Molecular Dissection of DAAM Function during Axon Growth in Drosophila Embryonic Neurons
Abstract
1. Introduction
2. Materials and Methods
2.1. Fly Stocks and Genetics
2.2. Molecular Biology
2.3. Protein Expression and GST Pull-Down
2.4. MT Co-Sedimentation
2.5. SDS-PAGE and Western Blot Analysis of Embryonic Lysates
2.6. Primary Neuronal Cultures and Immunohistochemistry
2.7. Schneider 2 Cell Cultures and Transfection
2.8. Microscopy and Image Analysis
2.9. Statistical Analysis and Figures
3. Results
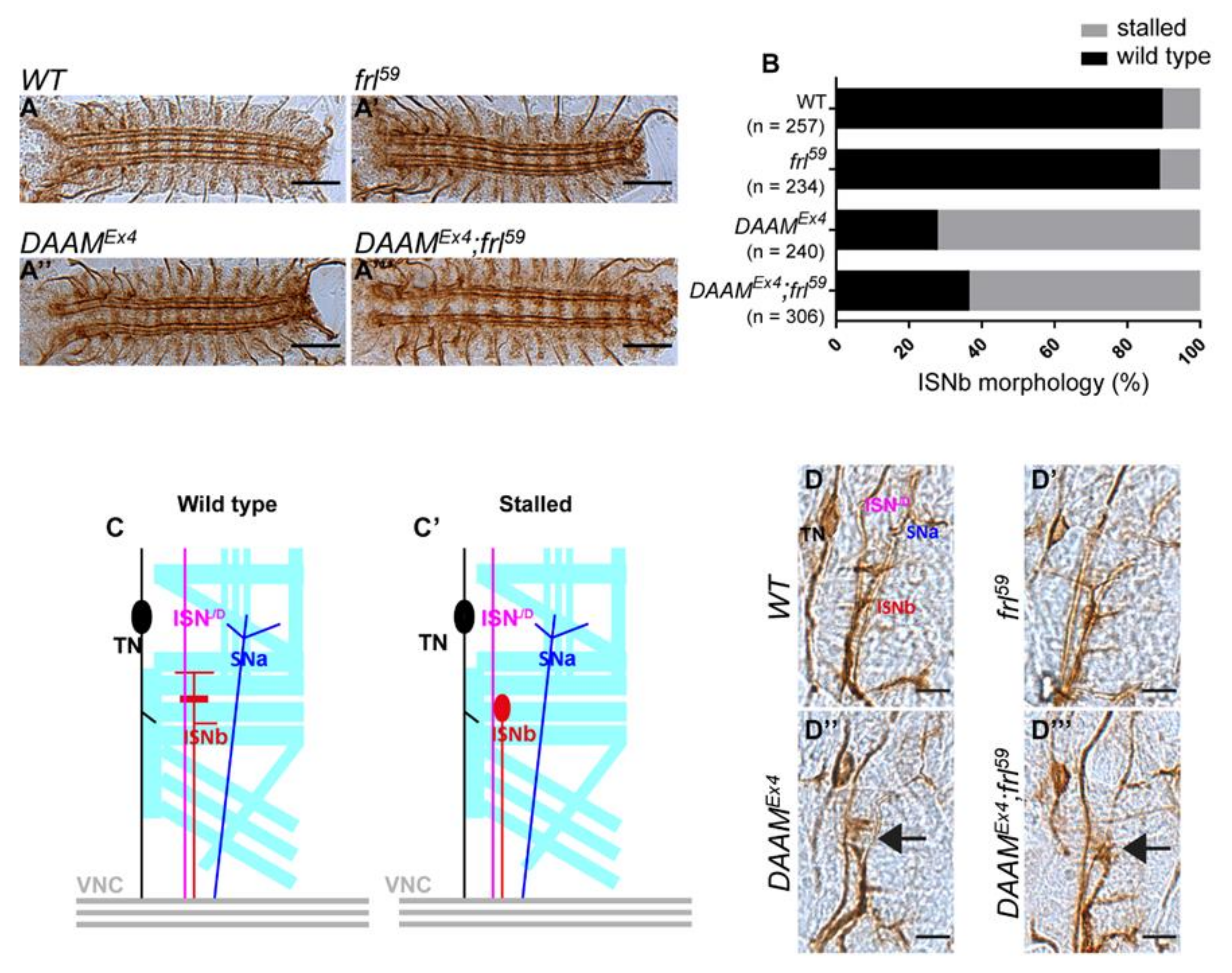
3.1. DAAM and frl Mutant Analysis in Primary Neuronal Cultures
3.2. Separation of Function Alleles of DAAM
3.3. The Actin-Processing Activity of DAAM Is Essential for Axon Growth
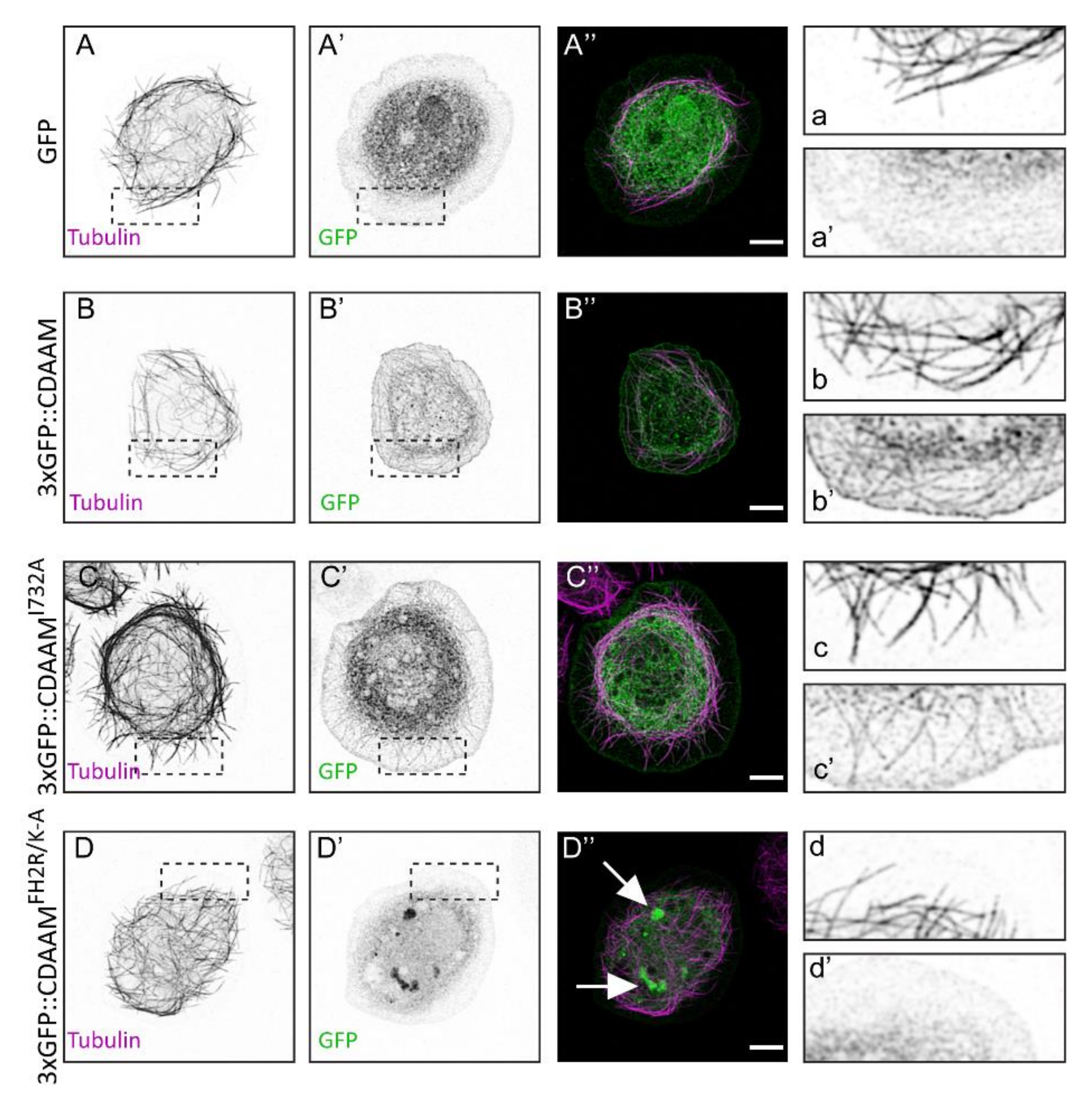
3.4. Direct MT-Binding of DAAM Is Not Essentially Required for Axon Growth
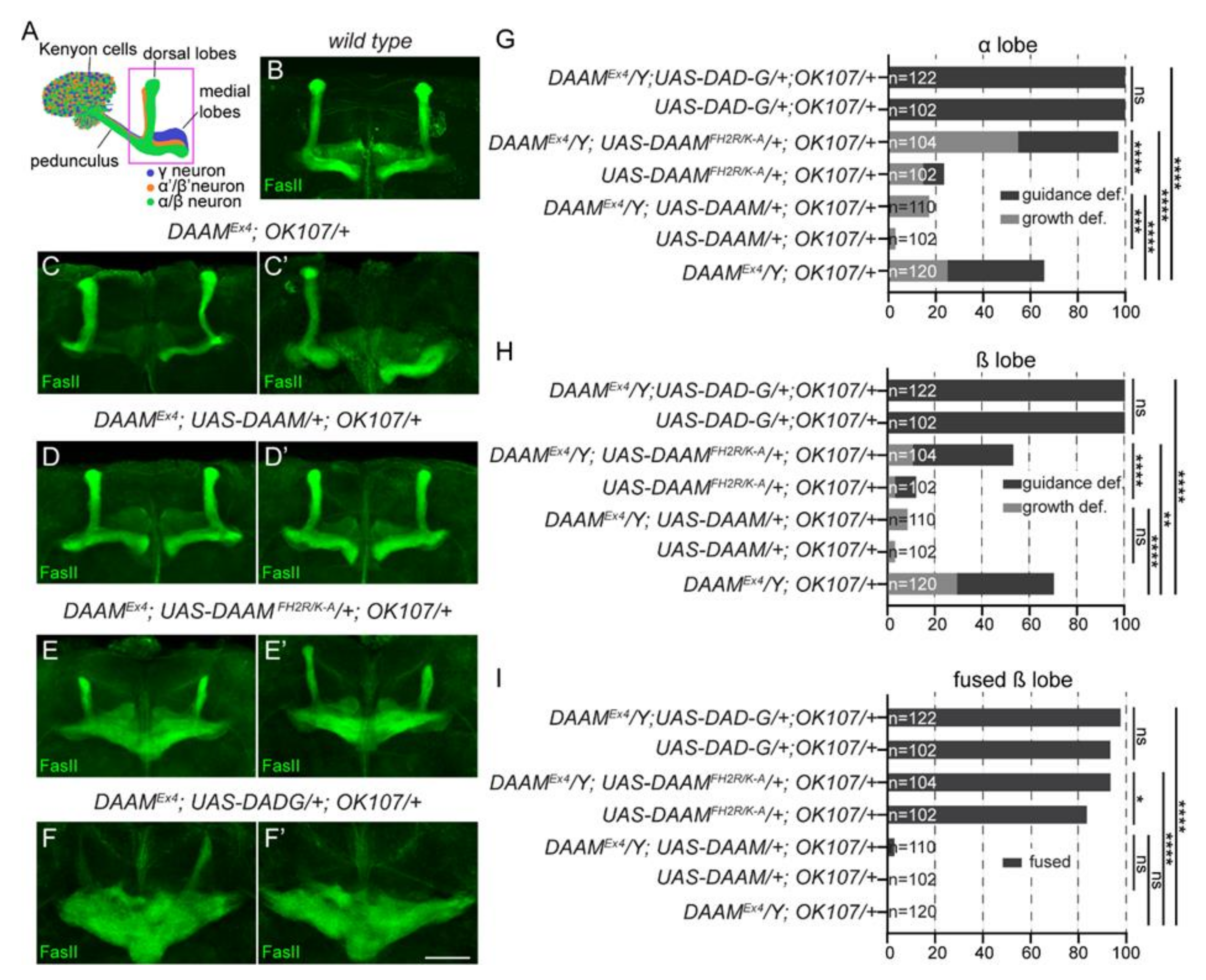
3.5. Overexpression of a Constitutively Active Form of DAAM
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coles, C.H.; Bradke, F. Coordinating Neuronal Actin–Microtubule Dynamics. Curr. Biol. 2015, 25, R677–R691. [Google Scholar] [CrossRef] [PubMed]
- Leterrier, C.; Dubey, P.; Roy, S. The Nano-Architecture of the Axonal Cytoskeleton. Nat. Rev. Neurosci. 2017, 18, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Araújo, S.J.; Tear, G. Axon Guidance Mechanisms and Molecules: Lessons from Invertebrates. Nat. Rev. Neurosci. 2003, 4, 910–922. [Google Scholar] [CrossRef] [PubMed]
- Pinto-Costa, R.; Sousa, M.M. Microtubules, Actin and Cytolinkers: How to Connect Cytoskeletons in the Neuronal Growth Cone. Neurosci. Lett. 2021, 747, 135693. [Google Scholar] [CrossRef] [PubMed]
- Dickson, B.J. Molecular Mechanisms of Axon Guidance. Science 2002, 298, 1959–1964. [Google Scholar] [CrossRef]
- Dent, E.W.; Gupton, S.L.; Gertler, F.B. The Growth Cone Cytoskeleton in Axon Outgrowth and Guidance. Cold Spring Harb. Perspect. Biol. 2011, 3, a001800. [Google Scholar] [CrossRef]
- Efimova, N.; Yang, C.; Chia, J.X.; Li, N.; Lengner, C.J.; Neufeld, K.L.; Svitkina, T.M. Branched Actin Networks Are Assembled on Microtubules by Adenomatous Polyposis Coli for Targeted Membrane Protrusion. J. Cell Biol. 2020, 219, e202003091. [Google Scholar] [CrossRef]
- Higgs, H.N.; Peterson, K.J. Phylogenetic Analysis of the Formin Homology 2 Domain. Mol. Biol. Cell 2005, 16, 1–13. [Google Scholar] [CrossRef]
- Paul, A.S.; Paul, A.; Pollard, T.D.; Pollard, T. The Role of the FH1 Domain and Profilin in Formin-Mediated Actin-Filament Elongation and Nucleation. Curr. Biol. 2008, 18, 9–19. [Google Scholar] [CrossRef]
- Paul, A.S.; Pollard, T.D. Review of the Mechanism of Processive Actin Filament Elongation by Formins. Cell Motil. Cytoskelet. 2009, 66, 606–617. [Google Scholar] [CrossRef]
- Seth, A.; Otomo, C.; Rosen, M.K. Autoinhibition Regulates Cellular Localization and Actin Assembly Activity of the Diaphanous-Related Formins FRLα and MDia1. J. Cell Biol. 2006, 174, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Otomo, T.; Tomchick, D.R.; Otomo, C.; Machius, M.; Rosen, M.K. Crystal Structure of the Formin MDia1 in Autoinhibited Conformation. PLoS ONE 2010, 5, e12896. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Cook, T.A.; Alberts, A.S.; Gundersen, G.G. MDia Mediates Rho-Regulated Formation and Orientation of Stable Microtubules. Nat. Cell Biol. 2001, 3, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Eng, C.H.; Schmoranzer, J.; Cabrera-Poch, N.; Morris, E.J.S.; Chen, M.; Wallar, B.J.; Alberts, A.S.; Gundersen, G.G. EB1 and APC Bind to MDia to Stabilize Microtubules Downstream of Rho and Promote Cell Migration. Nat. Cell Biol. 2004, 6, 820–830. [Google Scholar] [CrossRef]
- Zhou, F.; Leder, P.; Martin, S.S. Formin-1 Protein Associates with Microtubules through a Peptide Domain Encoded by Exon-2. Exp. Cell Res. 2006, 312, 1119–1126. [Google Scholar] [CrossRef]
- Young, K.G.; Thurston, S.F.; Copeland, S.; Smallwood, C.; Copeland, J.W. INF1 Is a Novel Microtubule-Associated Formin. MBoC 2008, 19, 5168–5180. [Google Scholar] [CrossRef]
- Bartolini, F.; Moseley, J.B.; Schmoranzer, J.; Cassimeris, L.; Goode, B.L.; Gundersen, G.G. The Formin MDia2 Stabilizes Microtubules Independently of Its Actin Nucleation Activity. J. Cell Biol. 2008, 181, 523–536. [Google Scholar] [CrossRef]
- Gaillard, J.; Ramabhadran, V.; Neumanne, E.; Gurel, P.; Blanchoin, L.; Vantard, M.; Higgs, H.N. Differential Interactions of the Formins INF2, MDia1, and MDia2 with Microtubules. Mol. Biol. Cell 2011, 22, 4575–4587. [Google Scholar] [CrossRef]
- Roth-Johnson, E.A.; Vizcarra, C.L.; Bois, J.S.; Quinlan, M.E. Interaction between Microtubules and the Drosophila Formin Cappuccino and Its Effect on Actin Assembly. J. Biol. Chem. 2014, 289, 4395–4404. [Google Scholar] [CrossRef]
- Bartolini, F.; Andres-Delgado, L.; Qu, X.; Nik, S.; Ramalingam, N.; Kremer, L.; Alonso, M.A.; Gundersen, G.G. An MDia1-INF2 Formin Activation Cascade Facilitated by IQGAP1 Regulates Stable Microtubules in Migrating Cells. Mol. Biol. Cell 2016, 27, 1797–1808. [Google Scholar] [CrossRef]
- Thurston, S.F.; Kulacz, W.A.; Shaikh, S.; Lee, J.M.; Copeland, J.W. The Ability to Induce Microtubule Acetylation Is a General Feature of Formin Proteins. PLoS ONE 2012, 7, e48041. [Google Scholar] [CrossRef] [PubMed]
- Szikora, S.; Foldi, I.; Tóth, K.; Migh, E.; Vig, A.; Bugyi, B.; Maléth, J.; Hegyi, P.; Kaltenecker, P.; Sanchez-Soriano, N.; et al. The Formin DAAM Is Required for Coordination of the Actin and Microtubule Cytoskeleton in Axonal Growth Cones. J. Cell Sci. 2017, 130, 2506–2519. [Google Scholar] [CrossRef] [PubMed]
- Kundu, T.; Dutta, P.; Nagar, D.; Maiti, S.; Ghose, A. Coupling of Dynamic Microtubules to F-Actin by Fmn2 Regulates Chemotaxis of Neuronal Growth Cones. J. Cell Sci. 2021, 134, jcs252916. [Google Scholar] [CrossRef] [PubMed]
- Lewkowicz, E.; Herit, F.; Le Clainche, C.; Bourdoncle, P.; Perez, F.; Niedergang, F. The Microtubule-Binding Protein CLIP-170 Coordinates MDia1 and Actin Reorganization during CR3-Mediated Phagocytosis. J. Cell Biol. 2008, 183, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Webb, R.L.; Zhou, M.-N.; McCartney, B.M. A Novel Role for an APC2-Diaphanous Complex in Regulating Actin Organization in Drosophila. Development 2009, 136, 1283–1293. [Google Scholar] [CrossRef]
- Henty-Ridilla, J.L.; Rankova, A.; Eskin, J.A.; Kenny, K.; Goode, B.L. Accelerated Actin Filament Polymerization from Microtubule plus Ends. Science 2016, 352, 1004–1009. [Google Scholar] [CrossRef]
- Dollar, G.; Gombos, R.; Barnett, A.A.; Sanchez Hernandez, D.; Maung, S.M.T.; Mihály, J.; Jenny, A. Unique and Overlapping Functions of Formins Frl and DAAM During Ommatidial Rotation and Neuronal Development in Drosophila. Genetics 2016, 202, 1135–1151. [Google Scholar] [CrossRef][Green Version]
- Gombos, R.; Migh, E.; Antal, O.; Mukherjee, A.; Jenny, A.; Mihály, J. The Formin DAAM Functions as Molecular Effector of the Planar Cell Polarity Pathway during Axonal Development in Drosophila. J. Neurosci. 2015, 35, 10154–10167. [Google Scholar] [CrossRef]
- Dehapiot, B.; Clément, R.; Alégot, H.; Gazsó-Gerhát, G.; Philippe, J.-M.; Lecuit, T. Assembly of a Persistent Apical Actin Network by the Formin Frl/Fmnl Tunes Epithelial Cell Deformability. Nat. Cell Biol. 2020, 22, 791–802. [Google Scholar] [CrossRef]
- Matusek, T.; Djiane, A.; Jankovics, F.; Brunner, D.; Mlodzik, M.; Mihály, J. The Drosophila Formin DAAM Regulates the Tracheal Cuticle Pattern through Organizing the Actin Cytoskeleton. Development 2006, 133, 957–966. [Google Scholar] [CrossRef]
- Migh, E.; Götz, T.; Földi, I.; Szikora, S.; Gombos, R.; Darula, Z.; Medzihradszky, K.F.; Maléth, J.; Hegyi, P.; Sigrist, S.; et al. Microtubule Organization in Presynaptic Boutons Relies on the Formin DAAM. Development 2018, 145, dev158519. [Google Scholar] [CrossRef] [PubMed]
- Barkó, S.; Bugyi, B.; Carlier, M.-F.; Gombos, R.; Matusek, T.; Mihály, J.; Nyitrai, M. Characterization of the Biochemical Properties and Biological Function of the Formin Homology Domains of Drosophila DAAM. J. Biol. Chem. 2010, 285, 13154–13169. [Google Scholar] [CrossRef] [PubMed]
- Fassier, C.; Fréal, A.; Gasmi, L.; Delphin, C.; Ten Martin, D.; De Gois, S.; Tambalo, M.; Bosc, C.; Mailly, P.; Revenu, C.; et al. Motor Axon Navigation Relies on Fidgetin-like 1-Driven Microtubule plus End Dynamics. J. Cell Biol. 2018, 217, 1719–1738. [Google Scholar] [CrossRef] [PubMed]
- Matusek, T.; Gombos, R.; Szécsényi, A.; Sánchez-Soriano, N.; Czibula, A.; Pataki, C.; Gedai, A.; Prokop, A.; Raskó, I.; Mihály, J. Formin Proteins of the DAAM Subfamily Play a Role during Axon Growth. J. Neurosci. 2008, 28, 13310–13319. [Google Scholar] [CrossRef]
- Sánchez-Soriano, N.; Gonçalves-Pimentel, C.; Beaven, R.; Haessler, U.; Ofner-Ziegenfuss, L.; Ballestrem, C.; Prokop, A. Drosophila Growth Cones: A Genetically Tractable Platform for the Analysis of Axonal Growth Dynamics. Dev. Neurobiol. 2010, 70, 58–71. [Google Scholar] [CrossRef]
- Xu, K.; Zhong, G.; Zhuang, X. Actin, Spectrin, and Associated Proteins Form a Periodic Cytoskeletal Structure in Axons. Science 2013, 339, 452–456. [Google Scholar] [CrossRef]
- Meijering, E.; Jacob, M.; Sarria, J.-C.F.; Steiner, P.; Hirling, H.; Unser, M. Design and Validation of a Tool for Neurite Tracing and Analysis in Fluorescence Microscopy Images. Cytom. Part A 2004, 58, 167–176. [Google Scholar] [CrossRef]
- Dent, E.W.; Callaway, J.L.; Szebenyi, G.; Baas, P.W.; Kalil, K. Reorganization and Movement of Microtubules in Axonal Growth Cones and Developing Interstitial Branches. J. Neurosci. 1999, 19, 8894–8908. [Google Scholar] [CrossRef]
- Tanaka, H.; Takasu, E.; Aigaki, T.; Kato, K.; Hayashi, S.; Nose, A. Formin3 Is Required for Assembly of the F-Actin Structure That Mediates Tracheal Fusion in Drosophila. Dev. Biol. 2004, 274, 413–425. [Google Scholar] [CrossRef]
- Xu, Y.; Moseley, J.B.; Sagot, I.; Poy, F.; Pellman, D.; Goode, B.L.; Eck, M.J. Crystal Structures of a Formin Homology-2 Domain Reveal a Tethered Dimer Architecture. Cell 2004, 116, 711–723. [Google Scholar] [CrossRef]
- Lu, J.; Meng, W.; Poy, F.; Maiti, S.; Goode, B.L.; Eck, M.J. Structure of the FH2 Domain of Daam1: Implications for Formin Regulation of Actin Assembly. J. Mol. Biol. 2007, 369, 1258–1269. [Google Scholar] [CrossRef] [PubMed]
- Breitsprecher, D.; Goode, B.L. Formins at a Glance. J. Cell Sci. 2013, 126, 1–7. [Google Scholar] [CrossRef]
- Vig, A.T.; Földi, I.; Szikora, S.; Migh, E.; Gombos, R.; Tóth, M.Á.; Huber, T.; Pintér, R.; Talián, G.C.; Mihály, J.; et al. The Activities of the C-Terminal Regions of the Formin Protein Disheveled-Associated Activator of Morphogenesis (DAAM) in Actin Dynamics. J. Biol. Chem. 2017, 292, 13566–13583. [Google Scholar] [CrossRef] [PubMed]
- Skube, S.B.; Chaverri, J.M.; Goodson, H.V. Effect of GFP Tags on the Localization of EB1 and EB1 Fragments in Vivo. Cytoskeleton 2010, 67, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kawabata Galbraith, K.; Kengaku, M. Multiple Roles of the Actin and Microtubule-Regulating Formins in the Developing Brain. Neurosci. Res. 2019, 138, 59–69. [Google Scholar] [CrossRef]
- Juanes, M.A.; Fees, C.P.; Hoeprich, G.J.; Jaiswal, R.; Goode, B.L. EB1 Directly Regulates APC-Mediated Actin Nucleation. Curr. Biol. 2020, 30, 4763–4772.e8. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Földi, I.; Tóth, K.; Gombos, R.; Gaszler, P.; Görög, P.; Zygouras, I.; Bugyi, B.; Mihály, J. Molecular Dissection of DAAM Function during Axon Growth in Drosophila Embryonic Neurons. Cells 2022, 11, 1487. https://doi.org/10.3390/cells11091487
Földi I, Tóth K, Gombos R, Gaszler P, Görög P, Zygouras I, Bugyi B, Mihály J. Molecular Dissection of DAAM Function during Axon Growth in Drosophila Embryonic Neurons. Cells. 2022; 11(9):1487. https://doi.org/10.3390/cells11091487
Chicago/Turabian StyleFöldi, István, Krisztina Tóth, Rita Gombos, Péter Gaszler, Péter Görög, Ioannis Zygouras, Beáta Bugyi, and József Mihály. 2022. "Molecular Dissection of DAAM Function during Axon Growth in Drosophila Embryonic Neurons" Cells 11, no. 9: 1487. https://doi.org/10.3390/cells11091487
APA StyleFöldi, I., Tóth, K., Gombos, R., Gaszler, P., Görög, P., Zygouras, I., Bugyi, B., & Mihály, J. (2022). Molecular Dissection of DAAM Function during Axon Growth in Drosophila Embryonic Neurons. Cells, 11(9), 1487. https://doi.org/10.3390/cells11091487







