The Role of CD36 in Cancer Progression and Its Value as a Therapeutic Target
Abstract
1. Introduction
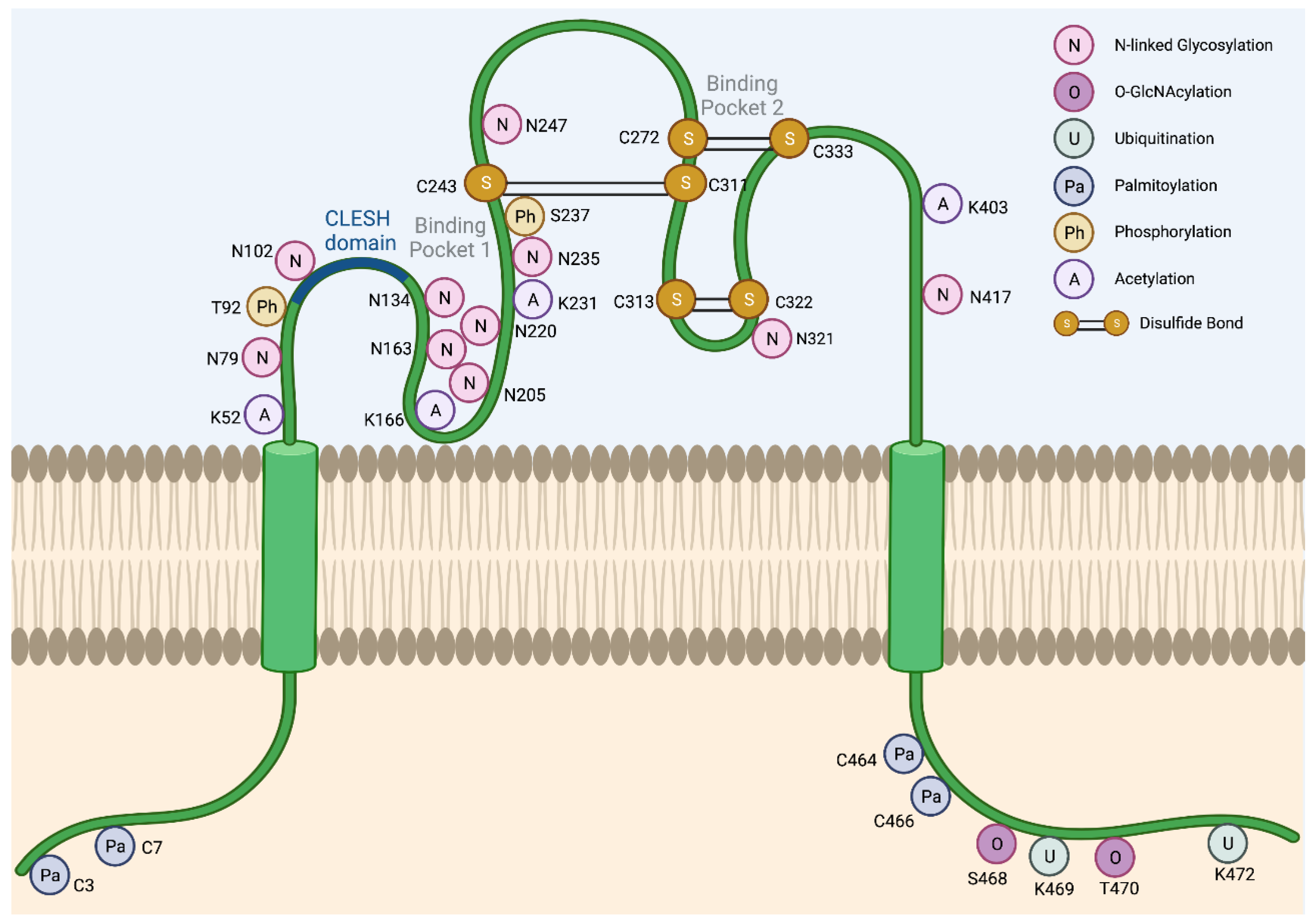
2. Regulation of CD36 Expression and Activity
2.1. Glycosylation
2.2. Ubiquitination
2.3. Palmitoylation
2.4. Other Post-Translational Modifications
2.5. Transcriptional Regulation of CD36
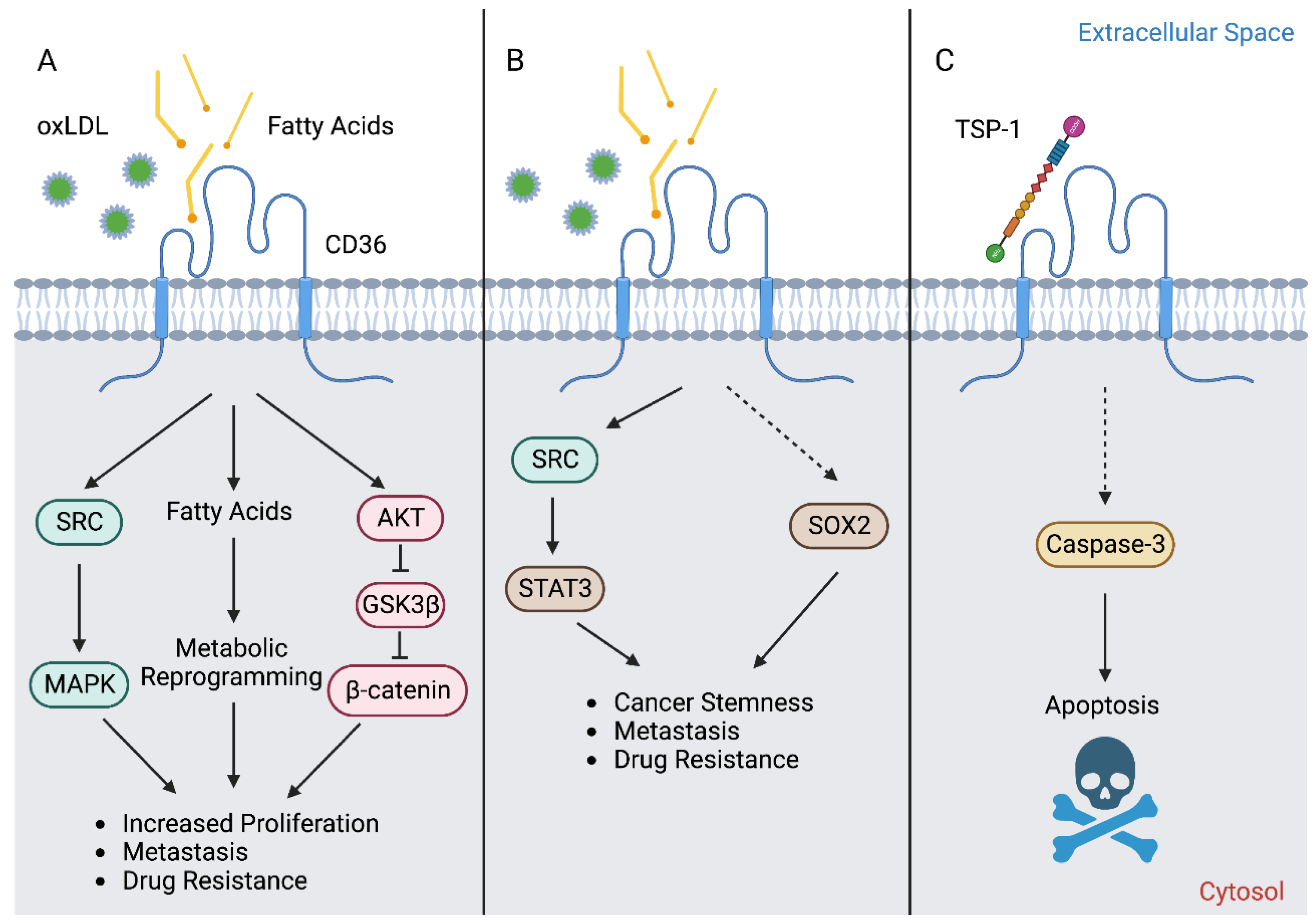
3. CD36 in Cancer
4. CD36 in Cancer Stemness and EMT
5. CD36 in Metastasis
6. CD36 in Cancer Drug Resistance
7. CD36 in Immune Evasion
8. CD36 as a Therapeutic Target
| Name of Compound | Disease Context | Description of Effect | References | |
|---|---|---|---|---|
| Antibodies | FA6.152 | Breast cancer, OSCC | Blocks all known functions of CD36, including interactions with TSP-1 and FA transporter properties. Anti-metastatic effects observed in vivo | [23,52] |
| JC63.1 | Breast, OSCC, Ovarian, Gastric cancers | Blocks FA and oxLDL uptake. Anti-metastatic effects observed in vivo | [14,23,52,54] | |
| Melanoma | Inhibits CD36 FA uptake on immunosuppressive Tregs and cytotoxic CD8+ T cells and restores anti-tumor immunity to TME | [56,73,81] | ||
| Peptides | Cyclic psap | Ovarian cancer | Induces expression of TSP-1 from MDSCs to promote apoptosis of cancer cells and endothelial cells | [75] |
| EP80317 | Cardiovascular diseases | Mitigates development of hypercholesterolemia and atherosclerosis. Also exhibits cardioprotective effects against myocardial ischemia and reperfusion injury | [109,110,111] | |
| VT1021 | Breast Glioblastoma, Ovarian, Pancreatic cancers | Induces expression of TSP-1 from MDSCs to activate CD36- and CD47-mediated apoptotic signaling in cancer cells and endothelial cells. Also increases CTL infiltration as well as M1:M2 macrophage ratios | [92,93] | |
| Small molecules | AP5258 and AP5156 | Diabetes | Protects against diabetic atherosclerosis, dyslipidemia, and insulin resistance | [106] |
| 2-methylthio-1,4-naphthoquinone (MTN) | Glioblastoma | Blocks growth of glioblastoma CSCs | [16] | |
| Development | Anti-angiogenic properties | [88] | ||
| Puerarin | Diabetes | Mitigates diabetic dyslipidemia by promoting FAO in skeletal muscle of diabetic rats | [107] | |
| Salvianolic acid (SAB) | Macrophages | Blocks macrophage uptake of oxLDL | [86] | |
| Obesity | Reduces visceral fat and improved insulin resistance in diet-induced obese mice | [87] | ||
| Synthetic FA analogs | FA-Linked Pt (IV)-Prodrugs | Ovarian cancer | Synthetic FA mimetic conjugated to cisplatin, selective drug uptake via elevated CD36 | [74] |
| Sulfo-N-succinimidyl oleate (SSO) | Breast, Cervical, Colorectal, Ovarian Cancers | Blocks uptake of long chain FAs and oxLDL | [17,52,53,61] | |
| Macrophages | Blocks uptake of long-chain FAs and oxLDL | [39,112] | ||
| Diabetes | Corrects cardiomyopathy observed in diabetic rat hearts | [85] |
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, J.; Li, Y. CD36 Tango in Cancer: Signaling Pathways and Functions. Theranostics 2019, 9, 4893–4908. [Google Scholar] [CrossRef] [PubMed]
- Clemetson, K.J.; Pfueller, S.L.; Luscher, E.F.; Jenkins, C.S. Isolation of the membrane glycoproteins of human blood platelets by lectin affinity chromatography. Biochim. Biophys. Acta 1977, 464, 493–508. [Google Scholar] [CrossRef] [PubMed]
- Tandon, N.N.; Hines, A.; Jamieson, G.A. Role of glycoprotein IV in collagen-induced platelet aggregation. Blood 1985, 66, 1148A. [Google Scholar]
- Shaw, S. Characterization of human leukocyte differentiation antigens. Immunol. Today 1987, 8, 1–3. [Google Scholar] [CrossRef]
- Asch, A.S.; Barnwell, J.; Silverstein, R.L.; Nachman, R.L. Isolation of the thrombospondin membrane receptor. J. Clin. Investig. 1987, 79, 1054–1061. [Google Scholar] [CrossRef]
- Tandon, N.N.; Lipsky, R.H.; Burgess, W.H.; Jamieson, G.A. Isolation and characterization of platelet glycoprotein IV (CD36). J. Biol. Chem. 1989, 264, 7570–7575. [Google Scholar] [CrossRef]
- Tandon, N.N.; Kralisz, U.; Jamieson, G.A. Identification of glycoprotein IV (CD36) as a primary receptor for platelet-collagen adhesion. J. Biol. Chem. 1989, 264, 7576–7583. [Google Scholar] [CrossRef]
- Endemann, G.; Stanton, L.W.; Madden, K.S.; Bryant, C.M.; White, R.T.; Protter, A.A. CD36 is a receptor for oxidized low density lipoprotein. J. Biol. Chem. 1993, 268, 11811–11816. [Google Scholar] [CrossRef]
- Abumrad, N.A.; El-Maghrabi, M.R.; Amri, E.Z.; Lopez, E.; Grimaldi, P.A. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36. J. Biol. Chem. 1993, 25, 17665–17668. [Google Scholar] [CrossRef]
- Silverstein, R.L.; Febbraio, M. CD36, a Scavenger Receptor Involved in Immunity, Metabolism, Angiogenesis, and Behavior. Sci. Signal. 2009, 2, re3. [Google Scholar] [CrossRef]
- Park, Y.M.; Febbraio, M.; Silverstein, R.L. CD36 Modulates Migration of Mouse and Human Macrophages in Response to Oxidized LDL and May Contribute to Macrophage Trapping in the Arterial Intima. J. Clin. Investig. 2009, 119, 136–145. [Google Scholar] [CrossRef]
- Stuart, L.M.; Bell, S.A.; Stewart, C.R.; Silver, J.M.; Richard, J.; Goss, J.L.; Tseng, A.A.; Zhang, A.; El Khoury, J.B.; Moore, K.J. CD36 Signals to the Actin Cytoskeleton and Regulates Microglial Migration via a P130Cas Complex. J. Biol. Chem. 2007, 282, 27392–27401. [Google Scholar] [CrossRef]
- Balaban, S.; Shearer, R.F.; Lee, L.S.; van Geldermalsen, M.; Schreuder, M.; Shtein, H.C.; Cairns, R.; Thomas, K.C.; Fazakerley, D.J.; Grewal, T.; et al. Adipocyte Lipolysis Links Obesity to Breast Cancer Growth: Adipocyte-Derived Fatty Acids Drive Breast Cancer Cell Proliferation and Migration. Cancer Metab. 2017, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Ladanyi, A.; Mukherjee, A.; Kenny, H.A.; Johnson, A.; Mitra, A.K.; Sundaresan, S.; Nieman, K.M.; Pascual, G.; Benitah, S.A.; Montag, A.; et al. Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene 2018, 37, 2285–2301. [Google Scholar] [CrossRef] [PubMed]
- Daquinag, A.C.; Gao, Z.; Fussell, C.; Immaraj, L.; Pasqualini, R.; Arap, W.; Akimzhanov, A.M.; Febbraio, M.; Kolonin, M.G. Fatty acid mobilization from adipose tissue is mediated by CD36 posttranslational modifications and intracellular trafficking. JCI Insight 2021, 6, e147057. [Google Scholar] [CrossRef] [PubMed]
- Hale, J.S.; Otvos, B.; Sinyuk, M.; Alvarado, A.G.; Hitomi, M.; Stoltz, K.; Wu, Q.; Flavahan, W.; Levison, B.; Johansen, M.J.; et al. Cancer Stem Cell-Specific Scavenger Receptor CD36 Drives Glioblastoma Progression. Stem Cells 2014, 32, 1746–1758. [Google Scholar] [CrossRef]
- Gyamfi, J.; Yeo, J.H.; Kwon, D.; Min, B.S.; Cha, Y.J.; Koo, J.S.; Jeong, J.; Lee, J.; Choi, J. Interaction between CD36 and FABP4 Modulates Adipocyte-Induced Fatty Acid Import and Metabolism in Breast Cancer. NPJ Breast Cancer 2021, 7, 129. [Google Scholar] [CrossRef]
- Zhang, L.; Billet, S.; Gonzales, G.; Rohena-Rivera, K.; Muranaka, H.; Chu, G.C.; Yang, Q.; Kim, H.; Bhowmick, N.A.; Smith, B. Fatty Acid Signaling Impacts Prostate Cancer Lineage Plasticity in an Autocrine and Paracrine Manner. Cancers 2022, 14, 3449. [Google Scholar] [CrossRef]
- Hoosdally, S.J.; Andress, E.J.; Wooding, C.; Martin, C.A.; Linton, K.J. The human scavenger receptor CD36: Glycosylation status and its role in trafficking and function. J. Biol. Chem. 2009, 284, 16277–16288. [Google Scholar] [CrossRef]
- Jiang, M.; Wu, N.; Xu, B.; Chu, Y.; Li, X.; Su, S.; Chen, D.; Li, W.; Shi, Y.; Gao, X.; et al. Fatty acid-induced CD36 expression via O-GlcNAcylation drives gastric cancer metastasis. Theranostics 2019, 9, 5359. [Google Scholar] [CrossRef]
- Nabeebaccus, A.A.; Zoccarato, A.; Hafstad, A.D.; Santos, C.X.; Aasum, E.; Brewer, A.C.; Zhang, M.; Beretta, M.; Yin, X.; West, J.A.; et al. Nox4 reprograms cardiac substrate metabolism via protein O-GlcNAcylation to enhance stress adaptation. JCI Insight 2017, 2, e96184. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Su, X.; El-Maghrabi, R.; Stahl, P.D.; Abumrad, N.A. Opposite regulation of CD36 ubiquitination by fatty acids and insulin: Effects on fatty acid uptake. J. Biol. Chem. 2008, 283, 13578–13585. [Google Scholar] [CrossRef] [PubMed]
- Pascual, G.; Avgustinova, A.; Mejetta, S.; Martín, M.; Castellanos, A.; Attolini, C.S.-O.; Berenuer, A.; Prats, N.; Toll, A.; Hueto, J.A.; et al. Targeting Metastasis-Initiating Cells through the Fatty Acid Receptor CD36. Nature 2017, 541, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.Y.; Stevens, M.V.; Akter, M.H.; Rusk, S.E.; Huang, R.J.; Cohen, A.; Noguchi, A.; Springer, D.; Bocharov, A.V.; Eggerman, T.L.; et al. Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. J. Clin. Investig. 2011, 121, 2037–2047. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Thundyil, J.; Lim, G.G.; Tng, T.J..; Yeow, S.Q.; Chai, C.; Yao, T.P.; Lim, K.L. Parkin regulates neuronal lipid homeostasis through SREBP2-lipoprotein lipase pathway—Implications for Parkinson’s disease. Hum. Mol. Genet. 2022, 32, 1466–1482. [Google Scholar] [CrossRef]
- Wu, W.; Wang, S.; Liu, Q.; Shan, T.; Wang, X.; Feng, J.; Wang, Y. AMPK facilitates intestinal long-chain fatty acid uptake by manipulating CD36 expression and translocation. FASEB J. 2020, 34, 4852–4869. [Google Scholar] [CrossRef]
- Xia, X.; Hu, T.; He, J.; Xu, Q.; Yu, C.; Liu, X.; Shao, Z.; Liao, Y.; Huang, H.; Liu, N. USP10 deletion inhibits macrophage-derived foam cell formation and cellular-oxidized low density lipoprotein uptake by promoting the degradation of CD36. Aging 2020, 12, 22892. [Google Scholar] [CrossRef]
- Zhang, F.; Xia, X.; Chai, R.; Xu, R.; Xu, Q.; Liu, M.; Chen, X.; Liu, B.; Liu, S.; Liu, N. Inhibition of USP14 suppresses the formation of foam cell by promoting CD36 degradation. J. Cell. Mol. Med. 2020, 24, 3292–3302. [Google Scholar] [CrossRef]
- Xia, X.; Xu, Q.; Liu, M.; Chen, X.; Liu, X.; He, J.; Hu, T.; Yu, C.; Huang, H.; Liu, S. Deubiquitination of CD36 by UCHL1 promotes foam cell formation. Cell Death Dis. 2020, 11, 636. [Google Scholar] [CrossRef]
- Tao, N.; Wagner, S.J.; Lublin, D.M. CD36 is palmitoylated on both N-and C-terminal cytoplasmic tails. J. Biol. Chem. 1996, 13, 22315–22320. [Google Scholar] [CrossRef]
- Thorne, R.F.; Ralston, K.J.; de Bock, C.E.; Mhaidat, N.M.; Zhang, X.D.; Boyd, A.W.; Burns, G.F. Palmitoylation of CD36/FAT regulates the rate of its post-transcriptional processing in the endoplasmic reticulum. Biochim. Biophys. Acta Mol Cell. Res. 2010, 1803, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- van Oort, M.M.; Drost, R.; Janβen, L.; Van Doorn, J.M.; Kerver, J.; Van der Horst, D.J.; Liuken, J.J.F.P.; Rodenburg, K.C.W. Each of the four intracellular cysteines of CD36 is essential for insulin-or AMP-activated protein kinase-induced CD36 translocation. Arch. Physiol. Biochem. 2014, 120, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hao, J.W.; Wang, X.; Guo, H.; Sun, H.H.; Lai, X.Y.; Liu, L.Y.; Zhu, M.; Wang, H.Y.; Li, Y.F. DHHC4 and DHHC5 facilitate fatty acid uptake by palmitoylating and targeting CD36 to the plasma membrane. Cell Rep. 2019, 26, 209–221. [Google Scholar] [CrossRef]
- Hao, J.W.; Wang, J.; Guo, H.; Zhao, Y.Y.; Sun, H.H.; Li, Y.F.; Lai, X.Y.; Zhao, N.; Wang, X.; Xie, C.; et al. CD36 facilitates fatty acid uptake by dynamic palmitoylation-regulated endocytosis. Nat. Commun. 2020, 11, 4765. [Google Scholar] [CrossRef] [PubMed]
- Asch, A.S.; Liu, I.; Briccetti, F.M.; Barnwell, J.W.; Kwakye-Berko, F.; Dokun, A.; Goldberger, J.; Pernambuco, M. Analysis of CD36 binding domains: Ligand specificity controlled by dephosphorylation of an ectodomain. Science 1993, 262, 1436–1440. [Google Scholar] [CrossRef] [PubMed]
- Hatmi, M.; Gavaret, J.M.; Elalamy, I.; Vargaftig, B.B.; Jacquemin, C. Evidence for cAMP-dependent platelet ectoprotein kinase activity that phosphorylates platelet glycoprotein IV (CD36). J. Biol. Chem. 1996, 271, 24776–24780. [Google Scholar] [CrossRef]
- Chu, L.Y.; Silverstein, R.L. CD36 ectodomain phosphorylation blocks thrombospondin-1 binding: Structure-function relationships and regulation by protein kinase C. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 760–767. [Google Scholar] [CrossRef]
- Guthmann, F.; Maehl, P.; Preiss, J.; Kolleck, I.; Rüstow, B. Ectoprotein kinase-mediated phosphorylation of FAT/CD36 regulates palmitate uptake by human platelets. Cell. Mol. Life Sci. 2002, 59, 1999–2003. [Google Scholar] [CrossRef]
- Kuda, O.; Pietka, T.A.; Demianova, Z.; Kudova, E.; Cvacka, J.; Kopecky, J.; Abumrad, N.A. Sulfo-N-Succinimidyl Oleate (SSO) Inhibits Fatty Acid Uptake and Signaling for Intracellular Calcium via Binding CD36 Lysine 164: SSO Also Inhibits Oxidized Low Density Lipoprotein Uptake by Macrophages. J. Biol. Chem. 2013, 288, 15547–15555. [Google Scholar] [CrossRef]
- Wang, R.; Tao, B.; Fan, Q.; Wang, S.; Chen, L.; Zhang, J.; Hao, Y.; Dong, S.; Wang, Z.; Wang, W.; et al. Fatty-acid receptor CD36 functions as a hydrogen sulfide-targeted receptor with its Cys333-Cys272 disulfide bond serving as a specific molecular switch to accelerate gastric cancer metastasis. EBioMedicine 2019, 45, 108–123. [Google Scholar] [CrossRef]
- Liang, Y.; Han, H.; Liu, L.; Duan, Y.; Yang, X.; Ma, C.; Zhu, Y.; Han, J.; Li, X.; Chen, Y. CD36 plays a critical role in proliferation, migration and tamoxifen-inhibited growth of ER-positive breast cancer cells. Oncogenesis 2018, 7, 98. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Febbraio, M.; Wada, T.; Zhai, Y.; Kuruba, R.; He, J.; Lee, J.H.; Khadem, S.; Ren, S.; Li, S.; et al. Hepatic fatty acid transporter Cd36 is a common target of LXR, PXR, and PPARγ in promoting steatosis. Gastroenterology 2008, 134, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Ghodsian, N.; Yeandle, A.; Hock, B.D.; Gieseg, S.P. CD36 down regulation by the macrophage antioxidant 7, 8-dihydroneopterin through modulation of PPAR-γ activity. Free Radic. Res. 2022, 56, 366–377. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Zou, C.; Shao, P.; Schaack, J.; Johnson, P.F.; Shao, J. Transcriptional regulation of fatty acid translocase/CD36 expression by CCAAT/enhancer-binding protein α. J. Biol. Chem. 2008, 283, 8788–8795. [Google Scholar] [CrossRef] [PubMed]
- Sp, N.; Kang, D.Y.; Kim, D.H.; Park, J.H.; Lee, H.G.; Kim, H.J.; Darvin, P.; Park, Y.M.; Yang, Y.K. Nobiletin inhibits CD36-dependent tumor angiogenesis, migration, invasion, and sphere formation through the Cd36/Stat3/Nf-Κb signaling axis. Nutrients 2018, 10, 772. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Lugea, A.; Zheng, L.; Gukovsky, I.; Edderkaoui, M.; Rozengurt, E.; Pandol, S.J. Protein kinase D1 mediates NF-κB activation induced by cholecystokinin and cholinergic signaling in pancreatic acinar cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 295, G1190–G1201. [Google Scholar] [CrossRef]
- Storz, P.; Toker, A. Protein kinase D mediates a stress-induced NF-κB activation and survival pathway. EMBO J. 2003, 22, 109–120. [Google Scholar] [CrossRef]
- Harikumar, K.B.; Kunnumakkara, A.B.; Ochi, N.; Tong, Z.; Deorukhkar, A.; Sung, B.; Kelland, L.; Jamieson, S.; Sutherland, R.; Raynham, T.; et al. A Novel Small-Molecule Inhibitor of Protein Kinase D Blocks Pancreatic Cancer Growth In vitro and In vivo. Mol. Cancer Ther. 2010, 9, 1136–1146. [Google Scholar] [CrossRef]
- Guo, Y.; Jiang, Y.; Rose, J.B.; Nagaraju, G.P.; Jaskula-Sztul, R.; Hjelmeland, A.B.; Beck, A.W.; Chen, H.; Ren, B. Protein Kinase D1 Signaling in Cancer Stem Cells with Epithelial-Mesenchymal Plasticity. Cells 2022, 11, 3885. [Google Scholar] [CrossRef]
- Kuemmerle, N.B.; Rysman, E.; Lombardo, P.S.; Flanagan, A.J.; Lipe, B.C.; Wells, W.A.; Pettus, J.R.; Froehlich, H.M.; Memoli, V.A.; Morganelli, P.M.; et al. Lipoprotein lipase links dietary fat to solid tumor cell proliferation. Mol. Cancer Ther. 2011, 10, 427–436. [Google Scholar] [CrossRef]
- Jiang, Y.; Guo, Y.; Hao, J.; Guenter, R.; Lathia, J.; Beck, A.W.; Hattaway, R.; Hurst, D.; Wang, Q.J.; Liu, Y.; et al. Development of an arteriolar niche and self-renewal of breast cancer stem cells by lysophosphatidic acid/protein kinase D signaling. Commun. Biol. 2021, 4, 780. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.W.; Wilkins, O.; Bang, S.; Ung, M.; Li, J.; An, J.; Del Genio, C.; Canfield, K.; DiRenzo, J.; Wells, W.; et al. CD36-Mediated Metabolic Rewiring of Breast Cancer Cells Promotes Resistance to HER2-Targeted Therapies. Cell Rep. 2019, 29, 3405–3420. [Google Scholar] [CrossRef] [PubMed]
- Drury, J.; Rychahou, P.G.; He, D.; Jafari, N.; Wang, C.; Lee, E.Y.; Weiss, H.L.; Evers, B.M.; Zaytseva, Y.Y. Inhibition of Fatty Acid Synthase Upregulates Expression of CD36 to Sustain Proliferation of Colorectal Cancer Cells. Front. Oncol. 2020, 10, 1185. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Fan, Z.; Wang, Z.; Dai, Q.; Xiang, Z.; Yuan, F.; Yan, M.; Zhu, Z.; Liu, B.; Li, C. CD36 mediates palmitate acid-induced metastasis of gastric cancer via AKT/GSK-3β/β-catenin pathway. J. Exp. Clin. Cancer Res. 2019, 38, 52. [Google Scholar] [CrossRef]
- Wang, J.; Wen, T.; Li, Z.; Che, X.; Gong, L.; Jiao, Z.; Qu, X.; Liu, Y. CD36 upregulates DEK transcription and promotes cell migration and invasion via GSK-3β/β-catenin-mediated epithelial-to-mesenchymal transition in gastric cancer. Aging 2021, 13, 1883. [Google Scholar] [CrossRef]
- Wang, H.; Franco, F.; Tsui, Y.C.; Xie, X.; Trefny, M.P.; Zappasodi, R.; Mohmood, S.R.; Fernández-García, J.; Tsai, C.H.; Schulze, I.; et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat. Immunol. 2020, 21, 298–308. [Google Scholar] [CrossRef]
- Kubo, M.; Gotoh, K.; Eguchi, H.; Kobayashi, S.; Iwagami, Y.; Tomimaru, Y.; Akita, H.; Asaoka, T.; Noda, T.; Takeda, Y.; et al. Impact of CD36 on chemoresistance in pancreatic ductal adenocarcinoma. Ann. Surg. Oncol. 2020, 27, 610–619. [Google Scholar] [CrossRef]
- Ye, H.; Adane, B.; Khan, N.; Sullivan, T.; Minhajuddin, M.; Gasparetto, M.; Stevens, B.; Pei, S.; Balys, M.; Ashton, J.M.; et al. Leukemic Stem Cells Evade Chemotherapy by Metabolic Adaptation to an Adipose Tissue Niche. Cell Stem Cell 2016, 19, 23–37. [Google Scholar] [CrossRef]
- Nath, A.; Li, I.; Roberts, L.R.; Chan, C. Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma. Sci. Rep. 2015, 5, 14752. [Google Scholar] [CrossRef]
- Deng, M.; Cai, X.; Long, L.; Xie, L.; Ma, H.; Zhou, Y.; Liu, S.; Zeng, S. CD36 promotes the epithelial–mesenchymal transition and metastasis in cervical cancer by interacting with TGF-β. J. Trans. Med. 2019, 17, 352. [Google Scholar] [CrossRef]
- Yang, P.; Su, C.; Luo, X.; Zeng, H.; Zhao, L.; Wei, L.; Zhang, X.; Varghese, Z.; Moorhead, J.F.; Chen, Y.; et al. Dietary oleic acid-induced CD36 promotes cervical cancer cell growth and metastasis via up-regulation Src/ERK pathway. Cancer Lett. 2018, 438, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Watt, M.J.; Clark, A.K.; Selth, L.A.; Haynes, V.R.; Lister, N.; Rebello, R.; Porter, L.H.; Niranjan, B.; Whitby, S.T.; Lo, J.; et al. Suppressing fatty acid uptake has therapeutic effects in preclinical models of prostate cancer. Sci. Transl. Med. 2019, 11, eaau5758. [Google Scholar] [CrossRef] [PubMed]
- ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef]
- Nath, A.; Chan, C. Genetic alterations in fatty acid transport and metabolism genes are associated with metastatic progression and poor prognosis of human cancers. Sci. Rep. 2016, 6, 18669. [Google Scholar] [CrossRef] [PubMed]
- Rae, C.; Fragkoulis, G.I.; Chalmers, A.J. Cytotoxicity and radiosensitizing activity of the fatty acid synthase inhibitor C75 is enhanced by blocking fatty acid uptake in prostate cancer cells. Adv. Radiat. Oncol. 2020, 5, 994–1005. [Google Scholar] [CrossRef]
- Frank, A.C.; Ebersberger, S.; Fink, A.F.; Lampe, S.; Weigert, A.; Schmid, T.; Ebersberger, I.; Syed, S.N.; Brüne, B. Apoptotic tumor cell-derived microRNA-375 uses CD36 to alter the tumor-associated macrophage phenotype. Nat. Commun. 2019, 10, 1135. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Lin, K.; Zhao, Y.; Wu, Q.; Chen, D.; Wang, J.; Liang, Y.; Li, J.; Hu, J.; Wang, H.; et al. Adipocytes fuel gastric cancer omental metastasis via PITPNC1-mediated fatty acid metabolic reprogramming. Theranostics 2018, 8, 5452. [Google Scholar] [CrossRef] [PubMed]
- Luanpitpong, S.; Janan, M.; Thumanu, K.; Poohadsuan, J.; Rodboon, N.; Klaihmon, P.; Issaragrisil, S. Deciphering the elevated lipid via CD36 in mantle cell lymphoma with bortezomib resistance using synchrotron-based Fourier transform infrared spectroscopy of single cells. Cancers 2019, 24, 576. [Google Scholar] [CrossRef]
- Landberg, N.; von Palffy, S.; Askmyr, M.; Lilljebjörn, H.; Sandén, C.; Rissler, M.; Mustjoki, S.; Hjorth-Hansen, H.; Richter, J.; Ågerstam, H.; et al. CD36 defines primitive chronic myeloid leukemia cells less responsive to imatinib but vulnerable to antibody-based therapeutic targeting. Haematologica 2018, 103, 447–455. [Google Scholar] [CrossRef]
- Luo, X.; Zheng, E.; Wei, L.; Zeng, H.; Qin, H.; Zhang, X.; Liao, M.; Chen, L.; Zhao, L.; Ruan, X.Z.; et al. The fatty acid receptor CD36 promotes HCC progression through activating Src/PI3K/AKT axis-dependent aerobic glycolysis. Cell Death Dis. 2021, 12, 328. [Google Scholar] [CrossRef]
- Yang, P.; Qin, H.; Li, Y.; Xiao, A.; Zheng, E.; Zeng, H.; Su, C.; Luo, X.; Lu, Q.; Liao, M.; et al. CD36-mediated metabolic crosstalk between tumor cells and macrophages affects liver metastasis. Nat. Commun. 2022, 13, 5782. [Google Scholar] [CrossRef] [PubMed]
- Aloia, A.; Müllhaupt, D.; Chabbert, C.D.; Eberhart, T.; Flückiger-Mangual, S.; Vukolic, A.; Eichhoff, O.; Irmisch, A.; Alexander, L.T.; Scibona, E.; et al. A Fatty Acid Oxidation-dependent Metabolic Shift Regulates the Adaptation of BRAF-mutated Melanoma to MAPK Inhibitors. Clin. Cancer Res. 2019, 25, 6852–6867. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Chaudhary, O.; Rodríguez-Morales, P.; Sun, X.; Chen, D.; Zappasodi, R.; Xu, Z.; Pinto, A.F.M.; Williams, A.; Schulze, I.; et al. Uptake of oxidized lipids by the scavenger receptor CD36 promotes lipid peroxidation and dysfunction in CD8+ T cells in tumors. Immunity 2021, 54, 1561–1577. [Google Scholar] [CrossRef] [PubMed]
- Jayawardhana, A.M.D.S.; Stilgenbauer, M.; Datta, P.; Qiu, Z.; Mckenzie, S.; Wang, H.; Bowers, D.; Kurokawa, M.; Zheng, Y.R. Fatty Acid-like Pt(IV) Prodrugs Overcome Cisplatin Resistance in Ovarian Cancer by Harnessing CD36. Chem. Commun. 2020, 56, 10706–10709. [Google Scholar] [CrossRef]
- Wang, S.; Blois, A.; El Rayes, T.; Liu, J.F.; Hirsch, M.S.; Gravdal, K.; Palakurthi, S.; Bielenberg, D.R.; Akslen, L.A.; Drapkin, R.; et al. Development of a prosaposin-derived therapeutic cyclic peptide that targets ovarian cancer via the tumor microenvironment. Sci. Transl. Med. 2016, 8, 329ra34. [Google Scholar] [CrossRef]
- Nieman, K.M.; Kenny, H.A.; Penicka, C.V.; Ladanyi, A.; Buell-Gutbrod, R.; Zillhardt, M.R.; Romero, I.L.; Carey, M.S.; Mills, G.B.; Hotamisligil, G.S.; et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011, 17, 1498–1503. [Google Scholar] [CrossRef]
- Ferraro, G.B.; Ali, A.; Luengo, A.; Kodack, D.P.; Deik, A.; Abbott, K.L.; Bezwada, D.; Blanc, L.; Prideaux, B.; Jin, X.; et al. Fatty Acid Synthesis Is Required for Breast Cancer Brain Metastasis. Nat. Cancer 2021, 2, 414–428. [Google Scholar] [CrossRef]
- Haderk, F.; Fernández-Méndez, C.; Čech, L.; Yu, J.; Meraz, I.; Olivas, V.; Rabago, D.B.; Kerr, D.L.; Gomez, C.; Allegakoen, D.V.; et al. A Focal Adhesion Kinase-YAP Signaling Axis Drives Drug Tolerant Persister Cells and Residual Disease in Lung Cancer. Biorxiv 2021. preprint. [Google Scholar] [CrossRef]
- Tian, Y.; Yang, B.; Qiu, W.; Hao, Y.; Zhang, Z.; Yang, B.; Li, N.; Cheng, S.; Lin, Z.; Rui, Y.C.; et al. ER-residential Nogo-B accelerates NAFLD-associated HCC mediated by metabolic reprogramming of oxLDL lipophagy. Nat. Commun. 2019, 10, 3391. [Google Scholar] [CrossRef]
- Hrdinova, T.; Toman, O.; Dresler, J.; Klimentova, J.; Salovska, B.; Pajer, P.; Bartos, O.; Polivkova, V.; Linhartova, J.; Polakova, K.M.; et al. Exosomes released by imatinib-resistant K562 cells contain specific membrane markers, IFITM3, CD146 and CD36 and increase the survival of imatinib-sensitive cells in the presence of imatinib. Int. J. Oncol. 2021, 58, 238–250. [Google Scholar] [CrossRef]
- Ma, X.; Xiao, L.; Liu, L.; Ye, L.; Su, P.; Bi, E.; Wang, Q.; Yang, M.; Qian, J.; Yi, Q. CD36-mediated ferroptosis dampens intratumoral CD8+ T cell effector function and impairs their antitumor ability. Cell Metab. 2021, 33, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Woo, M.S.; Yang, J.; Beltran, C.; Cho, S. Cell surface CD36 protein in monocyte/macrophage contributes to phagocytosis during the resolution phase of ischemic stroke in mice. J. Biol. Chem. 2016, 291, 23654–23661. [Google Scholar] [CrossRef] [PubMed]
- Pennathur, S.; Pasichnyk, K.; Bahrami, N.M.; Zeng, L.; Febbraio, M.; Yamaguchi, I.; Okamura, D.M. The macrophage phagocytic receptor CD36 promotes fibrogenic pathways on removal of apoptotic cells during chronic kidney injury. Am. J. Pathol. 2015, 185, 2232–2245. [Google Scholar] [CrossRef] [PubMed]
- Cannarile, M.A.; Weisser, M.; Jacob, W.; Jegg, A.-M.; Ries, C.H.; Rüttinger, D. Colony-Stimulating Factor 1 Receptor (CSF1R) Inhibitors in Cancer Therapy. J. Immunother. Cancer 2017, 5, 53. [Google Scholar] [CrossRef] [PubMed]
- Mansor, L.S.; Sousa Fialho, M.D.; Yea, G.; Coumans, W.A.; West, J.A.; Kerr, M.; Carr, C.A.; Luiken, J.J.P.; Glatz, J.F.C.; Evans, R.D.; et al. Inhibition of sarcolemmal FAT/CD36 by sulfo-N-succinimidyl oleate rapidly corrects metabolism and restores function in the diabetic heart following hypoxia/reoxygenation. Cardiovasc. Res. 2017, 113, 737–748. [Google Scholar] [CrossRef]
- Bao, Y.; Wang, L.; Xu, Y.; Yang, Y.; Wang, L.; Si, S.; Cho, S.; Hong, B. Salvianolic acid B inhibits macrophage uptake of modified low density lipoprotein (mLDL) in a scavenger receptor CD36-dependent manner. Atherosclerosis 2012, 223, 152–159. [Google Scholar] [CrossRef]
- Yang, J.; Park, K.W.; Cho, S. Inhibition of the CD36 receptor reduces visceral fat accumulation and improves insulin resistance in obese mice carrying the BDNF-Val66Met variant. J. Biol. Chem. 2018, 293, 13338–13348. [Google Scholar] [CrossRef]
- Müller, W.E.; Thakur, N.L.; Ushijima, H.; Thakur, A.N.; Krasko, A.; Le Pennec, G.; Indap, M.M.; Perovic-Ottstadt, S.; Schröder, H.C.; Lang, G.; et al. Matrix-mediated canal formation in primmorphs from the sponge Suberites domuncula involves the expression of a CD36 receptor-ligand system. J. Cell Sci. 2004, 117, 2579–2590. [Google Scholar] [CrossRef]
- Jiménez, B.; Volpert, O.V.; Crawford, S.E.; Febbraio, M.; Silverstein, R.L.; Bouck, N. Signals leading to apoptosis-dependent inhibition of neovascularization by thrombospondin-1. Nat. Med. 2000, 6, 41–48. [Google Scholar] [CrossRef]
- Ren, B.; Song, K.; Parangi, S.; Jin, T.; Ye, M.; Humphreys, R.; Duquette, M.; Zhang, X.; Benhaga, N.; Lawler, J.; et al. A double hit to kill tumor and endothelial cells by TRAIL and antiangiogenic 3TSR. Cancer Res. 2009, 69, 3856–3865. [Google Scholar] [CrossRef]
- Matuszewska, K.; Ten Kortenaar, S.; Pereira, M.; Santry, L.A.; Petrik, D.; Lo, K.M.; Bridle, B.W.; Wootton, S.K.; Lawler, S.; Petrik, J. Addition of an Fc-IgG induces receptor clustering and increases the in vitro efficacy and in vivo anti-tumor properties of the thrombospondin-1 type I repeats (3TSR) in a mouse model of advanced stage ovarian cancer. Gynecol. Oncol. 2022, 164, 154–169. [Google Scholar] [CrossRef] [PubMed]
- Vigeo Therapeutics News Release. Vigeo Therapeutics Advances VT1021 Into Phase 2–3 Registrational Study for Glioblastoma. Available online: https://vigeotherapeutics.com/news/vigeo-therapeutics-advances-vt1021-into-phase-2-3-registrational-study-for-glioblastoma (accessed on 1 September 2022).
- Mahalingam, D.; Harb, W.; Patnaik, A.; Ulahannan, S.; Mahdi, H.; Ahluwalia, M.; Patel, M.; Dowlati, A.; Bullock, A.; Wen, P.; et al. 374 A First-in-Human Phase ½ Open Label Trial Evaluating the Safety, Pharmacology, and Preliminary Efficacy of VT1021 in Subjects with Advanced Solid Tumors. J. Immunother. Cancer 2020, 8, A228. [Google Scholar] [CrossRef]
- Hoekstra, R.; de Vos, F.Y.; Eskens, F.A.; Gietema, J.A.; van der Gaast, A.; Groen, H.J.; Knight, R.A.; Carr, R.A.; Humerickhouse, R.A.; Verweij, J.; et al. Phase I safety, pharmacokinetic, and pharmacodynamic study of the thrombospondin-1–mimetic angiogenesis inhibitor ABT-510 in patients with advanced cancer. J. Clin. Oncol. 2005, 23, 5188–5197. [Google Scholar] [CrossRef] [PubMed]
- Gietema, J.A.; Hoekstra, R.; de Vos, F.Y.; Uges, D.R.; van der Gaast, A.; Groen, H.J.; Loos, W.J.; Knight, R.A.; Carr, R.A.; Humerickhouse, R.A.; et al. A phase I study assessing the safety and pharmacokinetics of the thrombospondin-1-mimetic angiogenesis inhibitor ABT-510 with gemcitabine and cisplatin in patients with solid tumors. Ann. Oncol. 2006, 17, 1320–1327. [Google Scholar] [CrossRef] [PubMed]
- Nabors, L.B.; Fiveash, J.B.; Markert, J.M.; Kekan, M.S.; Gillespie, G.Y.; Huang, Z.; Johnson, M.J.; Meleth, S.; Kuo, H.; Gladson, C.L.; et al. A phase 1 trial of ABT-510 concurrent with standard chemoradiation for patients with newly diagnosed glioblastoma. Arch. Neurol. 2010, 67, 313–319. [Google Scholar] [CrossRef]
- Markovic, S.; Suman, V.; Rao, R.; Creagan, E.; Maples, W.; Kaur, J.; Erickson, L.A.; Pitot, H.C.; Croghan, G.A.; McWilliams, R.R.; et al. A phase II study of ABT-510 for the treatment of metastatic melanoma. Am. J. Clin. Oncol. 2007, 30, 303–309. [Google Scholar] [CrossRef]
- Ebbinghaus, S.; Hussain, M.; Tannir, N.; Gordon, M.; Desai, A.A.; Knight, R.A.; Humerickhouse, R.A.; Qian, J.; Gordon, G.B.; Figlin, R. Phase 2 study of ABT-510 in patients with previously untreated advanced renal cell carcinoma. Clin. Cancer Res. 2007, 13, 6689–6695. [Google Scholar] [CrossRef]
- Baker, L.H.; Rowinsky, E.K.; Mendelson, D.; Humerickhouse, R.A.; Knight, R.A.; Qian, J.; Carr, R.A.; Gordon, G.B.; Demetri, G.D. Randomized, phase II study of the thrombospondin-1-mimetic angiogenesis inhibitor ABT-510 in patients with advanced soft tissue sarcoma. J. Clin. Oncol. 2008, 26, 5583–5588. [Google Scholar] [CrossRef]
- Volpert, O.V.; Zaichuk, T.; Zhou, W.; Reiher, F.; Ferguson, T.A.; Stuart, P.M.; Amin, M.; Bouck, N.P. Inducer-stimulated Fas targets activated endothelium for destruction by anti-angiogenic thrombospondin-1 and pigment epithelium–derived factor. Nat. Med. 2002, 8, 349–357. [Google Scholar] [CrossRef]
- Yang, M.; Liang, E.; Mao, L.; Chong, B.H.; Li, C. Thrombospondin-1 induces apoptosis in megakaryocytic leukemia via CD36 and caspase-3 signaling. Blood 2016, 128, 5209. [Google Scholar] [CrossRef]
- Dolgin, E. A Drug to Block Fat Intake and Combat Cancer Spread. Available online: https://www.nature.com/articles/d41586-021-01667-8 (accessed on 11 September 2022).
- Febbraio, M.; Abumrad, N.A.; Hajjar, D.P.; Sharma, K.; Cheng, W.; Pearce, S.F.; Silverstein, R.L. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J. Biol. Chem. 1999, 274, 19055–19062. [Google Scholar] [CrossRef]
- Yamashita, S.; Hirano, K.I.; Kuwasako, T.; Janabi, M.; Toyama, Y.; Ishigami, M.; Sakai, N. Physiological and pathological roles of a multi-ligand receptor CD36 in atherogenesis; insights from CD36-deficient patients. Mol. Cell. Biochem. 2007, 299, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Miyaoka, K.; Kuwasako, T.; Hirano, K.I.; Nozaki, S.; Yamashita, S.; Matsuzawa, Y. CD36 deficiency associated with insulin resistance. Lancet 2001, 357, 686–687. [Google Scholar] [CrossRef] [PubMed]
- Geloen, A.; Helin, L.; Geeraert, B.; Malaud, E.; Holvoet, P.; Marguerie, G. CD36 inhibitors reduce postprandial hypertriglyceridemia and protect against diabetic dyslipidemia and atherosclerosis. PLoS ONE 2012, 7, e37633. [Google Scholar] [CrossRef]
- Chen, X.F.; Wang, L.; Wu, Y.Z.; Song, S.Y.; Min, H.Y.; Yang, Y.; He, X.; Liang, Q.; Yi, L.; Wang, Y.; et al. Effect of puerarin in promoting fatty acid oxidation by increasing mitochondrial oxidative capacity and biogenesis in skeletal muscle in diabetic rats. Nutr. Diabetes 2018, 8, 1. [Google Scholar] [CrossRef]
- Wilson, C.G.; Tran, J.L.; Erion, D.M.; Vera, N.B.; Febbraio, M.; Weiss, E.J. Hepatocyte-specific disruption of CD36 attenuates fatty liver and improves insulin sensitivity in HFD-fed mice. Endocrinology 2016, 157, 570–585. [Google Scholar] [CrossRef] [PubMed]
- Marleau, S.; Harb, D.; Bujold, K.; Avallone, R.; Iken, K.; Wang, Y.; Demers, A.; Sirois, M.G.; Febbraio, M.; Silverstein, R.L.; et al. EP 80317, a ligand of the CD36 scavenger receptor, protects apolipoprotein E-deficient mice from developing atherosclerotic lesions. FASEB 2005, 19, 1869–1871. [Google Scholar] [CrossRef]
- Bujold, K.; Mellal, K.; Zoccal, K.F.; Rhainds, D.; Brissette, L.; Febbraio, M.; Marleau, S.; Ong, H. EP 80317, a CD36 selective ligand, promotes reverse cholesterol transport in apolipoprotein E-deficient mice. Atherosclerosis 2013, 229, 408–414. [Google Scholar] [CrossRef]
- Bessi, V.L.; Labbé, S.M.; Huynh, D.N.; Menard, L.; Jossart, C.; Febbraio, M.; Guérin, B.; Bentourkia, M.; Lecomte, R.; Carpentier, A.C.; et al. EP 80317, a selective CD36 ligand, shows cardioprotective effects against post-ischaemic myocardial damage in mice. Cardiovasc. Res. 2012, 96, 99–108. [Google Scholar] [CrossRef]
- Nicholls, H.T.; Kowalski, G.; Kennedy, D.J.; Risis, S.; Zaffino, L.A.; Watson, N.; Kanellakis, P.; Watt, M.J.; Bobik, A.; Bonen, A.; et al. Hematopoietic cell–restricted deletion of CD36 reduces high-fat diet–induced macrophage infiltration and improves insulin signaling in adipose tissue. Diabetes 2011, 60, 1100–1110. [Google Scholar] [CrossRef]
- Ookhtens, M.U.; Kannan, R.A.; Lyon, I.R.; Baker, N.O. Liver and adipose tissue contributions to newly formed fatty acids in an ascites tumor. Am. J. Physiol. 1984, 247, R146–R153. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.H.; Fowle-Grider, R.; Mahieu, N.G.; Liu, G.Y.; Chen, Y.J.; Wang, R.; Singh, M.; Potter, G.S.; Gross, R.W.; Schaefer, J.; et al. Exogenous fatty acids are the preferred source of membrane lipids in proliferating fibroblasts. Cell Chem. Biol. 2016, 23, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Corbet, C.; Pinto, A.; Martherus, R.; de Jesus, J.P.; Polet, F.; Feron, O. Acidosis drives the reprogramming of fatty acid metabolism in cancer cells through changes in mitochondrial and histone acetylation. Cell Metab. 2016, 24, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Hangauer, M.J.; Viswanathan, V.S.; Ryan, M.J.; Bole, D.; Eaton, J.K.; Matov, A.; Galeas, J.; Dhruv, H.D.; Berens, M.E.; Schreiber, S.L.; et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 2017, 551, 247–250. [Google Scholar] [CrossRef]
- Oren, Y.; Tsabar, M.; Cuoco, M.S.; Amir-Zilberstein, L.; Cabanos, H.F.; Hütter, J.C.; Hu, B.; Thakore, P.I.; Tabaka, M.; Fulco, C.P.; et al. Cycling cancer persister cells arise from lineages with distinct programs. Nature 2021, 596, 576–582. [Google Scholar] [CrossRef]
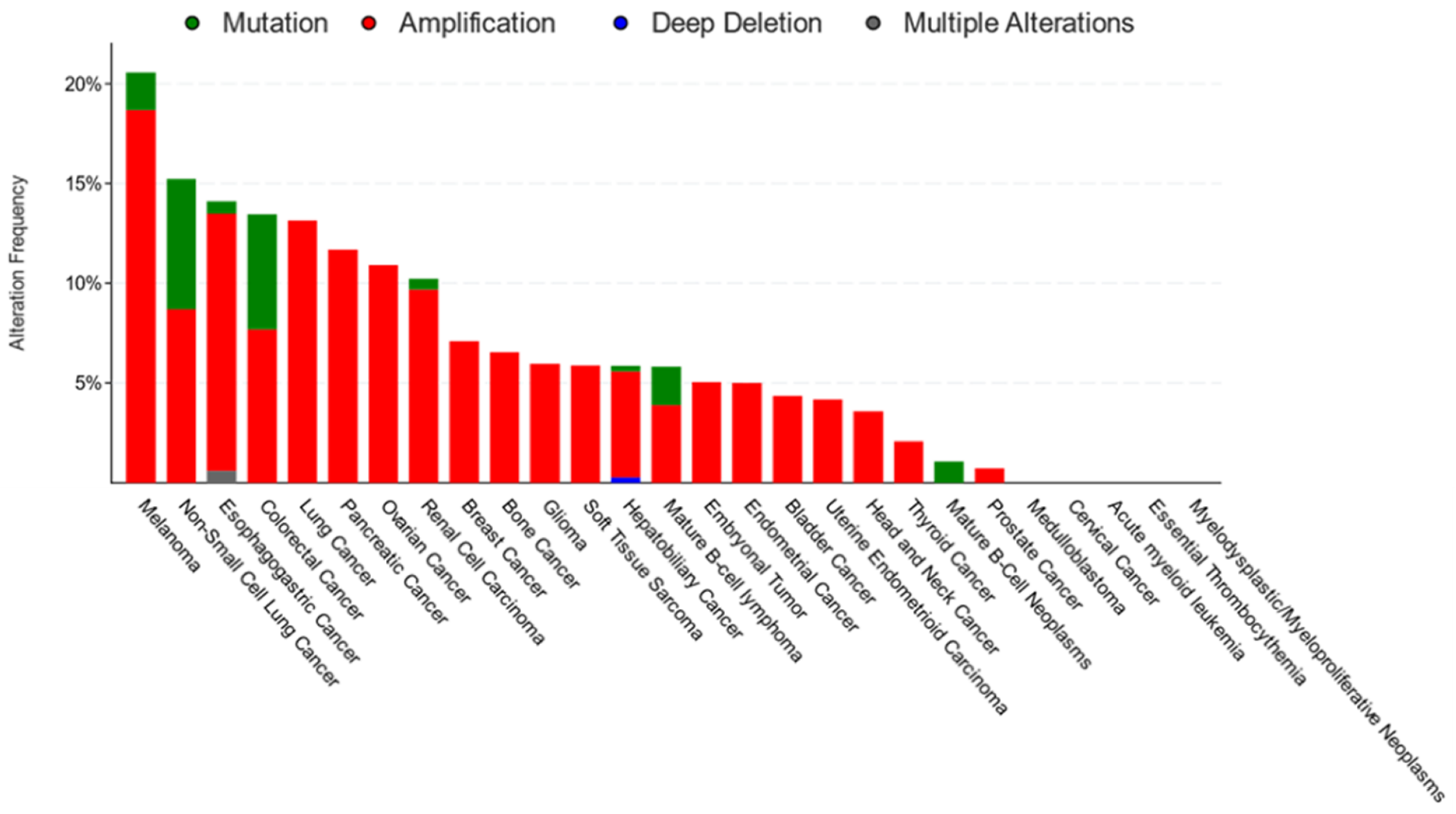
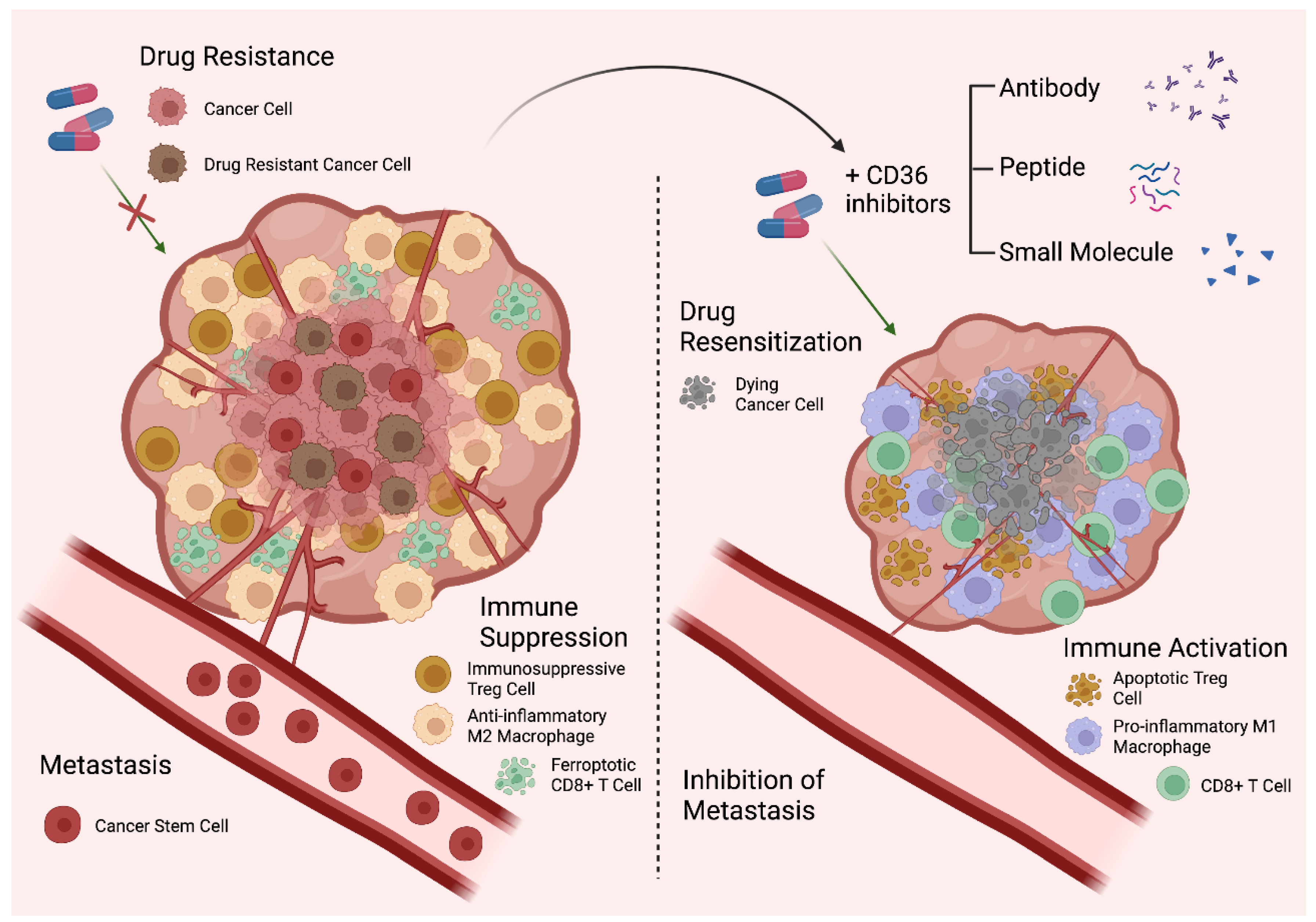
| Cancer | Cell Type That Expresses CD36 | Main Effects of CD36 | References |
|---|---|---|---|
| Brain | CSCs | Stemness marker, promotes tumor initiating ability and self-renewal | [16] |
| Breast | Cancer Cells, CSCs | FA transporter, drug resistance, stemness marker, promotes EMT, metabolic reprogramming | [17,41,50,51,52] |
| TAMs | Phagocytosis of apoptotic cells, recruitment to the TME, immune evasion | [66] | |
| Cervical | Cancer Cells | FA transporter, tumor growth, metastasis, promoted EMT | [60,61] |
| Colorectal | Cancer Cells | FA transporter, metastasis | [53] |
| Gastric | Cancer Cells | FA transporter, metastasis, metabolic reprogramming | [20,40,54,55,67] |
| Leukemia | Cancer Cells, CSCs | FA transporter, drug resistance, metastasis | [58,68,69] |
| Liposarcoma | Cancer Cells | FA transporter | [50] |
| Liver | Cancer Cells, CSCs, | Stemness marker, promoted EMT, metabolic reprogramming | [59,70] |
| TAMs | Immune evasion, metastasis | [71] | |
| Melanoma | Cancer Cells | Drug resistance | [72] |
| Intratumoral Tregs, CD8+ T Cells | FA transporter, immune evasion | [56,73] | |
| OSCC | CSCs | FA transporter, metastasis | [23] |
| Ovarian | Cancer Cells | TSP-1 receptor, FA transporter, drug resistance, metastasis | [14,74,75] |
| Pancreatic | Cancer Cells | Drug resistance | [57] |
| Prostate | Cancer Cells | FA transporter | [50,62,65] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, W.W.; Zuppe, H.T.; Kurokawa, M. The Role of CD36 in Cancer Progression and Its Value as a Therapeutic Target. Cells 2023, 12, 1605. https://doi.org/10.3390/cells12121605
Feng WW, Zuppe HT, Kurokawa M. The Role of CD36 in Cancer Progression and Its Value as a Therapeutic Target. Cells. 2023; 12(12):1605. https://doi.org/10.3390/cells12121605
Chicago/Turabian StyleFeng, William W., Hannah T. Zuppe, and Manabu Kurokawa. 2023. "The Role of CD36 in Cancer Progression and Its Value as a Therapeutic Target" Cells 12, no. 12: 1605. https://doi.org/10.3390/cells12121605
APA StyleFeng, W. W., Zuppe, H. T., & Kurokawa, M. (2023). The Role of CD36 in Cancer Progression and Its Value as a Therapeutic Target. Cells, 12(12), 1605. https://doi.org/10.3390/cells12121605








