Analysis of Plasma Proteins Involved in Inflammation, Immune Response/Complement System, and Blood Coagulation upon Admission of COVID-19 Patients to Hospital May Help to Predict the Prognosis of the Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Aspects
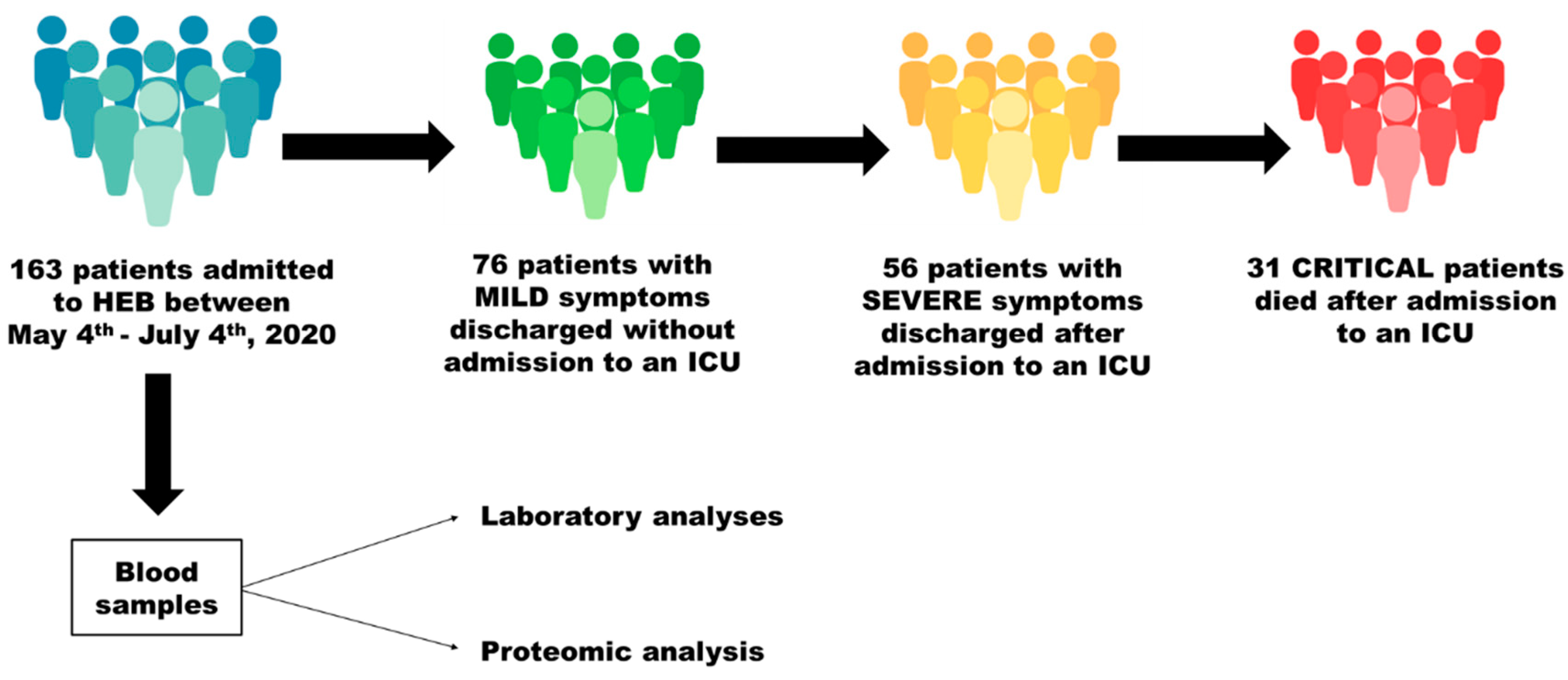
2.2. Study Design and Patients
2.3. Comparisons and Sampling
2.4. Preparation of the Plasma Samples for Proteomic Analysis
2.5. Proteomic Analysis
2.6. Statistical Analysis
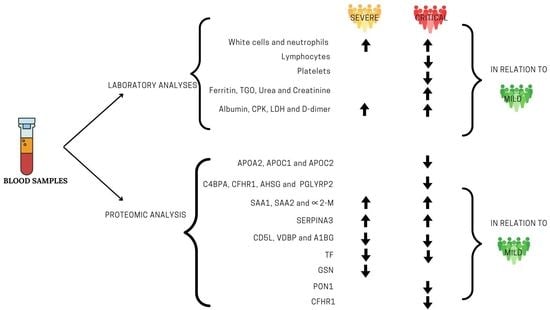
3. Results
3.1. Characterization of the Patients Included in the Study
3.2. Laboratory Findings
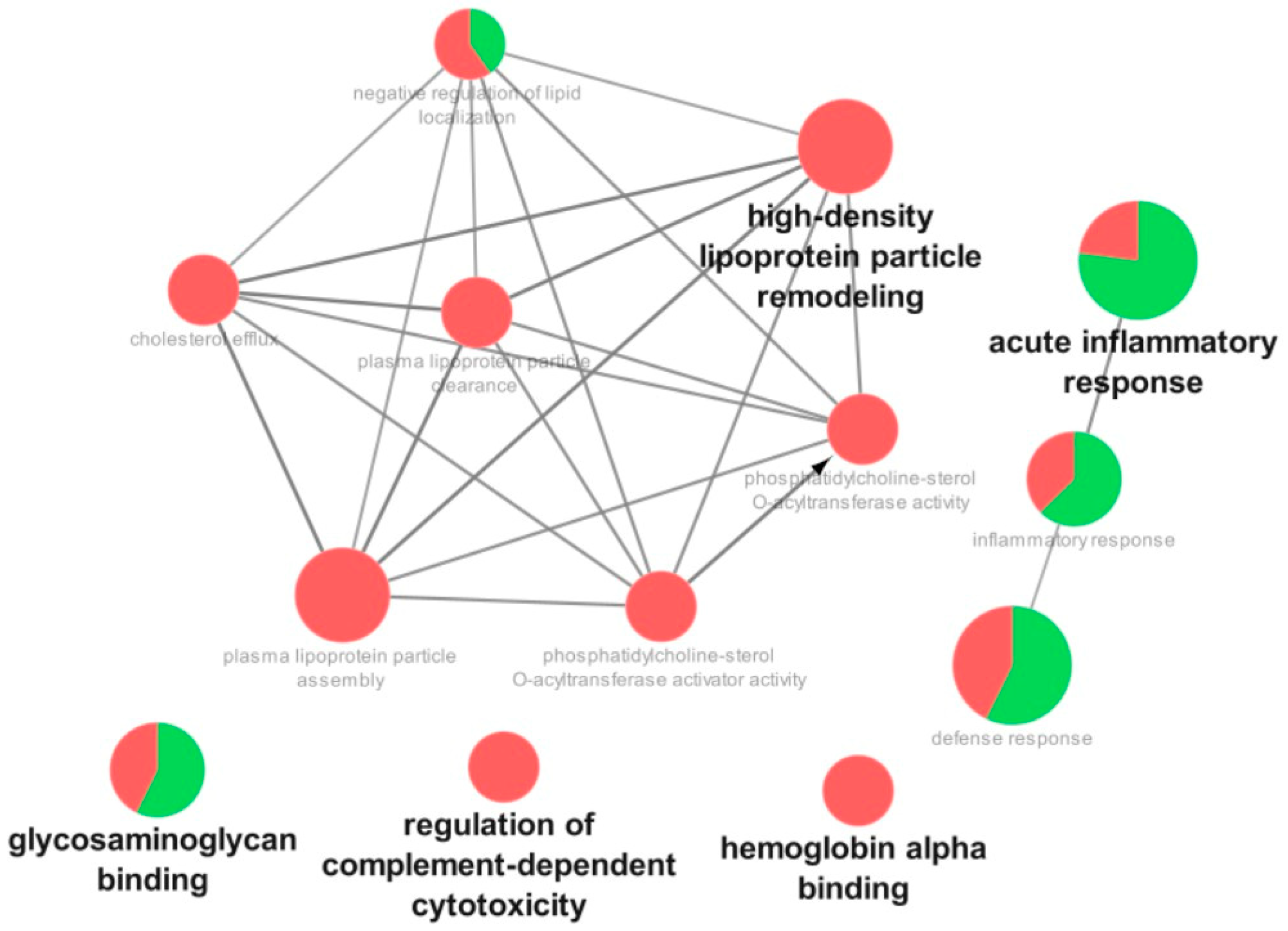
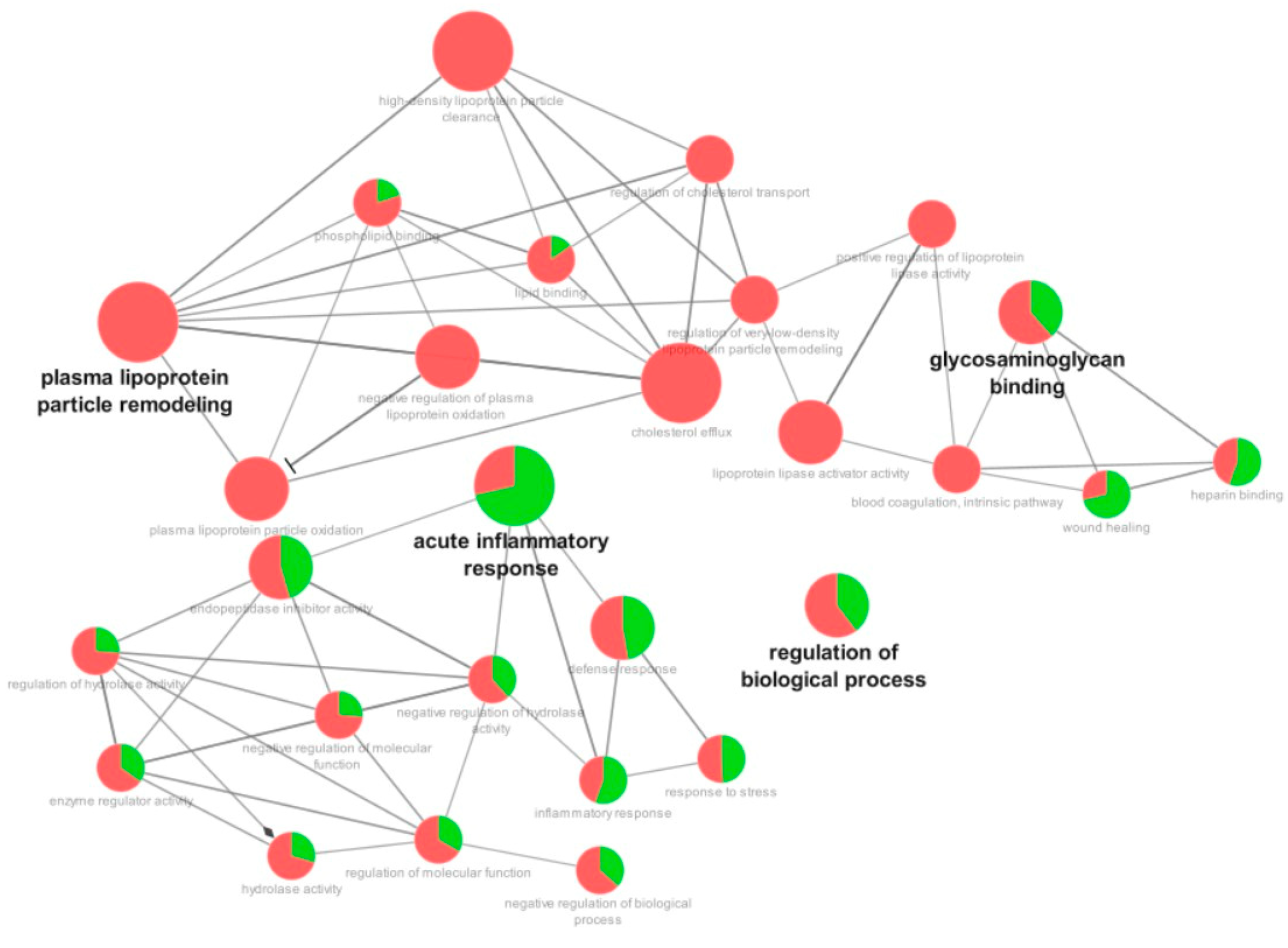
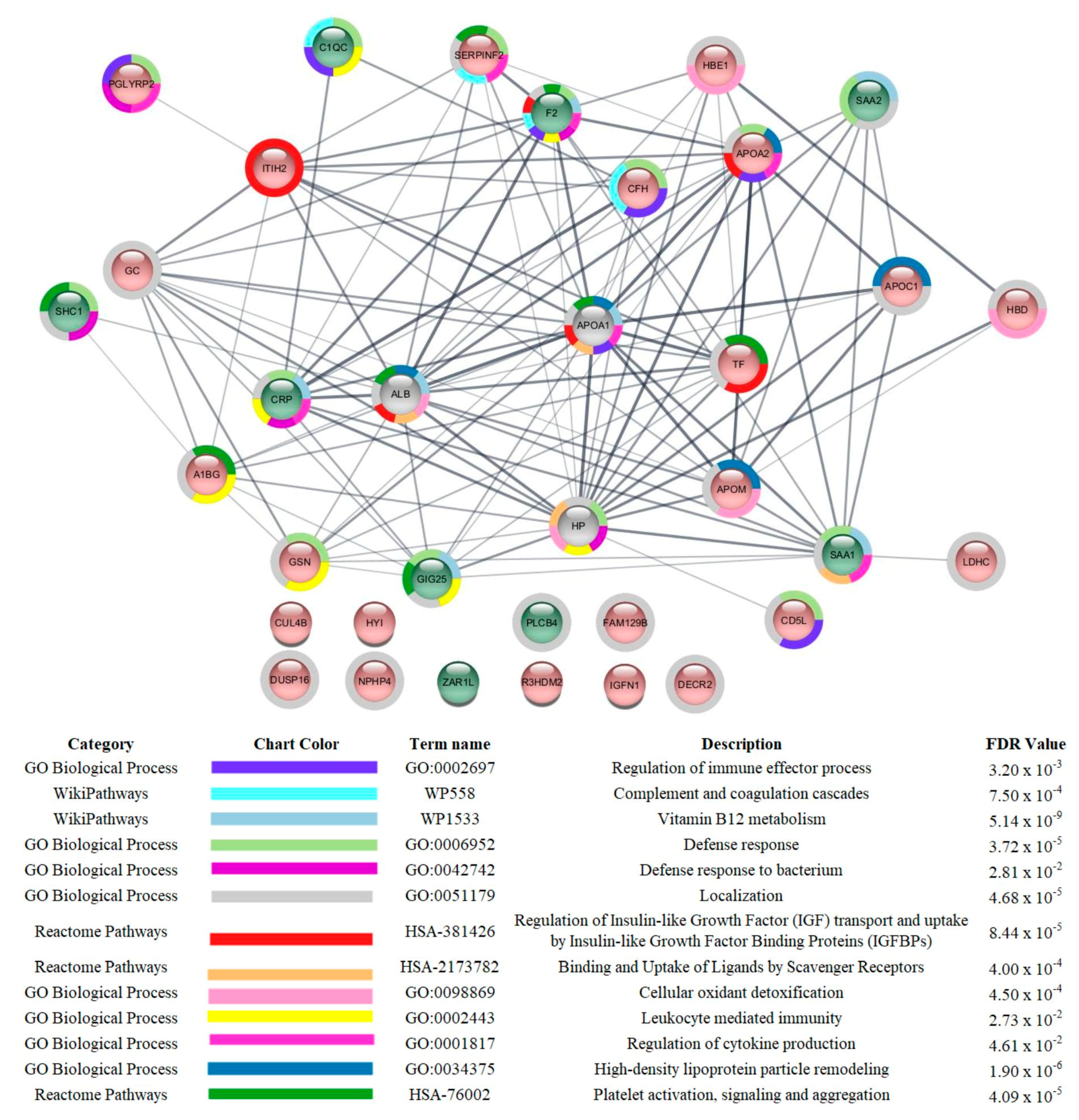
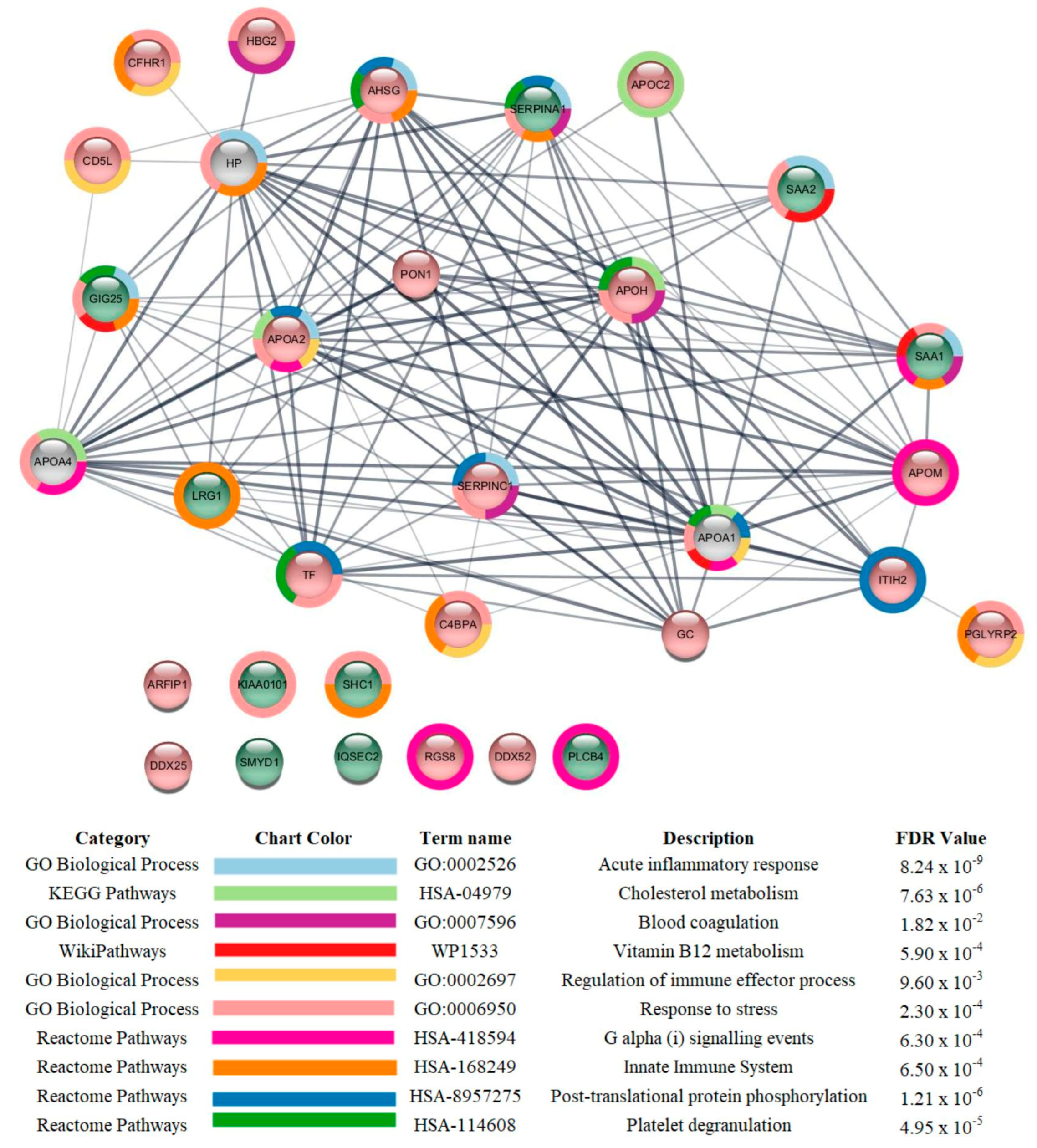
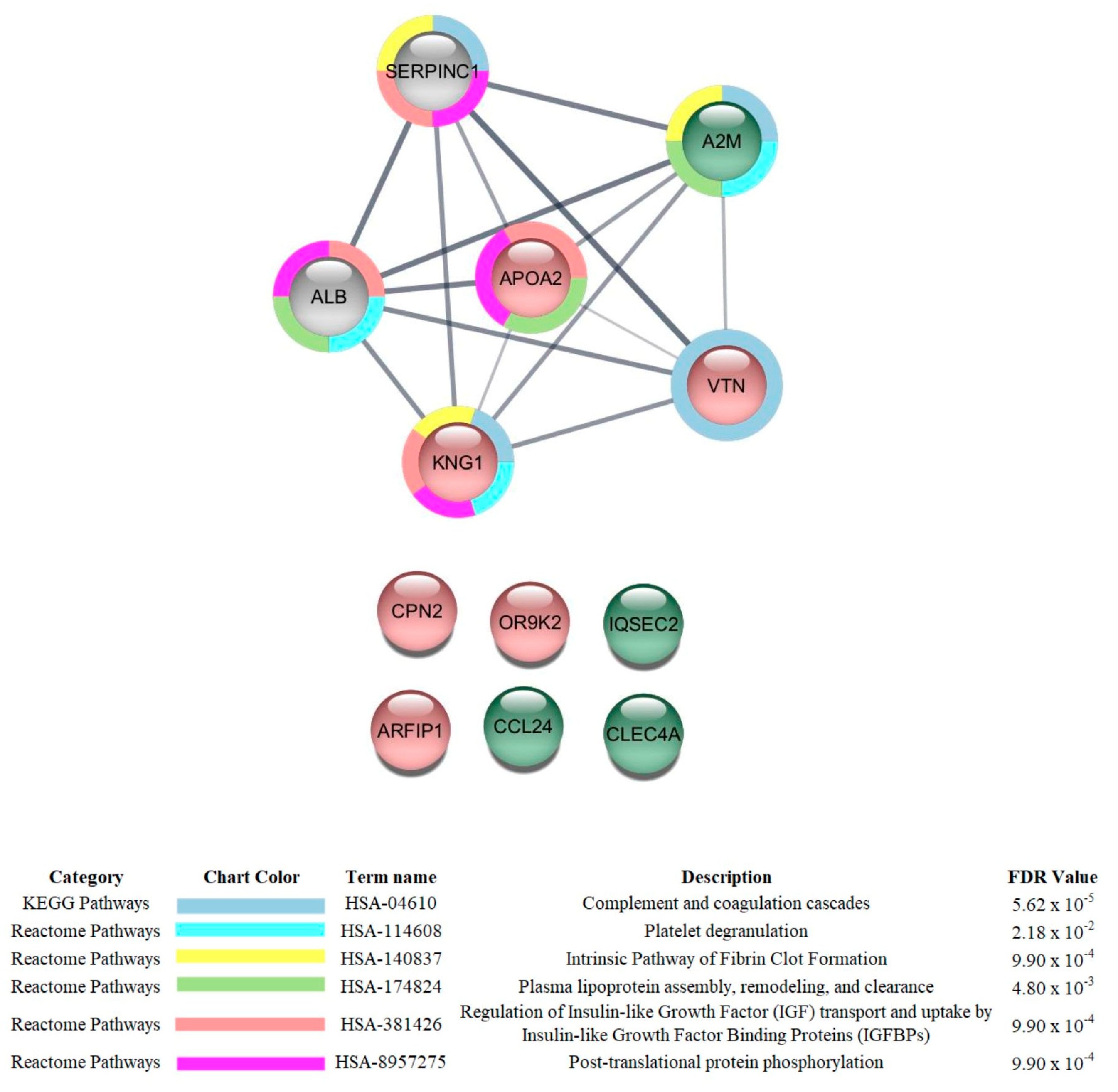
3.3. Proteomic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Weekly Epidemiological Update on COVID-19. 2022. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---16-november-2022 (accessed on 16 November 2022).
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Tracking SARS-CoV-2 Variants. 2022. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 15 December 2022).
- Grasselli, G.; Zangrillo, A.; Zanella, A.; Antonelli, M.; Cabrini, L.; Castelli, A.; Cereda, D.; Coluccello, A.; Foti, G.; Fumagalli, R.; et al. Baseline Characteristics and Outcomes of 1591 Patients Infected with SARS-CoV-2 Admitted to ICUs of the Lombardy Region, Italy. JAMA 2020, 323, 1574–1581. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Fan, Y.; Chen, M.; Wu, X.; Zhang, L.; He, T.; Wang, H.; Wan, J.; Wang, X.; Lu, Z. Cardiovascular Implications of Fatal Outcomes of Patients with Coronavirus Disease 2019 (COVID-19). JAMA Cardiol. 2020, 5, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Jin, H.; Wang, M.; Hu, Y.; Chen, S.; He, Q.; Chang, J.; Hong, C.; Zhou, Y.; Wang, D.; et al. Neurologic Manifestations of Hospitalized Patients with Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020, 77, 683–690. [Google Scholar] [CrossRef]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Thevarajan, I.; Nguyen, T.H.O.; Koutsakos, M.; Druce, J.; Caly, L.; van de Sandt, C.E.; Jia, X.; Nicholson, S.; Catton, M.; Cowie, B.; et al. Breadth of concomitant immune responses prior to patient recovery: A case report of non-severe COVID-19. Nat. Med. 2020, 26, 453–455. [Google Scholar] [CrossRef]
- Kattan, M.; Ji, X.; Milinovich, A.; Adegboye, A.; Duggal, A.; Dweik, R.; Khouli, H.; Gordon, S.; Young, J.; Jehi, L. An Algorithm for Classifying Patients Most Likely to Develop Severe Coronavirus Disease 2019 Illness. Crit. Care Explor. 2020, 2, e0300. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, X.; Li, Y.; Chen, H.; Chen, T.; Su, N.; Huang, F.; Zhou, J.; Zhang, B.; Yan, F.; et al. Clinical Course and Outcomes of 344 Intensive Care Patients with COVID-19. Am. J. Respir. Crit. Care Med. 2020, 201, 1430–1434. [Google Scholar] [CrossRef]
- Lee, P.Y.; Osman, J.; Low, T.Y.; Jamal, R. Plasma/serum proteomics: Depletion strategies for reducing high-abundance proteins for biomarker discovery. Bioanalysis 2019, 11, 1799–1812. [Google Scholar] [CrossRef]
- Lazari, L.C.; Ghilardi, F.R.; Rosa-Fernandes, L.; Assis, D.M.; Nicolau, J.C.; Santiago, V.F.; Dalcoquio, T.F.; Angeli, C.B.; Bertolin, A.J.; Marinho, C.R.; et al. Prognostic accuracy of MALDI-TOF mass spectrometric analysis of plasma in COVID-19. Life Sci. Alliance 2021, 4, e202000946. [Google Scholar] [CrossRef]
- Messner, C.B.; Demichev, V.; Wendisch, D.; Michalick, L.; White, M.; Freiwald, A.; Textoris-Taube, K.; Vernardis, S.I.; Egger, A.S.; Kreidl, M.; et al. Ultra-High-Throughput Clinical Proteomics Reveals Classifiers of COVID-19 Infection. Cell Syst. 2020, 11, 11–24 e14. [Google Scholar] [CrossRef] [PubMed]
- Overmyer, K.A.; Shishkova, E.; Miller, I.J.; Balnis, J.; Bernstein, M.N.; Peters-Clarke, T.M.; Meyer, J.G.; Quan, Q.; Muehlbauer, L.K.; Trujillo, E.A.; et al. Large-Scale Multi-omic Analysis of COVID-19 Severity. Cell Syst. 2020, 12, 23–40.e7. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15. [Google Scholar] [CrossRef] [PubMed]
- Filbin, M.R.; Mehta, A.; Schneider, A.M.; Kays, K.R.; Guess, J.R.; Gentili, M.; Fenyves, B.G.; Charland, N.C.; Gonye, A.L.K.; Gushterova, I.; et al. Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions. Cell Rep. Med. 2021, 2, 100287. [Google Scholar] [CrossRef]
- Shu, T.; Ning, W.; Wu, D.; Xu, J.; Han, Q.; Huang, M.; Zou, X.; Yang, Q.; Yuan, Y.; Bie, Y.; et al. Plasma Proteomics Identify Biomarkers and Pathogenesis of COVID-19. Immunity 2020, 53, 1108–1122.e5. [Google Scholar] [CrossRef]
- Ciccosanti, F.; Antonioli, M.; Sacchi, A.; Notari, S.; Farina, A.; Beccacece, A.; Fusto, M.; Vergori, A.; D’Offizi, G.; Taglietti, F.; et al. Proteomic analysis identifies a signature of disease severity in the plasma of COVID-19 pneumonia patients associated to neutrophil, platelet and complement activation. Clin. Proteom. 2022, 19, 38. [Google Scholar] [CrossRef]
- Mohammed, Y.; Goodlett, D.R.; Cheng, M.P.; Vinh, D.C.; Lee, T.C.; McGeer, A.; Sweet, D.; Tran, K.; Lee, T.; Murthy, S.; et al. Longitudinal Plasma Proteomics Analysis Reveals Novel Candidate Biomarkers in Acute COVID-19. J. Proteome Res. 2022, 21, 975–992. [Google Scholar] [CrossRef]
- Zhong, W.; Altay, O.; Arif, M.; Edfors, F.; Doganay, L.; Mardinoglu, A.; Uhlen, M.; Fagerberg, L. Next generation plasma proteome profiling of COVID-19 patients with mild to moderate symptoms. EBioMedicine 2021, 74, 103723. [Google Scholar] [CrossRef]
- Garcia, S.; Silva-Costa, L.C.; Reis-de-Oliveira, G.; Guest, P.C.; Baldasso, P.A.; Cassoli, J.S.; Martins-de-Souza, D. Identifying Biomarker Candidates in the Blood Plasma or Serum Proteome. Adv. Exp. Med. Biol. 2017, 974, 193–203. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Lima Leite, A.; Gualiume Vaz Madureira Lobo, J.; Barbosa da Silva Pereira, H.A.; Silva Fernandes, M.; Martini, T.; Zucki, F.; Sumida, D.H.; Rigalli, A.; Buzalaf, M.A.R. Proteomic analysis of gastrocnemius muscle in rats with streptozotocin-induced diabetes and chronically exposed to fluoride. PLoS ONE 2014, 9, e106646. [Google Scholar] [CrossRef] [PubMed]
- Kaeuffer, C.; Le Hyaric, C.; Fabacher, T.; Mootien, J.; Dervieux, B.; Ruch, Y.; Hugerot, A.; Zhu, Y.J.; Pointurier, V.; Clere-Jehl, R.; et al. Clinical characteristics and risk factors associated with severe COVID-19: Prospective analysis of 1,045 hospitalised cases in North-Eastern France, March 2020. Eurosurveillance 2020, 25, 2000895. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Kong, H.; Zheng, X.Z.; Li, X.Y.; Ma, J.; Zhang, H.; Wang, D.X.; Li, H.C.; Liu, X.M. Early predictive factors of progression from severe type to critical ill type in patients with Coronavirus Disease 2019: A retrospective cohort study. PLoS ONE 2020, 15, e0243195. [Google Scholar] [CrossRef]
- Jin, J.M.; Bai, P.; He, W.; Wu, F.; Liu, X.F.; Han, D.M.; Liu, S.; Yang, J.K. Gender Differences in Patients with COVID-19: Focus on Severity and Mortality. Front. Public Health 2020, 8, 152. [Google Scholar] [CrossRef]
- Krieger, N.; Chen, J.T.; Waterman, P.D. Excess mortality in men and women in Massachusetts during the COVID-19 pandemic. Lancet 2020, 395, 1829. [Google Scholar] [CrossRef]
- Chen, I.Y.; Moriyama, M.; Chang, M.F.; Ichinohe, T. Severe Acute Respiratory Syndrome Coronavirus Viroporin 3a Activates the NLRP3 Inflammasome. Front. Microbiol. 2019, 10, 50. [Google Scholar] [CrossRef]
- Suriawinata, E.; Mehta, K.J. Iron and iron-related proteins in COVID-19. Clin. Exp. Med. 2022, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Burugu, H.R.; Kandi, V.; Kutikuppala, L.V.S.; Suvvari, T.K. Activities of Serum Ferritin and Treatment Outcomes Among COVID-19 Patients Treated with Vitamin C and Dexamethasone: An Uncontrolled Single-Center Observational Study. Cureus 2020, 12, e11442. [Google Scholar] [CrossRef]
- Cao, P.; Wu, Y.; Wu, S.; Wu, T.; Zhang, Q.; Zhang, R.; Wang, Z.; Zhang, Y. Elevated serum ferritin level effectively discriminates severity illness and liver injury of coronavirus disease 2019 pneumonia. Biomarkers 2020, 26, 207–212. [Google Scholar] [CrossRef]
- Topcu, H.; Arik, Y.E. The Importance of D-Dimer, Ferritin, CRP and Lymphocyte Values in Determining Mortality in COVID-19 Disease in Turkey. Clin. Lab. 2022, 68, 2274–2280. [Google Scholar] [CrossRef]
- Gurusamy, E.; Mahalakshmi, S.; Kaarthikeyan, G.; Ramadevi, K.; Arumugam, P.; Gayathri, M.S. Biochemical predictors for SARS-CoV-2 severity. Bioinformation 2021, 17, 834–839. [Google Scholar] [CrossRef]
- Abbaspour, N.; Hurrell, R.; Kelishadi, R. Review on iron and its importance for human health. J. Res. Med. Sci. 2014, 19, 164–174. [Google Scholar]
- Bellmann-Weiler, R.; Lanser, L.; Barket, R.; Rangger, L.; Schapfl, A.; Schaber, M.; Fritsche, G.; Woll, E.; Weiss, G. Prevalence and Predictive Value of Anemia and Dysregulated Iron Homeostasis in Patients with COVID-19 Infection. J. Clin. Med. 2020, 9, 2429. [Google Scholar] [CrossRef]
- Tanaka, S.; Couret, D.; Tran-Dinh, A.; Duranteau, J.; Montravers, P.; Schwendeman, A.; Meilhac, O. High-density lipoproteins during sepsis: From bench to bedside. Crit. Care 2020, 24, 134. [Google Scholar] [CrossRef]
- Ulloque-Badaracco, J.R.; Hernandez-Bustamante, E.A.; Herrera-Anazco, P.; Benites-Zapata, V.A. Prognostic value of apolipoproteins in COVID-19 patients: A systematic review and meta-analysis. Travel. Med. Infect. Dis. 2021, 44, 102200. [Google Scholar] [CrossRef]
- Pasrija, R.; Naime, M. The deregulated immune reaction and cytokines release storm (CRS) in COVID-19 disease. Int. Immunopharmacol. 2020, 90, 107225. [Google Scholar] [CrossRef]
- Frieman, M.; Heise, M.; Baric, R. SARS coronavirus and innate immunity. Virus Res. 2008, 133, 101–112. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, H.; Fan, L.; Fang, M.; He, X.; Lu, B.; Pang, Z. CLEC4s as Potential Therapeutic Targets in Hepatocellular Carcinoma Microenvironment. Front. Cell Dev. Biol. 2021, 9, 681372. [Google Scholar] [CrossRef]
- Baker, J.R.; Mahdi, M.; Nicolau, D.V., Jr.; Ramakrishnan, S.; Barnes, P.J.; Simpson, J.L.; Cass, S.P.; Russell, R.E.K.; Donnelly, L.E.; Bafadhel, M. Early Th2 inflammation in the upper respiratory mucosa as a predictor of severe COVID-19 and modulation by early treatment with inhaled corticosteroids: A mechanistic analysis. Lancet Respir. Med. 2022, 10, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Maisonnasse, P.; Poynard, T.; Sakka, M.; Akhavan, S.; Marlin, R.; Peta, V.; Deckmyn, O.; Ghedira, N.B.; Ngo, Y.; Rudler, M.; et al. Validation of the Performance of A1HPV6, a Triage Blood Test for the Early Diagnosis and Prognosis of SARS-CoV-2 Infection. Gastro Hep Adv. 2022, 1, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Seitz, R.; Gurtler, L.; Schramm, W. Thromboinflammation in COVID-19: Can α2-macroglobulin help to control the fire? J. Thromb. Haemost. 2021, 19, 351–354. [Google Scholar] [CrossRef]
- Cui, T.; Miao, G.; Jin, X.; Yu, H.; Zhang, Z.; Xu, L.; Wu, Y.; Qu, G.; Liu, G.; Zheng, Y.; et al. The adverse inflammatory response of tobacco smoking in COVID-19 patients: Biomarkers from proteomics and metabolomics. J. Breath. Res. 2022, 16, 046002. [Google Scholar] [CrossRef]
- Souza Junior, D.R.; Silva, A.R.M.; Rosa-Fernandes, L.; Reis, L.R.; Alexandria, G.; Bhosale, S.D.; Ghilardi, F.R.; Dalcoquio, T.F.; Bertolin, A.J.; Nicolau, J.C.; et al. HDL proteome remodeling associates with COVID-19 severity. J. Clin. Lipidol. 2021, 15, 796–804. [Google Scholar] [CrossRef]
- Barnes, B.J.; Adrover, J.M.; Baxter-Stoltzfus, A.; Borczuk, A.; Cools-Lartigue, J.; Crawford, J.M.; Dassler-Plenker, J.; Guerci, P.; Huynh, C.; Knight, J.S.; et al. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 2020, 217, e20200652. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Yalavarthi, S.; Shi, H.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.; Weber, A.; Barnes, B.J.; Egeblad, M.; et al. Neutrophil extracellular traps (NETs) as markers of disease severity in COVID-19. medRxiv 2020, preprint. [Google Scholar] [CrossRef]
- Becker, R.C. COVID-19 update: COVID-19-associated coagulopathy. J. Thromb. Thrombolysis 2020, 50, 54–67. [Google Scholar] [CrossRef]
- Isakadze, N.; Engels, M.C.; Beer, D.; McClellan, R.; Yanek, L.R.; Mondaloo, B.; Hays, A.G.; Metkus, T.S.; Calkins, H.; Barth, A.S. C-reactive Protein Elevation Is Associated with QTc Interval Prolongation in Patients Hospitalized with COVID-19. Front. Cardiovasc. Med. 2022, 9, 866146. [Google Scholar] [CrossRef]
- Sanchez-Moral, L.; Rafols, N.; Martori, C.; Paul, T.; Tellez, E.; Sarrias, M.R. Multifaceted Roles of CD5L in Infectious and Sterile Inflammation. Int. J. Mol. Sci. 2021, 22, 4076. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Favaloro, E.J. D-dimer is Associated with Severity of Coronavirus Disease 2019: A Pooled Analysis. Thromb. Haemost. 2020, 120, 876–878. [Google Scholar] [CrossRef]
- Tang, N.; Li, D.; Wang, X.; Sun, Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020, 18, 844–847. [Google Scholar] [CrossRef]
- Magro, C.; Mulvey, J.J.; Berlin, D.; Nuovo, G.; Salvatore, S.; Harp, J.; Baxter-Stoltzfus, A.; Laurence, J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: A report of five cases. Transl. Res. 2020, 220, 1–13. [Google Scholar] [CrossRef]
- Wang, Z.M.; Li, X.; Cocklin, R.R.; Wang, M.; Wang, M.; Fukase, K.; Inamura, S.; Kusumoto, S.; Gupta, D.; Dziarski, R. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J. Biol. Chem. 2003, 278, 49044–49052. [Google Scholar] [CrossRef] [PubMed]
- Duygu, F.; Tekin Koruk, S.; Aksoy, N. Serum paraoxonase and arylesterase activities in various forms of hepatitis B virus infection. J. Clin. Lab. Anal. 2011, 25, 311–316. [Google Scholar] [CrossRef]
- Cava, C.; Bertoli, G.; Castiglioni, I. In Silico Discovery of Candidate Drugs against COVID-19. Viruses 2020, 12, 404. [Google Scholar] [CrossRef]
- Asteris, P.G.; Gavriilaki, E.; Touloumenidou, T.; Koravou, E.E.; Koutra, M.; Papayanni, P.G.; Pouleres, A.; Karali, V.; Lemonis, M.E.; Mamou, A.; et al. Genetic prediction of ICU hospitalization and mortality in COVID-19 patients using artificial neural networks. J. Cell. Mol. Med. 2022, 26, 1445–1455. [Google Scholar] [CrossRef]
- Vollmy, F.; van den Toorn, H.; Zenezini Chiozzi, R.; Zucchetti, O.; Papi, A.; Volta, C.A.; Marracino, L.; Vieceli Dalla Sega, F.; Fortini, F.; Demichev, V.; et al. A serum proteome signature to predict mortality in severe COVID-19 patients. Life Sci. Alliance 2021, 4, e202101099. [Google Scholar] [CrossRef] [PubMed]
- Suvarna, K.; Biswas, D.; Pai, M.G.J.; Acharjee, A.; Bankar, R.; Palanivel, V.; Salkar, A.; Verma, A.; Mukherjee, A.; Choudhury, M.; et al. Proteomics and Machine Learning Approaches Reveal a Set of Prognostic Markers for COVID-19 Severity with Drug Repurposing Potential. Front. Physiol. 2021, 12, 652799. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | Mild | Severe | Critical |
|---|---|---|---|
| Median age, years (95% CI) | 51.0 (48.8–56.7) a | 56.5 (51.8–60.4) a | 73.0 (63.7–72.7) b |
| Female (n, %) | 41, 53.9% | 30, 53.6% | 11, 35.5% |
| Male (n, %) | 35, 46.1% | 26, 46.4% | 20, 64.5% |
| Comorbidity | |||
| Hypertension | 31.6% | 49.1% | 64.5% |
| Diabetes | 21.1% | 41.8% | 32.3% |
| Cardiovascular disease | 10.5% | 10.9% | 9.7% |
| Obesity | 22.4% | 14.5% | 3.2% |
| COPD | 5.3% | 10.9% | 12.9% |
| Cancer | 2.6% | 5.5% | 3.2% |
| Nephropathy | 3.9% | 5.5% | 16.1% |
| Hepatic disease | 1.3% | 0 | 0 |
| Stroke | 5.3% | 1.8% | 6.5% |
| Autoimmune disease | 0 | 0 | 6.5% |
| Characteristics | Mild | Severe | Critical | p |
|---|---|---|---|---|
| Full blood counts | ||||
| Red blood cells, ×106/mm3 | 4.43 ± 0.58 a | 4.24 ± 0.69 a | 4.12 ± 0.79 a | 0.058 ** |
| White blood cells, /mm3 | 5930 (5565–6949) a | 8050 (7090–9090) b | 8020 (7349–12024) b | 0.001 ** |
| Neutrophil, /mm3 | 4112 (4086–5707) a | 6018 (5553–7628) b | 5849 (5613 –9889) b | 0.005 * |
| Lymphocyte, /mm3 | 954 (986–1397) a | 870 (817–1353) a,b | 529 (458–1255) b | 0.003 * |
| Hemoglobin, g/dL | 13.0 (12.5–13.8) a | 12.8 (12.2–13.2) a | 12.5 (11.5–13.2) a | 0.217 * |
| Eosinophil, /mm3 | 0 (15.4–51.8) a | 0 (11.7–104.6) a | 0 (7.7–61.2) a | 0.848 * |
| Platelets, ×103/mm3 | 220 (210–245) a | 220 (207–268) a | 176 (154–245) b | 0.011 * |
| Biochemical tests | ||||
| Ferritin, µg/L | 417 (511–796) a | 631 (664–1100) a | 931 (883–1474) b | 0.003 * |
| Albumin, g/dL | 3.60 (3.42–3.82) a | 3.20 (3.17–3.41) b | 3.1 (2.82–3.23) b | <0.001 * |
| TGO, U/L | 29.5 (31.5–50.0) a | 37.0 (36.7–57.1) a | 46.0(40.9–81.5) b | 0.014 * |
| TGP, U/L | 29.0 (33.5–51.1) a | 36.0 (37.6–65.6) a | 31.0 (27.8–61.8) a | 0.789 * |
| CPK, U/L | 75 (85–138) a | 120 (173–344) b | 124 (140–865) b | 0.001 * |
| Ure a, mg/dL | 32.8 (34.3–50.6) a | 33.8 (35.1–54.5) a | 44.0 (43.3–60.9) b | 0.007 * |
| Creatinine, mg/dL | 0.80 (1.00–1.99) a | 0.80 (0.86–1.35) a | 1.20 (1.19–4.60) b | 0.001 * |
| PCR, mg/L | 46.0 (60.3–93.7) a | 120.6 (92.5–140.0) b | 102.0 (86.5–151.9) a,b | 0.004 * |
| LDH, U/L | 213 (221–263) a | 314 (285–363) b | 402 (290–531) b | <0.001 * |
| D–dimer, mg/L | 0.73 (0.83–1.32) a | 1.11 (1.55–2.62) b | 2.14 (1.66–6.43) b | <0.001 * |
| Plasma Proteomic Findings | Implications for the Course of COVID-19 According to the Literature | Potential Therapies |
|---|---|---|
| GSN levels were reduced in severe patients compared to those with mild symptoms | Calcium-binding protein that scavenges circulating filamentous actin, thus possessing anti-inflammatory properties. Reduced levels of GELS have been shown in serum [13,15] and plasma [14] of COVID-19 patients with worse outcomes. | GSN supplementation has been suggested as a potential therapy for COVID-19 [13], and a clinical trial of recombinant plasma from GSN is currently being conducted (NCT04358406). |
| PON1 levels were reduced in critical patients compared to those with mild symptoms | This enzyme possesses aryadialkylphosphatase activity, as it is involved in the protection of low-density lipoproteins against oxidative damage and the formation of atheroma [40]. It is also important for the innate immune response [56]. In a recent study involving in silico discovery of candidate drugs against COVID-19, it was reported that genes correlated with ACE2 are enriched in aryadialkylphosphatase activity [57]. | Increasing the activity of PON1. |
| CFHR1 levels were reduced in critical patients compared to those with mild symptoms | Involved in complement regulation. The variant rs414628 found in CFHR1 was associated with severe COVID-19 in adult Caucasian patients [58] | Regulation of CFHR1. |
| AHSG levels were reduced in critical patients compared to those with mild symptoms | Promotes endocytosis and possesses opsonic properties. This protein was reported to be increased in the serum of survivor COVID-19 patients admitted to the respiratory and ICU because of respiratory failure [59]. | Increasing AHSG levels. |
| SERPINA3 levels were increased in critical and severe patients compared to those with mild symptoms | Inhibits neutrophil cathepsin G and mast cell chymase, both of which can convert angiotensin-1 to the active angiotensin-2. The levels of this protein were reported to be reduced in the serum of survivor COVID-19 patients admitted to the respiratory ward and ICU because of respiratory failure [59] and in the plasma of non-severe compared to severe patients [60] | Inhibition of SERPINA3 |
| Plasma Proteomic Findings | Implications for the Course of COVID-19 According to the Literature |
|---|---|
| TF levels were reduced in critical and severe patients compared to those with mild symptoms | High ferritin and low transferrin levels are associated with increased risk for ICU admission and the need for mechanical ventilation in COVID-19 patients [35]. |
| APOA1 *, APOA2, APOC1, and APOC2 levels were reduced in critical and/or severe patients compared to those with mild symptoms | Apolipoproteins transport cholesterol from peripheral tissues back to the liver, performing cardioprotective, antiapoptotic, antioxidant, anti-inflammatory, antithrombotic, and anti-infectious functions [36]. Adequate levels of APOA1 were related to protection against mortality in patients hospitalized for COVID-19 [37]. |
| CLEC4 levels were increased in critical patients compared to those with severe symptoms | CLEC4 is a C-type lectin receptor that, once triggered by an antigen, is internalized by clathrin-dependent endocytosis and delivers its antigenic cargo into the antigen presentation pathway, thereby promoting expansion of CD8+ T cells and high production of IFN-γ and TNFα. Functional analysis revealed the potential role of CLEC4A in viral infection, including that of COVID-19 [41]. |
| CCL24 levels were increased in critical patients compared to those with severe symptoms | Chemotactic for resting T-lymphocytes and eosinophils. COVID-19 patients that will clinically deteriorate have a blunted IFN and an exaggerated CCL24 airway response [42]. |
| SAA1 and SAA2 were increased in critical and severe patients compared to those with mild symptoms | SAAs are acute-phase proteins that have been suggested to be predictors of COVID-19 severity [45,46]. |
| CFHR1 levels were reduced in critical patients compared to those with mild symptoms | Involved in complement regulation. The variant rs414628 in CFHR1 was associated with severe COVID-19 in adult Caucasian patients [58] |
| AHSG levels were reduced in critical patients compared to those with mild symptoms | Promotes endocytosis and possesses opsonic properties. This protein was reported to be increased in the serum of survivor COVID-19 patients admitted to the respiratory and ICU because of respiratory failure [59]. |
| SERPINA3 levels were increased in critical and severe patients compared to those with mild symptoms | Inhibits neutrophil cathepsin G and mast cell chymase, both of which can convert angiotensin-1 to the active angiotensin-2. The levels of this protein were reported to be reduced in the serum of survivor COVID-19 patients admitted to the respiratory and ICU because of respiratory failure [59] and in the plasma of non-severe compared to severe patients [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
di Flora, D.C.; Dionizio, A.; Pereira, H.A.B.S.; Garbieri, T.F.; Grizzo, L.T.; Dionisio, T.J.; Leite, A.d.L.; Silva-Costa, L.C.; Buzalaf, N.R.; Reis, F.N.; et al. Analysis of Plasma Proteins Involved in Inflammation, Immune Response/Complement System, and Blood Coagulation upon Admission of COVID-19 Patients to Hospital May Help to Predict the Prognosis of the Disease. Cells 2023, 12, 1601. https://doi.org/10.3390/cells12121601
di Flora DC, Dionizio A, Pereira HABS, Garbieri TF, Grizzo LT, Dionisio TJ, Leite AdL, Silva-Costa LC, Buzalaf NR, Reis FN, et al. Analysis of Plasma Proteins Involved in Inflammation, Immune Response/Complement System, and Blood Coagulation upon Admission of COVID-19 Patients to Hospital May Help to Predict the Prognosis of the Disease. Cells. 2023; 12(12):1601. https://doi.org/10.3390/cells12121601
Chicago/Turabian Styledi Flora, Daniele Castro, Aline Dionizio, Heloisa Aparecida Barbosa Silva Pereira, Thais Francini Garbieri, Larissa Tercilia Grizzo, Thiago José Dionisio, Aline de Lima Leite, Licia C. Silva-Costa, Nathalia Rabelo Buzalaf, Fernanda Navas Reis, and et al. 2023. "Analysis of Plasma Proteins Involved in Inflammation, Immune Response/Complement System, and Blood Coagulation upon Admission of COVID-19 Patients to Hospital May Help to Predict the Prognosis of the Disease" Cells 12, no. 12: 1601. https://doi.org/10.3390/cells12121601
APA Styledi Flora, D. C., Dionizio, A., Pereira, H. A. B. S., Garbieri, T. F., Grizzo, L. T., Dionisio, T. J., Leite, A. d. L., Silva-Costa, L. C., Buzalaf, N. R., Reis, F. N., Pereira, V. B. R., Rosa, D. M. C., dos Santos, C. F., & Buzalaf, M. A. R. (2023). Analysis of Plasma Proteins Involved in Inflammation, Immune Response/Complement System, and Blood Coagulation upon Admission of COVID-19 Patients to Hospital May Help to Predict the Prognosis of the Disease. Cells, 12(12), 1601. https://doi.org/10.3390/cells12121601