) supplemented with a protease inhibitor cocktail. Crude synaptosome preparations were obtained essentially as described in [37] with an extra centrifugation step added after collecting the first pellet. To study the RNA distribution, we collected at least 400 flies of each genotype. The flies were frozen in liquid nitrogen, vortexed, and sifted through two 800 and 380 µm sieves to separate heads from bodies. The 0.32 M sucrose homogenization buffer was additionally supplemented with a ribonuclease inhibitor (Ribolock, Thermo Fisher Scientific, Waltham, MA, USA, catalog number EO0382).
2.7. Semiquantitative Western Blot Analysis
To assess the distribution of the proteins of interest in Drosophila nervous cells, we used the strategy described in [32]. Twenty fly heads of each genotype were taken for analysis.
2.8. Confocal Microscopy
Stained samples of fly brains were scanned and imaged under a Leica STELLARIS 5 (Wetzlar, Germany) confocal microscope. We used a 40× oil objective with a numerical aperture of 1.3. Images with a frame size of 2048 × 2048 pixels and a z resolution of 1 µm were taken at a scan speed of 400. At least four brains were imaged for each genotype, and selected images were processed using LazX software (Leica Application Suite X, version 3.7.25997.6., Leica Microsystems, Wetzlar, Germany). Average intensity values in the marked brain regions were determined using Fiji 1.53t software.
2.9. Quantification and Statistical Analysis
Experimental data were statistically analyzed with GraphPad Prism 8.0.2. software. Graphs presented in the figures were plotted using the same software. Columns display the mean values; error bars display the standard deviation. Two-tailed Student’s t-test was used to compare the relative mRNA and protein concentrations between flies with different genotypes. Asterisks in the graphs indicate p-value < 0.05; ns, nonsignificant differences.
3. Results
3.1. Generation of Fly Strains with Replacements of Deleted Part of orb2 3′UTR
In previous studies, we generated a 4.5 kb deletion of sequences encoding two of the larger orb2 3′UTRs [35]. As indicated in Figure 1, the largest orb2 3′UTR has 37 CPE-like elements, while the orb2R deletions have only four putative CPE elements, which are located closer to the stop codon for Orb2B (hereinafter referred to as Orb2) protein. The deletion does not remove sequences from the 3′UTR sequences of the mRNA encoding the Orb2A isoform, nor does it remove sequences from the shortest orb2B mRNA, orb2-RB (see Figure 1). Analysis of the starting orb2R deletion showed that Orb2 protein levels in the synaptic zone of neurons were reduced, while the levels in neuronal cell bodies were equivalent to those in WT. We also found that the formation of long-term memory (LTM) in the orb2R mutant was significantly impaired [32].
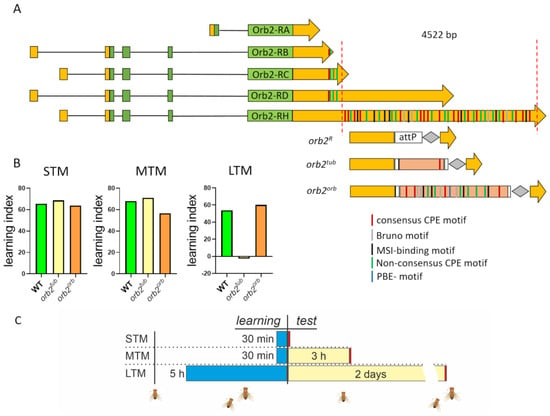
Figure 1.
Structure of the orb2 locus and replacements used in the study. (A) Structure of orb2 transcripts. The coding regions are shown with green boxes; the 5′- and 3′UTRs, with yellow boxes. The 3′UTR part deleted in the orb2R strain is shown with red dashed lines. The structure of the modified orb2 locus is given below. The remaining part of the 3′UTR (yellow), the attP site used for integration, the integrated sequences, and the LoxP site (gray rhombus) are also shown. Several regulatory motifs are marked with strips of different colors (see detail in text). (B) Learning indices calculated for short-term (STM), mid-term (MTM), and long-term (LTM) memory. STM and MTM of the orb2tub and orb2orb fly strains were the same as in the WT strain; orb2tub flies showed a significant reduction in LTM formation. (C) Behavioral assessment timeline. The blue bars indicate the learning time, the yellow bars indicate the time before the test, and the red bars indicate the test of the courtship behavior.
To provide further evidence that the proper localization/regulation of orb2 mRNA is critical for LTM formation, we replaced the deletion with two different UTR sequences, the αTub84B tubulin gene 3′UTR, which contains a single consensus CPE sequence, and the 3′UTR that belongs to the orb, another CPEB gene, and contains eight CPEs [34,38]. The full sequences are presented in Materials and Methods. We chose these two test UTRs because in the orb2R deletion, the remaining UTR sequences (see Figure 1A) had only four CPE-like elements and we supposed that among other things, the number of CPE sequences likely played a critical role in the localization of the orb2 mRNA and/or its efficient translation. In addition, other regulatory motifs were found in the 3′UTR of the orb2 mRNA, namely, the bird-box motif, Bruno motifs, and Musashi binding elements (MSI) [39,40]. All of them were also present in the orb 3′UTR. A Pumilio binding motif (PBE) is additionally found in the 3′UTR of the orb mRNA [41].
After phiC31-mediated integration of the UTR sequences in the attP site, the dsRed marker was removed. The final fly strains used in further experiments were designated as orb2tub and orb2orb (Figure 1B).
3.2. Introduction of Different Sequences in orb2R Allele Does Not Affect Short-Term and Mid-Term Memory Formation but Affects Long-Term Memory
The CIs of orb2tub and orb2orb adult males were assessed to evaluate their ability to form short-term (STM) and mid-term memory (MTM) (Figure S1). Figure 1B summarizes the calculated LIs. As expected, the CIs of orb2tub flies were similar to the CIs of WT flies. The same picture was observed in orb2orb flies—i.e., their STM and MTM formation was normal (Figure 1B). Thus, we found that different sequences of the 3′UTR of the orb2 mRNA did not affect the STM and MTM formation, as has been shown for the initial orb2R fly strain.
Next, we studied how different 3′UTR sequences affect the LTM formation when added to the orb2-coding region. The αTub84B 3′UTR did not restore the LTM formation, and orb2tub flies showed the same CIs as naive males 2 days after training. In contrast, orb2orb flies showed a significant reduction in CI just after training and retained this reduction 2 days after training. Thus, a replacement of the 3′UTR sequence deleted from the orb2 gene with the orb 3′UTR sequence leads to complete recovery of the LTM formation.
3.3. Different 3′UTR Sequences Used to Replace Deletion of orb2 3′UTR Do Not Influence mRNA Level but Affect Protein Level of Orb2 in Total Fly Head Extracts
To better understand why LTM formation in orb2tub and orb2orb males is different, we assessed how insertions into the orb2R allele affect the orb2 mRNA level. The relative orb2 mRNA level mRNA was calculated by the ΔΔCt method with gapdh2 used as a reference gene. The results are presented in Figure 2A. We found that the orb2 mRNA level did not differ from the WT level in both the orb2tub and orb2orb fly strains. The same result was observed in the orb2R strain [32]. Thus, the insertion of an additional sequence in place of the deleted part of the orb2 gene does not change the relative production or the stability of the orb2 mRNA.
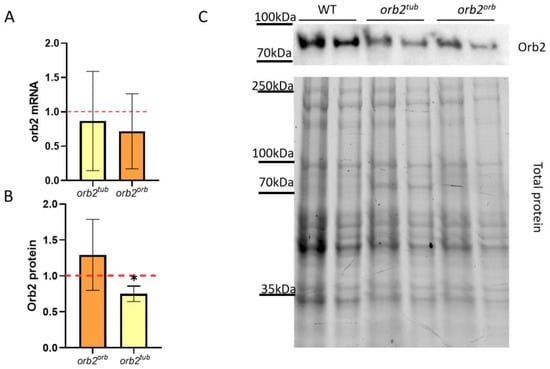
Figure 2.
Effects of different sequences inserted in the orb2 3′UTR on orb2 gene expression. (A) Relative levels of the orb2 mRNA detected in heads of orb2tub and orb2orb flies by qPCR. A red dashed line presents the orb2 mRNA level in WT flies, which was used for normalization. Columns show the mean mRNA levels detected in orb2tub and orb2orb (t-test, n = 6, SD is shown). (B) Relative levels of the Orb2 protein in total head extracts from orb2tub and orb2orb flies. A red dashed line presents the Orb2 protein level in WT flies, which was used for normalization. The mean Orb2 protein level was found to decrease in orb2tub flies (n = 6, SD is shown, asterisk shows p < 0.05), while orb2orb flies had the same Orb2 protein level as WT flies. (C) Western blot analysis of head lysates from orb2tub and orb2orb flies with antibodies against the Orb2 protein (top). Total protein staining (bottom) was used for normalization in a twofold serial dilution.
Next, the Orb2 protein level was assessed in total fly head extracts of the orb2tub and orb2orb fly strains. Chemiluminescent signals from staining with antibodies against the Orb2 protein were normalized to the total protein in the sample. We found that orb2orb flies had the same Orb2 protein level as WT flies. In contrast, orb2tub flies showed a 34% decrease in Orb2 protein level as compared with WT flies (Figure 3B). The results demonstrate that the orb 3′UTR sequence restored the total Orb2 protein amount in the fly brain, while the tubulin gene 3′UTR did not elevate the Orb2 protein amount to the WT level. The orb2tub and initial orb2R strains showed a similar level of decreasing of Orb2 protein amount.
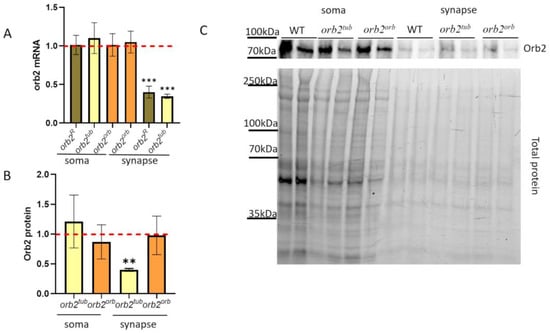
Figure 3.
Subcellular distribution of the orb2 mRNA and protein in neurons. (A) Relative levels of the orb2 mRNA were detected in fractions of neurons of the orb2R, orb2tub, and orb2orb fly strains by qPCR. A red dashed line presents the orb2 mRNA level in WT flies, which was used for normalization. Columns show the mean mRNA levels detected in orb2R, orb2tub, and orb2orb (t-test, n = 8, SD is shown). Asterisks show significant changes (p < 0.001) in the orb2 mRNA level in the synaptic fractions of the orb2R and orb2tub strains. (B) Relative mean levels of the Orb2 protein in the soma and synaptic fractions from orb2tub and orb2orb flies. A red dashed line presents the Orb2 protein level in WT flies, which was used for normalization. Flies of the orb2tub strain showed a decrease in Orb2 protein amount (n = 6, SD is shown, asterisks show p-value < 0.01), while orb2orb flies had the same Orb2 protein level as WT flies. (C) The top panel shows an example of staining the fractions from different fly strains with anti-Orb2 antibodies. Each sample was examined in a twofold dilution as well. The bottom panel shows the total protein.
3.4. 3′UTR of orb2 Gene Is Essential for Intracellular Transport of orb2 mRNA
The next question to study was how the sequences introduced into the orb2 3′UTR influence the orb2 mRNA intracellular distribution. We applied the previously used biochemical approach to separate the soma and synaptic regions [32]. The results are presented in Figure 3A. We did not find any difference in the orb2 mRNA level from the WT in the soma fraction of neurons in both the orb2tub and the orb2orb fly strains. The synaptic fraction of the orb2orb strain was similar to the WT in the amount of the orb2 mRNA. In contrast, the orb2tub synaptic fraction displayed a nearly threefold reduction in orb2 mRNA compared with the WT. We checked the distribution of the orb2 mRNA in the initial orb2R fly train. The amount of the orb2 mRNA in the soma of neurons was found to be the same as in the WT, whereas a decrease in the orb2 mRNA level was observed in the synaptic fraction, similar to the orb2tub strain. Thus, we can conclude that the orb2 3′UTR sequence is essential for localizing the orb2 mRNA to the synaptic compartment of the nerve cell.
3.5. 3′UTR of orb2 Gene Is Essential for Orb2 Protein Distribution
To analyze the Orb2 protein distribution in neurons of the orb2tub and orb2orb fly strains, we prepared the fractions of cell bodies and synaptosomes. We found that the Orb2 protein amount in the cell bodies of the strains did not differ from that in WT flies. In the synaptosome fraction, the Orb2 protein amount in orb2orb flies did not differ from that in WT flies, whereas orb2tub flies showed a more than twofold reduction in the Orb2 amount (Figure 3A). Thus, the distribution of the Orb2 protein among the fractions in orb2tub flies was similar to that reported previously for orb2R flies [32]. The results have led us to the conclusion that the 3′UTR of the orb2 gene is essential for the proper localization of the orb2 mRNA and protein to synapses.
3.6. Sequence in 3′UTR of orb2 Is Important for mRNA Localization to Neuron Outgrowths
To more precisely study the mRNA distribution in the fly brain for different orb2 alleles, in situ hybridization with Stellaris probes was performed using whole-mount brain samples. The orb2 mRNA was abundantly present in neuropile regions in WT flies (Figure 4A, Figures S2 and S3). We also found the orb2 mRNA in the soma of neurons, as has been shown before [23]. When studying orb2R fly brains, we found that staining was decreased in neuropile regions and that the signal from the orb2 probe was distributed evenly in different parts of the brain as compared with the WT (Figure 4A, Figures S2 and S3). We quantified the signal intensities of the orb2 mRNA in the neuropile and soma regions and then calculated their ratio. The relative intensity of the orb2 mRNA in the neuropile regions is significantly reduced in orb2R mutants (Figure 4B).
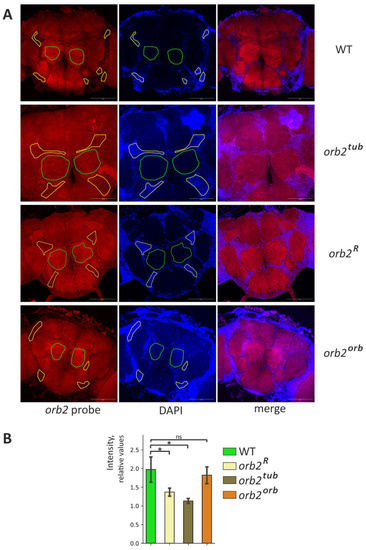
Figure 4.
Distribution of orb2 mRNA in the fly brain. (A) Staining of whole-mount brain samples with anti-orb2 probes (red channel) and staining of nuclei with DAPI (blue channel). The same exposure was used to take images in the cases of the WT and orb2 mutants. The neuropile regions are marked with a green line. The areas marked by the yellow line are the neuronal body regions. Scale bar, 100 µm. (B) The mean intensity of the neuropile regions relative to the neuronal nucleus regions in the indicated fly lines (Mann-Whitney test, n = 4, SD is shown, asterisk shows p-value < 0.05, ns is nonsignificant). The relative intensity of neuropile staining is reduced in the orb2R and orb2tub preparations.
A hybridization pattern similar to the orb2R pattern was observed in orb2tub samples. Regions enriched in axons were poorly stained with the orb2 probe (Figure 4A, Figures S2 and S3). In contrast, a recovery of the orb2 mRNA distribution observed in WT brains was detected in orb2orb brains. This is confirmed by a quantitative assessment of staining intensity (Figure 4B).
An interesting feature of the orb2 mRNA distribution was additionally observed in the neuron soma. A perinuclear ring with several bright speckles per cell was formed in the region of the calyx (dendritic input for Kenyon cells) by the orb2 mRNA (Figures S4 and S5). The speckles were clearly observed in WT flies. Sites of a high local concentration of the orb2 mRNA in the soma may be assumed to act as sites of RNP assembly. The same picture was found in orb2orb brains. In orb2R and orb2tub flies, the perinuclear ring was preserved, but the speckles were hardly distinct from other orb2 mRNA fractions localized in the soma. In addition, the orb2 mRNA was distributed more evenly between the cell soma and the calyx in orb2R and orb2tub brains and the background staining was stronger compared with the WT.
The changes in orb2 mRNA distribution observed in orb2R and orb2tub could be attributed to changes in brain structures. To check this assumption, brain samples were stained with antibodies against Synapsin, which is a marker of neuropile regions. The results are presented in Figure 5. We did not find any obvious change in brain organization of the orb2R, orb2tub, and orb2orb strains compared with the WT.
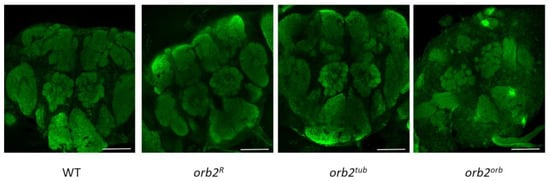
Figure 5.
The brain structure of the studied mutants. Staining with antibodies against synapsin. The brain structure in the strains orb2R, orb2tub, and orb2orb is indistinguishable from that in WT brains. Scale bar, 50 µm.
3.7. Distribution of mRNAs and Proteins of Potential Orb2 Targets in Nerve Cells
We have previously found that the Csp and Pyd proteins, whose mRNAs are potential targets of the Orb2 protein, elevate their representation in synapses of orb2R flies. csp encodes a chaperone associated with synaptic vesicles, and pyd encodes a protein involved in cell adhesion, which is important for mushroom body formation. The mRNAs encoding the proteins are thought to be regulatory targets of Orb2. One explanation for the increased amount of Csp and Pyd proteins in the synaptic fraction is that Csp and Pyd mRNAs are transported more efficiently and/or stabilized in the orb2R deletion mutant, resulting in increased total mRNA levels encoding these two proteins. Alternatively, both mRNAs may be subject to translational repression by Orb2. In this case, a decrease in the level of Orb2 in the synapse would result in an increase in the translation of csp and pyd mRNA.
To explore these possibilities, we used qPCR to measure the amount of csp and pyd mRNA in soma and synapse fractions in the head obtained from orb2R, orb2tub, and orb2orb flies. As shown in Figure 6A, the relative level and distribution in the soma and synaptic fractions were the same as in WT.
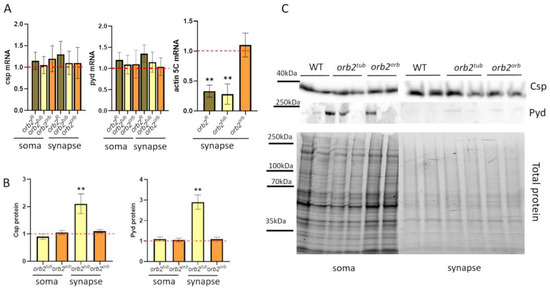
Figure 6.
Distribution of potential Orb2 targets in nerve cells. (A) Relative mean levels of the csp and pyd mRNAs in the soma and synaptic fractions of the fly strains. A red dashed line presents the mRNA level in WT flies, which was used for normalization. Error bars show SD; there was no significant change in mRNA levels compared with the WT. Relative mean levels of actin5C mRNA in a synaptic fraction of studied strains (n = 10, Student t-test, asterisks show p-value < 0.01). (B) Relative mean levels of the Csp and Pyd proteins in the soma and synaptic fractions in the indicated strains (n = 10, Student’s t-test, asterisks show that p-value < 0.01). A red dashed line presents the protein level in WT flies, which was used for normalization. (C) Example of staining with antibodies against the Csp and Pyd proteins (top) and total protein for normalization (bottom). Nerve cell fractions were obtained from WT, orb2tub, and orb2orb brain samples.
Next, we examined the distribution of Csp and Pyd proteins in the soma and synaptic fractions. In the orb2orb sample, the levels of the proteins were the same as in the WT flies. However, in orb2tub samples, the levels of the proteins were elevated in the synaptic fraction compared with the WT (Figure 6B). Similar results have been obtained previously for the orb2R deletion [32].
Changes in distribution were also explored for the actin5C mRNA in the fly strains because the mRNA has been shown to provide a potential target for the Orb2 protein [42,43]. We found that its amount significantly decreased (almost three times as compared with WT flies) in the synaptic fraction in the orb2tub strain and was almost the same as in WT flies in the orb2orb fly strain (Figure 6A).
4. Discussion
One of the most important discoveries in neurosciences is that the synapse, rather than a single neuron, is an elementary unit for memory formation [44]. Presynaptic and postsynaptic termini contain unique sets of molecules for the transmission and reception of nerve impulses. A mechanism to target key proteins to the pre- and postsynaptic termini relies on mRNA transport followed by local translation [7,9]. Perhaps for this reason, many neuronal mRNAs have extended 3′UTRs compared with mRNAs encoding the same proteins in non-neuronal tissues [45].
Previously, we have reported the isolation and characterization of a large deletion (4.5 kb) that removes most of the sequence for two large orb2 3′UTRs [32]. The longest of the orb2 3′UTRs (orb2-RH) has 37 canonical and noncanonical CPE motifs, and all but 4 of these are preserved in flies with our orb2R deletion. The deletion does not appear to impact the translation of the orb2 mRNAs in the neuronal cell body, and the levels of the Orb2 protein in this cell compartment are similar to that in the WT. On the other hand, the deletion reduces the level of the orb2 mRNA in synaptic fractions by more than half. A concomitant reduction in Orb2 protein in the synaptic fraction accompanies this reduction in mRNA.
In the studies reported here, we used a 3′UTR replacement strategy to rescue the orb2R 3′UTR deletion. We tested the 3′UTRs derived from two different fly genes, the αTub84B tubulin gene and the orb gene for the other Drosophila CPEB protein. The former has a single CPE, while the latter has eight canonical CPEs. The αTub84B 3′UTR does not rescue the localization of the orb2 mRNA to the synaptic fractions. Moreover, like in the starting orb2R deletion mutant, Orb2 protein levels in the synaptic fraction are reduced, while the protein amount in the neuronal cell body is similar to the WT. Consistent with the idea that localization of the orb2 mRNA to synaptic fractions is important for the orb2 function, orb2tub flies exhibit phenotypes similar to those observed for orb2R. These include elevated levels of the Csp and Pyd proteins in the synaptic fraction and pronounced deficits in LTM formation. The suggestion is supported by our analysis of the orb 3′UTR replacement of orb2orb. This orb 3′UTR replacement appears to fully rescue the phenotype of the orb2 3′UTR deletion. Both orb2 mRNA and protein levels in the synaptic fraction are equivalent to the WT, as are the levels of the Csp and Pyd proteins. In addition, there is no obvious deficit in LTM formation.
Orb is known to be essential for LTM formation [33]. It is also known that the orb 3′UTR is essential for the proper localization of the Orb protein in the oocyte [38] through an autoregulatory loop. Additionally, the orb 3′UTR contains the BRE, MSI, and PBE motifs, as well as binding sites for the Pumilio protein, which could be essential for proper translation activation [39,40,41]. Fragments of similarity between the orb and orb2 3′UTRs are not long enough, but all of them contain multiple CPE sequences. Based on this observation, we assume that the number of CPE sequences in the orb2 3′UTR together with other cis-regulatory elements can somehow influence the Orb2 protein amount in synapses.
A model that could explain our results is as follows: The UTR sequences remaining in orb2R transcripts lack signals critical for neuronal transport, and the levels of the orb2 mRNA in the synaptic fractions are consequently reduced. The same would be true for the orb2tub replacement. The levels of the Orb2 protein are reduced in both orb2R and orb2tub, and this reduction is responsible for elevated translation of the csp and pyd mRNAs in the synaptic fraction. One would suppose that the failure to properly regulate translation in the synaptic fraction is responsible for deficits in LTM formation as well. In the case of Csp and Pyd, the protein levels increase rather than decrease. Thus, Orb2 appears to repress, rather than activate, the translation of the mRNAs encoding these two proteins. Since both mRNAs have CPEs and bind with the Orb2 protein in tissue culture cells [43], it is quite possible that Orb2 regulates their translation directly. While CPEBs are known to activate translation by promoting polyadenylation, there are contexts and mRNAs whose translation is negatively regulated by CPEBs. It will clearly be of interest to determine whether the translation of other potential Orb2 targets in neurons is repressed rather than activated.
While the distribution of the csp and pyd mRNAs in orb2R and orb2tub cell bodies and synaptic fractions is similar to the WT, this is not true for the actin5C mRNA. The mRNA level of the actin5C mRNA is significantly reduced in the synaptic fraction as are the levels of the orb2 mRNAs in orb2R and orb2tub flies. This finding raises the possibility that Orb2 may be involved in the transport of a subset of neuronal mRNAs. It will be of interest to examine the distribution of other neuronal mRNAs in orb2R and orb2tub.
Thus, our results allow us to assume a new function for the 3′UTR of the orb2 mRNA. In Figure 7, we summarize our hypothesis. The 3′UTR of the orb2 mRNA mediates the interaction between Orb2 proteins in the soma of a neuron. Multiple copies of connected Orb2 proteins form an RNP complex, which can more efficiently bind to transport proteins and be transported to synapses. Once in the synapse, the Orb2 protein regulates the local translation of its target mRNAs. We suppose that further studies will provide new insights into the roles of long 3′UTRs in neuronal cell functions and their participation in intracellular transport.
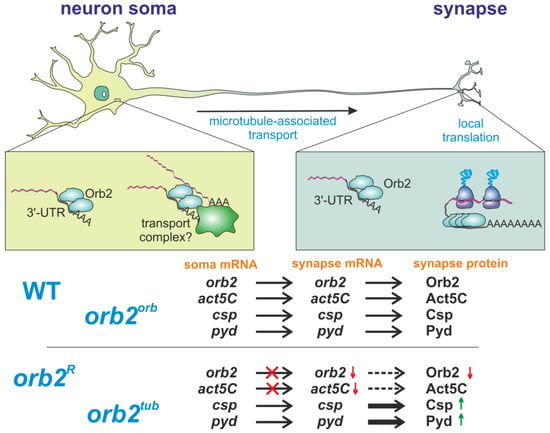
Figure 7.
A model of the orb2 role in neurons. The Orb2 protein participates in two major processes, intracellular transport in neurons and local translation in synapses. In the soma, Orb2 binds to the 3′UTRs of its target transcripts and participates in the formation of transport-competent mRNP. This mechanism is valid for several transcripts—e.g., Orb2 is required for intracellular transport of its own mRNA and the act5C mRNA to synapses, but not for transport of the csp and pyd transcripts. The orb2R and orb2tub mutants thus show lower synaptic levels of the orb2 mRNA and Orb2 protein. In synapses, Orb2 participates in regulating translation. The orb2R and orb2tub mutants consequently show higher levels of csp and pyd mRNA translation in synapses. The orb2orb mutant shows complete rescue of the described molecular defects.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells12131717/s1, Figure S1: Courtship indices of the studied fly lines; Figures S2 and S3: Brain preparations of the WT, orb2R, orb2tub, and orb2orb lines; Figures S4 and S5: Brain preparations of the WT, orb2R, orb2tub, and orb2orb lines.
Author Contributions
Conceptualization, E.N.K., P.S. and Y.V.S.; methodology, E.N.K., A.T., Z.M.K. and E.V.T.; validation, E.N.K., R.A.G. and M.Z.; formal analysis E.V.T., R.V.D. and Y.V.S.; investigation, E.N.K., E.V.T., R.A.G., R.V.D., A.T. and M.Z.; resources, M.Z., P.S. and Y.V.S.; writing—original draft preparation, E.N.K. and Y.V.S.; writing—review and editing, E.N.K., E.V.S.-P., M.Z., P.S. and Y.V.S.; visualization, R.V.D. and A.T.; supervision, Y.V.S.; project administration, Y.V.S.; funding acquisition, M.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation (grant 18-74-10051-Π). Part of this work (Section 3.6., the confocal microscopy imaging) was supported by grant 075-15-2019-1661 from the Ministry of Science and Higher Education of the Russian Federation.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
3′UTR, 3′-untranslated region; CPE, cytoplasmic polyadenylation element; CPEB, cytoplasmic polyadenylation element binding protein; STM, short-term memory; MTM, mid-term memory; LTM, long-term memory; CI, courtship index; LI, learning index.
References
- Barr, J.; Charania, S.; Gilmutdinov, R.; Yakovlev, K.; Shidlovskii, Y.; Schedl, P. The CPEB translational regulator, Orb, functions together with Par proteins to polarize the Drosophila oocyte. PLoS Genet. 2019, 15, e1008012. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Dumelie, J.G.; Li, X.; Cheng, M.H.; Yang, Z.; Laver, J.D.; Siddiqui, N.U.; Westwood, J.T.; Morris, Q.; Lipshitz, H.D.; et al. Global regulation of mRNA translation and stability in the early Drosophila embryo by the Smaug RNA-binding protein. Genome Biol. 2014, 15, R4. [Google Scholar] [CrossRef] [PubMed]
- Chia, W.; Somers, W.G.; Wang, H. Drosophila neuroblast asymmetric divisions: Cell cycle regulators, asymmetric protein localization, and tumorigenesis. J. Cell Biol. 2008, 180, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Singer, R.H.; Yoon, Y.J. The travels of mRNAs in neurons: Do they know where they are going? Curr. Opin. Neurobiol. 2019, 57, 110–116. [Google Scholar] [CrossRef]
- Harris, K.P.; Littleton, J.T. Transmission, Development, and Plasticity of Synapses. Genetics 2015, 201, 345–375. [Google Scholar] [CrossRef]
- Holt, C.E.; Martin, K.C.; Schuman, E.M. Local translation in neurons: Visualization and function. Nat. Struct. Mol. Biol. 2019, 26, 557–566. [Google Scholar] [CrossRef]
- Li, L.; Yu, J.; Ji, S.J. Axonal mRNA localization and translation: Local events with broad roles. Cell. Mol. Life Sci. CMLS 2021, 78, 7379–7395. [Google Scholar] [CrossRef]
- Si, K.; Giustetto, M.; Etkin, A.; Hsu, R.; Janisiewicz, A.M.; Miniaci, M.C.; Kim, J.H.; Zhu, H.; Kandel, E.R. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell 2003, 115, 893–904. [Google Scholar] [CrossRef]
- Wang, D.O.; Martin, K.C.; Zukin, R.S. Spatially restricting gene expression by local translation at synapses. Trends Neurosci. 2010, 33, 173–182. [Google Scholar] [CrossRef]
- Kandel, E.R.; Dudai, Y.; Mayford, M.R. The molecular and systems biology of memory. Cell 2014, 157, 163–186. [Google Scholar] [CrossRef]
- Ivshina, M.; Lasko, P.; Richter, J.D. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu. Rev. Cell Dev. Biol. 2014, 30, 393–415. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Sartain, C.V.; Pleiss, J.A.; Wolfner, M.F. Cytoplasmic polyadenylation is a major mRNA regulator during oogenesis and egg activation in Drosophila. Dev. Biol. 2013, 383, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Mendez, R.; Richter, J.D. Translational control by CPEB: A means to the end. Nat. Rev. Mol. Cell Biol. 2001, 2, 521–529. [Google Scholar] [CrossRef]
- Huang, Y.S.; Carson, J.H.; Barbarese, E.; Richter, J.D. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003, 17, 638–653. [Google Scholar] [CrossRef]
- Oe, S.; Yoneda, Y. Cytoplasmic polyadenylation element-like sequences are involved in dendritic targeting of BDNF mRNA in hippocampal neurons. FEBS Lett. 2010, 584, 3424–3430. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Culver, B.P.; Baj, G.; Tongiorgi, E.; Chao, M.V.; Tanese, N. Localization of BDNF mRNA with the Huntington’s disease protein in rat brain. Mol. Neurodegener. 2010, 5, 22. [Google Scholar] [CrossRef]
- Lee, S.; Chen, Y.C.; Gillen, A.E.; Taliaferro, J.M.; Deplancke, B.; Li, H.; Lai, E.C. Diverse cell-specific patterns of alternative polyadenylation in Drosophila. Nat. Commun. 2022, 13, 5372. [Google Scholar] [CrossRef] [PubMed]
- Crerar, H.; Scott-Solomon, E.; Bodkin-Clarke, C.; Andreassi, C.; Hazbon, M.; Logie, E.; Cano-Jaimez, M.; Gaspari, M.; Kuruvilla, R.; Riccio, A. Regulation of NGF Signaling by an Axonal Untranslated mRNA. Neuron 2019, 102, 553–563.e558. [Google Scholar] [CrossRef]
- Ma, W.; Mayr, C. A Membraneless Organelle Associated with the Endoplasmic Reticulum Enables 3’UTR-Mediated Protein-Protein Interactions. Cell 2018, 175, 1492–1506.e1419. [Google Scholar] [CrossRef]
- Kocabas, A.; Duarte, T.; Kumar, S.; Hynes, M.A. Widespread Differential Expression of Coding Region and 3’ UTR Sequences in Neurons and Other Tissues. Neuron 2015, 88, 1149–1156. [Google Scholar] [CrossRef]
- Malka, Y.; Steiman-Shimony, A.; Rosenthal, E.; Argaman, L.; Cohen-Daniel, L.; Arbib, E.; Margalit, H.; Kaplan, T.; Berger, M. Post-transcriptional 3´-UTR cleavage of mRNA transcripts generates thousands of stable uncapped autonomous RNA fragments. Nat. Commun. 2017, 8, 2029. [Google Scholar] [CrossRef] [PubMed]
- Hafer, N.; Xu, S.; Bhat, K.M.; Schedl, P. The Drosophila CPEB protein Orb2 has a novel expression pattern and is important for asymmetric cell division and nervous system function. Genetics 2011, 189, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Keleman, K.; Krüttner, S.; Alenius, M.; Dickson, B.J. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat. Neurosci. 2007, 10, 1587–1593. [Google Scholar] [CrossRef] [PubMed]
- Krüttner, S.; Stepien, B.; Noordermeer, J.N.; Mommaas, M.A.; Mechtler, K.; Dickson, B.J.; Keleman, K. Drosophila CPEB Orb2A mediates memory independent of Its RNA-binding domain. Neuron 2012, 76, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.R.; Li, L.; Pérez-Sánchez, C.; Saraf, A.; Florens, L.; Slaughter, B.D.; Unruh, J.R.; Si, K. Amyloidogenic Oligomerization Transforms Drosophila Orb2 from a Translation Repressor to an Activator. Cell 2015, 163, 1468–1483. [Google Scholar] [CrossRef] [PubMed]
- Krüttner, S.; Traunmüller, L.; Dag, U.; Jandrasits, K.; Stepien, B.; Iyer, N.; Fradkin, L.G.; Noordermeer, J.N.; Mensh, B.D.; Keleman, K. Synaptic Orb2A Bridges Memory Acquisition and Late Memory Consolidation in Drosophila. Cell Rep. 2015, 11, 1953–1965. [Google Scholar] [CrossRef]
- Bai, Y.; Suzuki, T. Activity-Dependent Synaptic Plasticity in Drosophila melanogaster. Front. Physiol. 2020, 11, 161. [Google Scholar] [CrossRef]
- Hirano, Y.; Ihara, K.; Masuda, T.; Yamamoto, T.; Iwata, I.; Takahashi, A.; Awata, H.; Nakamura, N.; Takakura, M.; Suzuki, Y.; et al. Shifting transcriptional machinery is required for long-term memory maintenance and modification in Drosophila mushroom bodies. Nat. Commun. 2016, 7, 13471. [Google Scholar] [CrossRef]
- Josselyn, S.A.; Tonegawa, S. Memory engrams: Recalling the past and imagining the future. Science 2020, 367, eaaw4325. [Google Scholar] [CrossRef]
- White-Grindley, E.; Li, L.; Mohammad Khan, R.; Ren, F.; Saraf, A.; Florens, L.; Si, K. Contribution of Orb2A stability in regulated amyloid-like oligomerization of Drosophila Orb2. PLoS Biol. 2014, 12, e1001786. [Google Scholar] [CrossRef]
- Hervás, R.; Li, L.; Majumdar, A.; Fernández-Ramírez Mdel, C.; Unruh, J.R.; Slaughter, B.D.; Galera-Prat, A.; Santana, E.; Suzuki, M.; Nagai, Y.; et al. Molecular Basis of Orb2 Amyloidogenesis and Blockade of Memory Consolidation. PLoS Biol. 2016, 14, e1002361. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, E.N.; Tokmatcheva, E.V.; Khrustaleva, A.M.; Grebenshchikov, E.S.; Deev, R.V.; Gilmutdinov, R.A.; Lebedeva, L.A.; Zhukova, M.; Savvateeva-Popova, E.V.; Schedl, P.; et al. Long-Term Memory Formation in Drosophila Depends on the 3’UTR of CPEB Gene orb2. Cells 2023, 12, 318. [Google Scholar] [CrossRef] [PubMed]
- Pai, T.P.; Chen, C.C.; Lin, H.H.; Chin, A.L.; Lai, J.S.; Lee, P.T.; Tully, T.; Chiang, A.S. Drosophila ORB protein in two mushroom body output neurons is necessary for long-term memory formation. Proc. Natl. Acad. Sci. USA 2013, 110, 7898–7903. [Google Scholar] [CrossRef] [PubMed]
- Lantz, V.; Schedl, P. Multiple cis-acting targeting sequences are required for orb mRNA localization during Drosophila oogenesis. Mol. Cell. Biol. 1994, 14, 2235–2242. [Google Scholar] [CrossRef] [PubMed]
- Gilmutdinov, R.; Kozlov, E.N.; Yakovlev, K.V.; Olenina, L.V.; Kotov, A.A.; Barr, J.; Zhukova, M.; Schedl, P.; Shidlovskii, Y.V. The 3’UTR of the Drosophila CPEB translation factor gene orb2 plays a crucial role in spermatogenesis. Development 2021, 148, dev198788. [Google Scholar] [CrossRef]
- Kamyshev, N.G.; Iliadi, K.G.; Bragina, J.V. Drosophila conditioned courtship: Two ways of testing memory. Learn. Mem. 1999, 6, 1–20. [Google Scholar] [CrossRef]
- Kamat, P.K.; Kalani, A.; Tyagi, N. Method and validation of synaptosomal preparation for isolation of synaptic membrane proteins from rat brain. MethodsX 2014, 1, 102–107. [Google Scholar] [CrossRef]
- Barr, J.; Gilmutdinov, R.; Wang, L.; Shidlovskii, Y.; Schedl, P. The Drosophila CPEB Protein Orb Specifies Oocyte Fate by a 3’UTR-Dependent Autoregulatory Loop. Genetics 2019, 213, 1431–1446. [Google Scholar] [CrossRef]
- Reveal, B.; Garcia, C.; Ellington, A.; Macdonald, P.M. Multiple RNA binding domains of Bruno confer recognition of diverse binding sites for translational repression. RNA Biol. 2011, 8, 1047–1060. [Google Scholar] [CrossRef]
- Zearfoss, N.R.; Deveau, L.M.; Clingman, C.C.; Schmidt, E.; Johnson, E.S.; Massi, F.; Ryder, S.P. A conserved three-nucleotide core motif defines Musashi RNA binding specificity. J. Biol. Chem. 2014, 289, 35530–35541. [Google Scholar] [CrossRef]
- Flora, P.; Wong-Deyrup, S.W.; Martin, E.T.; Palumbo, R.J.; Nasrallah, M.; Oligney, A.; Blatt, P.; Patel, D.; Fuchs, G.; Rangan, P. Sequential Regulation of Maternal mRNAs through a Conserved cis-Acting Element in Their 3’ UTRs. Cell Rep. 2018, 25, 3828–3843.e3829. [Google Scholar] [CrossRef] [PubMed]
- Mastushita-Sakai, T.; White-Grindley, E.; Samuelson, J.; Seidel, C.; Si, K. Drosophila Orb2 targets genes involved in neuronal growth, synapse formation, and protein turnover. Proc. Natl. Acad. Sci. USA 2010, 107, 11987–11992. [Google Scholar] [CrossRef]
- Stepien, B.K.; Oppitz, C.; Gerlach, D.; Dag, U.; Novatchkova, M.; Krüttner, S.; Stark, A.; Keleman, K. RNA-binding profiles of Drosophila CPEB proteins Orb and Orb2. Proc. Natl. Acad. Sci. USA 2016, 113, E7030–E7038. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.C.; Casadio, A.; Zhu, H.; Yaping, E.; Rose, J.C.; Chen, M.; Bailey, C.H.; Kandel, E.R. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: A function for local protein synthesis in memory storage. Cell 1997, 91, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Sanfilippo, P.; Wen, J.; Lai, E.C. Landscape and evolution of tissue-specific alternative polyadenylation across Drosophila species. Genome Biol. 2017, 18, 229. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).